Abstract
The alphavirus nucleocapsid core is formed through the energetic contributions of multiple noncovalent interactions mediated by the capsid protein. This protein consists of a poorly conserved N-terminal region of unknown function and a C-terminal conserved autoprotease domain with a major role in virion formation. In this study, an 18-amino-acid conserved region, predicted to fold into an α-helix (helix I) and embedded in a low-complexity sequence enriched with basic and Pro residues, has been identified in the N-terminal region of the alphavirus capsid proteins. In Sindbis virus, helix I spans residues 38 to 55 and contains three conserved leucine residues, L38, L45, and L52, conforming to the heptad amino acid organization evident in leucine zipper proteins. Helix I consists of an N-terminally truncated heptad and two complete heptad repeats with β-branched residues and conserved leucine residues occupying the a and d positions of the helix, respectively. Complete or partial deletion of helix I, or single-site substitutions at the conserved leucine residues (L45 and L52), caused a significant decrease in virus replication. The mutant viruses were more sensitive to elevated temperature than wild-type virus. These mutant viruses also failed to accumulate cores in the cytoplasm of infected cells, although they did not have defects in protein translation or processing. Analysis of these mutants using an in vitro assembly system indicated that the majority were defective in core particle assembly. Furthermore, mutant proteins showed a trans-dominant negative phenotype in in vitro assembly reactions involving mutant and wild-type proteins. We propose that helix I plays a central role in the assembly of nucleocapsid cores through coiled coil interactions. These interactions may stabilize subviral intermediates formed through the interactions of the C-terminal domain of the capsid protein and the genomic RNA and contribute to the stability of the virion.
Alphaviruses are members of the Togaviridae family and have a well-defined virion organization. Their genome consists of a single-stranded positive-sense RNA of ∼12 kb (38). This genomic RNA is packaged within an icosahedral T=4 virus particle that contains (i) an internal nucleocapsid core (NC) surrounded by a host-derived lipid bilayer and (ii) a glycoprotein shell consisting of two transmembrane proteins, E1 and E2 (4, 26, 31). The assembly of the multilayered alphaviruses has been studied extensively both in vivo and in vitro (39, 40, 43) and is driven by energetic contributions of multiple repeating noncovalent interactions. The assembly of subviral intermediates occurs in distinct cellular compartments, as the NCs are formed in the cytoplasm and the two glycoproteins associate to form heterodimers and undergo processing and maturation in the endoplasmic reticulum and Golgi. This glycoprotein processing is complex, although discrete intermediates in the process have been identified (3, 28, 33). At the plasma membrane, glycoprotein spikes, each consisting of a trimer of E1-E2 heterodimers, can be found. These spikes oligomerize at the cell surface due to lateral interactions between the glycoproteins (9). Independently, the genomic RNA is encapsidated by the association of 240 capsid protein (CP) monomers in the cytoplasm to form the NC. These preformed NCs interact with the viral glycoproteins at the plasma membrane. The energy derived from the NC-glycoprotein interaction presumably drives the budding of the mature virus from the infected cell (21).
A combination of X-ray crystallographic data from the CPs of Sindbis virus (SINV) and Semliki Forest virus (SFV) and cryoelectron microscopy reconstructions of Ross River virus and SFV have highlighted possible residues in the C-terminal protease domain (residues 114 to 264, SINV numbering) that may be involved in intermolecular interactions during particle assembly (4, 6, 7, 26). However, these structural analyses have not provided information concerning the N-terminal region of the CP (residues 1 to 106). Biochemical and molecular genetic studies on SINV CPs have suggested that residues 76 to 132 contain important determinants that are involved in genomic RNA recognition (14, 30, 45). These residues have also been suggested to be involved in the formation of nucleic acid-bound CP dimers, based on cross-linking experiments in an in vitro core assembly system (40). Due to their highly positively charged character, residues 1 to 80 of the CP have been implicated in nonspecific interaction with the genomic RNA (43). In addition, deletion analysis of large segments of the N terminus of the SFV CP has indicated that the region between residues 11 and 63 (SFV numbering) is important for NC assembly (11), but the structural basis of this result has not been clarified.
In this report we describe a theoretical identification of a conserved stretch of 18 amino acids in the N-terminal region of the alphavirus CPs with predicted properties of an amphipathic coiled coil α-helix (helix I) facilitating CP oligomerization. A panel of mutants in helix I of the SINV CP has been characterized with respect to virus growth properties and virion assembly, as well as in vitro assembly reactions of the NC. The results obtained suggests that helix I contributes significantly to NC formation and stability, although it is not absolutely required for virus particle formation. Thus, helix I is a newly described structural determinant involved in alphavirus core assembly.
MATERIALS AND METHODS
Computer sequence analyses.
Amino acid sequences were derived from the protein database maintained at the National Center for Biotechnological Information (NCBI), National Institutes of Health (NIH). Sequence alignments were produced using the ClustalX program (42). These alignments were sent as input for the PhD program (34) to predict secondary structure. Proteins were also analyzed for the presence of potential coiled coil regions using the Coils program (25).
Viruses and cells.
All viruses were grown at 37°C in BHK-15 cells that were propagated in Eagle minimal essential medium (MEM) supplemented with 10% fetal bovine serum, unless otherwise noted. The construction and isolation of the wild-type parental virus Toto71 are described below. The full-length plasmid pToto71 was used for all mutageneses unless otherwise indicated.
Construction of pToto71.
To facilitate the rapid introduction of mutations, four new restriction sites were introduced into the cDNA region encoding helix I of the SINV CP. Although these restriction sites changed the nucleotide sequence of the cDNA, they preserved the amino acid sequence of the protein. Unique EcoRI and HindIII restriction sites from an M13mp18-derived clone of SINV (pSM810) (30) were used to shuttle the region of the CP nucleotides ([nt] 7334 to 9120) into pGEM3Zf(+) (Promega, Madison, Wis.). The resulting plasmid, pGEMCAP, and its derivatives were used in QuikChange site-directed mutagenesis (Stratagene, La Jolla, Calif.) to introduce four restriction sites (NgoMIV, MfeI, BssHII, and MluI) into the cDNA region encoding the helix. These sites correspond to nt 7742, 7776, 7795, and 7818 (SINV numbering), respectively. Following mutagenesis, unique BstEII (nt 7472) and XbaI (nt 8529) sites flanking the CP coding sequence were used to shuttle the DNA from the pGEMCAP derivative into the full-length SINV cDNA plasmid, pToto50 (32). The resulting plasmid was designated pToto71.
Construction of mutants.
Full-length cDNA clones containing deletions in the CP are designated pCPΔ(deleted residues), with Δ indicating a deletion. Proteins expressed from these clones are designated CPΔ(deleted residues). pCPΔ(35–58) was constructed by digestion of pToto71 with NgoMIV (nt 7742) followed by the fill-in of overhanging ends with the Klenow fragment of DNA polymerase. This fragment was subsequently digested with SpeI (nt 5262), and the resulting 2,480-bp fragment was gel purified. Similarly, pToto71 was digested with MluI (nt 7818) and blunt ended with Klenow enzyme. This fragment was then digested with SpeI (nt 5262), and the large 11,306-bp fragment was purified. The two fragments were ligated using standard protocols (35) to produce pCPΔ(35–58). pCPΔ(45–50) was constructed in a similar manner, using the MfeI (nt 7776) and BssHII (nt 7795) restriction sites in pToto71. However, to achieve this deletion, a three-fragment ligation was required due to the presence of multiple MfeI and BssHII restriction sites in pToto71. Therefore, the following fragments were prepared and ligated: a 734-bp fragment from BssHII-blunt (nt 7795) to XbaI (nt 8529), a 12,805-bp fragment from XbaI (nt 8529) to BstEII (nt 7472), and a 304-bp fragment from BstEII (nt 7472) to MfeI-blunt (nt 7776) to produce pCPΔ(45–50). pCPΔ(39–43), pCPΔ(59–63), and pCPΔ(71–75) were constructed by Kunkel oligonucleotide-directed mutagenesis as previously described (18, 29).
Full-length cDNA clones containing substitutions in the CP are designated pCP(original amino acid, position, substituted amino acid). Mutations at L45 were constructed by PCR-mediated mutagenesis using a 3′ oligonucleotide primer containing the desired mutation. This primer, which spanned nt 7767 to 7802 of pToto71, included the BssHII site at nt 7795. The 5′ primer spanned nt 7467 to 7484 of pToto71 and included the BstEII site at nt 7472. Together, these primers were used to amplify a 333-bp DNA fragment from pToto71 using Pfu DNA polymerase (Stratagene). This fragment was digested with BstEII and BssHII and then ligated to a 792-bp fragment previously digested with BssHII (nt 7795) and XbaI (nt 8529) from pToto71 and a 12,737-bp fragment digested with XbaI (nt 8529) and BstEII (nt 7472), also from pToto71. Prior to ligation, all fragments were purified following digestion using a QIAX II gel extraction kit (Qiagen, Valencia, Calif.). CP(L52D) was constructed by Kunkel oligonucleotide-directed mutagenesis as previously described (30).
The full-length cDNA clones containing the mutations were sequenced in the vicinity of the introduced changes, using Thermo Sequenase radiolabeled terminator cycle sequencing (Amersham Pharmacia Biotech, Piscataway, N.J.). Full-length plasmids were linearized with SacI; RNA was transcribed in vitro from these clones and transfected into BHK cells by using DEAE-dextran as previously described (18). Stocks of mutant virus were prepared following two rounds of plaque purification. Cytoplasmic RNA was isolated from each of the mutants; following reverse transcription-PCR, the products were sequenced to confirm the presence of the mutation.
Pulse-chase analysis of cytoplasmic extracts.
Cells (2.5 × 107) were electroporated with 10 μg of in vitro-transcribed RNA according to previously established protocols (22, 30). At 6 h postelectroporation, the cells were starved for methionine and treated with actinomycin D (1 μg/ml, final concentration). At 6.5 h postelectroporation, the cells were pulsed with [35S]methionine (50 μCi/ml; ICN Biomedicals, Costa Mesa, Calif.) for 15 min. The samples were then washed with MEM containing 100× unlabeled methionine and overlaid with MEM containing 5% dialyzed fetal bovine serum and 10× unlabeled methionine. The cells were incubated for an additional 30 or 60 min at 37°C, at which time cytoplasmic extracts were prepared from 2.6 × 106 cells and examined by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to previously established protocols (30). These extracts were also immunoprecipitated with anti-SINV CP antibodies.
Thermal inactivation studies.
Thermal inactivation studies were carried out as described elsewhere (37). Virus stocks were diluted in 1× phosphate-buffered saline supplemented with Ca2+, Mg2+, and 1% fetal bovine serum, to give a final concentration of 1,000 PFU/ml. Then 500 μl of virus was incubated at 56°C for up to 20 min. At time zero and every 5 min thereafter, 30 μl of sample was removed and added to 270 μl of the same buffer on ice. Following the last time point, virus titers were determined by plaque assay on BHK cells.
Accumulation of NCs.
NC accumulation studies were carried out as previously described (23, 30). Briefly, 10 μg of in vitro-transcribed RNA was electroporated into ∼2.5 × 107 BHK cells and then incubated at 37°C. At 5 h postelectroporation, cells were treated with actinomycin D to a final concentration of 1 μg/ml. Six hours postelectroporation, replicating RNA was labeled with [5,6-3H]uridine (40 μCi/ml; Amersham Pharmacia Biotech, Piscataway, N.J.). Cytoplasmic extracts from 2.6 × 106 cells were harvested at 12 h postelectroporation and layered onto a 22% freeze-thaw sucrose gradient, which was centrifuged at 32,000 rpm in an SW41 rotor (Beckman, Palo Alto, Calif.) for 2.5 h. The gradients were fractionated using an ECONO system (Bio-Rad, Melville, N.Y.) into 600-μl aliquots, and radioactivity from 50 μl of each fraction was counted in 8 ml of Cytoscint liquid scintillant (ICN Biomedicals, Costa Mesa, Calif.). Immunofluorescence was also carried out on these samples to determine electroporation efficiency as previously described (29).
In vitro assembly.
SINV CPs used for in vitro experiments are identified by the abbreviation CPΔ(deleted) or CP(original amino acid, position, substituted amino acid). The full-length wild type, as well as the mutant proteins used in these assays, starts at residue 19 and ends at the ultimate amino acid of the CP, residue 264. Proteins for the wild type and mutants were expressed and purified according to previously established protocols; in vitro assembly was also carried out as described previously (40). Briefly, equal volumes of wild-type or mutant CPs (400 μg/ml) were incubated with a 48-base nonspecific oligonucleotide (240 μg/ml) and incubated at room temperature for 30 min. Assembly of core-like particles (CLPs) was assayed by agarose gel, sucrose gradient sedimentation, and negative-stain electron microscopy. Inhibition experiments were performed by incubation of wild-type and mutant CPs at various wild type/mutant molar ratios between 1:0.2 and 1:5, followed by the addition of the 48-base oligonucleotide to maintain the same molar ratio of protein to nucleic acid as used in the wild-type assembly reactions (1:1). These reaction mixtures were then incubated at room temperature for 30 min and analyzed for CLP formation as described above.
RESULTS
A putative α-helix in the alphavirus CP.
Structural studies of the alphavirus CP have revealed a conserved chymotrypsin-like fold in the C-terminal half of the protein (residues 114 to 264 of the SINV CP) (7). Using a combination of the available X-ray crystal structures and the cryoelectron microscopy image reconstructions, Cheng et al. suggested a unique orientation of this domain into the capsomeres present in the NC structure (4). Additional crystallographic studies have shown that residues 106 to 113 of the SINV CP are involved in intermolecular contacts between adjacent molecules in the crystal lattice, although this arrangement was not seen in the fit obtained by the cryoelectron microscopy reconstructions (5, 21). Despite numerous attempts, the structural organization of the remaining N-terminal residues has not been resolved. It has been suggested that the bulk of this sequence resides on the interior of the NC, neutralizing the negative charge of RNA through its basic residues (43).
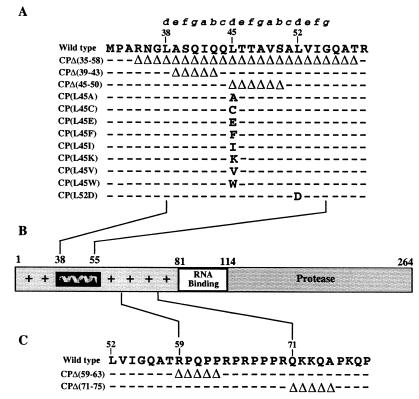
We have performed a comparative sequence analysis of the N-terminal region of the CP from a representative set of alphaviruses and identified a conserved 18-amino-acid region embedded in a low-complexity sequence enriched with basic and Pro residues. This conserved region occupies a variable position in different alphaviruses, being located between residues 38 and 55 in the SINV CP (Fig. 1A). A characteristic feature of this region is a periodical heptad organization of three leucine residues, L38, L45, and L52, the first of which is conserved among the New World alphaviruses and the two others among all alphaviruses. A similar periodicity of conserved leucine residues was previously found in α-helix leucine zipper motifs. These motifs have a high propensity to oligomerize through coiled coil interactions (24, 25). In leucine zippers and other coiled coil proteins, residues of each heptad unit are labeled from a to g, with leucine occupying the d position. The leucine at the d position along with hydrophobic residues at the a position form a coiled coil interface. Likewise, this CP N-terminal conserved region is strongly predicted to adopt an α-helix conformation (34), has a significant coiled coil potential (25), and thus can be labeled according to the coiled coil convention (Fig. 1A). The alphavirus helix (helix I) consists of an N-terminally truncated heptad and two complete heptads. A wheel model of helix I predicts that it has a hydrophobic face formed by leucine residues at the d position and β-branched residues at the a position (Fig. 1B). The opposite face of helix I is formed primarily by noncharged polar residues which are predicted to be solvent exposed. The structural similarity between the alphavirus helix I and characterized coiled coil domains suggest that helix I possesses properties typical of the coiled coil class proteins. In the context of the alphavirus life cycle, the expected ability to mediate oligomerization would make helix I central to the process of virus assembly and/or disassembly. To investigate the possible physiological roles of helix I and its conserved leucine residues experimentally, we have characterized a panel of SINV CP helix I mutants in a number of in vivo assays as well as in vitro assembly reactions of the NC.
FIG. 1.
Helix I in the alphavirus CP. (A) Sequence alignment of amino acid residues in helix I from indicated alphaviruses. Letters above the alignment identify amino acid positions in a coiled coil helix. Invariant residues are marked by asterisks; residues that are identical to the SINV amino acids are represented by dashes. (B) Coiled coil representation of helix I of SINV. The invariant leucine residues exclusively occupy the d position of the helix, and β-branched residues occupy the a position. Abbreviations (NCBI protein numbers): SINV (130579), Sindbis virus; AURAV (4240569), Aura virus; EEEV (130557), eastern equine encephalitis virus; WEEV (130581), western equine encephalitis virus; VEEV (130559), Venezuelan equine encephalitis; SPDV (4808420), salmon pancreatic disease virus; RRV (130571), Ross River virus; SFV (130577), Semliki Forest virus; CHIKV (576465), Chikungunya virus; ONNV (130568), O'nyong-nyong virus. Numbering corresponds to amino acid residues of the SINV CP.
Deletions in helix I compromise SINV replication.
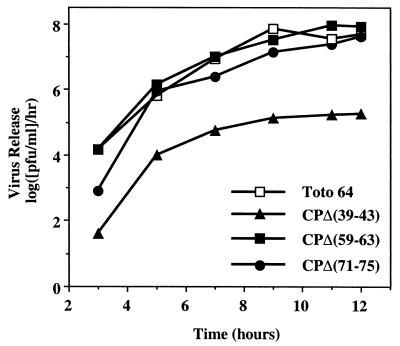
Previous studies of SFV showed that deletion of amino acids 66 to 77 (SINV numbering) of the CP did not affect virus replication, whereas deletion of amino acids 10 to 63 caused severe replication defects (11). To investigate whether the SINV CP could accommodate such changes, we have characterized five deletion mutants in the N-terminal region of SINV CP: three mutants with deletions of amino acids within helix I [pCPΔ(35–58), pCP(Δ39–43), pCPΔ(45–50)], and two mutants with deletions of amino acids downstream of helix I [pCPΔ(59–63) and pCPΔ(71–75)]. Viruses rescued from mutants CPΔ(59–63) and CPΔ(71–75) had plaque phenotypes in BHK cells 48 h posttransfection identical to that of the wild-type parental virus, Toto64 (Table 1). One of the mutants in helix I, pCPΔ(39–43), had a small plaque phenotype; the others [pCPΔ(35–58) and pCPΔ(45–50)] had very small plaque phenotypes that reverted rapidly during growth (Table 1 and data not shown). Results of one-step growth analysis of CPΔ(59–63) and CPΔ(71–75) were similar to those for the wild-type virus, whereas CPΔ(39–43) exhibited significantly less virus release (Fig. 2). Previous studies by Owen and Kuhn (30) and by Frolov and coworkers (12) had established that deletions in the CP could be made that had little or no effect on NC or virus assembly yet had profound effects on infectious virus released. In the work by Frolov and colleagues, a deletion in the N-terminal domain of the Ross River virus CP resulted in particles that were released but lacked RNA (12). In the work by Owen and Kuhn, a deletion was constructed in the CP that permitted RNA packaging, but specificity was lost, resulting in nonspecific encapsidation of RNA (30). Therefore, it was possible that in these studies, the decrease in infectious virus released was the result of defective particles and not the result of a decrease in particle assembly. To examine this possibility, equivalent amounts of infectious virus from CPΔ(39–43) and Toto71 were examined by immunoblotting using an anti-CP antiserum. The results indicate that equivalent levels of infectious virus are the result of equivalent numbers of CPs and presumably virus particles (data not shown). Hence, there is no change in the particle-to-PFU ratio for the CPΔ(39–43) virus. These results suggested that residues within the proposed helix I are involved in SINV assembly. Due to the extreme defect in replication, insufficient amounts of the CPΔ(35–58) and CPΔ(45–50) viruses were available for further characterization.
TABLE 1.
Deletions in helix I of the CP
| Genotype | Plaque phenotypea | NC accumulation (%)b |
|---|---|---|
| Wild type | lp | 100 |
| CPΔ(71–75) | lp | 100 |
| CPΔ(59–63) | lp | 85 |
| CPΔ(39–43) | sp | <2 |
| CPΔ(45–50) | vsp | <2 |
| CPΔ(35–58) | vsp | <2 |
Determined by plaque assay in BHK cells 48 h posttransfection. lp, large plaque (3.0 to 3.5 mm); sp, small plaque (1.5 to 2.5 mm); vsp, very small plaque (<1.5 mm).
Determined at 12 h after electroporation of in vitro-transcribed RNA.
FIG. 2.
One-step growth analysis of SINV CP deletion mutants in BHK cells. BHK cells were infected with the indicated virus at a multiplicity of infection of 1. Media were replaced every 30 min for the first 2 h and then every hour for 12 h. Supernatant was collected at the indicated times, and released virus was assayed by titration on BHK cell monolayers at 37°C. The results represent data obtained from a single experiment. The wild-type strain is Toto64.
Two out of the five deletion mutants characterized above [pCPΔ(35–58) and pCPΔ(45–50)] were generated using a newly constructed genetic background designated pToto71. It was produced from the original pToto50 full-length cDNA clone of SINV (32) by engineering four new restriction sites to facilitate mutagenesis in the helix I coding region. The nucleotide changes introduced into pToto71 (compared to pToto50) did not affect the open reading frame, the size of plaques recovered after transfection, or the kinetics of virus growth (data not shown). This suggests that pToto71 was phenotypically indistinguishable from the parental virus. pToto71 was used to generate all but one of the mutations in helix I described below.
Conserved leucines in helix I are important for SINV replication.
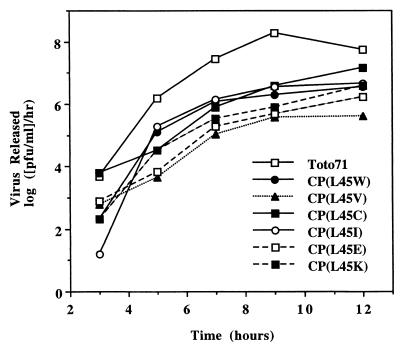
To gain further insights into the function of helix I, we extended our study to characterize a set of point mutations in helix I. Previous studies of various leucine zipper proteins have shown that leucine in the d position of an amphipathic helix is critical for determining the oligomerization properties of coiled coils (15) and that it contributes more than other hydrophobic residues to stabilizing these oligomeric protein structures (16, 27). To determine whether similar properties were associated with helix I residues, mutations were introduced at the conserved L45 and L52 residues occupying the d positions in helix I according to our model (Fig. 1). L52 was probed with one substitution (L52D), and L45 was replaced with amino acids varying in hydrophobicity and side chain geometry (Fig. 3). Following transfection of the mutant RNAs into BHK cells and subsequent rescue of mutant viruses, their plaque phenotypes and replication efficiencies were compared to those of the Toto71 wild-type virus (Table 2 and Fig. 4). As shown in Table 2, the results indicated that L45 mutants with hydrophobic amino acids such as Trp, Ile, or Cys displayed mid-sized plaque phenotypes; however, CP(L45V) displayed a small plaque phenotype in BHK cells, and its yield was reduced nearly 100-fold in the one-step growth analysis. CP(L45F) and CP(L45A) also displayed small plaque phenotypes and produced even less virus, with amounts insufficient for one-step growth analysis (Table 2 and data not shown). The L45 as well as the L52 mutant, carrying charged amino acids [CP(L45K), CP(L45E), and CP(L52D), respectively), displayed small plaque phenotypes and moderate growth defects when analyzed by one-step growth experiments (Fig. 4 and data not shown). The particle-to-PFU ratio for these mutants was also examined by immunoblotting as described above; in all cases, no significant change in the ratio was observed (data not shown). Collectively, the above results strongly indicate that L45 and L52 are important determinants of SINV growth, in accordance with their roles predicted by the model.
FIG. 3.
(A) Amino acid deletions and substitutions in helix I. The wild-type sequence corresponds to residues 32 to 59 of the SINV CP. Deletions (▵) and substitutions are indicated. Dashes indicate the wild-type sequence. Mutations were generated using the new restriction sites in the pToto71 cDNA clone of SINV as described in Materials and Methods. (B) Schematic diagram of the SINV CP. Residues 1 to 80 have a high degree of positive charge and are implicated in nonspecific recognition of the RNA, with the exception of residues 38 to 55, which are uncharged and form helix I. Residues 81 to 113 contain important determinants that are involved in specific recognition of the genomic RNA. Residues 114 to 264 form the protease domain and are also involved in capsomeric contacts as well as contacts with the cytoplasmic domain of E2. (C) Amino acid deletions outside helix I. Mutations were constructed by oligonucleotide-directed mutagenesis in the pToto63 cDNA clone of SINV as previously described (30).
TABLE 2.
Substitutions in helix I of the CP
| Genotype | Plaque phenotypea | NC accumulation (%)b |
|---|---|---|
| Wild type | lp | 100 |
| CP(L45W) | mp | 35 |
| CP(L45I) | mp | <2 |
| CP(L45C) | mp | <2 |
| CP(L45K) | sp | <2 |
| CP(L45E) | sp | <2 |
| CP(L45F) | sp | <2 |
| CP(L45A) | sp | <2 |
| CP(L45V) | sp | <2 |
| CP(L52D) | sp | <2 |
Determined by plaque assay in BHK cells 48 h posttransfection. lp, large plaque (3.0 to 3.5 mm); mp, medium plaque (2.5 to 3.0 mm); sp, small plaque (1.5 to 2.5 mm).
Determined at 12 h after electroporation of in vitro-transcribed RNA.
FIG. 4.
One-step growth analysis of SINV CP substitution mutants in BHK cells. BHK cells were infected with the indicated virus at a multiplicity of infection of 1. Media were replaced every 30 min for the first 2 h and then every hour for 12 h. Supernatant was collected at the indicated times, and released virus was assayed by titration on BHK cell monolayers at 37°C. The data represent averages of two independent experiments. The wild-type strain is Toto71.
Mutations in helix I disrupt NC assembly.
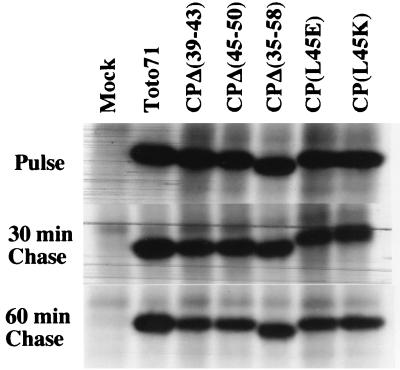
Previous studies showed that CP, along with the other structural proteins, is not essential for virus RNA translation and replication (47). Accordingly, our model predicts that the compromised SINV replication observed in the helix I mutants must be caused by a defect(s) in virion assembly and/or disassembly. To evaluate this prediction, we examined the protein synthesis and processing patterns of five of the mutants tested, three deletion mutants [pCPΔ(35–58), pCPΔ(39–43), pCPΔ(45–50)] and two point mutants [CP(L45E) and CP(L45K)]. In vitro-transcribed RNA from plasmids containing the mutations were electroporated into BHK cells; at 6.5 h postelectroporation, cells were labeled with [35S]methionine for 15 min and then chased for either 30 or 60 min in unlabeled medium. Cytoplasmic cell extracts from these mutants were prepared and analyzed by SDS-PAGE and autoradiography (Fig. 5). As expected, the mutants showed no significant defect in protein synthesis or proteolytic processing, although there was less CP in each of the mutants than in the wild type at the 60-min chase time point. Impairment of NC accumulation by mutations in helix I leads to the accumulation of free CP whose half-life may be different from that of the bound form. The observed difference in the amount of CP in the wild-type virus and helix I mutants (Fig. 6; see above) might thus be explained by an accelerated turnover of the mutant CPs.
FIG. 5.
Pulse-chase analysis of a subset of SINV mutants. BHK cells were electroporated with 10 μg of in vitro-transcribed RNA and incubated at 37°C. At 6 h postelectroporation, the cells were starved for methionine and treated with actinomycin D to a final concentration of 1 μg/ml. At 6.5 h postelectroporation, the cells were pulsed with [35S]methionine (50 μCi/ml) for 15 min. The samples were then washed with MEM–100× methionine and overlaid with MEM containing 5% dialyzed fetal bovine serum and 10× methionine. The cells were incubated for an additional 30 or 60 min at 37°C, at which time cytoplasmic extracts were prepared and examined by SDS-PAGE followed by autoradiography.
FIG. 6.
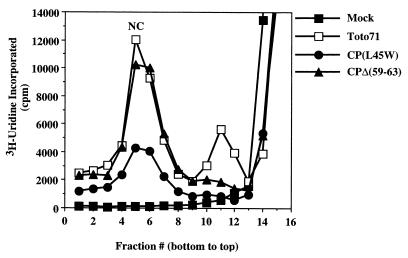
Accumulation of NC in the cytoplasm of a subset of mutants. In vitro-transcribed RNA from mutants and wild type were electroporated into BHK cells and incubated at 37°C. Five hours posttransfection, actinomycin D was added to the cells to a final concentration of 1 μg/ml, to block host cell transcription. At 6 h posttransfection, replicating RNA was labeled with [5,6-3H]uridine (40 μCi/ml). Cytoplasmic extracts of the infected cells were prepared at 12 h posttransfection and layered onto a 25% freeze-thaw sucrose gradient. The [3H]uridine profile of intracellular nucleocapsids following sucrose gradient centrifugation fractionation and scintillation counting is shown.
Alphavirus assembly is a multistep process which involves NC assembly in the cytoplasm of an infected cell prior to budding from the plasma membrane. The amount of accumulated NC in vivo can be monitored by sucrose gradient sedimentation (23, 30). We used this assay to characterize the helix I mutants. As shown in Fig. 6 and Tables 1 and 2, an accumulation of NCs was observed for the wild-type virus and deletion mutants outside helix I [CΔ(59–63) and CΔ(71–75)] but not for deletion or point mutants within helix I. The only exception was the CP(L45W) mutant, which showed a reduced level of NC accumulation but still produced approximately 35% of the wild-type level. Western blot analysis of cytoplasmic extracts following NC accumulation assays confirmed that the helix I mutants had no visible defects in protein translation or processing (data not shown). These results imply that helix I functions in the assembly of the SINV NC.
Single-site mutations in helix I diminish the stability of the virus particle.
The defect in NC accumulation observed in the helix I mutants suggested that the stability of the NCs could also be reduced in those mutants. This stability was analyzed by assaying the sensitivity of SINV to incubations at 56°C for short periods up to 20 min. As shown in Fig. 7, the helix I mutants were less stable at 56°C than the wild-type virus, with the surviving fraction of the mutants being 0.5 log to 1 log lower than for Toto71. Some of the curves appear biphasic, suggesting a two-step defect in stability. However, no pattern of amino acid substitutions responsible for this behavior is apparent. Similar results were obtained for the deletion mutants within helix I, whereas deletion mutants outside of the helix had stabilities similar to wild type (data not shown). These observations are consistent with the results of previous analyses (see above) and support our hypothesis. This effect of capsid mutations on glycoprotein function has previously been shown by the work of Burge and Pfefferkorn (2) and the study by Lee and coworkers (20), which suggest intimate cross talk between the inner NC and the outer glycoproteins.
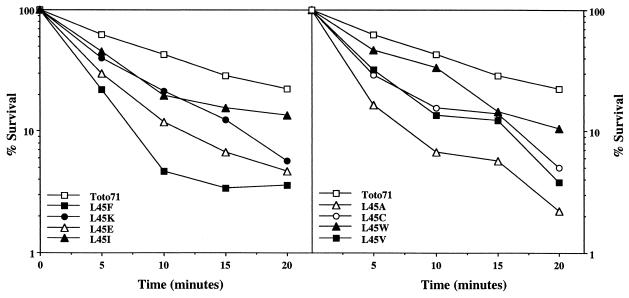
FIG. 7.
Thermal inactivation of SINV mutants. Thermal stability of mutant virus particles was determined by incubation of 500 PFU (diluted in phosphate-buffered saline supplemented with Ca2+ and Mg2+) at 56°C for the times indicated. Aliquots were taken at the indicated time points, and virus titers were determined by plaque assay on BHK cell monolayers. The results represent averages of three independent experiments. The data were divided into two panels to improve clarity.
The mutations in helix I confer a trans-dominant negative phenotype to in vitro assembly of core particles.
To study the helix I mutants in more detail, we analyzed a subset of these mutants by using a well-established in vitro assembly system for CLPs. CPs corresponding to residues 19 to 264 from CPΔ(45–50), CPΔ(35–58), CP(L45E), CP(L45W), and CP(L52D) were expressed in Escherichia coli according to previously established protocols (40). Assembly reactions were carried out with mutant CP (400 μg/ml) and oligonucleotide (240 μg/ml) and assayed by methods previously described for the wild-type protein (40, 44). As indicated in Table 3, all of the mutants tested except CP(L45W) failed to assemble CLPs in vitro. CP(L45W) assembled CLPs that sedimented similarly to wild type and were similar in morphology to wild-type CLPs when examined by negative-stain electron microscopy. However, many of the CP(L45W) particles appeared disrupted, presumably due to instability. The lack of CLP assembly observed for the majority of the mutants in helix I further supported the in vivo observations suggesting that mutants in helix I were defective in core assembly.
TABLE 3.
In vitro assembly of CP mutants
| Protein | Assembly | Inhibition of assemblya |
|---|---|---|
| Wild type | Yes | |
| CP(L45W) | Yes | NDb |
| CP(L45E) | No | Yes |
| CP(L52D) | No | Yes |
| CPΔ(45–50) | No | Yes |
| CPΔ(35–58) | No | Yes |
Determined in the presence of CP(19–264).
ND, not determined.
The effect of mutant CP on wild-type particle assembly was also analyzed (Table 3). Wild-type and mutant CPs were mixed at various molar ratios and incubated at room temperature for 30 min; then oligonucleotide was added to the mixture, maintaining the protein/nucleic acid ratio required for efficient assembly (40). The reaction mixtures were incubated at room temperature for an additional 30 min and assayed as described above. Wild-type CLP assembly was inhibited by mutant CP when present at a wild-type/mutant protein molar ratio of 1:1 or greater, with a majority of the protein and nucleic acid remaining trapped in the well when these reactions were analyzed on agarose gels. This was not observed in the wild-type assembly reactions. It is not clear whether inhibition of CLP assembly is caused by the mutant CP incorporating into wild-type CLPs, rendering them unstable, or by the mutant protein trapping intermediates in the assembly of CLPs. At present, no trapped intermediates have been identified.
DISCUSSION
The alphavirus CP contains helix I, a leucine zipper-type determinant of core assembly.
Alphavirus virions are complex oligomeric structures formed by multiple interactions between homologous and heterologous subunits (38). In this report, we provide molecular genetic and biochemical evidence that the N-terminal region of the CP contains a newly recognized structural determinant of alphavirus assembly. This determinant, helix I, is likely to stabilize the NC through coiled coil interactions between capsid monomers. The predicted hydrophobic intersubunit interface composed of conserved leucines and β-branched hydrophobic residues mediates the intersubunit contacts. This type of oligomer stabilization is characteristic of leucine zipper motifs, first observed in the bZIP class of transcription factors (19). Besides numerous cellular proteins, leucine zippers were previously implicated in virion formation in simian virus 40 and hepadnaviruses.
Leucine zippers consist of a series of heptad repeats (abcdefg)n that form an amphipathic α-helix. Leucines occupy the d position of the heptad ∼80% of the time; other hydrophobic residues including leucine usually occupy the a position. Together, the a and d positions define a hydrophobic interface involved in the oligomerization of coiled coil proteins (15, 19). In simian virus 40, two types of C-terminal interactions between VP1 proteins are present. In one type, the short C helix of VP1 mediates leucine zipper-type contacts between the β-β′ VP1 monomers through the interactions of isoleucine 301, leucine 304, and leucine 308 (13, 36). In the hepadnaviruses, four hydrophobic residues in the CP form two heptad repeats, with a glycine residue separating the two repeats into the a4b and a5 helices. Alanine scanning mutagenesis has shown that these residues are important for capsid assembly (46, 48). Studies on peptides derived from transcriptional factor GCN4 have demonstrated that when the a position is predominantly occupied by β-branched residues and the d position is occupied by leucines, as observed in helix I of the SINV CP, the formation of dimeric parallel coiled coils is strongly favored (15).
Mutant phenotypes are compatible with the helix I model.
We probed SINV helix I with deletion and point mutations and monitored their impact on virus assembly in in vivo and in vitro assays. Although only a few mutants were characterized in every assay used, results obtained consistently revealed a deleterious effect of the mutations on SINV assembly.
Mutants with a deletion of the entire helix I [CPΔ(35–58)] or a deletion of six amino acids spanning the first and second full heptad repeats [CPΔ(45–50)] were most detrimental to virus assembly. In contrast, point mutations and deletion of five amino acids spanning the truncated and first heptad repeats [CPΔ(39–43)] of helix I had visible but less pronounced effects on virus production. At present, the phenotypic difference between the deletion mutants CPΔ(39–43) and CPΔ(45–50) can be explained by the helix I model. First, the severity of the CPΔ(45–50) phenotype compared to the CPΔ(39–43) phenotype could be attributed to the deletion of an invariant residue, L45, which is deleted in the first but not the second mutant. Another, not mutually exclusive explanation is that the Δ39–43 deletion was better suited than the Δ45–50 deletion for heptad reorganization that must occur when residues are removed through deletion. Specifically, deletion of residues 39 to 43 would place T47 at the a position and S50 at the d position in CPΔ(39–43). In turn, deletion of residues 45 to 50 would place A51 at the d position and G55 at the a position of the helix in CPΔ(45–50). A pair of threonine and serine residues compared to a pair of glycine and alanine residues has a larger van der Waals radius and thus may be better suited to fill the void volume of the hydrophobic interface between CPs.
In an attempt to obtain a phenotype intermediate between those of the deletion mutants and the wild type, L45 was replaced with residues varying in hydrophobicity and van der Waals radii. These mutants displayed 50- to 100-fold less virus yield than the wild-type virus and were defective in NC assembly. Yet interestingly, there was a wide tolerance for diverse amino acids at this position. For instance, one-step growth analyses of these mutants indicated that tryptophan, isoleucine, and cysteine were well tolerated at this position, whereas phenylalanine, alanine, and valine caused more severe defects in replication; charged residues such as glutamate and lysine yielded an intermediate phenotype. Variant CP(L52D) had a phenotype similar to that of the CP(L45E) mutant. Analysis of in vivo NC assembly indicated that only the tryptophan mutant was able to form cytoplasmic cores, albeit only to 35% of wild-type levels, indicating that it was still compromised in NC stability.
In vitro assembly of a subset of mutants in helix I was also carried out so that NC assembly of these mutants could be studied as an isolated process without the interference of other cellular and viral processes. Among the mutants tested (Table 3), only CP(L45W) assembled CLPs in vitro, in agreement with results obtained in vivo. Furthermore, CLPs from CP(L45W) were identical in morphology to wild-type CLPs with the exception that a majority of them were disrupted in the negative-stained electron micrographs. This is possibly due to the lower stability of the cores formed by CP(L45W) compared to the wild type. The in vitro data further confirmed that mutations within helix I affected NC assembly.
Leucine 45 tolerance to substitutions and rescue by glycoproteins.
Analysis of mutants within helix I indicated that although leucine is the preferred residue at position 45, helix I has some flexibility in its interactions involving position 45. This permits the formation of unique interactions involving helix I that maintain functionality at the expense of stability. This hypothesis is supported by the fact that all mutants at position 45 were less stable than the wild type in thermal inactivation studies. Although these mutants were compromised in particle stability, as observed by the inability to form stable NC cores in the cytoplasm, they still released a limited number of virus particles. Therefore, it is possible that viruses with mutations within helix I require NCs to assemble in conjunction with the glycoproteins to provide added stability for particle assembly. Similar suggestions have been made in studies of SFV mutants, where deletion of residues CPΔ(105–118) and CPΔ(111–118) (SFV numbering) disrupted cytoplasmic core assembly but only mildly affected the release of virus particles (10). Likewise, the release of virus particles in the SINV deletion mutants may be the result of NCs that assembled in the presence of the glycoproteins that provided sufficient stability for particle formation to occur at the plasma membrane. This suggests the possibility of second-site suppressors that could map to glycoprotein E2. Based on the observed phenotypes of the helix I mutants, this is unlikely to have occurred in these stocks, although a search for revertants is in progress.
Role for helix I in alphavirus assembly.
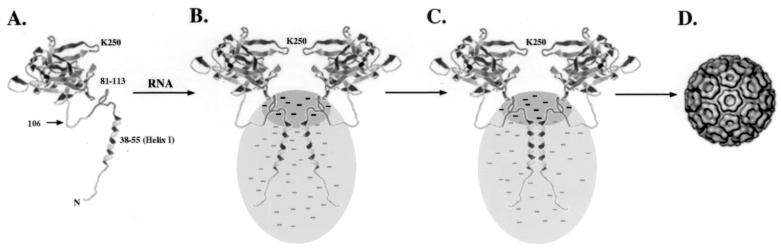
The analysis of helix I has suggested that it is important for NC assembly, perhaps functioning to stabilize CP-CP interactions during and after the assembly of the NC. Based on these data, we propose a model for the involvement of helix I in the assembly pathway of the NC (Fig. 8). In the organization of the alphavirus NC, residues 114 to 264 of the CP are proposed to lie within the capsomeres that project off the surface of the NC (4). The N-terminal 113 residues have been implicated in forming contacts between these capsomeres and must also project toward the interior of the particle and interact with the genomic RNA. As shown in Fig. 8A and B, it is proposed that the initial step in NC assembly involves the formation of nucleic acid-dependent CP dimers (41). This dimerization occurs through contacts that occur in the C-terminal two-thirds of the CP and involves CP-CP contacts that occur within residues 81 to 264, and CP-RNA contacts that occur through CP residues 81 to 113. This conclusion is supported by extensive in vivo and in vitro analyses that have demonstrated that residues 81 to 113 are important for specific recognition of the genomic RNA (14, 30, 45) and that in the presence of nucleic acid, residues 81 to 264 are capable of forming CLPs in vitro (T. L. Tellinghuisen, R. Perera, and R. J. Kuhn, submitted for publication). Yet, it has also been shown that these CLPs are unstable structures and need to be stabilized by a covalent chemical cross-link in order to be isolated. This suggests that the N-terminal one-third of the CP must play a crucial role in stabilizing these dimeric assembly intermediates. As shown in Fig. 8C, this additional stability can be afforded by coiled coil interactions mediated by helix I. This is supported by recent studies that have demonstrated that chemical cross-linking of lysine 250 of the CP could rescue the formation of CLPs using assembly-defective mutant proteins. Specifically, a mutant within helix I, CP(L52D), that was unable to form NCs was rescued and assembled into NCs when the dimer intermediates were stabilized through cross-linking (Tellinghuisen et al., submitted). These data strongly suggest that the chemical cross-link substitutes for helix I and that the helix functions in stabilizing dimer CP intermediates that proceed on to form NCs, which are then sufficiently stable to interact with the glycoproteins and bud out of the cell.
FIG. 8.
A model for coiled coil interactions in alphavirus assembly. (A) Monomer of SINV CP. Structural information is available only for residues 106 to 264 (upper portion of monomer). The remaining residues are shown as a random coil, and residues 38 to 55 are shown as an α-helix. (B) In the presence of RNA (grey area with negative charges), the CP forms nucleic acid-bound dimers initiated by interactions through residues 81 to 264. These dimers are transient but can be isolated in vitro from assembly defective CPs by chemical cross-linking methods (41). (C) In the absence of a chemical cross-link, coiled coil interactions mediated through helix I stabilize the nucleic-acid bound dimers; 120 copies of these dimers further oligomerize to form the NC. (D) Assembled NC structure observed from cryoelectron microscopy of intact virus particles.
In coiled coil helices, the e and g positions are commonly occupied by charged amino acids that form stabilizing salt bridges and confer dimerization specificity to the coiled coil (1, 8). Particularly, experimental data on bZIP proteins have suggested that the most favored amino acids at these positions are lysine, arginine, and glutamine (17). The rationale for this preference is that the methylene groups present in these long side chains increase the energetic contributions by the van der Waals forces and hydrophobic effects as they pack against the hydrophobic core of the helix. Surprisingly, among the alphaviruses, SINV and salmon pancreatic disease virus have no charged residues at the e and g positions of the helix, and the rest of the alphaviruses only have a limited number of charged amino acids at these positions. The significance of this amino acid distribution as well as the role of the hydrophilic face in helix I remain to be determined. Future studies should also clarify the role(s) of the absolutely conserved glutamine residue of helix I (SINV Q41), which is predicted to be solvent exposed.
ACKNOWLEDGMENTS
We acknowledge Suchetana Mukhopadhyay for help with data analysis and critical reading of the manuscript and Cyndy North for technical assistance. Critical discussions with Michael Rossmann, Thomas Smith, Cynthia Stauffacher, Christopher Jones, and Sergei Pletnev are also gratefully acknowledged. A.E.G. is grateful to Michael Rossmann for encouragement and hospitality during his stay at Purdue.
This research was supported by Public Health Service grant GM56279 from the National Institutes of Health. Additional funding from the Lucille Markey Foundation for structural studies is acknowledged. T.L.T. was supported in part by NIH biophysics training grant GM98296. A.E.G. (together with Michael G. Rossmann) was funded by a Cooperation in Applied Science and Technology Award from the National Academy of Sciences and with federal funds from the National Cancer Institute, NIH, under contract NO1-CO-56000.
REFERENCES
- 1.Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 2.Burge B W, Pfefferkorn E R. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology. 1966;30:204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- 3.Carleton M, Brown D T. Disulfide bridge-mediated folding of Sindbis virus glycoproteins. J Virol. 1996;70:5541–5547. doi: 10.1128/jvi.70.8.5541-5547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi H-K, Lee S, Zhang Y-P, McKinney B R, Wengler G, Rossmann M G, Kuhn R J. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J Mol Biol. 1996;262:151–167. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 6.Choi H-K, Lu G, Lee S, Wengler G, Rossmann M G. The structure of Semliki Forest virus core protein. Proteins Struct Funct Genet. 1997;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Choi H K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature (London) 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen C, Parry D A. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 9.Ekstrom M, Liuestrom P, Garoff H. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 1994;13:1058–1064. doi: 10.1002/j.1460-2075.1994.tb06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsell K, Suomalainen M, Garoff H. Structure-function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J Virol. 1995;69:1556–1563. doi: 10.1128/jvi.69.3.1556-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolov I, Frolova E, Schlesinger S. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J Virol. 1997;71:2819–2829. doi: 10.1128/jvi.71.4.2819-2829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia R L, Liddington R C. Structural biology of polymaviruses. In: Chui W, Burnett R M, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 187–208. [Google Scholar]
- 14.Geigenmüller-Gnirke U, Nitschko H, Schlesinger S. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J Virol. 1993;67:1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 16.Hodges R S, Zhou N E, Kay C M. Synthetic model proteins: contribution of hydrophobic residues and disulfide bonds to protein stability. Peptide Res. 1990;3:123–137. [PubMed] [Google Scholar]
- 17.Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn R J, Hong Z, Strauss J H. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landshultz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Ricker P D, Brown D T. The configuration of Sindbis virus envelope proteins is stabilized by the nucleocapsid protein. Virology. 1994;204:471–474. doi: 10.1006/viro.1994.1557. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Owen K E, Choi H K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 22.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez S, Yao J-S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 25.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 26.Mancini E J, Clarke M, Gowen B E, Rutten T, Fuller S D. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 27.Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 28.Mulvey M, Brown D T. Assembly of the Sindbis virus spike protein complex. Virology. 1996;219:125–132. doi: 10.1006/viro.1996.0229. [DOI] [PubMed] [Google Scholar]
- 29.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 30.Owen K E, Kuhn R J. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice C M, Strauss J H. Association of Sindbis virion glycoproteins and their precursors. J Mol Biol. 1982;154:325–348. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Bridging the protein sequence-structure gap by structure predictions. Annu Rev Biophys Biomol Struct. 1996;25:113–136. doi: 10.1146/annurev.bb.25.060196.000553. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 37.Strauss E G, Lenches E M, Strauss J H. Mutants of Sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology. 1976;74:154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- 38.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss J H, Strauss E G, Kuhn R J. Budding of alphaviruses. Trends Microbiol. 1995;3:346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 40.Tellinghuisen T L, Hamburger A E, Fisher B R, Ostendorp R, Kuhn R J. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tellinghuisen T L, Kuhn R J. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J Virol. 2000;74:4302–4309. doi: 10.1128/jvi.74.9.4302-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL:X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wengler G. The mode of assembly of alphavirus cores implies a mechanism for the disassembly of the cores in the early stages of infection. Arch Virol. 1987;94:1–14. doi: 10.1007/BF01313721. [DOI] [PubMed] [Google Scholar]
- 44.Wengler G, Boege U, Wengler G, Bischoff H, Wahn K. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology. 1982;118:401–410. doi: 10.1016/0042-6822(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 45.Wengler G, Würkner D, Wengler G. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191:880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 46.Wynne S A, Crowther R A, Leslie A G. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 47.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]
- 48.Yu M, Miller R H, Emerson S, Purcell R H. A hydrophobic heptad repeat of the core protein of woodchuck hepatitis virus is required for capsid assembly. J Virol. 1996;70:7085–7091. doi: 10.1128/jvi.70.10.7085-7091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]