Summary
High oleic oil is an important industrial feedstock that has been one of the main targets for oil improvement in a number of oil crops. Crambe (Crambe abyssinica) is a dedicated oilseed crop, suitable for industrial oil production. In this study, we down‐regulated the crambe fatty acid desaturase ( FAD ) and fatty acid elongase ( FAE ) genes for creating high oleic seed oil. We first cloned the crambe CaFAD2, CaFAD3 and CaFAE1 genes. Multiple copies of each of these genes were isolated, and the highly homologous sequences were used to make RNAi constructs. These constructs were first tested in Arabidopsis, which led to the elevated oleic or linoleic levels depending on the genes targeted, indicating that the RNAi constructs were effective in regulating the expression of the target genes in nonidentical but closely related species. Furthermore, down‐regulation of CaFAD2 and CaFAE1 in crambe with the FAD2– FAE1 RNAi vector resulted in even more significant increase in oleic acid level in the seed oil with up to 80% compared to 13% for wild type. The high oleic trait has been stable in subsequent five generations and the GM line grew normally in greenhouse. This work has demonstrated the great potential of producing high oleic oil in crambe, thus contributing to its development into an oil crop platform for industrial oil production.
Keywords: novel oil crop, renewable industrial oil, RNAi silencing, metabolic engineering, CaFAE1, CaFAD2
Introduction
Seed oil in general is composed of saturated and unsaturated fatty acids (FAs), while unsaturated FAs consist of monounsaturated FAs (MUFAs, mainly oleic acid, 18:1) and polyunsaturated FAs (PUFAs, mainly linoleic acid, 18:2, and linolenic acid, 18:3). Vegetable oils with a high content of monoenoic oleic acid (18:1) are of interest for both food and industrial purposes. This is due to the high heat and oxidative stability of oleic acid and makes high oleic (HO) oils suitable to be used directly in long‐life deep‐frying applications for food industry without the need for hydrogenation. Hydrogenation could lead to the conversion of naturally occurring cis‐FAs to trans‐FAs that are increasingly recognized as having cholesterol‐raising properties with increased risk of cardiovascular diseases (Mozaffarian et al., 2006). For industrial purpose, the high percentage of PUFAs in seed oil makes it highly prone to oxidation (Graef et al., 2009; Tat et al., 2007), while a more ideal biodiesel blend or a bio‐based lubricant should have high proportions of oleic acid that has greater oxidative stability than PUFAs and better cold flow properties than saturated FAs (e.g. 16:0 and 18:0, Durrett et al., 2008). Finally, oleic acid with double bond at delta‐9 position (mid‐chain) can also be converted into a range of derivatives that can be used as industrial FA feedstock.
Genetic improvement of seed oil with HO trait could be achieved through different breeding approaches. Traditional breeding utilizing natural variants or induced mutants as materials has been predominantly used to improve some oilseed crops, for example HO rapeseed (Auld et al., 1992), HO peanut (Norden et al., 1987) and HO sunflower (Soldatov, 1976). However, the selection process of traditional breeding is more time‐consuming, and the selection efficiency is often highly limited by the available genetic resources, and mutants having increased oleic acid levels are often associated with undesirable agronomic traits. Genetic engineering provides more precise and effective method for manipulating FA compositions in oilseed; for example, molecular techniques such as antisense and cosuppression have been used to down‐regulate the activity of fatty acid desaturase enzymes that control the synthesis of the major seed oil FAs (Kinney, 1994; Stoutjesdijk et al., 2000, 2002). However, the antisense and cosuppression strategies used in these cases have variable and unpredictable effectiveness and generally require the production of large populations of transgenic plants to obtain an acceptable number of lines exhibiting sufficient degrees of target gene suppression (Knutzon et al., 1992; Kinney, 1994). RNA interference (RNAi) technique, through a widespread silencing mechanism, has already been demonstrated to be more efficient than either antisense or cosuppression in terms of both higher frequency of transgenics showing silencing and generally higher degrees of silencing and has been successfully applied in a number of crops (Liu et al., 2002; Sheng et al., 2006; Zaplin et al., 2013).
FAD2 is a key gene encoding an ER membrane‐bound FA desaturase 2 (FAD2), which catalyses the conversion of oleic acid to linoleic acid (Okuley et al., 1994). FAD3 is another key gene for important enzyme converting 18:2 to 18:3 (Browse et al., 1993), while FAE1 encodes an enzyme responsible for the carbon chain elongation for synthesis of very long‐chain FAs (Millar and Kunst, 1997). These genes are considered to be priority targets for genetic manipulation towards lowering the PUFA content and increasing the oleic acid content. Up to now, a number of HO oilseed crops have been mainly developed through down‐regulating FAD2; for example, RNAi suppression of FAD2‐1 in cotton seeds clearly increased the oleic acid content and decreased the linoleic acid content (Liu et al., 2002). Double FAD2–FAE1 RNAi suppression to increase oleic acid content in seed oil has also been reported in camelina (Nguyen et al., 2013) and rapeseed (Peng et al., 2010).
Crambe (Crambe abyssinica Hochst.), a member of the Brassicaceae family, is a suitable oilseed crop for genetic engineering for industrial purposes (Li et al., 2012) as it does not outcross with any existing food oilseed crops, thereby eliminating the problem of gene flow into such crops. The crambe seed oil contains 55–60% erucic acid that makes the oil nonedible, despite its potential application as a starting material for developing other industrial oil products, such as long‐chain wax esters. Genetic modification of crambe has been reported (Guan et al., 2013; Li et al., 2012) to increase erucic acid levels in the seed oil. Down‐regulating FAD2 alone in crambe led to the increased oleic acid content up to 30% in the seed oil compared with 15% in the wild type (WT) and the erucic acid level up to 73% when combined with overexpression of the LdLPAAT and BnFAE1 genes (Li et al., 2012).
In this study, we used CaFAD2 or CaFAD3 partial coding sequences fused to CaFAE1 either partial coding sequence or 3′‐UTR under the control of a seed‐specific truncated napin promoter (Fp1) to construct the RNAi vectors. We first tested their ability for silencing the target genes in Arabidopsis and confirmed that these RNAi constructs were effective in affecting FA desaturation and elongation in seed oil, even though the target sequences were not completely identical. The CaFAD2–CaFAE1 RNAi vector was further used to silence crambe CaFAD2 and CaFAE1 genes to increase the level of oleic acid in crambe seed oil. We obtained transgenic crambe lines showing significantly increased contents of oleic acid, accompanied by down‐regulation of endogenous FAD2 and FAE1 genes.
Results
Multicopy gene families of CaFAE1, CaFAD2 and CaFAD3 in crambe
Using one‐step RT‐PCR, partial coding sequences of CaFAE1 (1009 bp), CaFAD2 (617 bp) and CaFAD3 (498 bp) were obtained from the total RNA. Through RACE amplification, multiple copies of CaFAE1, CaFAD2 and CaFAD3 sequences were identified and each of them showed high homology to known plant FAE1, FAD2 and FAD3 genes (data not shown). During the course of this work, other groups had also isolated multiple copies of CaFAD2 sequences (Cheng et al., 2013a). One FAE1 homologue from crambe was also previously cloned and characterized (Mietkewska et al., 2007). We chose part of the sequences among the above amplified regions, 617 bases for CaFAD2 (accession KP987201), 444 bases for CaFAD3 (accession KP987202), 734 bases for CaFAE1 (nt 1‐734 of accession KP987203), for the construction of RNAi‐silencing constructs. Alignment of these sequences with those from other Brassica species such as B. napus, B. juncea, A. thaliana and the recently published crambe sequences showed high homology between these sequences (Figure S1).
Southern blot analysis with genomic DNA isolated from C. abyssinica probed with CaFAD2, CaFAD3 or CaFAE1 coding sequence fragments confirmed the existence of multicopies of the three genes (Figure 1). This is in agreement with the fact that we have cloned multiple representatives of these gene families in crambe.
Figure 1.
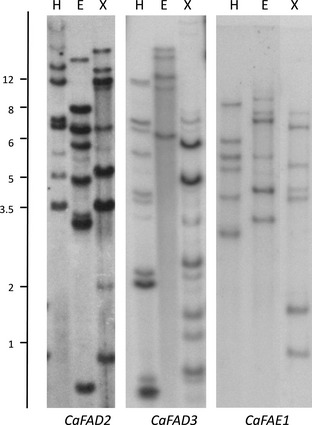
Southern analysis of Crambe abyssinica with probes CaFAD2, CaFAD3 and CaFAE1. The genomic DNA was digested with HindIII (H), Eco RI (E) and XbaI (X). The numbers on the right side of the figure indicate the sizes (kb) of the DNA bands.
Fusion PCR
The above‐selected RNAi‐silencing target regions were fused by PCR for preparing the constructs. The CaFAD2–CaFAE1 fusion fragment (1351 bp) consisted of the partial coding sequences (CDSs) of CaFAD2 and CaFAE1, while the CaFAD3–CaFAE1 fusion fragment (1178 bp) contained the partial CDSs of CaFAD3 and CaFAE1. Moreover, the 113‐bp‐long 3′‐UTR of CaFAE1 (nt 991‐1103 of accession KP987203) was also used for targeting sequence in other two constructs and fused to the partial sequence of CaFAD2 or CaFAD3 by PCR, resulting in either CaFAD2–CaFAE1‐U fusion fragment (730 bp) or CaFAD3–CaFAE1‐U fusion fragment (557 bp). Comparison of the sequences for these regions was shown in the supplementary data (Figure S2). Each of the gene fusion fragments was cloned into the binary vector pXZP401, generating RNAi gene‐silencing constructs with inverted repeat driven by the Fp1 promoter (Stalberg et al., 1993).
Cross‐species gene silencing of A. thaliana FAD2, FAD3 and FAE1 with C. abyssinica sequences
To test the functionality of the RNAi gene‐silencing constructs, and to test whether these crambe gene constructs could silence the genes in the closely related A. thaliana, the constructs were transformed in A. thaliana ecotype Columbia. The expression of RNAi gene‐silencing constructs containing CaFAD2–CaFAE1 or CaFAD2–CaFAE1‐U substantially decreased the 20:1 and 18:2 levels in the Arabidopsis seed oil (Figure 2). The 20:1 level was decreased from 21% in WT to 5% in the transgenic lines, with both the CaFAE1 coding sequence and 3′‐UTR, when combined with CaFAD2. The 18:2 level was decreased from 27% in WT to 6% or 9% in the transgenic lines with the CaFAE1 CDS or 3′‐UTR, respectively. In contrast, the 18:1 level was increased from 12% to 50% or 48%, respectively (Figure 2b,c).
Figure 2.
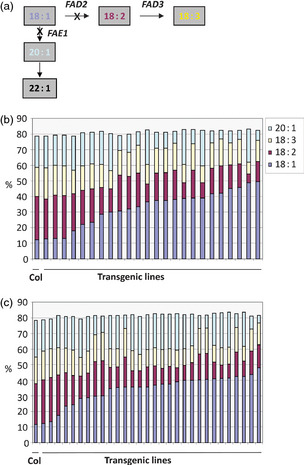
Silencing Arabidopsis genes with C. abyssinica CaFAD2 and CaFAE1 sequences resulted in high oleic acid in Arabidopsis seed. (a) Gene‐silencing targets in simplified representation of fatty acid metabolism pathway. (b) Major fatty acids in Arabidopsis WT (Col) or transgenic seeds transformed with RNAi‐silencing construct containing invert repeated CaFAD2 and CaFAE1 coding sequences. (c) Major fatty acids in Arabidopsis WT (Col) or transgenic seeds transformed with RNAi‐silencing construct containing invert repeated CaFAD2 coding sequence and CaFAE1 3′‐UTR sequence.
Similar to the above silencing effect, the expression of RNAi gene‐silencing constructs containing CaFAD3–CaFAE1 or CaFAD3–CaFAE1‐U substantially decreased the 18:3 level in the Arabidopsis seed oils. The 18:3 level was decreased from 19% in WT to 10% or 5% in the transgenic lines, with CaFAE1 CDS or 3′‐UTR, respectively. The 20:1 level was decreased from 21% in WT to 9% in the transgenic lines with CaFAE1 coding sequence. However, there was no significant decrease in 20:1 level in CaFAD3–CaFAE1‐U lines. In contrast, 18:2 level was increased from 28% in WT up to 45% or 46%, respectively, while the 18:1 level increased slightly, up to 22% (Figure 3).
Figure 3.
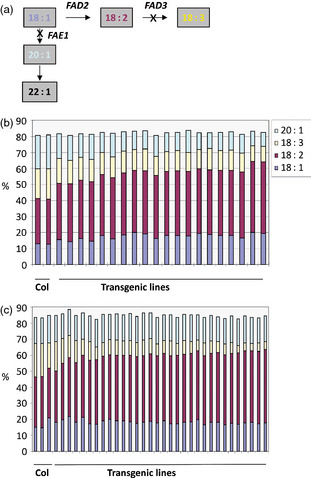
Silencing Arabidopsis genes with C. abyssinica CaFAD3 and CaFAE1 sequences resulted in high linoleic acid in Arabidopsis seed. (a) Gene‐silencing targets in simplified representation of fatty acid metabolism pathway. (b) Major fatty acids in Arabidopsis WT (Col) or transgenic seeds transformed with RNAi‐silencing construct containing invert repeated CaFAD3 and CaFAE1 coding sequences. (c) Major fatty acids in Arabidopsis WT (Col) or transgenic seeds transformed with RNAi‐silencing construct containing invert repeated CaFAD3 coding sequence and CaFAE1 3′‐UTR sequence.
Silencing FAD2 and FAE1 in crambe
To evaluate the silencing effect of the above RNAi constructs in crambe for obtaining ultrahigh oleic oil content, the CaFAD2–CaFAE1 RNAi‐silencing vector was introduced into crambe. Six transgenic lines were recovered and produced T1 seeds, of which five lines showed an altered seed oil composition with the average level of 18:1 ranging from 17.3% to 66.1% compared with 14.5% in WT. The best line P1 with the oleic acid content of 66.1% was further analysed using the half‐seed technique with over 50 seeds. The results showed that the 18:1 content was around 80% in two‐third of the seeds analysed, while the 18:1 content was 20–30% in the remaining seeds. This line was then followed up in the subsequent generations.
Twenty T1 half‐seeds from the line P1 with 18:1 content around 80% were planted in the greenhouse to produce T2 seeds. FA analysis showed a large segregation among the T2 seeds. Among twenty T1 progenies, one plant (P1‐8) produced seeds with the average 18:1 content of 76.4%, but some seeds contained more than 80% of 18:1 (Table 1).
Table 1.
Segregation of 18:1 content in seed oil from different generations of transgenic crambe with CaFAD2–CaFAE1 RNAi
| Generation | No of lines or individuals | 18:1 level (%) | No of plants at various 18:1 levels (%)** | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean* | Max | Min | <20 | 20‐39 | 40‐59 | 60‐74 | ≥75*** | ||
| T1 | 6 | 31.8 ± 19.0 | 66.1 | 13.8 | 2 | 3 | 0 | 1 | 1 |
| T2 | 20 | 34.1 ± 15.6 | 76.4 | 14.8 | 2 | 13 | 4 | 1 | 2 |
| T3 | 40 | 50.0 ± 14.3 | 75.6 | 24.2 | 0 | 12 | 15 | 13 | 6 |
| T4 | 16 | 69.1 ± 7.3 | 76.7 | 54.9 | 0 | 0 | 2 | 14 | 8 |
| T5 | 16 | 68.0 ± 9.3 | 77.9 | 54.1 | 0 | 0 | 4 | 12 | 10 |
*Mean value of all seeds from each generation. T1: 10 seeds for each line/plant, T2‐T5: 5 seeds for each line/plant. **Mean value of seeds from each line/plant. ***The plants having seeds with 18:1 content equal to or more than 75%.
Forty T2 progenies were generated from the T2 seeds possessing 18:1 content around 80% as shown in Table 1. Fifteen progenies had mean values of 18:1 between 40% and 59% and 13 progenies had the 18:1 content more than 60% in the T3 seeds. For the best progeny (P1‐8‐29), some seeds had 18:1 content more than 80%. Sixteen progenies each were analysed from T4 and T5 seeds. Among them, the majority of seeds contained the 18:1 content higher than 60%, and more individual progenies contained 18:1 content higher than 75% in the seed oil compared to earlier generations, suggesting that the stable transgenic lines with high 18:1 content have been generated after intensive selection of several generations (Table 1). In summary, the results shown in Table 1 indicate clearly that the high oleic trait has been stabilizing over the generations as shown by an increased percentage of individuals having high 18:1 level with each generation. This indicates the stable silencing effect of the target genes by RNAi.
Changes of 18:1, 18:2 and 18:3 in different generations of the best transgenic lines
The pedigree of the best transgenic line and the contents of 18:1, 18:2 and 18:3 in the seed oil from different generations are listed in Table 2. A clear segregation of 18:1 was observed in P1, but the variation became smaller in the subsequent generations, indicating a stable silencing effect of FAD2–FAE1 RNAi was maintained over generations. The transgenic seeds with 18:1 content higher than 80% could be produced in nearly all the following generations, indicating the stable silencing effect of the target genes by RNAi. The 18:2 level in the transgenic seeds was decreased compared with WT as expected, while no obvious change was observed for the 18:3 content (Table 2).
Table 2.
The 18:1, 18:2 and 18:3 levels (%) of seed fatty acids from different generations of selected transgenic crambe lines with CaFAD2–CaFAE1 RNAi
| Line | Seed no. | 18:1 | 18:2 | 18:3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Max | Min | Mean | Max | Min | Mean | Max | Min | ||
| Wild type | 20 | 14.5 ± 1.3 | 16.4 | 12.0 | 7.6 ± 0.6 | 9.3 | 6.7 | 6.2 ± 0.6 | 7.6 | 5.2 |
| P1 | 30 | 66.1 ± 28.7 | 84.3 | 12.9 | 4.1 ± 2.9 | 10.4 | 1.7 | 3.6 ± 0.7 | 4.7 | 2.8 |
| P1‐8 | 50 | 75.1 ± 3.3 | 80.9 | 58.0 | 3.6 ± 0.7 | 4.5 | 2.6 | 5.1 ± 1.0 | 7.3 | 3.7 |
| P1‐8‐29 | 80 | 75.1 ± 5.4 | 82.2 | 61.6 | 4.2 ± 1.2 | 7.4 | 2.8 | 5.4 ± 0.9 | 7.4 | 4.1 |
| P1‐8‐29‐34 | 60 | 76.1 ± 3.3 | 79.9 | 66.4 | 4.3 ± 1.2 | 7.4 | 2.9 | 6.9 ± 0.9 | 9.1 | 5.3 |
| P1‐8‐29‐34‐4 | 30 | 75.9 ± 6.6 | 80.0 | 55.0 | 4.2 ± 1.5 | 8.6 | 2.5 | 5.6 ± 0.7 | 7.2 | 4.3 |
Changes of fatty acid composition in the seed oil of transgenic crambe lines
We also analysed the FA composition in the seeds of the best transgenic crambe line (Table 3). The contents of 16:0, 18:0, 18:1 and 20:1 were increased in the transgenic line compared to WT, especially for 18:1 which reached up to 84.3% compared with 14.5% in WT. The other FAs were decreased in the transgenic seeds, except for 20:0 that remained unchanged. The most significant decrease was found in 22:1 level, which was decreased to 0.3% compared to 59.5% in WT. Two other T1 progenies from this line (P1‐6 and P1‐13) are also shown in Table 3, as their performance represented the most transgenic lines in the selection. For these lines, although the content of 18:1 was increased and 22:1 level was decreased, the contents of 18:2 and 20:1 were also increased, probably due to inefficient down‐regulation of the FAD2–FAE1 RNAi (see Figure 4 for gene expression analysis).
Table 3.
Oil composition (%) of some selected seeds with high 18:1 content from different generations of transgenic crambe line with CaFAD2–CaFAE1 RNAia
| Line/seed | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 20:2 | 22:0 | 22:1 | 24:0 | 24:1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 1.5 | 0.7 | 14.5 | 7.5 | 6.2 | 0.7 | 2.0 | 0.7 | 1.8 | 59.5 | 0.5 | 1.2 |
| P1 | 3.1 | 1.9 | 84.3 | 2.1 | 3.6 | 0.6 | 2.2 | 0.6 | 0.3 | 0.5 | 0.2 | 0.1 |
| P1‐8 | 4.3 | 3.0 | 79.9 | 3.0 | 4.5 | 0.9 | 2.4 | 0.3 | 0.3 | 0.4 | 0.3 | 0.2 |
| P1‐8‐29 | 3.9 | 2.1 | 79.7 | 2.8 | 4.1 | 0.7 | 2.8 | 0.2 | 0.2 | 0.5 | 0.2 | 0.2 |
| P1‐8‐29‐34 | 4.2 | 1.9 | 79.9 | 2.9 | 6.3 | 0.6 | 2.4 | 0.2 | 0.2 | 0.3 | 0.1 | 0.2 |
| P1‐8‐29‐34‐4 | 4.1 | 2.3 | 80.0 | 2.5 | 4.3 | 0.6 | 2.4 | 0.4 | 0.2 | 0.4 | 0.1 | 0.2 |
| P1‐6 | 2.8 | 1.1 | 44.0 | 7.0 | 3.2 | 0.6 | 10.3 | 0.3 | 0.4 | 27.5 | 0.2 | 0.7 |
| P1‐13 | 3.2 | 1.0 | 24.9 | 10.4 | 4.4 | 0.7 | 8.1 | 0.9 | 1.0 | 41.7 | 0.7 | 1.2 |
Some minor components are not shown in the table.
Figure 4.
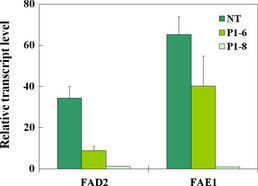
Relative transcript levels of FAD2 and FAE1 genes in the seeds from two transgenic crambe lines and untransformed control (NT). The 18:1 content of NT is 14.5%. The 18:1 contents of transgenic lines P1‐6 and P1‐8 are 44.0% and 76.4%, respectively. The result is from six different samples and each sample has 6–7 seeds. Error bars represent SE.
The expression of the target genes in the transgenic crambe lines
qRT‐PCR analysis was carried out on two progenies (P1‐6 and P1‐8) for the expression levels of two targeted genes (CaFAD2 and CaFAE1) as these two progenies had different 18:1 levels. The RNA levels of the targeted endogenous genes were negatively associated with the levels of 18:1 in the developing transgenic seeds. As shown in Figure 4, P1‐8 that had higher 18:1 level showed lower expression levels of FAD2 and FAE1 than P1‐6 (Figure 4).
Discussion
Crambe (C. abyssinica) is an allohexaploid species (2n = 6x = 90). Cheng et al. (2013a) recently reported seven copies of FAD2 cloned from C. abyssinica cv. Galactica. Both our gene cloning efforts from developing embryo and Southern analysis further confirmed the existence of multiple copies of CaFAD2, possibly even more than seven copies, although there might be also difference between two cultivars, that is Nigeria vs Galactica. In addition, CaFAD3 and CaFAE1 genes had also multiple copies, with likely six copies for CaFAE1. Small FAD2 or FAD3 gene families were also discovered in other plants, although only a single FAD2 or FAD3 gene exists in Arabidopsis. For example, two FAD2 sequences were isolated from flax (Khadake et al., 2009), three distinct family members in soya bean (Li et al., 2007), sunflower (Martínez‐Rivas et al., 2001) and Camelina sativa (Kang et al., 2011), and five members in cotton (Liu et al., 1999). A super large FAD2 gene family with at least 11 members was isolated from safflower (Cao et al., 2013). Recently, we also isolated a FAD2 gene family with four members along with two copies of FAD2‐like hydroxylase gene FAH12 (Zhou et al., 2013). These suggest the complicated regulation exists in plant FA desaturation and elongation on one hand, and maybe also gene redundancy on the other hand. It is intriguing how these divergent gene family members with similar fundamental properties carry out the specific functions.
Although the detailed expression profile of each member in the CaFAD2, CaFAD3 and CaFAE1 gene families was not investigated in this study, a subset of members may play a key role in determining the FA profile in crambe seeds. In fact, two members of CaFAD2 were highly up‐regulated during the seed development (Cheng et al., 2013a). Regardless, the high homology between different members of each gene family makes it feasible to silence all gene family members expressed in seed.
Both A. thaliana and C. abyssinia are members of Brassicaceae family. Their FAE1, FAD2 and FAD3 sequences shared high homology between two species (Figure S1). To quickly test the functionality of the RNAi gene‐silencing constructs containing crambe sequences, the crambe RNAi constructs were first transformed into Arabidopsis. The results showed that all four constructs were effective to alternate the Arabidopsis seed FA composition, that is elevating the 18:1 or 18:2 level. This suggests that the cross‐species gene silencing was effective by the closely related sequences. Surprisingly, even the RNAi CaFAE1 3′‐UTR construct had similar but a slightly lower effect than the coding sequence construct in alternating the Arabidopsis seed FA profile (Figures 2 and 3). Comparison of A. thaliana and C. abyssinia FAE1 3′‐UTR sequences revealed that they were more divergent than the coding sequences, and there was only one small region of 25 bases (Complementary Figure S2) with 3 nucleotides mismatch. This region was shorter than 21‐mer that is considered as small interfering RNA to break up the ‘perfect complementary’ target mRNA with RNA‐induced silencing complex (RISC) (Tijsterman et al., 2002). Nevertheless, this study demonstrated that the gene silencing could be effective by targeting one specific gene with the same gene sequence from different but closely related species, even with more divergent 3′‐UTR, which could lead to a similar or slightly lower effect on gene silencing.
Unlike Arabidopsis, WT crambe seeds have relative low amounts of 18:2 and 18:3. Most of 18:1 is elongated to 20:1 then further to 22:1 by FA elongase. Attempt to silence the crambe CaFAD2 genes alone had only resulted in about 30% of 18:1, much of it was still elongated to 22:1 (Cheng et al., 2013b; Li et al., 2012). In this study, we have shown that the simultaneous silence of CaFAD2 and CaFAE1 in crambe was more effective in elevating the 18:1 level with up to 80% in the seed oil, by blocking the desaturation from 18:1 to 18:2 as well as elongation from 18:1 to 22:1. The 22:1 level was greatly reduced from 60% in WT to less than 0.5% in the transgenic seeds, with reduced 18:2 and 18:3 levels as well. The high oleic line grew normally under greenhouse conditions. Although only one line among these six transgenic lines had very high 18:1 oleic acid level, considering the fact that there were multiple copies of CaFAD2 and CaFAE1 in crambe genome, the approach to silence CaFAD2 and CaFAE1 simultaneously has proved to be an efficient way compared to generating mutations in each copy of CaFAD2 and CaFAE1. In addition, the ultrahigh 18:1 level was a stable trait in the five subsequent generations tested.
In the engineered crambe lines, the saturated FAs 16:0 and 18:0 were also increased (Table 3) and the reason for this was unclear. When FAD2 and/or FAE1 are down‐regulated, the levels of 16:0 and 18:0 have been reported to be either reduced or increased depending on species. For instance, silencing of FAD2 in cotton resulted in a reduction of 16:0 and no change of 18:0, accompanied by an increase of 18:1 (Liu et al., 2002). In flax, a slight increase of 16:0 was detected in the FAD2‐silenced lines compared to WT (Chen et al., 2015). One possible explanation for the increased saturated FAs in the silenced crambe lines could be due to the substrate preference of acyltransferases. In WT crambe, the acyltransferases might prefer assembling more 22:1 into TAG, thus leading to about 60% of 22:1 accumulation in the seed oil. However, the significant reduction of 22:1 in the silenced crambe lines might have increased the chance for the acyltransferases to assemble more 16:0 and 18:0 into TAG, in addition to 18:1.
The efficiency of RNAi silencing by the crambe target sequences was lower in Arabidopsis than in crambe. For instance, silencing FAD2 and FAE1 in Arabidopsis resulted in a fourfold increase in oleic acid compared to fivefold increase in crambe. This was likely due to the imperfect match of the target genes in Arabidopsis with the crambe sequences. This was especially true for the silencing of FAE1 in Arabidopsis where the 20:1 levels were quite variable. When the CaFAE1 UTR sequence was used in combination with CaFAD2, the 20:1 level was decreased down to 5% in the transgenic Arabidopsis line compared to 21% in WT (Figure 2c). However, when the CaFAE1 UTR sequence was used in combination with CaFAD3, there was no significant difference in 20:1 level in Arabidopsis (Figure 3c). Meanwhile, the combination of CaFAE1 coding sequence with CaFAD3 resulted in the decrease of 20:1 from 21% to 9% (Figure 3b). Multiple copies of FAD2 and FAE1 sequences in crambe would be also targeted by the RNAi construct at the same time due to the high‐sequence homology. In a recent report, as high as 80% of oleic acid level was achieved in flax by silencing both copies of FAD2 genes in a single construct, compared to 12% in WT (Chen et al., 2015).
An alternative approach for elevating oleic acid level in crambe would be to generate FAD2 and FAE1 double mutants. Examples of FAD2 mutants in oil crops with high 18:1 levels have been reported (Liu, et al., 2013; Pham et al., 2010; Schuppert et al., 2006). Recently, Cheng et al. (2014) successfully combined EMS mutagenesis and Illumina sequencing to detect mutation in crambe FAD2 genes. Generation of single mutants in individual FAD2 or FAE1 genes could allow the functional analysis of different members of the same gene family and production of non‐GM genotypes with high oleic oil by crossing different mutants, thus avoiding the expensive cost of deregulation of GMO. However, it would be tedious to combine the mutations from all the individual members of gene families, especially in the case of crambe, in which both FAD2 and FAE1 have been shown to be multiple gene copies. The RNAi approach would be more effective to target multiple gene families in the same event to have maximal level of gene silencing, as shown in this study.
Nevertheless, we have shown that the RNAi constructs using either crambe CaFAD2 and CaFAE1 or CaFAD3 and CaFAE1 genes could generate high oleic acid in Arabidopsis seed oil. RNAi gene silencing targeting the crambe CaFAD2 and CaFAE1 genes could generate ultrahigh oleic acid level up to 80% in the crambe seed oil.
Experimental procedures
Plant material and in vitro growing conditions
Crambe abyssinica. cv. Nigeria was used for gene isolation. Different vegetative tissues from 35‐day‐old plant, as well as 1‐ to 4‐week‐old embryos after pollination, were harvested. C. abyssinica cv. Galactica was used for transformation. All in vitro materials were grown in a climate chamber with 16‐h photoperiod, 33 μmol/m2/s light intensity and 25 ⁄ 18 o C temperature (day/night).
DNA and RNA isolation
Genomic DNA was extracted from leaves and purified with CsCl gradient ultra centrifugation. Total RNA was isolated from roots, stems, petioles, leaves and 1‐, 2‐, 3‐ and 4‐week‐old embryo using RNeasy Plant Mini kit (Qiagen, Hilden, Germany).
PCR primers
To clone crambe FAE1 and FAD2 genes, PCR primers were designed based on the known sequence or targeting for the conserved region. CaFAE1 sequence was cloned using primers FAE1‐S1 and FAE1‐A1, designed based on CaFAE1 (AY793549) and Brassica napus BnFAE1 (EU543283). Primers FAD2‐S1 and FAD2‐A1 for CaFAD2 were designed based on the highly homology region of FAD2 from B. napus (AAF78778, AAS92240, AAT02411), Crambe hispanica (ABF70302, ABF70303), C. kralikii (ABF70313, ABF70314), C. maritime (ABF70315, ABF70316), Arabidopsis thaliana (AAA32782), Gossypium hirsutum (CAA71199) and Glycine max (P48630). Primers FAD3‐S3 and FAD3‐A3 for CaFAD3 were designed based on the conserved amino acid regions of FAD3 from A. thaliana (P48623), B. napus (AY599884), G. max (P48625), Vigna radiata (P32291), Linum usitatissimum (ABA02172, ABA02173), Nicotiana tabacum (P48626) and Perilla frutescens (AAD15744). 5′‐ and 3′‐RACE primers were designed according to the cloned sequences. PCR primers used for making the fusion construct of CaFAD2 or CaFAD3 coding sequences (underlined) with CaFAE1 coding region or 3′‐UTR were the same as for the gene‐silencing vector. Primer sequences used in this study are listed in Table S1.
Cloning of genes
The cDNAs were synthesized from the total RNA of embryos mentioned above using one‐step Superscript II RT Taq polymerase (Invitrogen, Carlsbad, CA). One‐step RT‐PCRs were carried out with the following program: 50 °C for 20 min, a predenaturation at 94 °C for 2 min and followed by 40 cycles of PC R consisting of 94 °C for 20 s; 60 °C for 30 s; and 70 °C for 1 min with a final extension at 70 °C for 10 min. The gel‐purified PCR products of expected sizes were cloned into the pGEM‐T Easy vector (Promega, Madison, WI). The sequences of inserts were determined with BigDyer 3.0 (Applied Biosystems, Inc. Foster City, CA) and used for designing the gene‐specific primers for rapid amplification of cDNA ends (RACE). RACE reactions were performed using the GeneRacer kit (Invitrogen) according to the manufacture's instruction, with 2 μg of the total RNA. The RACE products were cloned into the pGEM‐T Easy vector and sequenced. Full length of cDNAs was re‐amplified with primers designed based on the RACE product sequences.
Southern blotting
About 5–8 μg of genomic DNA was digested by EcoRI, HindIII or XbaI. The restriction fragments were separated on a 0.8% (w/v) agrose gel and transferred onto a Hybond‐N+ nylon membrane (Amersham, Buckinghamshire, UK). The hybridization was carried out following the standard procedure (Sambrook, 2000), with the [α‐32P] dCTP‐labelled probes of CaFAD2, CaFAD3 or CaFAE1 gene fragments isolated above.
Full‐length gene cloning and sequencing
The full‐length sequence of FAD3 was cloned using GeneRacer cDNA (Invitrogen). Other genes were cloned using the cDNA reverse transcription by superscript III RNase H RT Kit (Invitrogen). PCRs were performed in a total volume of 50 μL containing 1 μL of cDNA, 10 pmol of each primer, 2.5 unit of Pfu‐Ultra (Stratagene), 5 μL of 2 mM dNTPs and 5 μL 10 × Pfu‐Ultra Buffer. Thermocycling consisted of an initial denaturation at 94 °C for 2 min, followed by 40 cycles of 94 °C for 30s, 70 °C to 56 °C for 30 s (annealing temperature decreased 1 °C per cycle from the 14 cycles, and 72 °C for 2 min, with the final extension of 2 min at 72 °C. The desired PCR products were purified by PCR Purification Kit (Qiagen), and added appropriate Hotstar mix, kept at 72 °C for 1 h, followed by ligation into the pGEM‐T Easy vector (Promega) for sequence confirmation.
Fusion of two target gene sequences into single construct by PCR
Two rounds of PCR were used to fuse two targeted gene sequences containing either CaFAD2 or CaFAD3 coding sequences with CaFAE1 coding region or 3′‐UTR. The antisense primer of first targeted gene contained extra 17–20 bp of 5′‐end of the second gene, while the sense primer of the second gene contained 17–20 bp of 3′‐end of the first gene (Table S1). Each of these two genes was amplified separately by combining its own gene‐specific primer (end primer) in first round PCR, led to the overlap region of 37–40 bp between two PCR products. The two PCR products were then mixed in the second round and re‐amplified only with two gene‐specific primers (end primers) used in the first round PCR. The fusion PCR products were cloned into pGEM‐T Easy (Promega) for sequence confirmation.
RNAi gene‐silencing vectors
Four gene fusion PCR products were created, namely FAD2–FAE1 and FAD3–FAE1 containing the coding sequences of the target genes, FAD2–FAE1‐U and FAD3–FAE1‐U, consisting of coding sequences for CaFAD2 and CaFAD3, but also 3′‐UTR CaFAE1. The gene fusion PCR products were cloned in the pGEM‐T Easy (Promega) and subcloned into pENTR11 (Invitrogen), and flanked by attL1 and attL2 sequences. The resulted entry plasmids were used for constructing RNAi gene‐silencing vectors by LR clonase (Invitrogen) with the binary vector pXZP401 (Zhou et al., 2013) containing two inverted copies of attR1‐ccdB gene‐attR2 driven by the seed‐specific napin promoter Fp1 (Stalberg et al., 1993) and separated by PKS and Cat‐1 introns.
Plant transformation
A. thaliana ecotype Columbia was used for transformation with Agrobacterium tumefaciens (AGL1 strain) carrying the various RNAi gene‐silencing constructs as previously described (Zhou et al., 2006). Crambe transformation was performed according to Li et al. (2010, 2011) with pXZP401_FAD2–FAE1 RNAi‐silencing vector targeting coding regions of CaFAD2 and CaGAE1. The plant selection marker was neomycin phosphotransferase II (NPTII).
Crambe growth conditions and plant management
The transgenic and wild‐type (WT) shoots of crambe were rooted according to Li et al. (2011), and the plantlets were then planted in the biotron (T1, T3 generations) or greenhouse (T2, T4 and T5 generations). The conditions in the biotron were 16‐h photoperiod, 250 μmol/m2/s1 light intensity, 21⁄18 oC temperature (day ⁄ night) and 60% humidity, and those in the greenhouse were 16‐h photoperiod and 21⁄13 o C temperature (day ⁄ night). The established plants were watered regularly with fertilizer (N: P: K = 21: 3: 10). The harvested seeds were kept at 4 °C for later use.
Quantitative Real‐Time PCR (qRT‐PCR) analysis
Total RNA was extracted from immature seeds of transgenic and WT crambe collected at 20–30 days after flowering using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer's instructions. Six to seven seeds were used in one sample with 6 biological replications for each line and four technical replicates for each qRT‐PCR. Residual genomic DNA was removed through DNase treatment using TURBO DNA‐free (Ambion, Austin, TX). First‐strand cDNA was synthesized according to Li et al. (2013) using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, St. Leon‐Rot, Germany). The cDNA was diluted 4 x, and 2 μL was used for 20‐μL qRT‐PCR using Applied Biosystems 7300 Real‐Time PCR System (Applied Biosystems, Foster City, CA) with Platinum SYBR Green qPCR SuperMix‐UDG module (Invitrogen, Carlsbad, CA). The PCR program was 50 °C for 2 min and 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Melting curve analysis was performed to confirm the product specificity. The primers for FAD2 and FAE1 were selected from several sets of primers tested in preliminary analyses. Gene expression was normalized to the Arabidopsis reference gene AtUBC21 (Table S1) as the genome sequence information of crambe is not available. The partial sequence of crambe ubiquitin gene has shown that the primers used for qRT‐PCR are homologous between Arabidopsis and crambe (Li et al., 2012).
Fatty acid composition analysis
For Arabidopsis, 2 mg of seeds was used to prepare the FA methyl ester (FAME), as previously described (Zhou et al., 2006). The FAMEs were analysed with Agilent N6890 gas chromatography (GC) as described previously (Zhou et al., 2011), using FAME standard C8‐C24 (Sigma) to calculate the weight percentage.
For crambe, the half‐seed technique was used for FA analysis according to Li et al. (2012). Briefly, the seeds were surface‐sterilized using 15% calcium hypochlorite for 20 min and rinsed thoroughly with sterile water before being placed on the germination medium (Li et al., 2010) in the climate chamber as stated above. After about 15 h, the seed coats were removed, and the larger outer cotyledon was excised for FA analysis according to the method described by Li et al. (2012), while the rest were maintained on the culture medium until planted in pots to harvest progeny seeds. The FA composition was analysed using Shimadzu GC‐17A gas chromatograph with WCOT Fused Silica CP‐Wax 58 with a FID detector (Shimadzu Corporation, Kyoto, Japan). Peaks were identified according to their retention time in comparison with a standard FAME mixture, and results are expressed as area percentage of oleic acid methyl esters in total detectable peak areas.
Statistical analysis
Data of oil analysis for crambe were statistically analysed with ANOVA and Tukey's test using the Statgraphics program.
Supporting information
Figure S1 Comparison of crambe gene fragments used in RNAi constructs with related sequences. A. CaFAD2 (accession KP987201); B, CaFAD3 (accession KP987202); C, CaFAE1 (accession KP987203). Each nucleotide sequence is followed by an accession number. The plant species are: At, Arabidopsis thaliana; Bn, Brassica napus; Ca, Crambe hispanica subsp. abyssinica; Cf, C. filiformis; Cg, C. glabrata; Ch, C. hispanica subsp. hispanica; Ck, C. kralikii; Cm, C. maritime (ABF70315, ABF70316); Lu, Linum usitatissimum. Sequences are aligned with Vector NTi V11 (Life Technologies).
Figure S2 Nucleotide sequences used for RNAi constructs and their comparison to Arabidopsis sequences. A, FAD2; B, FAD3, C, FAE1 coding sequence; D, FAE1 3′‐UTR sequence.
Acknowledgements
We wish to thank Clive Hurlstone, Cheryl Blundell and Bei Dong at CSIRO for technical support. This study is a part of EC FP7 ICON project and we thank Prof. Sten Stymne for his great support and discussions for this project. D. Mei was supported by Australian Centre for International Agricultural Research (ACIAR) R & D Program. We also wish to thank Vinnova, FORMAS, SLU, Swedish Foundation for Strategic Research, Royal Physiographic Society in Lund and Einar och Inga Nilsson's Foundation for financial support to the crambe project.
References
- Auld, D.L. , Heikkinen, M.K. , Erickson, D.A. , Sernyk, J.L. and Romero, J.E. (1992) Rapeseed mutants with reduced levels of polyunsaturated fatty‐acids and increased levels of oleic‐acid. Crop Sci. 32, 657–662. [Google Scholar]
- Browse, J. , McConn, M. , James, D. Jr and Miquel, M. (1993) Mutants of Arabidopsis deficient in the synthesis of alpha‐linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351. [PubMed] [Google Scholar]
- Cao, S.J. , Zhou, X.R. , Wood, C.C. , Green, A.G. , Singh, S.P. , Liu, L.X. and Liu, Q. (2013) A large and functionally diverse family of FAD2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhou, X.‐R. , Zhang, Z.‐J. , Dribnenki, P. , Singh, S. and Green, A. (2015) Development of high oleic oil crop platform in flax through RNAi‐mediated multiple FAD2 gene silencing. Plant Cell Rep. 34, 643–653, DOI 10.1007/s00299-015-1737-5. [DOI] [PubMed] [Google Scholar]
- Cheng, J. , Salentijn, E.M.J. , Huang, B. , Krens, F.A. , Dechesne, A.C. , Visser, R.G.F. and Loo, E.N. (2013a) Isolation and characterization of the omega‐6 fatty acid desaturase (FAD2) gene family in the allohexaploid oil seed crop Crambe abyssinica Hochst. Mol. Breed. 32, 517–531. [Google Scholar]
- Cheng, J. , Zhu, L.‐H. , Salentijn, E.M.J. , Huang, B. , Gruber, J. , Dechesne, A. , Krens, F.A. , Qi, W. , Visser, R.G.F. and van Loo, E.N. (2013b) Functional analysis of the omega‐6 fatty acid desaturase (CaFAD2) gene family of the oil seed crop Crambe abyssinica . BMC Plant Biol. 13, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. , Salentijn, E.M.J. , Huang, B. , Denneboom, C. , Qi, W. , Dechesne, A.C. , Krens, F.A. , Visser, R.G.F. and van Loo, E.N. (2014) Detection of induced mutations in CaFAD2 genes by next‐generation sequencing leading to the production of improved oil composition in Crambe abyssinica. Plant Biotechnol. J., doi: 10.1111/pbi.12269. [DOI] [PubMed] [Google Scholar]
- Durrett, T.P. , Benning, C. and Ohlrogge, J. (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. [DOI] [PubMed] [Google Scholar]
- Graef, G. , LaVallee, B.J. , Tenopir, P. , Tat, M. , Schweiger, B. , Kinney, A.J. , Van Gerpen, J.H. and Clemente, T.E. (2009) A high‐oleic‐acid and low‐palmitic‐acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnol. J. 7, 411–421. [DOI] [PubMed] [Google Scholar]
- Guan, R. , Lager, I. , Li, X. , Stymne, S. and Zhu, L.H. (2013) Bottlenecks in erucic acid accumulation in genetically engineered ultrahigh erucic acid Crambe abyssinica . Plant Biotechnol. J. 12, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.L. , Snapp, A.R. and Lu, C.F. (2011) Identification of three genes encoding microsomal oleate desaturases (FAD2) from the oilseed crop Camelina sativa . Plant Physiol. Biochem. 49, 223–229. [DOI] [PubMed] [Google Scholar]
- Khadake, R.M. , Ranjekar, P.K. and Harsulkar, A.M. (2009) Cloning of a Novel Omega‐6 Desaturase from Flax (Linum usitatissimum L.) and Its Functional Analysis in Saccharomyces cerevisiae . Mol. Biotechnol. 42, 168–174. [DOI] [PubMed] [Google Scholar]
- Kinney, A.J. (1994) Genetic modification of the storage lipids of plants. Curr. Opin. Biotechnol. 5, 144–151. [Google Scholar]
- Knutzon, D.S. , Thompson, G.A. , Radke, S.E. , Johnson, W.B. , Knauf, V.C. and Krid, J.C. (1992) Modification of Brassica seed oil by antisense expression of a stearoyl‐acyl carrier protein desaturase gene. Proc Natl Acad Sci U S A. 89, 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L.Y. , Wang, X.L. , Gai, J.Y. and Yu, D.Y. (2007) Molecular cloning and characterization of a novel microsomal oleate desaturase gene from soybean. J. Plant Physiol. 164, 1516–1526. [DOI] [PubMed] [Google Scholar]
- Li, X.Y. , Ahlman, A. , Yan, X.F. , Lindgren, H. and Zhu, L.H. (2010) Genetic transformation of the oilseed crop Crambe abyssinica . Plant Cell Tiss. Organ Cult. 100, 149–156. [Google Scholar]
- Li, X.Y. , Ahlman, A. , Lindgren, H. and Zhu, L.H. (2011) Highly efficient in vitro regeneration of the industrial oilseed crop Crambe abyssinica . Ind. Crops Prod. 33, 170–175. [Google Scholar]
- Li, X.Y. , van Loo, E.N. , Gruber, J. , Fan, J. , Guan, R. , Frentzen, M. , Stymne, S. and Zhu, L.H. (2012) Development of ultra‐high erucic acid oil in the industrial oil crop Crambe abyssinica . Plant Biotechnol. J. 10, 862–870. [DOI] [PubMed] [Google Scholar]
- Li, X.Y. , Fan, J. , Gruber, J. , Guan, R. , Frentzen, M. and Zhu, L.H. (2013) Efficient selection and evaluation of transgenic lines of Crambe abyssinica . Front. Plant Sci. 4, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Singh, S.P. , Brubaker, C.L. , Sharp, P.J. , Green, A.G. and Marshall, D.R. (1999) Molecular cloning and expression of a cDNA encoding a microsomal omega‐6 fatty acid desaturase from cotton (Gossypium hirsutum). Aust. J. Plant Physiol. 26, 101–106. [Google Scholar]
- Liu, Q. , Singh, S.P. and Green, A.G. (2002) High‐stearic and High‐oleic cottonseed oils produced by hairpin RNA‐mediated post‐transcriptional gene silencing. Plant Physiol. 129, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Cao, S. , Zhou, X.‐R. , Wood, C. , Green, A. and Singh, S. (2013) Nonsense‐mediated mRNA degradation of CtFAD2‐1 and development of a perfect molecular marker for olol mutation in high oleic safflower (Carthamus tinctoris L.). Theor. Appl. Genet. 126, 2219–2231. [DOI] [PubMed] [Google Scholar]
- Martínez‐Rivas, J. , Sperling, P. , Lühs, W. and Heinz, E. (2001) Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal‐type and high‐oleic varieties of sunflower (Helianthus annuus L.). Mol. Breed. 8, 159–168. [Google Scholar]
- Mietkiewska, E. , Brost, J.M. , Giblin, E.M. , Barton, D.L. and Taylor, D.C. (2007) Cloning and functional characterization of the fatty acid elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv. Prophet. Plant. Biotechnol. J. 5, 636–645. [DOI] [PubMed] [Google Scholar]
- Millar, A.A. and Kunst, L. (1997) Very‐long‐chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. [DOI] [PubMed] [Google Scholar]
- Mozaffarian, D. , Katan, M.B. , Ascherio, A. , Stampfer, M.J. and Willett, W.C. (2006) Medical progress ‐Trans fatty acids and cardiovascular disease. N. Engl. J. Med. 354, 1601–1613. [DOI] [PubMed] [Google Scholar]
- Nguyen, H.T. , Silva, J.E. , Podicheti, R. , Macrander, J. , Yang, W. , Nazarenus, T.J. , Nam, J.‐W. , Jaworski, J.G. , Lu, C. , Scheffler, B.E. , Mockaitis, K. and Cahoon, E.B. (2013) Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotechnol. J. 11, 759–769. [DOI] [PubMed] [Google Scholar]
- Norden, A.J. , Gorbet, D.W. , Knauft, D.A. and Young, C.T. (1987) Variability in oil quality among peanut genotypes on the Florida breeding program. Peanut Sci. 14, 7–11. [Google Scholar]
- Okuley, J. , Lightner, J. , Feldmann, K. , Yadav, N. , Lark, E. and Browse, J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell, 6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Q. , Hu, Y. , Wei, R. , Zhang, Y. , Guan, C.Y. , Ruan, Y. and Liu, C.L. (2010) Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. [DOI] [PubMed] [Google Scholar]
- Pham, A.T. , Lee, J.D. , Shannon, J.G. and Bilyeu, K.D. (2010) Mutant alleles of FAD2‐1A and FAD2‐1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 10, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. (2000) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schuppert, G.F. , Tang, S. , Slabaugh, M.B. and Knapp, S.J. (2006) The sunflower high‐oleic mutant Ol carries variable tandem repeats of FAD2‐1, a seed‐specific oleoyl‐phosphatidyl choline desaturase. Mol. Breed. 17, 241–256. [Google Scholar]
- Sheng, Z.W. , Fen, L.Y. , Wu, Z.S. and Bao, X.X. (2006) Obtaining new germplast of Brassica napus with high oleic acid content by RNA interference and marker‐free transformation of Fad2 gene. J. Plant Physiol. Mol. Biol. 32, 665–671. [PubMed] [Google Scholar]
- Soldatov, K.I. (1976) Chemical mutagenesis in sunflower breeding. Proceedings of the Seventh International Sunflower Association, Vlaardingen, The Netherlands, pp. 352–357. Toowoomba, Australia: International Sunflower Association. [Google Scholar]
- Stalberg, K. , Ellerstrom, M. , Josefsson, L.G. and Rask, L. (1993) Deletion analysis of a 2S seed storage protein promoter of Brassica napus in transgenic tobacco. Plant Mol. Biol. 23, 671–683. [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk, P.A. , Hurlestone, C. , Singh, S.P. and Green, A.G. (2000) High‐oleic acid Australian Brassica napus and B. juncea varieties produced by co‐suppression of endogenous Δ12‐desaturases. Biochem. Soc. Trans. 28, 938–940. [PubMed] [Google Scholar]
- Stoutjesdijk, P.A. , Singh, S.P. , Liu, Q. , Hurlstone, C.J. , Waterhouse, P.A. and Green, A.G. (2002) hpRNA‐mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 129, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tat, M.E. , Wang, P.S. , Van Gerpen, J.H. and Clemente, T.E. (2007) Exhaust emissions from an engine fueled with biodiesel from high‐oleic soybeans. J. Am. Oil Chem. Soc. 84, 865–869. [Google Scholar]
- Tijsterman, M. , Ketting, R.F. and Plasterk, R.H.A. (2002) The genetics of RNA silencing. Annu. Rev. Genet. 36, 489–519. [DOI] [PubMed] [Google Scholar]
- Zaplin, E.S. , Liu, Q. , Li, Z.Y. , Butardo, V.M. , Blanchard, C.L. and Rahman, S. (2013) Production of high oleic rice grains by suppressing the expression of the OsFAD2‐1 gene. Funct. Plant Biol. 40, 996–1004. [DOI] [PubMed] [Google Scholar]
- Zhou, X.‐R. , Singh, S. , Liu, Q. and Green, A. (2006) Combined transgenic expression of Δ12‐desaturase and Δ12‐epoxygenase in high linoleic acid seeds leads to increased accumulation of vernolic acid. Funct. Plant Biol. 33, 585–592. [DOI] [PubMed] [Google Scholar]
- Zhou, X.‐R. , Green, A.G. and Singh, S.P. (2011) Caenorhabditis elegans delta 12‐desaturase FAT‐2 is a bifunctional desaturase able to desaturate a diverse range of fatty acid substrates at the Δ12 and Δ15 positions. J. Biol. Chem. 286, 43644–43650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.‐R. , Singh, S. and Green, A. (2013) Characterisation of the FAD2 gene family from Hiptage benghalensis: A ricinoleic acid accumulating plant. Phytochemistry, 92, 42–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison of crambe gene fragments used in RNAi constructs with related sequences. A. CaFAD2 (accession KP987201); B, CaFAD3 (accession KP987202); C, CaFAE1 (accession KP987203). Each nucleotide sequence is followed by an accession number. The plant species are: At, Arabidopsis thaliana; Bn, Brassica napus; Ca, Crambe hispanica subsp. abyssinica; Cf, C. filiformis; Cg, C. glabrata; Ch, C. hispanica subsp. hispanica; Ck, C. kralikii; Cm, C. maritime (ABF70315, ABF70316); Lu, Linum usitatissimum. Sequences are aligned with Vector NTi V11 (Life Technologies).
Figure S2 Nucleotide sequences used for RNAi constructs and their comparison to Arabidopsis sequences. A, FAD2; B, FAD3, C, FAE1 coding sequence; D, FAE1 3′‐UTR sequence.


