ABSTRACT
Archaea are widespread in the environment and play fundamental roles in diverse ecosystems; however, characterization of their unique biology requires advanced tools. This is particularly challenging when characterizing gene function. Here, we generate randomly barcoded transposon libraries in the model methanogenic archaeon Methanococcus maripaludis and use high-throughput growth methods to conduct fitness assays (RB-TnSeq) across over 100 unique growth conditions. Using our approach, we identified new genes involved in nutrient utilization and response to oxidative stress. We identified novel genes for the usage of diverse nitrogen sources in M. maripaludis including a putative regulator of alanine deamination and molybdate transporters important for nitrogen fixation. Furthermore, leveraging the fitness data, we inferred that M. maripaludis can utilize additional nitrogen sources including ʟ-glutamine, ᴅ-glucuronamide, and adenosine. Under autotrophic growth conditions, we identified a gene encoding a domain of unknown function (DUF166) that is important for fitness and hypothesize that it has an accessory role in carbon dioxide assimilation. Finally, comparing fitness costs of oxygen versus sulfite stress, we identified a previously uncharacterized class of dissimilatory sulfite reductase-like proteins (Dsr-LP; group IIId) that is important during growth in the presence of sulfite. When overexpressed, Dsr-LP conferred sulfite resistance and enabled use of sulfite as the sole sulfur source. The high-throughput approach employed here allowed for generation of a large-scale data set that can be used as a resource to further understand gene function and metabolism in the archaeal domain.
IMPORTANCE
Archaea are widespread in the environment, yet basic aspects of their biology remain underexplored. To address this, we apply randomly barcoded transposon libraries (RB-TnSeq) to the model archaeon Methanococcus maripaludis. RB-TnSeq coupled with high-throughput growth assays across over 100 unique conditions identified roles for previously uncharacterized genes, including several encoding proteins with domains of unknown function (DUFs). We also expand on our understanding of carbon and nitrogen metabolism and characterize a group IIId dissimilatory sulfite reductase-like protein as a functional sulfite reductase. This data set encompasses a wide range of additional conditions including stress, nitrogen fixation, amino acid supplementation, and autotrophy, thus providing an extensive data set for the archaeal community to mine for characterizing additional genes of unknown function.
KEYWORDS: archaea, transposons, metabolism
INTRODUCTION
Archaea are abundant in diverse habitats and play essential roles in ecosystem function (1) such as carbon and nitrogen cycling. Despite their ubiquity, basic aspects of archaeal physiology are still uncharacterized. To address these gaps, high-throughput methods are needed to rapidly assign putative gene function and characterize metabolic pathways.
The study of model organisms is key to overcoming the challenges associated with characterizing archaeal physiology. Methanococcus maripaludis is a model (2) with well-established genetic tools for understanding archaeal biology, methanogenesis, and diazotrophy. However, even in the relatively small genome (1.7 Mb) of this well-studied organism, ~35% of encoded proteins have RefSeq annotations of either “hypothetical protein” or “membrane protein” or belonging to a family rather than having specific functional assignments.
In the past decade, the development of randomly barcoded transposon site sequencing (RB-TnSeq) has facilitated the rapid characterization of gene function for a variety of cellular processes in diverse microorganisms (3, 4). RB-TnSeq is a variation of TnSeq where unique DNA sequences, or barcodes, are inserted alongside transposons. Thus, RB-TnSeq can be used to conduct comparable experiments to those classically done with TnSeq, in which mutant fitness in a control condition is compared to that of an experimental condition, to identify genes with significant phenotypes. This can be done at a fraction of the effort due to the simplicity of barcode sequencing (BarSeq). Previous work has used classical transposon site sequencing to characterize the essential genes in M. maripaludis strain S2 (5). However, this library was challenging to use to characterize gene fitness for non-essential genes in different growth environments in high throughput. To overcome this challenge, we developed the RB-TnSeq library that takes advantage of barcode sequencing for inexpensive detection of condition-specific phenotypes.
We generated RB-TnSeq libraries for two M. maripaludis strains, S2 and JJ. Libraries were generated in two strains to account for a range of genotypes. The genomes of JJ and S2 are 95% identical (two-way average nucleotide identity) (6, 7); however, they have major phenotypic differences. For example, strain JJ forms biofilms and can take up DNA via a natural transformation pathway in laboratory conditions while strain S2 exhibits more reliable growth in nitrogen fixing conditions. We also report methods for high-throughput cultivation and fitness profiling of these libraries to generate 536 genome-wide fitness assays representing 112 unique growth conditions, with replicates. Fitness assays were conducted across a range of autotrophic, diazotrophic, nutrient rich, and stress conditions with either H2 or formate as the electron donor. From this large data set, we identified novel genes of importance for carbon, nitrogen, and sulfur metabolism.
RESULTS
Generation of RB-TnSeq libraries
We generated barcoded transposon libraries in both M. maripaludis strain S2 and strain JJ wild-type backgrounds using a barcoded variant of a mariner transposon. In this study, we focus on the analysis and discussion of the M. maripaludis strain S2 library as strain S2 is the model used for many aspects of archaeal biology and metabolism including determination of essential genes (5), diazotrophy (8–10), and H2 and formate oxidation for methanogenesis (11–13). Data from M. maripaludis strain JJ fitness assays were used to confirm the results from M. maripaludis strain S2 when feasible.
Since M. maripaludis S2 is a polyploid (14), we observed insertions in essential genes. The insertion frequency of essential genes was previously analyzed by Sarmiento et al. using Tn5 mutagenesis (5). RB-TnSeq normalization approaches assume that most genes will not have a change in fitness in most conditions, which is not the case for essential genes. Thus, we excluded the previously identified essential genes from our analysis to streamline normalization and focus on conditionally essential phenotypes. The list of essential genes excluded is in the supplemental data set, and insertion data for the excluded genes are available on Figshare. See Materials and Methods for further detail.
For M. maripaludis strain S2, we used Tn-Seq to identify 38,887 unique barcodes that mapped ≥10 times and linked them to 29,150 unique insertion sites distributed across the genome (Fig. S1). Of these barcoded insertions, 21,269 were within the central 10–90% of a gene accounting for insertions in 1,615 of the 1,741 predicted protein-coding genes. Among the genes with transposon insertions, the average number of strains per gene is 13.1, with a median of 8 strains, and a mean coverage of 284.41 reads per million mapped reads (Table 1). For strain JJ, we identified 81,398 unique barcodes at 50,146 insertion locations resulting in insertions in 1,503 of the 1,796 predicted protein-coding genes (Table 1).
TABLE 1.
RB-TnSeq library quality statistics for M. maripaludis strains S2 and JJ
| Feature | Strain S2 | Strain JJ |
|---|---|---|
| Unique barcodes | 38,887 | 81,398 |
| Insertion locations | 29,150 | 50,146 |
| No. of protein-coding genes hit/total genes | 1,615/1,741 | 1,503/1,796 |
| Mean (median) no. of strains per protein-coding gene | 13.1 (8.0) | 20.0 (31.0) |
To take advantage of the throughput and simplicity of RB-TnSeq, we normalized the basal cultivation medium and optimized growth in high throughput (Fig. S2 and S3). Through incubation of 1 mL cultures in 96-well deep-well blocks in high-pressure incubation vessels, we were able to conduct fitness assays in over 100 unique conditions with biological replicates. Our goal with using this approach was to conduct assays with each library in both H2 and formate oxidizing conditions in parallel. The only past high-throughput growth assay in M. maripaludis was conducted in formate oxidizing conditions (15).
In an RB-TnSeq assay, each gene’s fitness is an estimate of the log2 fold change of the abundance in mutants of that gene during growth. It is calculated by comparing the read counts from the experimental sample (after growth) to the initial "Time 0" sample used for inoculation. Because it is a log2 change, gene fitness should not be affected by variations in the number of insertions per gene or in the initial abundance of insertions. To compute gene fitness values, we first calculated the fitness of each strain as the normalized log2 fold change in barcode counts. Then, we calculated a weighted average of the fitness values for relevant strains for each gene and normalized gene fitness by chromosomal position (3). This normalized value is referred to as the fitness value henceforth. We also calculated a t-like test statistic that gives an estimate of how significant a genes measurement is, with |t| > 4 being statistically significant (3).
Between the two strains and the 536 genome-wide fitness assays performed, we identified 189 genes with specific phenotypes. Specific phenotypes are defined as genes that have a strong phenotype (|fitness| > 1, |t| > 5) in a small subset of conditions tested (<5% conditions) and lack a phenotype (|fitness| < 1) in other conditions. Henceforth, we aim to highlight the phenotypes that we found the most impactful and/or surprising and highlight the utility that we hope this data set will provide to the archaeal research community.
Identification of genes important for autotrophic growth
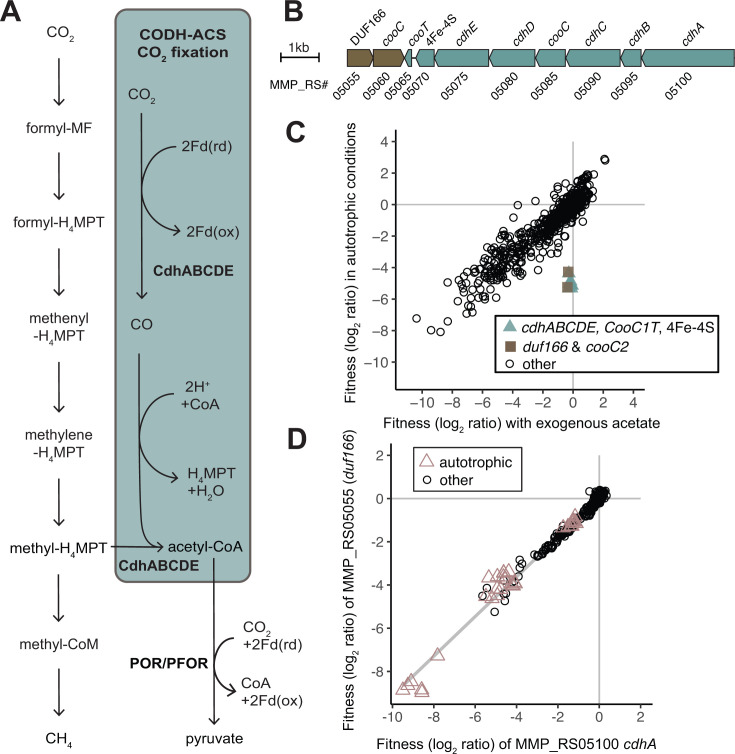
M. maripaludis can fix CO2 to acetyl-CoA via the Wood-Ljungdahl pathway. The key enzyme complex for autotrophic growth is the carbon monoxide (CO) dehydrogenase and acetyl-CoA synthase (CODH-ACS) encoded by an operon cdhABCDE spanning genes MMP_RS05100 to MMP_RS05065. CODH-ACS synthesizes the carbonyl group of acetyl-CoA via H2 and ferredoxin-dependent CO2 reduction to CO and derives the methyl group of acetyl-CoA from an intermediate of methanogenesis (Fig. 1, panel A). To determine if there were additional encoded functions essential to CO2 fixation, we conducted fitness assays under autotrophic growth conditions. The reproducibility of fitness assays in autotrophic H2 oxidizing conditions, as quantified by the linear Pearson's correlation, was r = 0.81–0.86).
Fig 1.
DUF166 encoded by MMP_RS05055 is important for fitness during autotrophic growth. (A) The Wood-Ljungdahl pathway for CO2 fixation, highlighted in blue, uses methyl-H4MPT from the methanogenic pathway (left) for the methyl group of acetyl-CoA. Acetyl-CoA can be further reduced to pyruvate by pyruvate ferredoxin oxidoreductase (POR/PFOR) and reduced ferredoxin (Fd). (B) Gene neighborhood of the CODH-ACS operon. (C) Comparison of gene fitness values for growth with exogenous acetate (10 mM) compared to autotrophic growth in H2 oxidizing conditions. Each value is the average of six autotrophic (Mc defined media) replicates and three replicates with exogenous acetate (Mc defined media + 10 mM acetate). (D) Cofitness of carbon monoxide dehydrogenase/acetyl-coenzyme synthase (CODH-ACS) subunit alpha (cdhA, MMP_RS05100) and DUF166 encoded by MMP_RS05055. Experiments identified as autotrophic conditions were performed in defined minimal media both with and without vitamins and either H2 or formate as the electron donor.
Insertions in a cluster of 10 genes had negative fitness specifically during autotrophic growth (Fig. 1B and C) in H2 oxidizing conditions. Autotrophic growth assays were conducted in media lacking fixed carbon (casamino acids or acetate). As expected, the CODH-ACS operon accounted for 8 of the 10 genes identified. The other two genes, MMP_RS05060 and MMP_RS05055, are encoded in an adjacent operon encoded on the opposite DNA strand (Fig. 1B) (16). MMP_RS05060 encodes a paralog of CooC, the CO dehydrogenase nickel insertion accessory protein. MMP_RS05055 encodes a protein with a domain of unknown function (DUF) DUF166 and is “cofit” with each gene encoded by the CODH-ACS operon and MMP_RS05060 (r = 0.94–0.99). Cofitness is defined as a linear Pearson correlation of fitness patterns (r > 0.8). The cofitness of insertions in genes encoding the CODH-ACS operon, DUF166, and MMP_RS05060 is conserved in strain JJ as well (MMJJ_RS00080 and MMJJ_RS00080; r = 0.96–0.99).
Both MMP_RS05060 and MMP_RS05085 (in the CODH-ACS gene cluster) are annotated to encode the CooC domain predicted to be involved in maturation of the nickel center in CODH and are strongly cofit with cdhA (r = 0.99). Both homologs contain the zinc CxC binding motif and conserved ADP binding residues. Previous work has shown that 63% of organisms that encode a CooC also encode a paralogous gene (AcsF) that is involved in delivery of the nickel to acetyl-CoA synthase (Acs) (17). CooC (MMP_RS05085) is 47% identical to the AcsF characterized in Carboxydothermus hydrogenoformans to catalyze the insertion of nickel into Acs (Fig. S4) (17). The M. maripaludis CooC (MMP_RS05060) is only 34% identical to the C. hydrogenoformans AcsF and 41% identical to the C. hydrogenoformans CooC1 (Fig. S4). Thus, we hypothesize that CooC (MMP_RS05085) is involved in the nickel insertion into CdhC (Acs) and that CooC (MMP_RS05060) is involved in CODH (CdhA) maturation.
Insertions in DUF166 occurred on both positive and negative strands, in both strains S2 and JJ, and have negative fitness values in autotrophic conditions (Fig. S5). However, to confirm that the fitness defect was not due to a polar effect on MMP_RS05060, we generated a gene knockout strain of duf166 in M. maripaludis strain S2 (Δduf166) that resulted in an inability to fix CO2. This defect was complemented when duf166 was expressed in trans (pLW40neo-duf166) (Fig. S5). This indicates that the defect is due to the importance of MMP_RS05055 in autotrophic conditions and not a polar effect, suggesting that its function is related to CODH-ACS carbon fixation. DUF166 co-occurs with a CooC homolog (always encoded in the same genomes) and is occasionally in a potential operon with CooC (12% of those genera) (18). Previous computational analysis of a Methanococcus jannaschii DUF166 (36% amino acid identity [AA ID] to MMP_RS05055) protein suggested that it may be a metal-dependent enzyme involved in a redox reaction (19). Taken together, we hypothesize that DUF166 is involved in the nickel cofactor maturation of CODH.
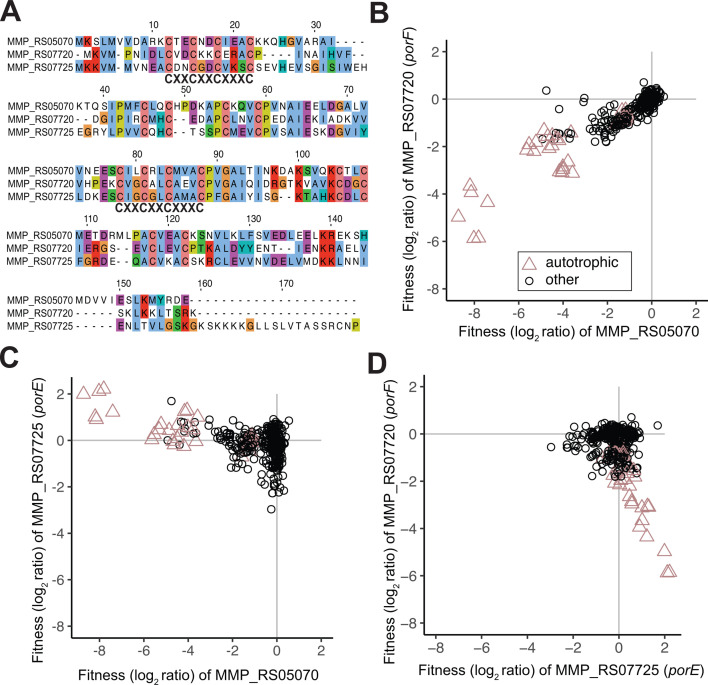
The CODH-ACS complex is predicted to receive electrons from a 4Fe-4S cluster containing protein (MMP_RS05070) encoded at the end of the CODH-ACS operon (20). MMP_RS05070 is homologous to the ferredoxins PorE (39% AA ID) and PorF (37% AA ID) (Fig. 2A), which are encoded in an operon with the pyruvate ferredoxin oxidoreductase (POR/PFOR). In addition to being cofit with the rest of the CODH-ACS operon, MMP_RS05070 is also cofit with porF (MMP_RS07720), but not porE (MMP_RS07725) (Fig. 2B–D). The cofitness of MMP_RS05070 and porF is conserved (r = 0.89) in M. maripaludis strain JJ orthologs (MMJJ_RS00065 and MMJJ_RS06635, respectively), indicating that they serve a role in similar metabolic context. Additionally, the importance of porF in autotrophic conditions further supports previous work done using M. maripaludis strain JJ indicating PorF as the dominant electron conduit to POR/PFOR for pyruvate biosynthesis (21). The lack of strong fitness defects in porE in autotrophic conditions indicates that additional experimentation is required to elucidate its role.
Fig 2.
Cofitness between 4Fe-4S encoding homologs is specific to PorF and the CODH-ACS 4Fe-4S encoded by MMP_RS05070. (A) Alignment of 4Fe-FS encoding MMP_RS05070, MMP_RS07720, and MMP_RS07725 with the two conserved 4Fe-4S motifs (CXXCXXCXXXC) labeled. Amino acids are colored using the Clustal X color scheme. (B) Cofitness of MMP_RS05070 and MMP_RS07720 (PorF). (C) Negative cofitness of MMP_RS05070 and MMP_RS07725 (PorE). (D) Negative cofitness of MMP_RS07725 (PorE) and MMP_RS07720 (PorF). Experiments identified as autotrophic conditions were performed in defined minimal media both with and without vitamins and either H2 or formate as the electron donor.
An alternate pathway of acetyl-CoA synthesis involves assimilation of acetate from the environment via an AMP-forming acetyl-CoA synthetase (20). Since growth in autotrophic conditions is slower compared to growth in the presence of exogenous acetate, we would expect insertions in the AMP-forming acetyl-CoA synthetase to have reduced fitness. However, insertions in AMP-forming acetyl-CoA synthetase (MMP_RS00845) have neutral fitness (|fitness| < 2) across all conditions in both strains S2 and JJ. Thus, we propose that the conversion of acetate to acetyl-CoA is genetically redundant among the AMP-forming acetyl-CoA synthetase, the ADP-dependent acetyl-CoA synthetase (Acd, MMP_RS01370), or possibly an acyl-CoA ligase (MMP_RS06550).
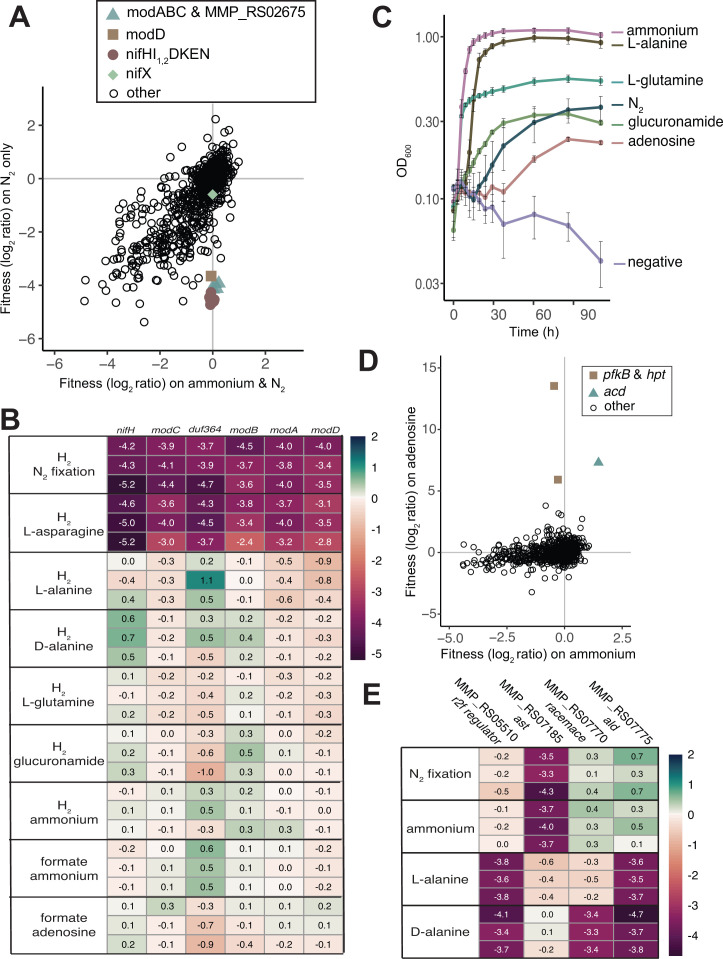
Fitness assays in nitrogen fixing conditions
M. maripaludis has been studied as a model for diazotrophic growth and the nitrogenase (nif) operon has been well characterized (8, 9, 22, 23). The reproducibility of fitness data across replicates in deep-well blocks in H2 oxidizing, N2 fixing conditions was evaluated by a linear Pearson's correlation, with r = 0.89–0.91. Insertions in genes in the nif operon were detrimental in conditions where N2 was the sole nitrogen source provided (Fig. 3A). These included the molybdate-dependent nitrogenase reductase (nifH, MMP_RS04440), nitrogenase regulators (nifI1,2, MMP_RS04445-MMP_RS04450), dinitrogenase (nifDK, MMP_RS04455-MMP_RS04460), and nifEN (MMP_RS04465-MMP_RS04470). Genes nifHI1,2DKEN are strongly cofit (r > 0.95). The last gene in the nif operon, nifX (MMP_RS04475), is non-essential in N2 fixing conditions as previously shown (8) (Fig. 3, panel A).
Fig 3.
Gene fitness for diazotrophic growth and growth on various nitrogen sources. (A) Comparison of fitness values between growth with ammonium and N2 as nitrogen sources versus N2 only (both in the presence of acetate and H2). MMP_RS02675 encodes a DUF364. Each value is the average of three replicate experiments. (B) Heatmap showing overview of fitness data for select genes strongly cofit with nifH across conditions with varying nitrogen sources added. N2 is present in all conditions. (C) Growth of M. maripaludis with various nitrogen sources. Averages and standard deviations are calculated from four replicate cultures. N2 is absent in all conditions except N2 fixing (blue circles). (D) Fitness for growth in formate oxidizing conditions with ammonium or adenosine added as a nitrogen source. Values are averages of three experiments. (E) Heatmap showing fitness data for genes of interest for alanine metabolism. N2 is present in all conditions.
While some archaea have V-nitrogenase and/or Fe-nitrogenase (24), archaeal nitrogen fixation is primarily a molybdenum-dependent process (25), and M. maripaludis only possess a Mo-nitrogenase. nifH also had strong cofitness with one of the three putative molybdate transporter gene clusters (26), namely, the operon composed of modABC (MMP_RS02685, MMP_RS02680, and MMP_RS02670) and MMP_RS02675 that encodes DUF364. Previous structural characterization and genome context analysis predicts that DUF364 has a role in heavy-metal chelation (27). In organisms that encode DUF346 homologs, its presence is conserved next to molybdate uptake genes (18). We hypothesize that it could have a role in chelating molybdate for biosynthesis of the molybdate cofactor (FeMoCo) required for nitrogen fixation. However, in M. maripaludis strain S2, insertions in MMP_RS02675 only occur on the positive strand (Fig. S6), and we cannot rule out polar effects on downstream genes in the same predicted operon. Additionally, there is not a homolog of MMP_RS02675 in strain JJ. Thus, additional investigation is required to determine if MMP_RS02675 is required for N2 fixation in strain S2.
MMP_RS02730 (modD) is also cofit with nifH and is associated with another molybdate transporter encoding gene cluster modABD (MMP_RS02720-MMP_RS02730) previously identified as an operon (26); however, recent transcriptional analysis of M. maripaludis (16) shows that modAB is transcribed independent of modD. modD encodes a putative ATPase and its role in molybdate transport is unclear. The modAB encoded upstream of modD is non-essential and had neutral fitness across all conditions (fitness values between +2 and −2). The third molybdate transporter gene cluster is also composed of two operons, modA MMP_RS01135 and modBC MMP_RS01140-MMP_RS01145 (16), is not cofit with the nif operon, and has weak fitness responses across conditions despite being encoded downstream of the nitrogen regulator (NrpR) operator consensus sequence GGAAN6TTCC (26). These results were unexpected considering that past proteomic analysis performed in similar growth medium showed a significant increase in ModA encoded by MMP_RS01135 in nitrogen limiting conditions (26). Thus, under the conditions we assayed, the protein abundance of molybdate transporter subunits is not determinative of their essentiality under N2 fixing conditions. However, more work is needed to tease apart the role of these molybdate transporter subunits in N2 fixation.
Identification of previously uncharacterized nitrogen sources
To our knowledge, N2, alanine, and ammonium are the only recognized nitrogen sources for M. maripaludis strain S2 (8, 10). We sought to identify additional nitrogen sources by screening 39 nitrogen-containing compounds in both formate and H2 oxidizing conditions, including 20 proteogenic amino acids as well as non-proteogenic amino acids, di-peptides, and nucleosides. To generate fitness data regardless of the substrate's potential use as a nitrogen source, growth was performed in the presence of N2. In formate oxidizing conditions, only cultures with ᴅ- or ʟ-alanine, ammonium, casamino acids, ʟ-glutamine, glucuronamide, or adenosine grew. Lack of growth in other conditions suggests that N2 fixation, a process that requires the H2-dependent reduction of ferredoxin, was poor in formate oxidizing conditions. The reproducibility of these experiments, as quantified by a linear Pearson's correlation, was r ≥ 0.89 except adenosine experiments, with r = 0.84–0.88.
We used the fitness of insertions in nifH to determine which of the nitrogen sources were utilized, based on an approach previously implemented in Desulfovibrio vulgaris BarSeq analysis (28). Insertions in nifH had neutral fitness in H2 oxidizing conditions, when alanine, ammonium, ʟ-glutamine, ᴅ-glucuronamide, or adenosine was provided as a nitrogen source, identifying ʟ-glutamine, ᴅ-glucuronamide, and adenosine as additional nitrogen sources for M. maripaludis (Fig. 3B). The use of these substrates as nitrogen sources is further supported by growth of M. maripaludis in medium with ʟ-glutamine, ᴅ-glucuronamide, or adenosine as the sole nitrogen source (no N2 in headspace) (Fig. 3C).
There were no specific phenotypes when ʟ-glutamine or ᴅ-glucuronamide was used as a nitrogen source suggesting that the gene(s) responsible for cleavage of the secondary amine is essential in M. maripaludis, that there are redundant genes for these activities, or that hydrolysis occurs outside the cell. The three domains of the large subunit of the bacterial glutamate synthase are encoded by three essential genes in M. maripaludis (MMP_RS00460-MMP_RS00470). No homolog of the small subunit of the bacterial glutamate synthase has been identified (20). The lack of fitness defects for transporters in conditions where ʟ-glutamate or ᴅ-glucuronamide is used as a nitrogen source suggests that there may be redundancy in uptake mechanisms or that the transporters are encoded by essential genes.
When adenosine was provided as a nitrogen source, we identified beneficial mutations (>2 log2FC) for several genes predicted to be involved in purine degradation and salvage (Fig. 3D). Most notably, inosine kinase (pfkB, MMP_RS02225) had an fitness value of +13.3–14.0 (|t| = 45–89) during growth with formate as the electron donor and adenosine and N2 as the sole nitrogen sources. We hypothesize that adenosine is deaminated to yield ammonium and inosine by adenosine deaminase MMP_RS07110, which is essential in M. maripaludis S2 (5). The inosine kinase phosphorylates inosine to IMP, costing an ATP. The other purine metabolism related gene with a strong positive selection was hypoxanthine phosphoribosyltransferase (hpt) MMP_RS00830 (fitness = 5.4–6.9, |t| = 28–36). Cofitness of inosine kinase and hpt orthologs is conserved in M. maripaludis strain JJ (MMJJ_RS03080 and MMJJ_RS04660; r = 0.90). We hypothesize that the strong positive fitness values of the inosine kinase is in part due to the ATP costs; however, further characterization to elucidate the mechanism of selection against both the inosine kinase and hpt is required.
There was also a strong positive selection for insertions in MMP_RS01370 (fitness = 6.8–7.9, |t| = 38–41), which is an (ADP)-forming acetyl-CoA synthetase (Acd) and hypothesized to be involved in an acetate switch (switch from acetate production to re-uptake in later growth phases) in M. maripaludis (29). However, acd being detrimental is not specific to adenosine utilizing conditions as many other conditions with acetate present and no adenosine have positive fitness values for strains with insertions in acd. For example, in formate oxidizing conditions with ammonium as the sole nitrogen source, acd had fitness values of 1.4–1.5 (|t| = 8.6–10.1). Thus, future work is required to understand why insertions in acd have positive fitness values in a variety of conditions.
In all amino acid free conditions, a gene (MMP_RS07185) annotated to encode an aspartate transaminase (Ast) had negative fitness except when alanine was provided (Fig. 3E). Ast encodes one of five potential type I aminotransferases that could be responsible for biosynthesis of alanine (30). Alternatively, ʟ-alanine dehydrogenase (ald, MMP_RS07775) is predicted to be responsible for both catabolism and biosynthesis of ʟ-alanine (31). However, insertions in ald do not have strong negative fitness in amino acid free conditions except for when ᴅ- or ʟ-alanine is provided as a nitrogen source without ammonium. Thus, we hypothesize that Ast is responsible for biosynthesis of ʟ-alanine and Ald is likely essential for catabolism.
An Rrf2 family transcriptional regulator (MMP_RS05510) is cofit (r = 0.87) with ald and has a negative fitness (fitness = −4.1 to −3.4, |t| = 6.8–11.3) when ᴅ- or ʟ-alanine is used as a nitrogen source (Fig. 3E). MMP_RS05510 does not have a strong response in other casamino free conditions, suggesting that it has a regulatory role in alanine catabolism. When ᴅ-alanine is provided as a nitrogen source, insertions in the alanine racemase (MMP_RS07770) have negative fitness (fitness = −3.4 to −3.3, |t| = 12.2–13.6) along with the ald (Fig. 3E), consistent with previous characterization in M. maripaludis (32).
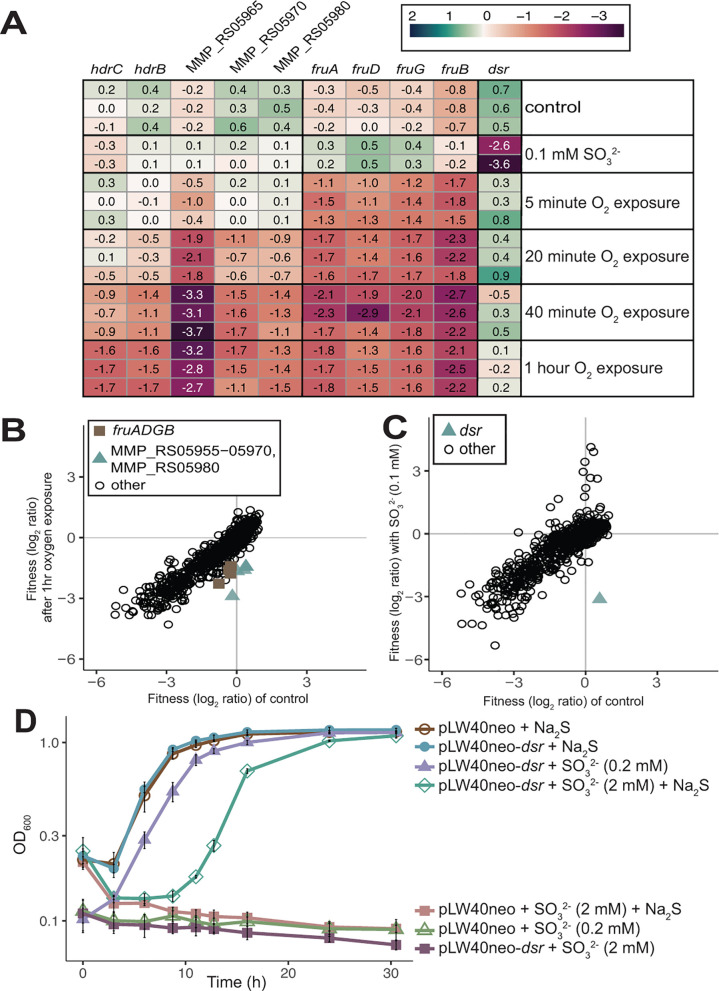
Oxygen and sulfite stress have distinct genome-wide fitness profiles
Methanogens are strict anaerobes and are therefore sensitive to a variety of oxidative stress. Sulfite is inhibitory to methanogenesis (33) and can be produced via an abiotic reaction when oxygen is introduced to sulfide-rich environments where methanogens are found (34). In vitro, enzymes like methyl-coenzyme M reductase (Mcr), an essential enzyme for methanogenesis, are irreversibly damaged by sulfite (35). However, oxygen itself can also irreversibly damage metalloenzymes, so we sought to determine if there were independent consequences of oxygen and sulfite toxicity. To separate these effects, cultures were washed with reductant- and sulfide-free media in anoxic conditions and then agitated in air for up to 1 hour (to introduce oxygen, but not sulfite). The oxidized culture was resuspended in anoxic media with reductant prior to the addition of sulfide, then grown in H2 oxidizing conditions. The sulfite stress cultures were not exposed to air and were grown with 0.1 mM sulfite in H2 oxidizing conditions. The reproducibility of fitness assays in these conditions, as determined by a linear Pearson's correlation, was r ≥ 0.90.
Increased importance of coenzyme reducing genes following oxygen exposure
There were two operons encoding genes with negative fitness in oxygen stress conditions but not control or sulfite stress conditions (Fig. 4A and B). The strongest negative fitness was for MMP_RS05965 (fitness after 40 minutes or 1 hour O2 exposure = −2.7 to −3.7, |t| = 3.7–4.5), which is annotated as a carboxymucolactone decarboxylase family protein and is encoded in a 10-gene operon (MMP_RS05955-MMP_RS06000). While other annotated carboxymucolactone decarboxylases have been characterized as disulfide reductases (MdrA) in Methanosarcina acetivorans, the protein encoded by M. maripaludis is only 30% identical to MdrA and is missing conserved cysteine residues (Fig. S7) (36). Thus, the function of MMP_RS05965 is unclear, but it is likely beneficial following oxygen exposure.
Fig 4.
Comparison of genes with strong phenotypes in oxygen versus sulfite stress. (A) Heatmap showing fitness of select genes across control, oxygen (O2), and sulfite (SO32−) stress conditions. (B) Comparison of fitness between control and oxygen exposed cultures. Values are replicates of triplicate cultures. (C) Comparison of fitness between control and sulfite containing cultures. Values are replicates of two cultures for sulfite and three replicates for control. (D) Growth of M. maripaludis with an empty vector (pLW40neo) or vector containing MMP_RS00450 (dsr) across SO32− stress conditions with or without sulfide (Na2S) added as a sulfur source. Values are averages of three replicates. Error bars represent standard deviation.
Additional genes in the operon with MMP_RS05965 have negative fitness values following oxygen exposure including those encoding heterodisulfide reductase (Hdr) subunits B and C; however, none of the fitness values for HdrC were significant (|t| > 4) (Fig. 4A and B) (HdrB fitness = −1.1 to −1.7, |t| = 3.7–7.2; HdrC fitness = −0.9 to −1.7, |t| = 1.3–2.5). The Hdr complex is responsible for the reversible reduction of the heterodisulfide CoM-S-S-CoB to coenzymes CoM-SH and CoB-SH. A second operon with negative fitness (MMP_RS07120-MMP_RS07135; fitness = −1.3 to −2.7, |t| = 3.1–12.7) encodes the selenocysteine containing coenzyme F420 hydrogenase (Fru), capable of using H2 to reduce F420, which is then used by other reductive enzymes in the reduction of CO2 to methane (37) (Fig. 4A and B). Since both operons encode genes involved in redox reactions, we hypothesize that mutants have decreased fitness due to the oxidation of the coenzymes and loss of reduced pools of coenzyme F420H2 and heterodisulfide required for methanogenesis.
Sulfite reductase enables detoxification and use of sulfite as sole sulfur source
For testing sulfite toxicity, the transposon library was grown with 0.1 mM sulfite or 10 mM thiosulfate, both with H2 as the electron donor. In sulfite (Fig. 4A and C) and thiosulfate (Fig. S8) stress conditions, strains with insertions in MMP_RS00450 had negative fitness (fitness = −4.1 to −2.6, |t| = 3.1–3.8) compared to controls (Fig. 4A and C). We hypothesize that the phenotype in thiosulfate conditions is due to the presence of sulfite in the thiosulfate stock. MMP_RS00450 encodes a gene predicted to be a group IIId dissimilatory sulfite reductase-like protein (Dsr) (38).
Sulfide is the only known sulfur source for M. maripaludis (39). However, some close relatives with a greater sulfite tolerance are able to use sulfite through reduction to sulfide by an F420-dependent sulfite reductase (Fsr) (35). Expression of the Fsr from Methanocaldococcus jannashii in M. maripaludis confers the ability to reduce sulfite to sulfide for both detoxification and use as a sulfur source (40). This sulfite reductase is a chimera of an N-terminal homolog of F420H2 oxidase (Fsr-N) and a C-terminal homolog of Dsr (Fsr-C). However, M. maripaludis does not contain this chimeric form of a sulfite reductase.
The M. maripaludis Dsr (MMP_RS00450) contains a siroheme and two 4Fe-4S clusters but no F420H2 oxidase (Fsr-N) fusion (38, 41). However, decreased mutant fitness for the native dsr in M. maripaludis fitness assays with sulfite and thiosulfate suggests that it still has a role in sulfite detoxification. We recombinantly expressed MMP_RS00450 under the control of the high expression promoter (phmv; histone promoter from Methanococcus voltae) (9) in M. maripaludis to investigate if increased expression of the native dsr was sufficient to confer increased sulfite resistance and use as the sole sulfur source (Fig. 4D). M. maripaludis with recombinantly expressed Dsr was able to use 0.2 mM sulfite as the sole sulfur source. The overexpression strain was also able to detoxify 2 mM sulfite but not as the sole sulfur source (Fig. 4D).
Sulfite and thiosulfate concentrations are typically in the low micromolar range or below in marine sediments (42). The Dsr-LP in M. maripaludis is functional as a sulfite reductase despite lacking the F420H2-oxidase domain, albeit at lower concentrations than that previously demonstrated (2 mM) with recombinant expression of M. jannashii Fsr (40). Since the F420H2-oxidase domain is lacking, we predict that the electron donor for sulfite reduction is either essential or redundant in M. maripaludis; however, further characterization is required to identify the electron donor for this reaction. Taken together, we hypothesize that Dsr-LP activity, while less robust than Fsr, may still be beneficial to Methanococcus spp. for the detoxification of sulfite under physiologically relevant conditions in anoxic marine environments.
Conclusion
By combining RB-TnSeq with high pressure incubations in deep-well blocks, we successfully assessed fitness of non-essential genes in the model archaeon M. maripaludis in over 100 unique growth conditions. This led to identifying several genes encoding proteins of unknown function as important in diverse cellular processes. Here, we demonstrate the essentiality of the DUF166 protein MMP_RS05055 for growth in autotrophic conditions. We speculate that it is involved in the maturation of CO dehydrogenase. Further, we were able to characterize three new nitrogen sources, a new sulfur source, and to identify different genes important for the response to oxygen or sulfite stress. Not only does this identification expand our understanding of the metabolic versatility of M. maripaludis but we hypothesize that it provides ground work for further development of M. maripaludis laboratory tools. We believe that the strong selection against the inosine kinase in conditions where adenosine is provided may be an opportunity to develop an additional selectable marker in M. maripaludis, which has only a handful of markers available. Determining that M. maripaludis can recover and grow following 1 hour of oxygen exposure (in the absence of sulfide/sulfite) may aid in the optimization of methods for some techniques to be performed aerobically followed by reduction for growth. Finally, the determination of a functional dsr in M. maripaludis provides novel insight to sulfur metabolism and is the first in vivo analysis of the function of a Dsr-LP group IIId protein as a sulfite reductase.
RB-TnSeq is particularly valuable when in silico gene functional assignments are incorrect or missing. The data presented here and available through the fitness browser (http://fit.genomics.lbl.gov/) are a valuable resource for further hypothesis generation with regard to gene function. We have highlighted a handful of findings from this data set, but there are 189 genes with specific phenotypes and additional cofitness and conserved cofitness patterns to be explored. As the first application to a member of the Archaea, this RB-TnSeq data set will empower future efforts to characterize novel aspects of archaeal physiology.
MATERIALS AND METHODS
Growth conditions
All M. maripaludis growth plates and deep-well blocks (Costar) were inoculated and harvested in an anaerobic chamber (Coy Laboratory Products; 2–3% H2, 10% CO2, N2 balanced atmosphere). Plates were incubated at 37°C with 10–20 mL of 25% Na2S on a paper towel in a falcon tube or plate as a sulfur source and under a pressurized atmosphere of 140 kPa H2:CO2 (80:20). For 96-well deep-well block experiments, anoxic H2O was added to the Na2S soaked paper towel each time the incubation vessels were opened to increase the humidity and reduce evaporation.
The 1,000× trace mineral solution used for library generation and growth was prepared by making 21 g/L Na3citrate·2H2O solution and adjusting the pH to 6.5, then in order 5 g/L MnSO4·2H2O, 1 g/L CoCl2·6H2O, 1 g/L ZnSO4·7H2O, 0.1 g/L CuSO4·5H2O, 0.1 g/L AlK(SO4)2, 0.1 g/L H3BO3, 1 g/L Na2MoO4·2H2O, 0.25 g/L NiCl2·6H2O, 2 g/L Na2SeO3, 0.1 g/L VCl3, and 0.033 g/L Na2WO4·2H2O was added. Aliquots (50 mL) of the trace mineral stock solution were bubbled under N2 for 20 minutes and stored in the dark in serum bottles sealed with butyl stoppers at 4°C.
For fitness assays and nitrogen fixation growth curves, 200 mM MOPS buffer (pH 7.2) was added to increase the buffering capacity and reduce the differences between formate and H2 growth medium. Trace minerals with SO42− anions were swapped for Cl− anion alternatives and all Mg was provided as MgCl2 to eliminate SO42− for future SO42− free experiments. Fitness assays to determine if there were genes with specific phenotypes in the medium with MOPS added or SO42− removed compared to the medium used to generate the library were performed (Fig. S2).
Balch tubes were pressurized with 280 kPa H2:CO2 (80:20) or 207 kPa N2:CO2 (80:20) with either H2 or formate oxidizing medium, with the exception of N2 fixation growth tubes that were pressurized to 103 kPa N2:CO2 then brought up to 280 kPa with H2:CO2 and repressurized with H2:CO2 daily.
Plasmid construction
All strains, constructs, and primers used can be found in Tables S1 and S2. For construction of the barcoded plasmid for generation of the RB-TnSeq libraries plasmid, pJJ605 (43) was digested with NotI at 37°C for 20 minutes then BstB1 was added and incubated at 65°C for an additional 20 minutes. The insertion fragment included DNA between the NotI and BstB1 restriction sites, the U1 U2 priming sites, and the 20 bp DNA barcode (3). Oligos 109-pLD26_insert1 and 110-pLD26_insert2 were used as a template DNA for amplification using 107-pLD26_F and 108-pLD26_R for 10 replicate reactions to minimize bottlenecks for barcode diversity (Table S2). PCR conditions were 98°C for 1 minute followed by 30 cycles of 98°C for 20 s, 56°C for 20 s, and 72°C for 30 s, followed by a final extension at 72°C for 5 minutes. The 10 reactions were then pooled and aliquoted into 10 equal volumes for purification.
The pLW40neo construct is a stable plasmid used for expression of genes under the control of the Methanococcus voltae histone promoter in M. maripaludis (9). The plasmid used for complementation of the ΔMMP_RS05055 (duf166) strain was generated by amplifying MMP_RS05055 using oligos 156-DUF166F and 157-DUF166R and Gibson assembling (44) the product into pLW40neo that had been digested with NsiI and AscI. Similarly, the gene encoding the dissimilatory sulfite reductase-like protein (dsr, MMP_RS00450) was PCR amplified using oligos 142-DsrF and 143-DsrR and Gibson assembled (40) into pLW40neo that had been digested with NsiI and AscI.
Digested pJJ605 and all PCR products were purified using Invitrogen PCR cleanup kit (REF K310001) per manufacturer's protocol. Plasmid and insert fragment(s) were ligated using Gibson assembly with NEBuilder HiFi DNA Assembly Master Mix (CAT E2621) per manufacturer's guidelines and electroporated into electrocompetent DH5alpha E. coli. For the barcoded pJJ605 construct (pLD026), electroporation reactions were pooled and recovered overnight, shaking at 37°C in 500 mL lysogeny broth with kanamycin (50 µg/mL) and ampicillin (50 µg/mL). E. coli containing pLW40neo-dsr was selected for on LB agar plates with ampicillin (50 µg/mL). All plasmids were purified using the Invitrogen PureLink HiPure Plasmid Filter Maxiprep Kit (REF K210016) per manufacturer's guidelines. All insertion fragments were amplified using Phusion HF polymerase. The pLD026 transposase sequence is available in the supplementary data set.
Transposon library generation and RB-TnSeq
Transposon library mutagenesis was performed following the optimized protocol described by Fonseca et al. (7, 43, 45) with slight differences in the starting culture preparation. Briefly, 16 overnight cultures in stationary phase (OD600 ~ 1.0) were supplemented with 1 mL fresh McCas and repressurized to 276 kPa H2:CO2 and incubated shaking at 37°C for 2 hours. While cultures were growing, the transposon reaction was set up in the reaction buffer with pLD026 and HimarI transposase then incubated at 28°C for 2 hours. Transposon reactions were transferred into wild-type M. maripaludis using the PEG transformation method for 16 replicate cultures (46) followed by a 4 hour outgrowth. The replicate transformations were pooled and transformants were selected on McCas agar medium with puromycin and grown at 37°C for 4 days in an anaerobic incubation vessel pressurized to 138 kPa H2:CO2. Plates were flooded with 2 mL of 20% glycerol McCas solution (5) and scraped into a sterile collection vessel then single use 1 mL aliquots were distributed into serum bottles and flash frozen with liquid nitrogen and stored at −80°C.
Genomic DNA was extracted using the Qiagen Blood and Tissue Kit (CAT 69504) per manufacturer's guidelines and sequenced at the University of Minnesota Genomics Center. The genomic locations and associated barcodes were determined using a previously described Tn-Seq protocol (3). We also mapped transposon insertions and linked them to their associated DNA barcodes using a two-step PCR approach (47) with a splinkerette adapter design (A M. Deutschbauer, unpublished data), described by Rubin et al. (48). In this approach, the splinkerette adapter replaces the Y-adapter we used previously (3) and greatly enriches transposon insertion junctions in the first round of PCR. In the second round of nested PCR, we add the Illumina adapter sequences. After bead-based cleanup, we sequenced the round two PCR products.
Fitness experiments were only included in subsequent analysis if they passed previously established quality metrics (3). First, we ensure that most of the non-essential genes are still represented in the experimental sample (median reads per gene at least 50). Second, as a measure of how consistent the fitness data is for each gene, we estimate the (normalized) fitness using only insertions within the first half of each gene (10–50%) or only within the second half of each gene (50–90%). We require that the Spearman rank correlation of those two sets of values be at least 0.1. This ensures that there is some biological consistency to the results. Third, as another measure of the consistency, we require that the median of the absolute differences between the fitness according to the first half of the gene and the fitness according to the second half of the gene be at most 0.5. Fourth, we require that the fitness values not be very correlated with the gene's GC content (Pearson correlation between −0.2 and +0.2), as a correlation with GC content can indicate artifacts due to PCR (3). Finally, as another check for artifacts from PCR, we require that the nearby genes that probably do not have related functions do not have correlated fitness values. Specifically, we compute the Pearson correlation between the fitness values of pairs of adjacent genes that are not on the same strand (and hence not in the same operon) and require this to be within −0.25 and +0.25 (3).
RB-TnSeq fitness assays
For reviving the library, a 1 mL aliquot was thawed in the anoxic chamber and inoculated into 50 mL McCas with sulfide in a serum bottle and pressurized to 138 kPa H2:CO2. Cultures were incubated overnight then centrifuged and resuspended in anoxic basal medium (unless otherwise indicated) twice at 4,000 rcf and resuspended in Basal Mc. The optical density (OD600) was determined on a Spectronic20, blanked with Basal Mc. After washing, for each experiment at least three times, zero samples, 1 mL each, were collected by centrifuging at 15,000 rpm in microcentrifuge tubes and stored at −80°C. The remaining culture was then diluted to an estimated OD600 of 0.04 and added to 2 × media for a starting density of 0.02 Abs. Cells were washed and resuspended in media type according to the assay being performed. The full list of experiments performed for both mutant libraries along with detailed metadata are available on Figshare (https://doi.org/10.6084/m9.figshare.24195057.v1). The full list of experiments discussed in the text and available on the fitness browser and the media used for each fitness assay for both strains S2 and JJ are also available in the supplementary data set.
Media and anoxic water used for deep-well blocks were prepared by bubbling them with N2:CO2 or N2 for 30 minutes before transferring to the anoxic chamber for 1–2 days before addition of reductant (DTT), sulfide (1 mM), and filter sterilization. When additional components were added to the media as indicated in Compound_1 and Compound_2 columns of the fitness assay description, they were filter sterilized and added in a 2 × concentration to 96-well deep-well blocks and covered with a AeraSeal gas permeable film (Millipore Sigma) then transferred into the anoxic chamber to degas for 2 days before inoculating with cells in 2 × media containing reductant and transferring plates to the incubation vessel. Growth of cultures was periodically checked by transferring 150 µL of culture to a Greiner 96-well microplate using an Avidien microPro 300 Benchtop pipettor. When possible, the OD600 was read inside an anoxic chamber using a Tecan Infinite 200 plate reader, then cultures were returned to the deep-well blocks.
Sulfite and oxidative stress assay
The sulfite stress assay cells were washed with McCas without DTT or sulfide. Culture was divided and resuspended in a 2× buffer that contained DTT and inoculated into deep-well blocks. Culture reserved for the oxygen stress assay was resuspended in media without DTT, removed from the anaerobic chamber, and incubated shaking in a flask at 37°C with foil loosely over the top. One milliliter of culture was collected after 5, 20, 40, and 60 minutes of oxygen exposure. The culture was centrifuged for 1 minute at 8,000 × g, and the supernatant was discarded and then cycled into the anoxic chamber where it degassed for 5 minutes before 2× media containing reductant (DTT) was used to resuspend the pellet. Cells remained in the 2× containing McCas for at least 10 minutes before being transferred to deep-well blocks containing sulfide.
Syntrophic fitness assays
Fitness assays conducted with Syntrophotalea carbinolica were performed as previously described except with the S2 barcoded transposon library (45).
Vitamin fitness assays
For vitamin dropout conditions, filter sterilized vitamin solutions were made lacking one of the vitamins for each vitamin present in the stock solutions (e.g., biotin dropout condition). A solution of all 20 amino acids (1 mM) each was added in place of casamino acids to ensure excess of all amino acids (formate McAA). However, there were few outliers from controls with complete vitamin solutions likely due to residual intracellular pools being sufficient to complement defects; thus, future studies will require serial transfers.
Essential genes in M. maripaludis strain S2
Essential genes have been previously characterized in M. maripaludis strain S2 using a derivative of the Tn5 transposon library (5). Similar to what was observed in the Tn5 transposon library, we discovered that insertions in many of these essential genes were present in the RB-TnSeq library, likely due to the polyploidy of M. maripaludis (14). We previously described a library of barcoded transposon mutants for Zymomonas mobilis ZM4 (49), which is also highly polyploid (50). In Z. mobilis, the insertions in essential genes contain both wild-type and mutated copies of the gene and are unstable (49).
Our data for M. maripaludis S2 show a similar pattern as in Z. mobilis: we obtained transposon mutants for most of the essential genes, and most of the insertions in essential genes have consistently low fitness. Low fitness may occur because some fractions of progeny lack a functional copy of the essential gene and are not viable. Alternatively, because there was no antibiotic selection, the mutated copy may be lost, and then barcode will no longer be detected: again, this would lead to low fitness values for that strain. Library outgrowth (T0) and fitness assays were performed in the absence of antibiotic selection. Previous work with M. maripaludis S2 demonstrated that insertions in non-essential genes persisted in the absence of antibiotic selection for ~30 generations in syntrophic conditions (45) and ~14 generations in axenic cultures (5) from the T0 population. Most fitness assays performed here were grown approximately 4–6 generations unless otherwise indicated (7–10 generations for subcultured fitness assays indicated as T2, or up to 20 generations for the syntrophic experiments).
The fitness data for essential genes complicate the normalization by chromosomal position, which assumes that most genes have little effect on fitness (3). Additionally, essential genes have a higher average GC content compared to non-essential genes (35% vs 33%, respectively), which impedes determination of the quality metric (gccor), which accounts for PCR-based spurious GC content correlations (3). Therefore, we removed fitness data for previously identified essential genes if they had consistent low fitness (median under −2) in a preliminary data set of 72 experiments. Of the 326 essential genes with fitness data, 300 were removed (these are listed in Supplemental Data Set and Figshare at https://doi.org/10.6084/m9.figshare.24195057.v1).
Data analysis
Gene fitness was calculated as described previously (3). Briefly, insertions in a gene were considered if they occurred within the middle 10–90% of the gene. Strain fitness values are calculated from the normalized log2 ratios of the abundance of the barcode following selection versus in the starting population before selection (Time0). Gene fitness is then calculated as the weighted average of the fitness of the individual mutant strains then normalized based on chromosome position to correct for variation in copy number along the chromosome using the running median along the chromosome (excluding essential genes values for strain S2). Finally, gene fitness is normalized so that the mode is zero. Specific phenotypes are identified for genes in an experiment where |fitness| > 1 and |t| > 5 with |fitness| < 1 in at least 95% of total experiments and that the fitness value of the outlier experiment(s) were more pronounced than most other experiments conducted [|fitness| > 95th percentile(|fitness|) +0.5] (4). Cofitness was calculated as the Pearson correlation coefficient (r) across all fitness assays for each pair of genes. Gene pairs were considered cofit if they had r > 0.8 and conserved cofitness for gene pairs included genes where cofitness is greater than 0.6 for the two genes and cofitness >0.6 for two orthologous genes.
ACKNOWLEDGMENTS
We thank Jon Badalamenti and Catalina Vega Hurtado for the preparation of the DNA libraries for TnSeq.
This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under grant number DE-SC0019148. This material was supported by the U.S. Department of Energy Graduate Student Research (SCGSR) program fellowship to L.A.D. The SCGSR program is administered by the Oak Ridge Institute for Science and Education for the U.S. Department of Energy under contract number DE‐SC0014664. Work by H.K.C., A.P.A., M.N.P., and A.M.D. was supported by ENIGMA-Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Science Focus Area Program at Lawrence Berkeley National Laboratory, and is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231.
Contributor Information
Adam M. Deutschbauer, Email: amdeutschbauer@lbl.gov.
Kyle C. Costa, Email: kcosta@umn.edu.
Sonja-Verena Albers, Albert-Ludwigs-Universitat Freiburg, Freiburg, Germany, USA.
DATA AVAILABILITY
Data for successful experiments are available via the Fitness Browser (http://fit.genomics.lbl.gov), which includes details of the experimental conditions, quality metrics for each experiment, per-strain fitness values, gene fitness scores, cofitness and conserved cofitness, and t values. All data from the fitness browser and tab-delimited files for M. maripaludis strains S2 and JJ are also archived on Figshare at https://doi.org/10.6084/m9.figshare.24195057.v1. In addition to being available from the fitness browser and archived on the Figshare, all growth media information and experiment metadata and quality information are available in the Supplemental Data Set. A list of M. maripaludis strain S2 essential genes excluded from the fitness analysis can be found in the Supplemental Data Set. All data from the Fitness Browser as of February 2024 are archived on Figshare at https://doi.org/10.6084/m9.figshare.25236931.v1.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.00781-24.
Supplemental figures and tables.
Additional metadata.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Offre P, Spang A, Schleper C. 2013. Archaea in biogeochemical cycles. Annu Rev Microbiol 67:437–457. doi: 10.1146/annurev-micro-092412-155614 [DOI] [PubMed] [Google Scholar]
- 2. Costa KC, Whitman WB. 2023. Model organisms to study methanogenesis, a uniquely archaeal metabolism. J Bacteriol 205:e0011523. doi: 10.1128/jb.00115-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0 [DOI] [PubMed] [Google Scholar]
- 5. Sarmiento F, Mrázek J, Whitman WB. 2013. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc Natl Acad Sci U S A 110:4726–4731. doi: 10.1073/pnas.1220225110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodriguez-R LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. doi: 10.7287/peerj.preprints.1900 [DOI]
- 7. Fonseca DR, Loppnow MB, Day LA, Kelsey EL, Abdul Halim MF, Costa KC. 2023. Random transposon mutagenesis identifies genes essential for transformation in Methanococcus maripaludis. Mol Genet Genomics 298:537–548. doi: 10.1007/s00438-023-01994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kessler PS, Leigh JA. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343–1351. doi: 10.1093/genetics/152.4.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dodsworth JA, Leigh JA. 2006. Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc Natl Acad Sci U S A 103:9779–9784. doi: 10.1073/pnas.0602278103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lie TJ, Leigh JA. 2002. Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source. J Bacteriol 184:5301–5306. doi: 10.1128/JB.184.19.5301-5306.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci U S A 107:11050–11055. doi: 10.1073/pnas.1003653107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costa KC, Lie TJ, Jacobs MA, Leigh JA. 2013. H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis. mBio 4:e00062-13. doi: 10.1128/mBio.00062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lohner ST, Deutzmann JS, Logan BE, Leigh J, Spormann AM. 2014. Hydrogenase-independent uptake and metabolism of electrons by the archaeon Methanococcus maripaludis. ISME J 8:1673–1681. doi: 10.1038/ismej.2014.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. 2011. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol 193:734–743. doi: 10.1128/JB.01016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weimar MR, Cheung J, Dey D, McSweeney C, Morrison M, Kobayashi Y, Whitman WB, Carbone V, Schofield LR, Ronimus RS, Cook GM. 2017. Development of multiwell-plate methods using pure cultures of methanogens to identify new inhibitors for suppressing ruminant methane emissions. Appl Environ Microbiol 83:e00396-17. doi: 10.1128/AEM.00396-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W, Ren D, Li Z, Yue L, Whitman WB, Dong X, Li J. 2023. Internal transcription termination widely regulates differential expression of operon-organized genes including ribosomal protein and RNA polymerase genes in an archaeon. Nucleic Acids Res 51:7851–7867. doi: 10.1093/nar/gkad575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregg CM, Goetzl S, Jeoung J-H, Dobbek H. 2016. AcsF catalyzes the ATP-dependent Insertion of nickel into the Ni,Ni-[4Fe4S] cluster of acetyl-CoA synthase. J Biol Chem 291:18129–18138. doi: 10.1074/jbc.M116.731638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Price MN, Arkin AP. 2024. A fast comparative genome browser for diverse bacteria and archaea. PLoS ONE 19:e0301871. doi: 10.1371/journal.pone.0301871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iyer LM, Aravind L, Bork P, Hofmann K, Mushegian AR, Zhulin IB, Koonin EV. 2001. Quod erat demonstrandum? The mystery of experimental validation of apparently erroneous computational analyses of protein sequences. Genome Biol 2:RESEARCH0051. doi: 10.1186/gb-2001-2-12-research0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, et al. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol 186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin W, Whitman WB. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch Microbiol 181:68–73. doi: 10.1007/s00203-003-0629-1 [DOI] [PubMed] [Google Scholar]
- 22. Kessler PS, Blank C, Leigh JA. 1998. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol 180:1504–1511. doi: 10.1128/JB.180.6.1504-1511.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lie TJ, Leigh JA. 2003. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol 47:235–246. doi: 10.1046/j.1365-2958.2003.03293.x [DOI] [PubMed] [Google Scholar]
- 24. Chanderban M, Hill CA, Dhamad AE, Lessner DJ. 2023. Expression of V-nitrogenase and Fe-nitrogenase in Methanosarcina acetivorans is controlled by molybdenum, fixed nitrogen, and the expression of mo-nitrogenase. Appl Environ Microbiol 89:e0103323. doi: 10.1128/aem.01033-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kessler PS, McLarnan J, Leigh JA. 1997. Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol 179:541–543. doi: 10.1128/jb.179.2.541-543.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia Q, Wang T, Hendrickson EL, Lie TJ, Hackett M, Leigh JA. 2009. Quantitative proteomics of nutrient limitation in the hydrogenotrophic methanogen Methanococcus maripaludis. BMC Microbiol 9:149. doi: 10.1186/1471-2180-9-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller MD, Aravind L, Bakolitsa C, Rife CL, Carlton D, Abdubek P, Astakhova T, Axelrod HL, Chiu HJ, Clayton T, et al. 2010. Structure of the first representative of Pfam family PF04016 (DUF364) reveals enolase and rossmann-like folds that combine to form a unique active site with a possible role in heavy-metal chelation. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1167–1173. doi: 10.1107/S1744309110007517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trotter VV, Shatsky M, Price MN, Juba TR, Zane GM, De León KB, Majumder EL-W, Gui Q, Ali R, Wetmore KM, Kuehl JV, Arkin AP, Wall JD, Deutschbauer AM, Chandonia J-M, Butland GP. 2023. Large-scale genetic characterization of the model sulfate-reducing bacterium, Desulfovibrio vulgaris Hildenborough. Front Microbiol 14:1095191. doi: 10.3389/fmicb.2023.1095191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vo CH, Goyal N, Karimi IA, Kraft M. 2020. First observation of an acetate switch in a methanogenic autotroph (Methanococcus maripaludis S2). Microbiol Insights 13:1178636120945300. doi: 10.1177/1178636120945300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saum R, Mingote A, Santos H, Müller V. 2009. A novel limb in the osmoregulatory network of Methanosarcina mazei Gö1: Nε-acetyl-β-lysine can be substituted by glutamate and alanine. Environ Microbiol 11:1056–1065. doi: 10.1111/j.1462-2920.2008.01826.x [DOI] [PubMed] [Google Scholar]
- 31. Goyal N, Zhou Z, Karimi IA. 2016. Metabolic processes of Methanococcus maripaludis and potential applications. Microb Cell Fact 15:107. doi: 10.1186/s12934-016-0500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore BC, Leigh JA. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol 187:972–979. doi: 10.1128/JB.187.3.972-979.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balderston WL, Payne WJ. 1976. Inhibition of methanogenesis in salt marsh sediments and whole-cell suspensions of methanogenic bacteria by nitrogen oxides. Appl Environ Microbiol 32:264–269. doi: 10.1128/aem.32.2.264-269.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cline JD, Richards FA. 1969. Oxygenation of hydrogen sulfide in seawater at constant salinity, temperature and pH. Environ Sci Technol 3:838–843. doi: 10.1021/es60032a004 [DOI] [Google Scholar]
- 35. Johnson EF, Mukhopadhyay B. 2005. A new type of sulfite reductase, a novel coenzyme F420-dependent enzyme, from the methanarchaeon Methanocaldococcus jannaschii. J Biol Chem 280:38776–38786. doi: 10.1074/jbc.M503492200 [DOI] [PubMed] [Google Scholar]
- 36. Lessner DJ, Ferry JG. 2007. The archaeon Methanosarcina acetivorans contains a protein disulfide reductase with an iron-sulfur cluster. J Bacteriol 189:7475–7484. doi: 10.1128/JB.00891-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendrickson EL, Leigh JA. 2008. Roles of coenzyme F420-reducing hydrogenases and hydrogen- and F420-dependent methylenetetrahydromethanopterin dehydrogenases in reduction of F420 and production of hydrogen during methanogenesis. J Bacteriol 190:4818–4821. doi: 10.1128/JB.00255-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Susanti D, Mukhopadhyay B. 2012. An intertwined evolutionary history of methanogenic archaea and sulfate reduction. PLoS One 7:e45313. doi: 10.1371/journal.pone.0045313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y, Sieprawska-Lupa M, Whitman WB, White RH. 2010. Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J Biol Chem 285:31923–31929. doi: 10.1074/jbc.M110.152447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnson EF, Mukhopadhyay B. 2008. Coenzyme F420-dependent sulfite reductase-enabled sulfite detoxification and use of sulfite as a sole sulfur source by Methanococcus maripaludis. Appl Environ Microbiol 74:3591–3595. doi: 10.1128/AEM.00098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jespersen M, Pierik AJ, Wagner T. 2023. Structures of the sulfite detoxifying F420-dependent enzyme from methanococcales. Nat Chem Biol 19:695–702. doi: 10.1038/s41589-022-01232-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zopfi J, Ferdelman TG, Fossing H. 2004. Distribution and fate of sulfur intermediates—sulfite, tetrathionate, thiosulfate, and elemental sulfur—in marine sediments. In Amend JP, Edwards KJ, Lyons TW (ed), Sulfur biogeochemistry - past and present. Geological Society of America. [Google Scholar]
- 43. Sattler C, Wolf S, Fersch J, Goetz S, Rother M. 2013. Random mutagenesis identifies factors involved in formate-dependent growth of the methanogenic archaeon Methanococcus maripaludis. Mol Genet Genomics 288:413–424. doi: 10.1007/s00438-013-0756-6 [DOI] [PubMed] [Google Scholar]
- 44. Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 45. Day LA, Kelsey EL, Fonseca DR, Costa KC. 2022. Interspecies formate exchange drives syntrophic growth of Syntrophotalea carbinolica and Methanococcus maripaludis. Appl Environ Microbiol 88:e0115922. doi: 10.1128/aem.01159-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tumbula DL, Makula RA, Whitman WB. 1994. Transformation of Methanococcus maripaludis and identification of a Pst I-like restriction system. FEMS Microbiol Lett 121:309–314. doi: 10.1111/j.1574-6968.1994.tb07118.x [DOI] [Google Scholar]
- 47. Potter CJ, Luo L. 2010. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One 5:e10168. doi: 10.1371/journal.pone.0010168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rubin BE, Diamond S, Cress BF, Crits-Christoph A, Lou YC, Borges AL, Shivram H, He C, Xu M, Zhou Z, Smith SJ, Rovinsky R, Smock DCJ, Tang K, Owens TK, Krishnappa N, Sachdeva R, Barrangou R, Deutschbauer AM, Banfield JF, Doudna JA. 2022. Species- and site-specific genome editing in complex bacterial communities. Nat Microbiol 7:34–47. doi: 10.1038/s41564-021-01014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skerker JM, Leon D, Price MN, Mar JS, Tarjan DR, Wetmore KM, Deutschbauer AM, Baumohl JK, Bauer S, Ibáñez AB, Mitchell VD, Wu CH, Hu P, Hazen T, Arkin AP. 2013. Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates. Mol Syst Biol 9:674. doi: 10.1038/msb.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuchino K, Wasser D, Soppa J. 2021. Genome copy number quantification revealed that the ethanologenic alpha-proteobacterium Zymomonas mobilis Is polyploid. Front Microbiol 12:705895. doi: 10.3389/fmicb.2021.705895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables.
Additional metadata.
Data Availability Statement
Data for successful experiments are available via the Fitness Browser (http://fit.genomics.lbl.gov), which includes details of the experimental conditions, quality metrics for each experiment, per-strain fitness values, gene fitness scores, cofitness and conserved cofitness, and t values. All data from the fitness browser and tab-delimited files for M. maripaludis strains S2 and JJ are also archived on Figshare at https://doi.org/10.6084/m9.figshare.24195057.v1. In addition to being available from the fitness browser and archived on the Figshare, all growth media information and experiment metadata and quality information are available in the Supplemental Data Set. A list of M. maripaludis strain S2 essential genes excluded from the fitness analysis can be found in the Supplemental Data Set. All data from the Fitness Browser as of February 2024 are archived on Figshare at https://doi.org/10.6084/m9.figshare.25236931.v1.