Abstract
Peach latent mosaic viroid (PLMVd) is a circular RNA pathogen that replicates in a DNA-independent fashion via a rolling circle mechanism. PLMVd has been shown to self-ligate in vitro primarily via the formation of 2′,5′-phosphodiester bonds; however, in vivo the occurrence and necessity of this nonenzymatic mechanism are not evident. Here, we unequivocally report the presence of 2′,5′-phosphodiester bonds at the ligation site of circular PLMVd strands isolated from infected peach leaves. These bonds serve to close the linear conformers (i.e., intermediates), yielding circular ones. Furthermore, these bonds are shown to stabilize the replicational circular templates, resulting in a significant advantage in terms of viroid viability. Although the mechanism responsible for the formation of these 2′,5′-phosphodiester bonds remains to be elucidated, a hypothesis describing in vivo nonenzymatic self-ligation is proposed. Most significantly, our results clearly show that 2′,5′-phosphodiester bonds are still present in nature and that they are of biological importance.
Viroids are the smallest nucleic acid-based pathogens known to date (see references 15 and 33 for reviews). They are small (∼300 nucleotides [nt]), single-stranded, circular RNAs that infect higher plants and cause significant losses in agriculture. Viroids have been classified into two groups (A and B) based primarily on whether or not they possess the five typical structural domains found in the group B viroids. Further division among the 27 group B members depends on the sequence and length of the conserved central regions (7, 14, 33). Viroids that do not possess any kind of sequence or structural similarity with group B viroids have been classified as belonging to group A. The latter group includes the avocado sunblotch viroid, the peach latent mosaic viroid (PLMVd), and the chrysanthenum chlorotic mottle viroid (7, 15). All group A viroids possess hammerhead self-cleaving motifs and are proposed to replicate in chloroplasts (8, 11, 23). This localization has received recent support from the identification of a chloroplastic RNA polymerase that has the ability to replicate avocado sunblotch viroid (26). In contrast, group B viroids are localized in the nucleus, and RNA polymerase II appears to be responsible for their replication (4, 18, 30).
In infected cells, viroids replicate in a DNA-independent manner via a rolling circle mechanism that follows either a symmetric or an asymmetric mode (6, 8, 33, 36). The replicational intermediates of PLMVd, a 338-nt group A viroid that is the causal agent of peach latent mosaic disease (19), were recently studied by Northern blot analysis (8). PLMVd has been shown to replicate in a symmetric mode involving the accumulation of both circular and linear monomeric strands of both polarities (see Fig. 1). No multimeric conformer (i.e., intermediate) has been detected, indicating that both strands self-cleave efficiently via their hammerhead sequences. Moreover, it has been observed that monomeric linear RNAs accumulate at a high level compared to circular conformers (8). The latter observation might be the result of a rather inefficient ligation step (i.e., circularization).
FIG. 1.
PLMVd rolling circle replication. The polymerization, hammerhead-mediated self-cleavage, and ligation steps are numbered 1 and 4, 2 and 5, and 3 and 6, respectively. The polarities of the strands are indicated in parentheses. The insets show schematic secondary structures for the PLMVd strands of both polarities according to a published previously model (9). For each strand, the nucleotide sequence and structure adopted at the ligation site are illustrated.
Based on in vitro experiments, two different ligation mechanisms were proposed for the conversion of monomeric linear PLMVd (L-PLMVd) strands into circular conformers (10, 22). Initially, a wheat germ RNA ligase was shown to catalyze the circularization of unit-length L-PLMVd transcripts, as has been observed for potato spindle tuber viroid (PSTVd) (5, 22). Such enzymatic ligation leaves a 2′-phosphomonoester 3′,5′-phosphodiester bond as its signature, as has been observed for two viroid-like satellite RNAs (21). More recently, PLMVd has been shown to self-ligate in vitro primarily via the formation of 2′,5′-phosphodiester bonds (i.e., >96%; see references 10 and 22). However, in vivo the occurrence and necessity of this nonenzymatic mechanism are not evident (15). In order to elucidate the PLMVd circularization step, we have investigated the nature of the phosphodiester bonds found at the ligation site of natural PLMVd strands isolated from infected cells. We report unequivocal evidence of the presence of 2′,5′-phosphodiester bonds at the ligation site of circular PLMVd (C-PLMVd) strands. These bonds serve to close the linear conformers, yielding circular ones. Furthermore, these bonds are shown to prevent self-cleavage of the replicational circular templates, resulting in a significant advantage in terms of viroid viability.
MATERIALS AND METHODS
RNA isolation.
RNA samples were isolated from leaves harvested from different peach cultivars either infected or not infected with PLMVd (see Table 1) using three previously described extraction methods (8, 17). The first procedure used an RNeasy plant minikit (Qiagen) to prepare RNA from 50 to 100 mg of tissue as specified by the manufacturer. This procedure was also performed with the addition of 5 mM EDTA to all buffers. The second procedure used was the Tris-EDTA extraction method (8). The third procedure used was a modification of that involving polyethylene glycol (PEG) precipitation (17). All RNA samples were quantified by UV spectrophotometry and electrophoresed on 1.3% agarose gels in order to assess their quality. Dried RNA samples were stored at −70°C.
TABLE 1.
Detection of the 2′,5′-linked dinucleotides
| Peach cultivar or transcript | Infected plantsa | Extraction procedure | PLMVd bands isolated by 2D PAGEb | C/U detectedc |
|---|---|---|---|---|
| Cultivars | ||||
| Siberian C | + | RNeasy | C | + |
| L | − | |||
| Tris-EDTA | C | + | ||
| − | RNeasy | C | − | |
| L | − | |||
| Redhaven | + | PEG precipitationd | C | + |
| L | − | |||
| Harrow Beauty | + | RNeasyd | C | + |
| L | − | |||
| Agua | − | PEG precipitation | C | − |
| L | − | |||
| Synthetic transcripts | ||||
| 3′,5′ linked | C | − | ||
| L | − | |||
| L + Ce | − | |||
| 2′,5′ linked | C | + | ||
| L | − |
+, infected; −, not infected.
L and C, L-PLMVd and C-PLMVd, respectively.
+, detected; −, not detected.
This experiment was repeated in the presence of 5 mM EDTA in all extraction buffers, and the results were similar.
A mixture of L- and C-PLMVd that was not gel purified but was treated directly.
Preparation of synthetic PLMVd transcripts and RNA probes.
The synthesis and purification of both the L-PLMVd and the C-PLMVd transcripts used were performed as described previously (2, 3, 10). Briefly, in vitro transcription was performed using recombinant plasmid pPD1, which possesses two tandemly repeated PLMVd sequences (plus and minus strands). During transcription, RNAs of both polarities and possessing hammerhead sequences are produced and self-cleave efficiently, yielding 338-nt monomeric transcripts (i.e., L-PLMVd). For random internal labeling, 50 μCi of [α-32P]UTP (3,000 Ci/mmol; Amersham Life Science) was added to the transcription reaction. After transcription, DNase (RNase-free) treatment, and precipitation of the nucleic acids, they were dissolved in 20 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA in 0.5 volume of stop buffer (0.3% [wt/vol] each bromophenol blue and xylene cyanol, 10 mM EDTA [pH 7.5], and 97.5% [vol/vol] deionized formamide) and electrophoresed through a 5% (wt/vol) polyacrylamide gel in 100 mM Tris-borate (pH 8.3)–1 mM EDTA–7 M urea buffer. Nonradioactive transcripts were detected by UV shadowing and radioactive ones were detected by autoradiography, and both were excised, eluted, precipitated, purified by passage through Sephadex G-50 spin columns (Amersham), lyophilized, resuspended in water, quantitated either by absorbance spectrophotometry at 260 nm or Cerenkov counting, and stored dry at −70°C. Both the plus- and the minus-strand-specific riboprobes used for Northern blot hybridization were synthesized and purified in an analogous manner, except that (i) we used a StripEZ transcription kit (Ambion) to obtain probes that can be stripped under mild washing conditions so as to permit multiple probings of the same membrane and (ii) the transcription reactions were performed in the presence of 50 μCi of [α-32P]GTP (3,000 Ci/mmol; Amersham) (8).
C-PLMVd transcripts which include exclusively 3′,5′-phosphodiester bonds (3′,5′-C-PLMVd) were synthesized by use of a procedure based on the circularly permuted RNA strategy described previously (2). C-PLMVd transcripts including a 2′,5′-phosphodiester bond at the ligation site (2′,5′-C-PLMVd) were synthesized by in vitro self-ligation of L-PLMVd as described previously 10, 22). L-PLMVd (∼500,000 cpm for radioactive transcripts or 1 to 10 μg for nonradioactive ones) was resuspended in a final volume of 15 μl of 4 mM Tris-HCl (pH 7.9)–100 mM MgCl2, incubated overnight at 16°C, ethanol precipitated, washed, and lyophilized. The resulting pellets were purified by 5% polyacrylamide gel electrophoresis (PAGE) as described above.
RNA fractionation by 2D PAGE.
RNA samples (5 μg) purified from peach tree leaves were mixed with trace amounts (<1 fmol; 500 cpm) of in vitro-synthesized 32P-labeled C-PLMVd in a volume of 30 μl of either water or 10 mM Tris-HCl (pH 8.0)–1 mM EDTA, and 10 μl of loading buffer (0.25% each xylene cyanol and bromophenol blue and 50% glycerol) was added. The resulting samples were then fractionated on a 5% two-dimensional (2D) polyacrylamide gel, with the first dimension being run under native conditions (4°C) and the second being run under denaturing conditions (50°C in the presence of 7 M urea) as described previously (29). Radiolabeled C- and L-PLMVd transcripts were detected by autoradiography, excised, eluted, precipitated, purified by passage through Sephadex G-50 spin columns, and stored dry at −70°C.
Analysis of phosphodiester bonds.
The RNA species isolated with the radiolabeled C-PLMVd and L-PLMVd spots from the 2D polyacrylamide gels were treated according to a method used to identify modified nucleosides (32). The RNA samples were resuspended in 10 μl of 10 mM ammonium acetate (pH 4.5) containing 0.05 U of RNase T2 (Gibco BRL; producing 3′ nucleoside monophosphates [NMP]) and were then incubated overnight at 37°C. The resulting mixtures were lyophilized, and the nucleotides (and dinucleotides) were 5′ end labeled using T4 polynucleotide kinase (Pharmacia) in the presence of [γ-32P]ATP (3,000 Ci/mmol; Amersham; producing 5′,3′-nucleoside diphosphates) in a total volume of 11 μl according to the manufacturer's recommendations (Pharmacia). The excess [γ-32P]ATP was chased using 0.008 U of yeast hexokinase (Sigma) in the presence of 22 nmol of glucose for 10 min at 37°C. Nonradioactive ATP (2.5 mM) was added, and the mixtures were incubated at 37°C for an additional 10 min. This latter step was then repeated. After the mixtures were cooled on ice, they were lyophilized and resuspended in 20 μl of ultrapure water. One-half (10 μl) of each of the resulting samples was treated with nuclease P1 by the addition of 10 μl of 150 mM ammonium acetate (pH 5.3) and 2 μg of nuclease P1 (Boehringer Mannheim Biochemicals; producing 5′ NMP). The reaction mixture was incubated at 37°C for 3 h and then analyzed by 2D thin-layer chromatography (TLC) on cellulose plates with a UV indicator (Mandell) using the solvent system described by Silberklang et al. (32). Alternatively, 2D TLC was run using the solvent system described by Nishimura (27). Nonradioactive mononucleotides and dinucleotides (including cytidylyl-2′,5′-uridine [C/U]) were purchased (Sigma) and were also fractionated. The resulting dried plates were analyzed by UV shadowing and autoradiography, and the spots were quantified with a PhosphorImager (Molecular Dynamics). The radioactive spot corresponding to the dinucleotide was recovered as described by Houssier et al. (20). Dinucleotides were digested by alkaline hydrolysis or were 5′ end labeled using polynucleotide kinase in the presence of [γ-32P]ATP, and the resulting samples were analyzed directly on polyethyleneimine-cellulose with a UV indicator (Mandell) using the solvent system described previously (37).
Self-cleavage assay.
The mixtures of RNA (i.e., extracted RNA and added radioactive PLMVd; ∼3,000 cpm) were resuspended in a volume of 9 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA and were then heated at 90°C for 1 min prior to snap cooling on ice for 30 s. Self-cleavage of the transcripts was initiated by the addition of 1 μl of 1 M MgCl2, and the reaction mixtures were incubated at 37°C for 15 to 30 min. The reactions were quenched by the addition of 0.5 volume of stop buffer, and the mixtures were stored on ice until being denatured for 2 min at 65°C and purified by denaturing 5% PAGE.
Northern blot hybridization.
RNA samples (5 μg) isolated from both healthy and PLMVd-infected Siberian C cultivar leaves were analyzed by Northern blot hybridization using radiolabeled probes as described previously (8). RNA samples from healthy leaves mixed with 0.5 ng of synthetic nonradioactive 3′,5′-C-PLMVd transcripts (of positive polarity) were also analyzed. The RNA samples were resuspended in 10 mM Tris-HCl (pH 7.5)–0.1 mM EDTA, self-cleavage experiments were performed, and the mixtures were subjected to 5% PAGE analysis as described above. Nucleic acid transfer to nylon filters (Hybond N+; Amersham), prehybridization, and hybridization were performed as described previously (8). The resulting filters were analyzed by autoradiography or were exposed to a phosphor screen. All blots were successfully hybridized with both the plus- and the minus-strand PLMVd probes.
RESULTS
Detection of 2′,5′-phosphodiester bonds at the ligation sites of PLMVd.
The processing of PLMVd concatameric intermediates is mediated by RNA self-catalytic hammerhead structures that produce linear monomeric strands with 2′,3′-cyclic phosphate and 5′-hydroxyl termini (3, 19). After self-cleavage, precise refolding is required in order to bring the two ends (i.e., 3′-cytosine and 5′-uridine) into the close proximity required for ligation (Fig. 1, insets). Although the in vivo ligation mechanism remains unknown, if self-ligation does occur, C/U dinucleotides should be found in C-PLMVd strands isolated from infected leaves. Since 2′,5′-phosphodiester bonds, but not 3′,5′-phosphodiester bonds, are resistant to both RNase T2 and nuclease P1 (10, 13), we should be able to simply verify whether or not self-ligation is a potential mechanism of in vivo circularization for PLMVd.
In order to detect the position of natural C-PLMVd in 2D PAGE, trace amounts of synthetic 32P-labeled C-PLMVd containing exclusively 3′,5′-phosphodiester bonds (2) were added to RNA samples isolated from infected leaves (Siberian C cultivar). Figure 2A shows that C-PLMVd migrates off the diagonal, while a trace of L-PLMVd, produced by self-cleavage, is barely detectable on the diagonal. The off-diagonal band (C-PLMVd) was excised from the gels, and the RNA was eluted. The eluted RNA was digested with RNase T2, and the digestion products were labeled at their 5′ ends with 32P using polynucleotide kinase prior to 2D TLC analysis (Fig. 2B and C). The four NMP and a species with a migration consistent with that of a commercial nonradioactive C/U dinucleotide linked by a 2′,5′ bond were detected in the infected samples but not in the RNA sample extracted from healthy (i.e., uninfected) leaves. Similar results were obtained using other solvent systems (data not shown).
FIG. 2.
(A) Autoradiogram of 2D-PAGE analysis. RNA from PLMVd-infected (Siberian C cultivar) peach leaves mixed with in vitro-synthesized 32P-labeled C-PLMVd was fractionated on a 2D 5% polyacrylamide gel. The C- and L-PLMVd transcripts are indicated by arrows. The diagonal (dotted line), 5S RNA, and rRNA were revealed by silver staining. XC, xylene cyanol. (B and C) Typical 2D TLC autoradiograms corresponding to hydrolysis of the RNA species isolated from healthy and infected peach leaves, respectively. Nonradioactive mono- and dinucleotides were also cofractionated and are identified. The directions of migration are shown, and the origin (o) is indicated. These autoradiograms were overexposed in order to allow for the detection of any trace products.
The C/U identity of this species was subsequently confirmed (data not shown) by the methods described previously for the in vitro self-ligation experiments (10). Specifically, in the absence of commercial nucleotides, the radioactive spot corresponding to the dinucleotide was recovered and digested by alkaline hydrolysis, thereby releasing the NMP. The resulting samples were analyzed by one-dimensional TLC to confirm the presence of the 5′ 32P-cytosine. Alternatively, the NMP released by alkaline hydrolysis were 5′ end labeled with T4 polynucleotide kinase. In this case, a 5′ 32P-cytosine and a 5′ 32P-uridine were detected, confirming the presence of the 3′ uridine. The chromatographic mobilities of these species (C/U, C, and U) in both 1D and 2D TLC analyses excluded the possibility that the spot was a 2′-phosphomonoester 3′,5′-phosphodiester bond, like that found in two viroid-like satellite RNAs (21). Moreover, the migration positions of all dinucleotides in 2D TLC analyses are well known and different. No other dinucleotide migrated to the same position as C/U.
Similar experiments were performed using either synthetic transcripts or various RNA samples isolated from both infected and uninfected peach leaves (Table 1). The dinucleotide was detected only in two cases: (i) in C-PLMVd isolated from infected leaves, regardless of the extraction method used or the particular cultivar, and (ii) in synthetic C-PLMVd prepared by in vitro self-ligation. No L-PLMVd species isolated from infected leaves was observed to possess a dinucleotide. Together, the latter result and the C/U identity of the dinucleotide confirm that 2′,5′-phosphodiester bonds originate from the ligation sites. Furthermore, the 2′,5′-phosphodiester bonds observed by TLC are not the result of subsequent self-cleavage and self-ligation of synthetic 3′,5′-C-PLMVd or of the self-ligation of natural L-PLMVd during the extraction procedure, as no dinucleotide was detected in experiments with this transcript alone (i.e., 3′,5′-C-PLMVd; Table 1); in addition, extraction performed in the presence of radioactive synthetic L-PLMVd did not result in the formation of C-PLMVd (data not shown). It is doubtful that the dinucleotide originated from another RNA species that comigrated with C-PLMVd strands because, if this were the case, we should have detected the dinucleotide in the experiments performed with the off-diagonal bands from healthy leaves. Furthermore, TLC analysis of synthetic 3′,5′-C-PLMVd in the presence of RNA isolated from healthy leaves always required a longer exposure (>10 times) in order to produce NMP intensities comparable to those derived from samples from infected leaves. This difference suggests that for the samples from healthy leaves, the NMP were derived exclusively from the added synthetic 32P-C-PLMVd; therefore, natural C-PLMVd strands are likely alone or almost alone in having this electrophoretic mobility.
Proportion of 2′,5′-phosphodiester bonds.
We estimate, by PhosphorImager quantification, that more than 88% of the C-PLMVd strands isolated from infected leaves contain a 2′,5′ bond (eight chromatograms were analyzed in total, with the percentage varying from 62 to 100%). Even allowing for imprecisions in the quantification of the different TLC spots, the predominance of the 2′,5′ isomer versus the 3′,5′ isomer at the ligation site appears unequivocal. However, this value may be inflated due to the self-cleavage of natural 3′,5′-C-PLMVd strands during the manipulations which produced the linear conformers seen on the 2D gels. In order to rule out this possibility, we performed two experiments. In one, EDTA was included (up to 5 mM) in the RNA extraction steps in order to prevent hammerhead self-cleavage of 3′,5′-C-PLMVd. No detectable differences were observed (Table 1). In the other, the addition of 32P-labeled synthetic C-PLMVd transcripts with either a 3′,5′- or a 2′,5′-phosphodiester bond at the ligation site to the extraction mixture (i.e., in the ground leaf powder) resulted in less than 1% self-cleavage, as determined by PAGE analysis (Fig. 3, lanes 2 and 7, respectively). The results were identical in the presence of additional 5 mM EDTA in all buffers used in the extraction procedures (Fig. 3, lanes 3 and 8), as well as when the synthetic transcripts were added to the leaves prior to extraction. Also, greater-than-one-unit-length L-PLMVd transcripts (e.g., a dimer) submitted to the same treatments showed less than 2% self-cleavage (data not shown).
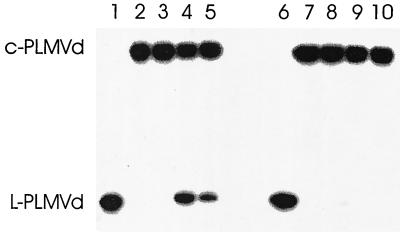
FIG. 3.
Autoradiogram of 5% PAGE analysis of self-cleavage experiments using either 2′,5′- or 3′,5′-C-PLMVd. Radioactive transcripts (C-PLMVd) were added to the ground leaf powder, and the RNA was extracted using the RNeasy plant minikit and incubated under self-cleavage conditions with or without prior heat denaturation and snap-cooling treatments. Lanes 1 and 6, untreated L-PLMVd (control); lanes 2 to 5 and 7 to 10, 3′,5′- and 2′,5′-C-PLMVd, respectively; lanes 2 and 7, C-PLMVd extracted and incubated under self-cleavage conditions without heat denaturation and snap-cooling steps; lanes 3 and 8, like lanes 2 and 7, except that the extraction was performed in the presence of additional 5 mM EDTA in all buffers; lanes 4 and 9, like lanes 2 and 7, except that the samples were heat denatured and snap-cooled prior to incubation under self-cleavage conditions; lanes 5 and 10, like lanes 4 and 9, except that the extraction was performed in the presence of additional 5 mM EDTA. Adjacent to the gel, the positions of the C-PLMVd and L-PLMVd transcripts are used as size references.
When equivalent samples were heat denatured and snap-cooled on ice and MgCl2 was added to a 100 mM final concentration prior to incubation at 37°C for 10 to 30 min (i.e., under optimal self-cleavage conditions), 3′,5′-C-PLMVd self-cleaved normally (i.e., ∼40%; Fig. 3, lane 4), showing that it had not lost its ability to self-cleave. In contrast, 2′,5′-C-PLMVd showed only a barely detectable level of self-cleavage (<0.5%; Fig. 3, lane 9), in agreement with the previous demonstration that 2′,5′-C-PLMVd transcripts (produced in vitro) are protected against self-cleavage (10). Finally, the mixture of synthetic transcripts and RNA samples extracted in the presence of 5 mM EDTA was also tested for self-cleavage activity. 3′,5′-C-PLMVd had a slightly reduced level of self-cleavage, most likely because the EDTA chelated a portion of the magnesium (Fig. 3, lane 5). For 2′,5′-C-PLMVd, no self-cleavage activity was detected (Fig. 3, lane 10). Most importantly, these controls demonstrated that the protocols used preserve the integrity of the RNA species, thereby confirming the predominance of 2′,5′-phosphodiester bonds.
The presence of a 2′,5′-phosphodiester bond prevents self-cleavage.
As mentioned above, it has previously been shown that 2′,5′-C-PLMVd transcripts (produced in vitro) are protected against self-cleavage (10). To verify if this property is shared by natural C-PLMVd, RNA extracted from infected leaves was incubated under self-cleavage conditions prior to Northern blot analysis using L-PLMVd strands of negative polarity as a probe (Fig. 4). While self-cleavage was observed with synthetic 3′,5′-C-PLMVd transcripts (Fig. 4, lanes 3 and 4), the ratio of L-PLMVd to C-PLMVd remained virtually identical for PLMVd isolated from infected leaves (lanes 5 and 6). Since the quantity of C-PLMVd is significantly smaller than that of L-PLMVd, it is not impossible that a small proportion self-cleaved without being detected by the method used. Identical results were obtained when positive-polarity L-PLMVd was used as a probe. These results are easily explained by the fact that the 2′-hydroxyl group adjacent to the hammerhead scissile phosphate is essential for the self-cleavage reaction (10); therefore, the presence of a 2′,5′-phosphodiester bond at this site prevents the self-cleavage of C-PLMVd and thus stabilizes this conformer. Moreover, the result in Fig. 4, lane 5, also shows that PLMVd strands accumulate predominantly as linear conformers rather than as circular ones and that no multimeric strands are detected. This result is in agreement with those published previously showing a larger proportion of linear conformers than of circular strands of PLMVd accumulated in infected cells (8). The accumulation of primarily L-PLMVd strands suggests that the ligation step is relatively inefficient.
FIG. 4.
Autoradiogram of a Northern blot with RNA samples incubated under both self-cleavage and non-self-cleavage conditions. Lane 1, synthetic, minus-strand, monomeric PLMVd without snap-cooling; lane 2, RNA sample from healthy leaves incubated without snap-cooling; lanes 3 and 4, RNA samples from healthy leaves mixed with 0.5 ng of synthetic 3′,5′-C-PLMVd transcripts (of positive polarity) and incubated without and with snap-cooling, respectively; lanes 5 and 6, RNA samples isolated from PLMVd-infected leaves either without or with prior self-cleavage, respectively. Adjacent to the gel, the positions of the C-PLMVd and L-PLMVd transcripts are used as size references, and ori indicates the origin. The panels including lanes 1 and 2 and lanes 5 and 6 were overexposed in order to allow for the detection of any trace products.
DISCUSSION
2′,5′-phosphodiester bonds in PLMVd.
Our results illustrate the presence of 2′,5′-phosphodiester bonds at the ligation site of C-PLMVd strands isolated from the leaves of infected peach plants. These data are in contrast to the report of the presence of a 2′-phosphomonoester 3′,5′-phosphodiester bond at the ligation site of two viroid-like satellite RNAs (21). In the latter study, the two viroid-like satellite RNAs were initially replicated in protoplasts from Nicotiana clevelandii in order to produce 32P-labeled RNA that was then purified by one-dimensional denaturing gel electrophoresis (i.e., fractionation strictly based on molecular weight). We wanted to use peach protoplasts but, unfortunately, this approach was not experimentally viable (data not shown). Consequently, RNA samples were isolated from infected peach leaves and purified by 2D electrophoresis involving fractionation based on molecular weight followed by one based on the conformation of the RNAs. As a result, the RNA species isolated from far off the diagonal in these gels correspond to circular 338-nt RNA molecules. The probability that these isolated RNA species might result from induction by viroid infection is infinitely small. The viroid infection modulates (i.e., results in the overexpression of) the expression of several genes compared to the normal state. However, most of these genes are expressed at a basal level; hence, if one produces an RNA with the same migration properties as circular PLMVd, we should be able to detect it in healthy samples. This was not the case (Table 1). Furthermore, the isolated RNA species were not a lariat because they would have produced a different junction after digestion (i.e., a three-way junction). Moreover, the possibility that a 2′,5′-linked oligo(A) might have comigrated with PLMVd is highly unlikely, as it has been shown that most oligonucleotides of this type are usually very small (i.e., ∼10 nt) (34). Also, it would be surprising for such an oligonucleotide to include a C/U dinucleotide and, more importantly, for this C/U dinucleotide to be the only 2′,5′-linked dinucleotide that it contains. Although the sequence context of this C/U dinucleotide is limited, the only conclusion that can be drawn is that it unequivocally originated from purified C-PLMVd. By itself, the demonstration that almost all C-PLMVd copies include the resistant bond constitutes additional evidence that PLMVd was the only RNA species found in the off-diagonal gel bands.
Since PLMVd multimeric strands do not accumulate in cells (8), the obligatory templates for replication are the circular conformers (i.e., C-PLMVd). Therefore, the 2′,5′-phosphodiester bonds most likely create these circular templates. The ability of reverse transcriptase to read through a 2′,5′ linkage has been estimated to be greater than 50% (10, 25), with the remaining cDNA synthesis being terminated. Therefore, the presence of a 2′,5′ linkage does not constitute an important obstacle to polymerase progression (10, 25); rather, it results in a significant advantage in terms of viroid viability, since it protects viroid integrity.
The mechanism responsible for the formation of these 2′,5′-phosphodiester bonds remains to be elucidated. One possibility is catalysis by a host 2′,5′ RNA ligase. Such an enzyme has been purified from Escherichia coli and demonstrated to ligate tRNA half-molecules in vitro, but the natural substrate(s) remains unidentified (1, 16). However, some observations suggest that the in vivo mechanism of ligation may be analogous to the nonenzymatic one observed in vitro (10). First, PLMVd accumulates predominantly and most likely replicates in chloroplasts (8); no chloroplastic gene encoding an RNA ligase has been identified, nor has such an activity been purified from chloroplasts (J. P. Perreault, personal communication). Second, self-ligation has minimal requirements other than the correct juxtaposition of the strand ends produced by hammerhead self-cleavage on a complementary strand (22). It has been shown that the self-ligation site is embedded in a very stable stem that is formed in solution and, most likely, also in vivo (9). Third, analysis of the sequence surrounding the ligation site suggests the existence of selective pressure in favor of the self-ligation mechanism, as it appears to be conserved (10). Thus, all self-ligation requirements are satisfied. Finally, in vitro self-ligation is relatively inefficient (i.e., ∼10%) (10), resulting in the accumulation of linear monomers. This effect correlates perfectly with the ratio of L- to C-PLMVd strands observed in vivo (i.e., >11:1) (8) (see also Fig. 3, lanes 5 and 6). Clearly, in vivo nonenzymatic self-ligation appears to be the most interesting hypothesis for PLMVd circularization. However, additional physical evidence in favor of this mechanism is required. Unfortunately, directed mutagenesis of the nucleotide(s) near the ligation site, in order to either reduce or abolish self-ligation in vivo, does not constitute an option, as it also inhibits the self-cleavage activity of the concatameric PLMVd strands. Nevertheless, if self-ligation, like self-cleavage, is indeed a part of PLMVd replication, then this process would be largely an RNA-based mechanism in which the only host component required is an RNA replicase.
Role of 2′,5′-phosphodiester bonds in nature.
RNA molecules are biological polymers composed of nucleotides covalently linked by phosphodiester bonds. Enzymes, such as RNA polymerases, that act on RNA use the ribose 3′-hydroxyl groups, instead of the 2′-hydroxyl groups, to create 3′,5′-phosphodiester bonds. There are some examples of 2′,5′-phosphodiester bonds in nature, but most of these are formed in RNA species that already possess a 3′,5′-phosphodiester bond and result in a “branched” 2′,5′ linkage like that observed in the lariats adopted by introns. Currently, the only “in-line” 2′,5′-phosphodiester bonds, which leave the 3′-hydroxyl group free, that have been retrieved from cells are the 2′,5′-oligoadenylates (see reference 34 for a review). Based on current knowledge, the discovery of any other in-line natural 2′,5′-phosphodiester bonds appear to be unlikely. In fact, the existence of these bonds has been essentially limited, so far, to in vitro experiments representative of prebiotic chemistry, in which the absence of the 3′,5′-phosphodiester bond has been attributed to the intrinsic lack of activity of the 3′-hydroxyl group (e.g., see references 24, 28, 31 and 35). For example, the nonenzymatic joining of both adenosine and short poly(A) oligonucleotides on poly(U) templates produces 97% 2′,5′-phosphodiester bonds (28, 35). Moreover, in vitro selection of RNA enzymes using pools of randomized sequences has led to the isolation of catalytic sequences that act as 2′,5′ RNA ligases (13).
The results reported here show that 2′,5′-phosphodiester bonds are still present in nature and that they are of biological importance. The finding of 2′,5′-phosphodiester bonds in an RNA species currently found in nature may have at least two consequences regarding molecular biology. On the one hand, since this type of phosphodiester bond is primarily associated with prebiotic chemistry, its presence in PLMVd supports the hypothesis that this viroid constitutes a model “relic” from the precellular world (12). On the other hand, regardless of the mechanism responsible for the production of the 2′,5′-phosphodiester bonds, it is tempting to speculate that these bonds are not restricted to PLMVd and that many other as-yet-unidentified RNA molecules possess this linkage. Since the requirements for self-ligation are minimal, this mechanism may be the source of the production of these bonds in various cellular RNAs. The 2′,5′-phosphodiester bonds could serve to stabilize the phosphate backbone at specific locations or even to create and modify an RNA molecule (i.e., RNA recombination). They have the potential to contribute significantly to biology.
ACKNOWLEDGMENTS
We thank D. A. Thompson for RNA samples and our laboratory colleagues for critical comments and helpful suggestions.
This research was supported by grants from the Natural Sciences and Engineering Research Council (NSERC, Canada) and Fonds pour la Formation des Chercheurs et l'Aide à la Recherche (FCAR; Québec, Québec, Canada) to J.-P.P. F.C. was the recipient of an NSERC studentship. J.-P.P. is a Medical Research Council (MRC, Canada) scholar.
REFERENCES
- 1.Arn E A, Abelson J N. The 2′,5′ RNA ligase of Escherichia coli. Proc Natl Acad Sci USA. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- 2.Beaudry D, Perreault J P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudry D, Bussière F, Lareau F, Lessard C, Perreault J P. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonfiglioli R G, Webb D R, Symons R H. Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J. 1996;9:457–465. [Google Scholar]
- 5.Branch A D, Robertson H D. Cell-free circularization of viroid progeny RNA by an RNA ligase from wheat germ. Science. 1982;217:1147–1149. doi: 10.1126/science.217.4565.1147. [DOI] [PubMed] [Google Scholar]
- 6.Branch A D, Robertson H D. A replication cycle for viroids and other small infectious RNAs. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 7.Bussière F, Lafontaine D, Perreault J P. Compilation and analysis of viroid and viroid-like RNA sequences. Nucleic Acids Res. 1996;24:1793–1798. doi: 10.1093/nar/24.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussière F, Lehoux J, Thompson D A, Skrzeczkowski L J, Perreault J P. Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J Virol. 1999;73:6353–6360. doi: 10.1128/jvi.73.8.6353-6360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bussière F, Ouellet J, Côté F, Lévesque D, Perreault J P. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J Virol. 2000;74:2647–2654. doi: 10.1128/jvi.74.6.2647-2654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Côté F, Perreault J P. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J Mol Biol. 1997;273:533–543. doi: 10.1006/jmbi.1997.1355. [DOI] [PubMed] [Google Scholar]
- 11.Daros J A, Marcos J F, Hernandez C, Flores R. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diener T O. Circular RNAs: relics of precellular evolution? Proc Natl Acad Sci USA. 1989;86:9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekland E H, Szostak J W, Bartel D P. Structurally complex and highly active RNA ligase derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 14.Flores R, Randles J W, Bar-Joseph M, Diener T O. A proposed scheme for viroid classification and nomenclature. Arch Virol. 1998;143:623–629. doi: 10.1007/s007050050318. [DOI] [PubMed] [Google Scholar]
- 15.Flores R, Navarro J A, Pena M, Navarro B, Ambros S, Vera A. Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group. Biol Chem. 1999;380:849–854. doi: 10.1515/BC.1999.104. [DOI] [PubMed] [Google Scholar]
- 16.Greer C L, Javor B, Abelson J. RNA ligase in bacteria: formation of a 2′,5′ linkage by an E. coli extract. Cell. 1983;33:899–906. doi: 10.1016/0092-8674(83)90032-6. [DOI] [PubMed] [Google Scholar]
- 17.Hadidi A, Giunchedi L, Schamloul A M, Poggi-Pollini C, Amer M A. Occurrence of peach latent mosaic viroid in stone fruits and its transmission with contaminated blades. Plant Dis. 1997;81:154–158. doi: 10.1094/PDIS.1997.81.2.154. [DOI] [PubMed] [Google Scholar]
- 18.Harders J, Lukacs N, Robert-Nicoud M, Jovin J M, Riesner D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989;8:3941–3949. doi: 10.1002/j.1460-2075.1989.tb08577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houssier C, Degee P, Nicoghosian K, Grosjean H. Effect of uridine dethiolation in the anticodon triplet of tRNA(Glu) on its association with tRNA(Phe) J Biomol Struct Dynam. 1988;5:1259–1266. doi: 10.1080/07391102.1988.10506468. [DOI] [PubMed] [Google Scholar]
- 21.Kiberstis P A, Haseloff J, Zimmern D. 2′-Phosphomonoester, 3′-5′-phosphodiester bond at a unique site in a circular viral RNA. EMBO J. 1985;4:817–822. doi: 10.1002/j.1460-2075.1985.tb03703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine D, Beaudry D, Marquis P, Perreault J P. Intra- and intermolecular nonenzymatic ligations occur within transcripts derived from the peach latent mosaic viroid. Virology. 1995;212:705–709. doi: 10.1006/viro.1995.1528. [DOI] [PubMed] [Google Scholar]
- 23.Lima M I, Fonseca M E N, Flores R, Kitajima E W. Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch Virol. 1994;13:385–390. doi: 10.1007/BF01379142. [DOI] [PubMed] [Google Scholar]
- 24.Lohrmann R, Orgel L E. Efficient catalysis of polycytidylic acid-directed oligoguanylate formation by Pb2+ J Mol Biol. 1980;142:555–567. doi: 10.1016/0022-2836(80)90263-6. [DOI] [PubMed] [Google Scholar]
- 25.Lorsch J R, Bartel D P, Szostak J W. Reverse transcriptase reads through a 2′,5′-linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 1995;23:2811–2814. doi: 10.1093/nar/23.15.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro J A, Vera A, Flores R. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology. 2000;268:218–225. doi: 10.1006/viro.1999.0161. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura S. Chromatographic mobilities of modified nucleotides. In: Schimmel P R, Söll D, Abelson J N, editors. Transfer RNA: structure, properties, and recognition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1979. pp. 547–552. [Google Scholar]
- 28.Renz M, Lohrmann R, Orgel L E. Catalysis for the polymerization of adenosine cyclic 2′,3′-phosphate on a poly (U) template. Biochim Biophys Acta. 1971;240:463–471. doi: 10.1016/0005-2787(71)90703-9. [DOI] [PubMed] [Google Scholar]
- 29.Roy G, Mercure S, Beuvon F, Perreault J P. Characterization of stable RNAs from the resected intestinal tissues of individuals with either Crohn's disease or ulcerative colitis. Biochem Cell Biol. 1997;75:789–794. doi: 10.1139/o97-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher J, Sänger H L, Riesner D. Subcellular localization of viroids in highly purified nuclei from tomato leaf tissue. EMBO J. 1983;2:1549–1555. doi: 10.1002/j.1460-2075.1983.tb01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharmeen L, Kuo M Y, Taylor J. Self-ligating RNA sequences on the antigenome of human hepatitis delta virus. J Virol. 1989;63:1428–1430. doi: 10.1128/jvi.63.3.1428-1430.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silberklang M, Gillum A M, RajBhandary U L. Use of in vitro32P labeling in the sequence analysis of non-radioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 33.Symons R H. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trujillo M A, Roux D, Fueri J P, Samuel D, Cailla H L, Rickenberg H V. The occurrence of 2′,5′ oligoadenylates in Escherichia coli. Eur J Biochem. 1987;169:167–173. doi: 10.1111/j.1432-1033.1987.tb13594.x. [DOI] [PubMed] [Google Scholar]
- 35.Usher D A, McHale A H. Nonenzymatic joining of oligoadenylates on a polyuridylic acid template. Science. 1976;192:53–54. doi: 10.1126/science.1257755. [DOI] [PubMed] [Google Scholar]
- 36.Warrilow D, Symons R H. Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch Virol. 1999;144:2367–2375. doi: 10.1007/s007050050650. [DOI] [PubMed] [Google Scholar]
- 37.Yang J H, Sklar P, Axel R, Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995;374:77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]