Abstract
Human cytomegalovirus (HCMV) exhibits a highly restricted host range. In this study, we sought to examine the relative significance of host and viral factors in activating early gene expression of the HCMV UL54 (DNA polymerase) promoter in murine cells. Appropriate activation of the UL54 promoter at early times is essential for viral DNA replication. To study how the HCMV UL54 promoter is activated in murine cells, a transgenesis system based on yeast artificial chromosomes (YACs) was established for HCMV. A 178-kb YAC, containing a subgenomic fragment of HCMV encompassing the majority of the unique long (UL) region, was constructed by homologous recombination in yeast. This HCMV YAC backbone is defective for viral growth and lacks the major immediate-early (IE) gene region, thus permitting the analysis of essential cis-acting sequences when complemented in trans. To quantitatively measure the level of gene expression, we generated HCMV YACs containing a luciferase reporter gene inserted downstream of either the UL54 promoter or, as a control for late gene expression, the UL86 promoter, which directs expression of the major capsid protein. To determine the early gene activation pathway, point mutations were introduced into the inverted repeat 1 (IR1) element of the UL54 promoter of the HCMV YAC. In the transgenesis experiments, HCMV YACs and derivatives generated in yeast were introduced into NIH 3T3 murine cells by polyethylene glycol-mediated fusion. We found that infection of YAC, but not plasmid, transgenic lines with HCMV was sufficient to fully recapitulate the UL54 expression program at early times of infection, indicating the importance of remote regulatory elements in influencing regulation of the UL54 promoter. Moreover, YACs containing a mutant IR1 in the UL54 promoter led to reduced (∼30-fold) reporter gene expression levels, indicating that HCMV major IE gene activation of the UL54 promoter is fully permissive in murine cells. In comparison with HCMV, infection of YAC transgenic NIH 3T3 lines with murine cytomegalovirus (MCMV) resulted in lower (more than one order of magnitude) efficiency in activating UL54 early gene expression. MCMV is therefore not able to fully activate HCMV early gene expression, indicating the significance of virus over host determinants in the cross-species activation of key early gene promoters. Finally, these studies show that YAC transgenesis can be a useful tool in functional analysis of viral proteins and control of gene expression for large viral genomes.
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that causes severe disease in newborns and immunosuppressed patients. The expression of HCMV genes upon infection is temporally regulated. The first genes expressed (immediate-early [IE] genes) are independent of any de novo protein synthesis and encode mainly either regulatory or immune-modulator factors (1, 11, 20). The regulatory IE genes, together with cellular host factors, coordinate the next level of gene expression (early [E] genes). The E genes are dependent on the viral IE proteins and contribute an essential source of factors, including viral DNA replication and repair enzymes and other nonstructural proteins. Late (L) genes are essentially expressed after the onset of viral DNA replication and contribute primarily to the assembly and morphogenesis of the virus (6).
Activation of the UL54 (DNA polymerase) promoter at early times is essential for DNA replication; accordingly, the UL54 promoter is one of the most intensely studied E gene promoters of HCMV. A number of factors involved in regulating viral E gene expression have been identified, these primarily being the major IE genes, IE1 and IE2 (UL122-3), and the UL36-38, IRS1-TRS1, and UL112-113 gene products. The precise control of temporal gene regulation in HCMV is not clearly understood. Analysis of the UL54 promoter and upstream region revealed that a single element (inverted repeat 1 [IR1]) is primarily responsible for the major IE-mediated activation of the promoter (13, 14). IE2 (IE86), a sequence-specific DNA binding protein, but not IE1 (IE72), is necessary and sufficient for activation of the UL54 promoter (24). IE2 apparently does not bind directly to IR1 but rather mediates the transcriptional activation of cellular factors bound to IR1. Recent studies have shown that the transcription factor Sp1 binds to IR1 and may potentially recruit IE86 to the UL54 promoter (18, 25). Thus, IE2 and probably the UL112-113 and IRS1-TRS1 gene products may directly or indirectly work as coactivators of the Sp1 protein on the IR1 element of the UL54 promoter. The function, if any, of coactivation in the transcription of UL54 is not known.
HCMV exhibits a highly restricted host range. This strict species specificity, the reason for which is not known, has hampered the development of suitable animal model systems for HCMV. Murine cells are known to be competent for infection by HCMV but are apparently restricted in the ability to replicate viral templates (10, 17). It has often been assumed that inappropriate levels of gene activation of key E genes, necessary for viral replication, may be in part responsible for the inability of the virus to replicate (5). Thus, an understanding of the species restriction of HCMV requires determining the relative importance of virus versus cellular factors in governing cross-species activation of its key viral E genes. A better understanding of the species restriction of CMV may lead to new ways of overcoming limitations in crossing the species barrier.
In this study, we used a novel genetic system based on yeast artificial chromosomes (YACs) to examine the transcriptional activation of the UL54 promoter in murine cells. The findings of our study show that optimal E gene activation of the UL54 promoter is fully permissible in murine cells and that murine cells contain host factors necessary for coordinating activation of HCMV E gene expression by the viral master regulator (IE2/IE86), indicating that the species restriction likely occurs downstream of HCMV E gene activation. Markedly, activation of UL54 was found to be strictly virus species restricted, showing the dominance of virus over host factors in the cross-species activation.
MATERIALS AND METHODS
Yeast, cells, and virus.
Saccharomyces cerevisiae strain YPH857 (Matα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 cyh2R) and derivatives were grown in standard media (9). Murine NIH 3T3 fibroblasts (ATCC CRL 1658) were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM glutamine, 100 U of penicillin per ml, 100 μg of gentamicin per ml, and 10% calf serum. Human foreskin fibroblasts (HFF cells) were cultured in DMEM as above, except with 10% fetal bovine serum. The AD169 and Towne strains of HCMV (ATCC VR-538 and VR-977, respectively) and the Smith strain of murine CMV (MCMV; ATCC VR-1399) were used.
Plasmid constructs.
pRML1 and pRML2 were used to create a series of right- and left-arm plasmids, respectively, of the YACs (23). pRML1+I2, the right-arm plasmid, was constructed in three steps. First, an EcoRI fragment from cosmid pCM1035 (22), containing the last 7,297 bp of the 3′ end of HCMV plus the first 3,967 bp of the 5′ end, was cloned into pBluescript-SK (Stratagene, La Jolla, Calif.), yielding pBS-SK-I1. This plasmid was digested with NotI and self-ligated, creating pBS-SK+I and -I2. The NotI-ClaI fragment from pBS-SK+I2 was purified and ligated to pRML1 (23) to construct pRML1+I2. This plasmid has 4,895 bp of the 5′ end of HCMV. pRML2(ura)+Ea, the left-arm plasmid, was constructed by cloning a 3,292-bp EcoRI fragment (nucleotide positions 175524 to 178816 in the HCMV genome; GenBank accession number X17403) from cosmid pCM1050 into pRML2 (22, 23). YPH857 has the appropriate auxotrophies to select for both YAC arm plasmids. To construct pRML2-Leu2-PUR, the PvuII-BamHI 1.4-kb DNA fragment from pPUR was inserted into the blunt-ended BamHI site of pRML2-Leu2 (Leu2 is a yeast protein involved in the synthesis of leucine, which complements the leu2 mutation of YPH857), generating pRML2/D. Then the yeast LEU2 gene was cloned in the place of URA3 as a HindIII DNA fragment. To develop a mutagenesis shuttle plasmid in which to clone the different promoters, we generated pLHN. This plasmid has the yeast HIS3 gene as a selectable marker in yeast, the neomycin resistance gene for selection in mammalian cells, and the luciferase gene as a reporter. The promoter sequence and the 3′ end of the targeted gene were both cloned, flanking the unique SmaI restriction site. pLHN.UL54wt was constructed as follows. A 445-bp DNA fragment from the UL54 promoter (nucleotide positions 80996 through 81441 of the HCMV genome), present in plasmid pPolCAT (24), and viral DNA fragment from the 3′ region of UL54 (nucleotide positions 77291 through 77516) were cloned into the multicloning site of pLHN, leaving a unique SmaI site between them. Digestion with SmaI directs recombination to the UL54 locus. pLHN.UL54IRM was constructed by cloning the mutated EcoRI-SmaI DNA fragment (see “IRM mutagenesis,” below) into EcoRI-SmaI-digested pLHN.UL54wt, replacing the wild-type (wt) fragment with the mutant one. PCR was used to test the correct integration of pLHN.UL54wt into the viral sequences of Y24P. Two oligonucleotides were used: YAC54.2 (TCGCCCTGGATATCGACCCGCT), complementary to a region of the UL54 promoter outside the one included in plasmid pLHN.UL54wt; and GLprimer2 (CTTTATGTTTTTGGCGTCTTCCA), complementary to the 5′ region of the luciferase gene. pLHN.UL86 was constructed as follows. A 432-bp DNA fragment, encompassing the UL86 promoter (nucleotide positions 128318 through 128750 of the HCMV genome), was cloned along with a 522-bp DNA fragment from the 3′-end fragment of the UL86 gene (nucleotide positions 124981 through 125503), with a SmaI site between them (Figure 2A). PCR was used to test the correct integration of pLHN.UL86 into the viral sequences of Y24P. The oligonucleotides used were YAC86.2 (GTAGCCGGAGACGGCGGTT), complementary to a region of the UL86 outside the one included in the plasmid pLHN.UL86, and GLprimer2.
FIG. 2.
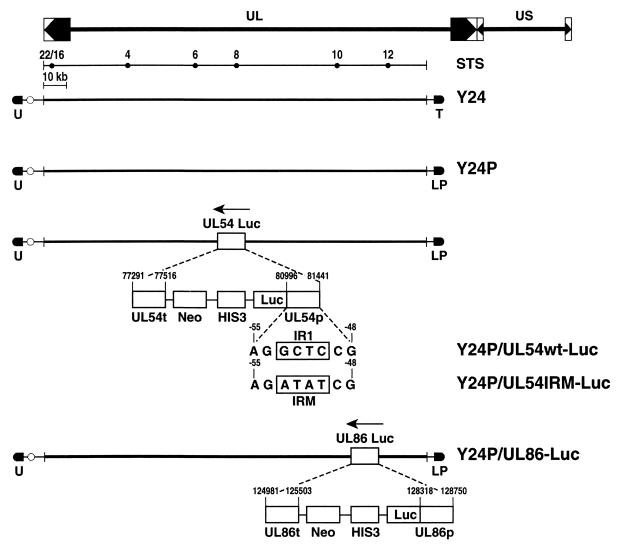
Structure of the HCMV genome, showing the UL and US (unique short) regions and the inverted and direct repetitive sequences. The STS diagram shows relative positions of the pairs of oligonucleotides used to check the integrity of HCMV. Five YACs are shown, with their corresponding mutations outlined. The selectable markers (U, URA3; T, TRP1; L, LEU2; P, puromycin resistance; Neo, neomycin resistance) are shown, as well as the positions of the HCMV sequences where the mutations were performed.
PFGE.
For the separation of yeast chromosomes, we used a Biometra Rotaphor R 23. The yeast DNA samples, agarose gels, and pulsed-field gel electrophoresis (PFGE) conditions were as directed by the manufacturer. To separate Y24, an artificial chromosome of approximately 180 kb, we used a 20-h run at 180 V, with 15-s intervals and a rotation angle of 120°.
STS analysis.
Specific pairs of primers were designed along the HCMV sequence, with an approximate spacing between them of 20 kb, to allow for checking the intactness of the viral genome. A subset of those sequence-tagged site (STS) pairs was used in this study see Fig. 2). PCR was performed using either yeast total DNA of a strain containing Y24 or DNA extracted from HCMV AD169 as a positive control. Each PCR mixture contained 100 ng of template DNA and 25 pmol of each oligonucleotide of the STS pair in a total volume of 25 μl, using standard PCR conditions. The reactions were performed as follows: 1 cycle at 94°C for 3 min, followed by 30 cycles of amplification for 30 s at 94°C, 1 min at 50°C, and 1 min at 72°C. An aliquot of the reaction mixture was loaded in a 1% agarose gel to check for the presence of the expected DNA fragment size. The pair S22 (oligonucleotides S225 [ATGAGGATCGCGACAG] and S223 [CGTTATCCGTTCCTCG]; positions 3809 and 4186, respectively, in the HCMV genome) amplifies a DNA fragment of 378 bp; the pair S04 (oligonucleotides S045 [AGATGGATTCGTGCAC] and S043 [ATCGATCTGGAGCACT]; positions 36489 and 36714, respectively) amplifies a DNA fragment of 226 bp; the pair S06 (oligonucleotides S065 [GGTCCGCAACTTCTGATCCA] and S063 [CAGATCAGTCCACAGGTTCT]; positions 66701 and 67183, respectively) amplifies a fragment of 483 bp; the pair S08 (oligonucleotides S085 [ACGCAGGTGAATATCC] and S083 [AGGTTATCGTCAAGCG]; positions 84721 and 85094, respectively) amplifies a DNA fragment of 374 bp; the pair S10 (oligonucleotides S105 [GATGGTGGAAATCGGA] and S103 [ATATCGCACCGATTGC]; positions 128975 and 129337, respectively) amplifies a DNA fragment of 363 bp; the pair S12 (oligonucleotides S125 [GTTGGCGTTGAGCACGTCTA] and S123 [AGCCGACAACCTGCTGCACT]; positions 149371 and 149770, respectively) amplifies a DNA fragment of 400 bp.
IRM mutagenesis.
The mutagenesis of IR1 in the UL54 promoter was done as previously described (13), using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) as instructed by the manufacturer. Briefly, 50 ng of pLHN.UL54wt DNA was mixed with the oligonucleotides IRM-A (GACACGTCGTTACAGATATCGCCTTCCTACGAGG) and IRM-B (CCTCGTAGGAAGGCGATATCTGTAACGACGTGTC), 125 ng of each, then incubated for 1 min at 95°C, and subjected to 16 PCR cycles (30 s at 95°C, 1 min at 55°C, and 15 min at 68°C). The reaction mixtures were digested with DpnI, and the digested DNA was transformed into Escherichia coli. Several clones were screened with the EcoRV site, a unique restriction site introduced with the IRM mutation. Positive clones of the new pLHN.UL54IRM plasmid were confirmed by sequencing.
PEG spheroplast fusion.
The method described by Julicher et al. (12) was followed. A yeast strain containing the YAC plasmid of interest was grown until it reached log phase (optical density at 600 nm of 0.6 to 0.8). The cells were washed twice with water and resuspended in SCE (1 M sorbitol, 0.1 M sodium citrate, 60 mM EDTA [pH 7.0]), 9 mM dithiothreitol, and 100 U of lyticase/ml. The protoplasts that formed were washed twice with SCE. Then 108 yeast spheroplasts were fused to 2 × 106 NIH 3T3 cells in 0.5 ml of 50% polyethylene glycol (PEG) 1500–10 mM CaCl2 for 2 min, washed with serum-free medium, and plated. After 48 h, neomycin was added at a concentration of 400 μg/ml; colonies appeared 2 weeks later.
Transfection and luciferase assay.
NIH 3T3 cells were grown in DMEM supplemented with 10% calf serum. Transfections were done in six-well plates, unless otherwise noted, by the calcium phosphate precipitation method (7). For transient transfections, cells were washed with phosphate-buffered saline 16 to 18 h after addition of the precipitate, then incubated for 24 h with 10% calf serum supplemented medium, and finally infected with either the Towne strain of HCMV or the Smith strain of MCMV in 3% calf serum. Stable transfections were done in the same way except that after the 24-h recovery in 10% calf serum medium, cells were selected on G418 (400 μg/ml; GIBCO BRL, Gaithersburg, Md.). Clones appeared 10 days after selection. Virus was adsorbed for 2 h prior to the addition of fresh overlay medium.
To measure luciferase activity, the Promega luciferase assay system was used as recommended by the manufacturer. Cells were washed twice with phosphate-buffered saline, and 200 μl of lysis buffer (provided by the manufacturer) was added to each well. Cells were scraped, transferred to an Eppendorf tube, freeze-thawed, and centrifuged for 2 min. The luciferase assays were done in a microplate luminometer (LB 96V; EG&G Berthold), using 100 μl of extract and injecting 100 μl of luciferase substrate, with 2-s delay and 20-s reading time. Luciferase activity is shown either as the absolute value in relative light units (RLU), as an average of at least two experiments done in triplicate, or as fold activation (absolute value divided by the value for corresponding mock-infected sample).
Reverse transcriptase-mediated PCR.
NIH 3T3 or HFF cells were infected with the Towne strain of HCMV at a multiplicity of infection (MOI) of 1. Total RNA was isolated at different times after infection by the RNAzol method (Tel-Test, Inc., Friendswood, Tex.) according to the manufacturer's protocol. RNA samples were treated with RNase-free DNase I for 15 min at room temperature, and the DNase was inactivated at 65°C for 15 min. The RNA was reverse transcribed using oligo(dT) primers at 42°C for 50 min, and reactions were terminated by heating at 70°C for 15 min. The reverse-transcribed products were treated with RNase H for 20 min at 37°C and amplified using specific primers. Primers IEP4BII (CAATACACTTCATCTCCTCGAAAGG) and IEP3C (CAACGAGAACCCCGAGAAAGATGTC) were used to amplify a 217-bp product within the HCMV ie1 gene (15), and primers RTUL54-2R (AAGCCGGCTCCAAGTGCAAGCGCC) and RTUL54-6F (CGTGTGCAACTACGAGGTAGCCGA) were used to amplify a 199-bp fragment within the HCMV UL54 gene. Primers TF-R and TF-F, designed to amplify a 601-bp product within the human tissue factor (TF) gene, and primers HPRT-R and HPRT-F, designed to amplify a 163-bp within the murine hypoxanthine phosphoribosyltransferase (HPRT) gene, have been previously described (2, 16). PCRs were performed under the following conditions: 1 cycle at 94°C for 3 min; 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; and 1 cycle at 72°C for 10 min. Control reactions carried out in the absence of reverse transcriptase were used to assess the specific detection of RNA. Amplified products were separated on a 1% agarose gel and visualized by ethidium bromide staining.
RESULTS
Plasmid-based analysis of HCMV UL54 promoter activity in NIH 3T3 cells, using transient expression assays and stable cell lines.
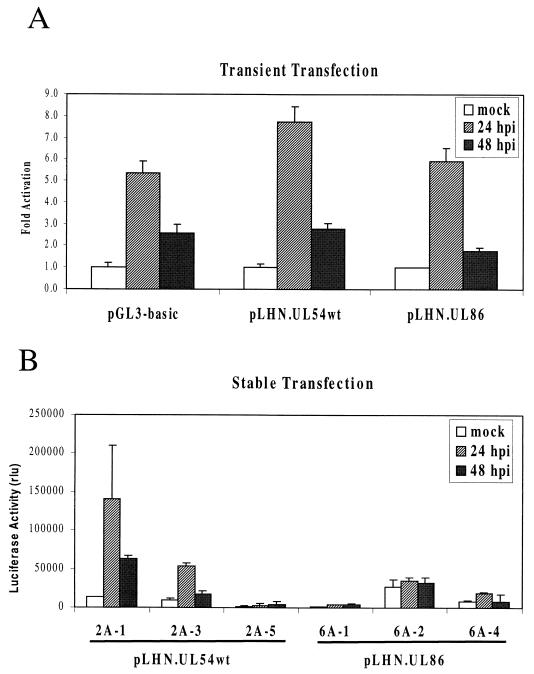
We first used a plasmid-based transient expression assay to investigate activation of the HCMV UL54 promoter by HCMV in murine cells. For these experiments, a 445-bp DNA fragment containing the UL54 promoter (nucleotide positions 80996 through 81441 in the HCMV genome) was cloned in plasmid pLHN, to produce pLHN.UL54wt (see Materials and Methods). We also cloned a 432-bp DNA fragment corresponding to the UL86 promoter (nucleotide positions 128318 through 128750 in the HCMV genome) into pLHN, deriving pLHN.UL86. In these recombinants, the UL54 and UL86 promoters drive the expression of the luciferase reporter gene. To test if the species restriction for HCMV in murine cells occurs at early times, each plasmid was transiently transfected into NIH 3T3 cells, which were then infected with HCMV, and luciferase activity was measured at different times postinfection. As shown in Fig. 1A, the infection caused a general activation of transcription, reflected by a 5-fold activation of the promoterless pGL3-basic plasmid 24 h postinfection (hpi) compared to the mock-infected sample, followed by a 2.5-fold activation at 48 hpi. Overall activation of the UL54 promoter was slightly (sixfold) higher but similar to that for the negative control (Fig. 1A). UL86 promoter-driven luciferase activity responded in a similar manner (Fig. 1A). When we tested the same plasmid in transient transfection assays in human HFF cells, the UL54 promoter was, as expected (13), specifically responsive to HCMV infection (data not shown). Thus, transient expression from isolated HCMV promoters is apparently not specifically responsive to HCMV infection in murine cells.
FIG. 1.
(A) Transient transfection assays using 1 μg of DNA of the plasmids described, followed by infection with HCMV at an MOI of 10. NIH 3T3 cells were lysed at different times postinfection or at the moment of infection (mock), and luciferase activity of the extracts was measured. Relative luciferase activities from triplicate measurements are shown. The RLU values at each time postinfection are normalized to the activity in the corresponding mock-infected cells. (B) Stably transfected clones were infected with HCMV at an MOI of 10, and samples were taken at different times postinfection. The average RLU values from at least two experiments performed in triplicate are shown. Clones 2A-1, 2A-3, and 2A-5 are three representative clones stably transfected with pLHN.UL54wt; 6A-1, 6A-2, and 6A-4 are representative clones stably transfected with pLHN.UL86.
To test if the nonresponsiveness of the HCMV promoters was due to the nature of the transient assay or to the inherent nonresponsiveness of the HCMV promoters in murine cells, we used the same plasmids to generate neomycin-resistant stable clones. Of the 12 neomycin-resistant UL54 promoter clones isolated, 4 showed some luciferase activity when infected with HCMV. The results for three representative clones are shown in Fig. 1B (2A clones); the luciferase activity values were variable, ranging from 3- to 10-fold peak activation at 24 hpi, which decreased by 48 hpi. The noninfected samples had some residual activity in the absence of infection, most likely influenced by the site of integration. To examine whether the effect of HCMV infection is specific, we next investigated the expression from UL86 (late promoter) stable cell lines containing plasmid pLHN.UL86. Since HCMV cannot replicate in murine cells, we expect the UL86 promoter not to be activated if regulation is specific, while a nonspecific enhancement would cause some expression to be detected upon infection. In these experiments, the UL86 neomycin-resistant clones (Fig. 1B, 6A clones) showed no significant activation of transcription either in the absence or in the presence of HCMV. Overall, we conclude that the UL54 promoter, when isolated from its natural context, is poorly activated by HCMV infection in murine cells.
Cloning and mutagenesis of an HCMV YAC.
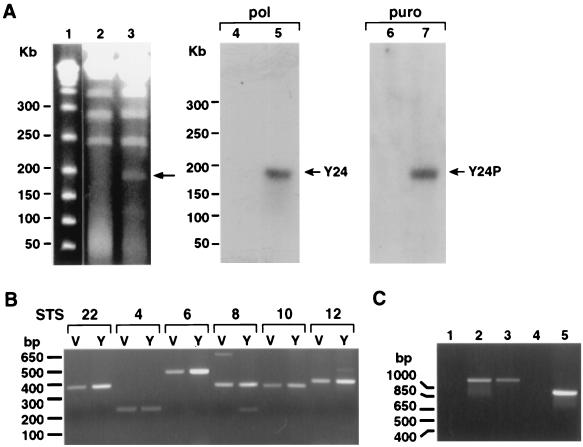
It is possible that the lack of a robust activation of UL54 was due to integration in host chromatin and/or the lack of its natural sequence context. To investigate the influence of cis-acting sequences on the activity of the UL54 promoter in its natural context, we constructed a YAC with a 178-kb HCMV DNA fragment, encompassing most of the UL (unique long) region of the viral genome (Fig. 2, Y24; for details, see Materials and Methods). This part of the HCMV genome lacks the major IE gene region and is expected to be defective for viral growth. Briefly, the centromeric plasmid pRML1+I2 contains a 4,895-bp DNA fragment from the 5′ end of HCMV genome. The noncentromeric plasmid pRML2(ura)+Ea has a 3,292-bp fragment from the 3′ end of the UL region. Both plasmids were cotransformed into yeast strain YPH857 with HCMV DNA (strain Towne). Different clones were obtained, and the integrity of the selected YAC (Y24) was verified by several methods. If recombination between the two plasmids and the viral DNA is as expected, a new chromosome of around 178 kb, encompassing the HCMV genome from the 5′ end to position 178816, should be generated. Figure 3A (left) shows results of PFGE of Y24 and of the parental strain YPH857. Y24 migrates as an approximately 180-kb extra chromosome in the stained gel. When the separated chromosomes were hybridized to an HCMV UL54-specific probe, only Y24 showed specific hybridization (Fig. 3A, middle).
FIG. 3.
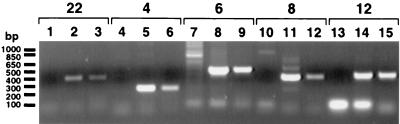
(A) PFGE to resolve different DNA samples: size markers (lane 1), YPH857 (lanes 2 and 4), Y24 (lanes 3, 5, and 6), and Y24P (lane 7). The gels were either stained with ethidium bromide (lanes 1 to 3) or hybridized to a UL54-specific probe (pol; lanes 4 and 5) or a puromycin-specific probe (puro; lanes 6 and 7). Sizes of the marker DNA fragments are shown at the left; arrows indicate bands corresponding to either Y24 or Y24P. (B) Agarose gel in which an aliquot of the STS PCR mixture was loaded. The lanes are marked V for AD169 DNA or Y for YAC DNA. The set numbers for the corresponding STS, as well as locations of the size markers, are indicated. (C) PCR to test the UL54-Luc and UL86-Luc mutations and their integration in the correct position in Y24P. The specific primers used were YAC54.2 and GLprimer2 (lanes 1 to 3), and YAC86.2 and GLprimer2 (lanes 4 and 5). The DNA samples are Y24P (lanes 1 and 4), Y24P/UL54wt-Luc (lane 2), Y24P/UL54IRM-Luc (lane 3), and Y24P/UL86-Luc (lane 5).
To further test the overall integrity of the viral DNA, we performed STS analysis with specific primer sets along the HCMV genome. DNA was prepared from a yeast strain harboring Y24, and PCR was performed using six different pairs of STS oligonucleotides specific to consecutive regions of HCMV (locations are shown in Fig. 2; see Materials and Methods for details). The results (Fig. 3B) show that all of the specific primer sets amplified DNA fragments of the expected sizes compared to the viral DNA, demonstrating the overall integrity of the 178,816 bp of viral sequence.
As we wanted to use the HCMV YAC for the analysis of essential cis-acting sequences in a transgenesis system, we next retrofitted the YAC for selection in mammalian cells. The first mutagenesis was targeted to the right arm of the YAC, to introduce the puromycin resistance gene, a mammalian selectable marker. The URA3 gene was replaced by LEU2 plus the puromycin resistance gene, using plasmid pRML2-Leu2-PUR digested with NotI (Materials and Methods and Fig. 2). This replacement is expected to provide a YAC with the same basic features on the noncentromeric arm but including the mammalian selectable marker puromycin. Recombinant yeast clones were selected in the corresponding medium (yeast synthetic medium lacking tryptophan and leucine) and analyzed by PFGE and Southern blot analysis. The expected 180-kb specific band appears in the stained gel; when the puromycin resistance gene was used as a probe in a Southern blot analysis, only the newly made Y24P YAC gave a specific signal of around 180 kb, indicating successful integration (Fig. 3A, right).
To investigate the effects of cis-acting sequences on the UL54 promoter in the context of the UL region, we constructed a YAC, Y24P/UL54wt-Luc, in which a luciferase reporter gene was inserted at position +20 of the UL54 promoter (Fig. 2). The strategy was as follows. SmaI-digested pLHN.UL54wt DNA, which contains the HIS3 gene for selection in yeast, was retrofitted into a strain containing Y24P. Integration of the plasmid in the YAC sequence would produce histidine prototrophs, in which the correct integration can be tested by PCR. Using specific primers, directed to the region adjacent to the UL54 promoter but not included in plasmid pLHN.UL54wt and to the luciferase gene, a specific DNA fragment of the expected size (1.1 kb) was amplified (Fig. 3C, lane 2). This new YAC has, besides the UL54 promoter fused to luciferase and integrated in its natural position in the HCMV genome, the neomycin resistance gene, which confers an additional marker for positive selection in mammalian cells (Fig. 2).
In addition, as a control for HCMV L gene expression, we constructed another YAC in which the luciferase gene was inserted downstream of the UL86 promoter. The strategy was very similar to the one used to generate the UL54-luciferase (UL54-Luc) fusion, but in this case we retrofitted plasmid pLHN.UL86, digested with SmaI, in a yeast strain harboring Y24P (Fig. 2). The recombinant clones were selected on medium lacking histidine; as described above, correct integration was confirmed by PCR using specific primers directed to the region neighboring the UL86 promoter and the luciferase gene (Fig. 3C, lanes 4 and 5).
Analysis of UL54 promoter activity by stable transfer (transgenesis) of the HCMV YAC to NIH 3T3 cells.
We next sought to examine the influence of the surrounding UL region on UL54 transcription activity by direct transfer of Y24P/UL54wt-Luc into NIH 3T3 fibroblasts. In these experiments, 108 yeast spheroplasts were fused to 2 × 106 mouse fibroblasts (see Materials and Methods). After a recovery period of 48 h, selection with neomycin (400 μg/ml) was applied and resistant clones were isolated.
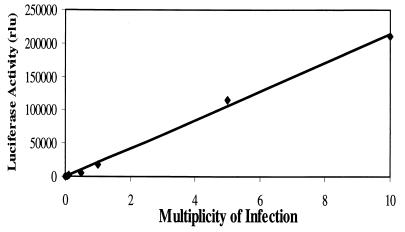
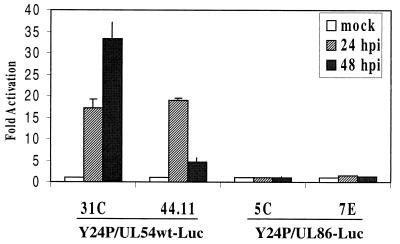
The nine neomycin-resistant clones selected were first tested for luciferase activity after infection with HCMV at an MOI of 10. Six of these clones showed some activity in the luciferase assays; three (clones 31C, 42.8, and 44.11) are shown in Fig. 4. UV-inactivated virus failed to elicit a response (data not shown), and the response of the UL54 promoter to infection in the YAC transgenic lines was proportional to the MOI (Fig. 5). To check the integrity of transferred YACs, we performed STS analysis using some of the specific pairs of oligonucleotides along the HCMV genome (see Materials and Methods). As shown in Fig. 6, PCRs using DNA extracted from NIH 3T3 cells did not amplify any DNA fragment of the expected size. However, DNA from both yeast strain Y24P and the transgenesis YAC clones showed amplified DNA fragments of the expected sizes, indicating that intact YACs had been successfully transferred to NIH 3T3 cells (Fig. 6). In marked contrast to the plasmid transgenic lines (Fig. 4A), the YAC-containing NIH 3T3 cells showed very low luciferase activity when mock infected; 24 h after infection with HCMV, they exhibited a very strong upregulation, which continued to increase up to 48 h (Fig. 4A). Furthermore, the absolute values of luciferase activity in the stable YAC clones were less variable and consistently higher than for the stable plasmid-containing cell lines. The basal reporter gene levels in mock-infected YAC transgenic cells are subject to a highly specific and very tight regulation. Comparison between the plasmid and the transgenesis cell lines showed very strong activation of the UL54 promoter upon HCMV infection, ranging from 170 at 24 hpi to 470-fold at 48 hpi in the YAC cell lines (Fig. 4B). This level of activation is much higher than in the stable plasmid clones, suggesting a positive influence of remote viral DNA sequences in addition to more stringent regulation prior to activation. It is noteworthy that the second peak of activation at 48 h is not observed with the plasmid transgenic lines.
FIG. 4.
(A) Y24P/UL54wt-Luc transgenic lines were infected with HCMV at an MOI of 10. Samples were harvested at the indicated times postinfection. Averages of at least two experiments done in triplicate are shown. As a negative control, Y24P/UL86-Luc clones 5C and 7E were used. (B) Fold activation of luciferase activity in transgenic plasmid pLHN.UL54wt lines 2A-1, 2A-3, and 2A-5 and in Y24P/UL54wt-Luc lines 31C, 42.8, and 44.11.
FIG. 5.
The stable NIH 3T3 line containing Y24P/UL54wt-Luc clone 44.11 was infected with HCMV at MOIs from 0.005 to 10. Cells were harvested at 48 hpi, and luciferase activity was measured. Each point represents the average of two independent infections.
FIG. 6.
STS analysis of YAC transgenic NIH 3T3 clones. PCRs were performed using specific primer sets designed along the HCMV genome (see Materials and Methods). DNA from NIH 3T3 cells (lanes 1, 4, 7, 10, and 13), yeast Y24P positive control (lanes 2, 5, 8, 11, and 14), or NIH 3T3 cells expressing Y24P/UL54wt-luc clone 44.11 (lanes 3, 6, 9, 12, and 15) was used, along with different sets of primers as indicated at the top.
Transgenesis experiments were also carried out with Y24P/UL86 in murine cells. The UL86 promoter is a true L promoter (4), being activated only after the onset of DNA replication. The transfer yielded three neomycin-resistant clones, and the presence of the YAC sequences was tested by Southern blotting and STS analysis (data not shown). None of these clones showed any luciferase activity before or after infection with HCMV (Fig. 4A, clones 5C and 7E). The lack of activation of UL86 transcription in the transgenesis experiments indicates the expected restriction in replication of HCMV in murine cells. Altogether, these results underscore the importance of sequence context in providing an optimal level of gene expression.
Analysis of the UL54 promoter activation pathway by HCMV in murine cells.
The primary pathway for early activation of the UL54 promoter is dependent on IE86-mediated activation via the IR1 element that binds the cellular transcription factor Sp1. To investigate the early activation pathway of HCMV UL54 in murine cells, we accordingly mutated the IR1 element present in the UL54 promoter (13). Figure 2 shows the point mutations introduced in the IR1 site, which has been previously shown to completely eliminate IE86 transactivation of the UL54 promoter (14).
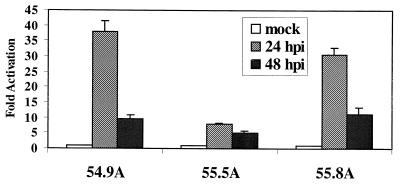
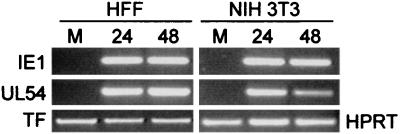
In agreement with previous studies, mutant plasmid pLHN.UL54IRM showed a much lower activation after transfection in HFF cells and infection with HCMV (around 17-fold reduction) compared to the wt counterpart (data not shown) (13). On the basis of this result, we next retrofitted plasmid pLHN.UL54IRM into Y24P, to study the effect of the IRM mutation in the UL54 promoter in the context of the UL region. Figure 2 shows a schematic of the recombinant generated in yeast (Y24P/UL54IRM-Luc), and its integrity was confirmed by PFGE, STS analysis, and Southern blot hybridization (not shown). We next performed transgenesis studies by spheroplast fusion of Y24P/UL54IRM-Luc to NIH 3T3 cells. The PEG fusion produced eight neomycin-resistant clones, three of which showed luciferase activity after infection with HCMV (Fig. 7). The profile of activation, with a peak at 24 h, was different from that for the UL54 wt promoter (compare Fig. 4B and 7), but most significantly, both the absolute values (not shown) and the extent of activation (10- to 30-fold [Fig. 7]), were significantly diminished. The remaining activity is most likely due to the ATF binding site positioned upstream of the IR-Sp1 element of the UL54 promoter (13). These results suggest that the pathway used for HCMV early activation of the UL54 promoter is conserved in murine cells and that the same or functionally similar cellular factors bind to the UL54 promoter. In agreement with this view, we observe IE1 and UL54 RNA upon infection of NIH 3T3 cells with HCMV (Fig. 8). From these experiments, we conclude that HCMV early activation of the UL54 promoter by IE transactivators is not restricted by host species-encoded cellular factors.
FIG. 7.
Stable Y24P/UL54IRM-Luc-containing NIH 3T3 clones were infected with HCMV at an MOI of 10. Samples were harvested at the indicated times, and luciferase activity determined as indicated in Materials and Methods. Each point represents the average of at least two independent experiments done in triplicate. Fold activation was calculated by dividing the absolute value by the mock-infected value. Results for three representative clones are shown.
FIG. 8.
Detection of HCMV IE1 and UL54 transcripts in 3T3 cells infected with HCMV. NIH 3T3 or HFF cells were mock infected (M) or infected with HCMV at an MOI of 1. Total RNA was harvested at different times (hours postinfection) indicated, treated with DNase, and reverse transcribed by using oligo(dT). PCRs were performed using primer sets specific for HCMV IE1 UL54, human TF (for HFF samples), and murine HPRT (for NIH 3T3 samples) as described in Materials and Methods. Amplified products were separated on 1% agarose gels and visualized by ethidium bromide staining. Amplified fragments obtained in the different reactions are shown. Sizes were as expected for each primer set (see Materials and Methods for details). Specific PCR-amplified products were not detected in control reactions in which reverse transcriptase was not added during the RNA reverse transcription reaction (data not shown).
Analysis of MCMV activation of the HCMV UL54 promoter in YAC transgenic clones.
In the experiments described above, we maintained a homologous promoter-virus relationship in a heterologous host cell background and demonstrated that the activation of E gene expression uses the same cellular pathway in both human and murine cells. Knowing that the cellular pathway is conserved between human and murine cells, we next sought to test the effect of changing the species origin of the virus. For these experiments we infected the YAC transgenic clones Y24P/UL54wt-Luc 31C and 44.11 with MCMV and examined their levels of activation of the UL54 promoter. In this scenario, the cell-virus relationship is homologous (both are murine), while the promoter-virus relationship is heterologous.
We infected the two Y24P/UL54wt-Luc clones with MCMV at an MOI of 10 and determined luciferase activity at 24 and 48 hpi. The maximum transcriptional activity by MCMV was 10-fold lower than the activation by HCMV (Fig. 4B and 9, clones 31C and 44.11). This lower activation implicates restriction at a viral trans-acting factor level, necessitating the concordance of species origin of both promoter and viral transactivators to recapitulate the activation profile of the UL54 promoter. As anticipated, the Y24P/UL86-Luc 5C and 7C clones were not activated by MCMV infection (Fig. 9). Taken together, these results suggest that there is a dominance of virus over host factors in determining the species specificity of E gene expression.
FIG. 9.
Y24P/UL54wt-Luc-containing NIH 3T3 clones were infected with MCMV at an MOI of 10. Samples were harvested at the indicated times, and luciferase activity measured. Each point represents the average of at least two independent experiments done in triplicate. Fold activation was calculated by dividing the absolute value by the mock-infected value. Results for two representative clones are shown.
DISCUSSION
CMV is a pathogen with a highly restricted spectrum of hosts. HCMV shows a very strong species specificity, being able to replicate only in human cells. HCMV can efficiently enter cells of other species, indicating that the species restriction is not at the level of viral binding or penetration (21). Recent studies have provided direct evidence that major IE expression determined by viral IE regulatory sequences is also not species restricted (3, 8). In the present study we show that an essential E gene activation pathway governed by HCMV major IE proteins is conserved between host species. However, we observed a marked divergence in the mechanism for this activation between viral species.
We used a YAC transgenesis approach to examine the species restriction checkpoint of an essential E gene activation pathway of HCMV in murine cells. The YAC vectors have advantages over other conventional vectors like plasmids and cosmids: they allow the manipulation of larger fragments of DNA, and the genetic manipulations can be accurately and easily performed in yeast. Toward this end, we designed a YAC vector encompassing most of the UL region of HCMV. This vector is replication defective, lacks the major IE region, and thus permits the analysis of essential cis-acting sequences within the UL region when complemented in trans. For the purpose of this study, we chose to study essential cis-acting sequences of the viral DNA polymerase (UL54) promoter complemented by HCMV infection in murine cells. The isolated UL54 promoter, in both transient and stable transfections of murine cells, showed a profile of activation not consistent with an E gene. Only when in the context of the UL region was the UL54 promoter able to recapitulate the early (48 hpi) gene activation described for human cells (13). To avoid variability, we used the same plasmids for the transfection assays and for the YAC mutagenesis. In the UL54wt plasmid stable clones, the basal activity in mock-infected cells was not as tightly regulated as in the YAC clones, and the extent and profile of activation were more random. This suggests the need for tight regulation of early events, and perhaps the involvement of long-range cis-acting sequences (sequence context) for optimal UL54 gene regulation. Indeed, the results of this study strongly support the suggestion that remote regulatory sequences participate in the control of expression of this essential E gene.
Members of the CMV family have evolved by cospeciation with their host and as a consequence have had sufficient time to acquire a significant degree of genetic drift (19). In particular, the genetics of HCMV speciation has led to nonviable replication in other host species. In this case we can assume that genetic changes that have been beneficial or neutral in its natural host background may well be deleterious in another host genetic background because of negative or inappropriate gene interactions that had not been screened by natural selection. For this reason, species-specific strains of CMV must have accumulated genetic changes that may be advantageous or neutral in its host species but lethal in other species. The results of this study suggest that the virus-host gene interaction pathway for E gene activation is viable between different host species but has significantly diverged between virus species. This strongly argues for coevolution of viral trans-acting factors and their viral target promoters. In agreement, mutation of the IR1 element of the HCMV UL54 promoter showed that the pathway of E activation is the same in human and murine cells. Thus, IE86-mediated activation of UL54 via IR1 is conserved in murine cells. In support, the primary host factor known to bind IR1 is Sp1, in which sequence homology between human and murine Sp1 is over 95%. In contrast, the MCMV transactivator IE3 not only exhibits more sequence differences (<40% identity) from its related HCMV IE86 counterpart but also is extremely inefficient at activating the HCMV UL54 promoter via IR1, indicating a divergence of mechanism of action. A prediction from these observations is that it would not be informative to study HCMV IE proteins in the context of MCMV infection. Consistent with this notion, we find that an IE3 mutant of MCMV is poorly complemented by HCMV infection in murine cells (A. Angulo and P. Ghazal, unpublished results).
In summary, we have used a YAC transgenesis system to explore critical host-virus gene interaction pathways in the cross-species activation of a key E gene promoter of HCMV, UL54. Our results show a clear dominance of virus factors (major IE proteins) for transactivation of the UL54 early promoter and evidence for a species specificity checkpoint at later times of infection. Whether the species barrier can be crossed remains an open question.
ACKNOWLEDGMENTS
We thank Forrest Spencer for plasmids pRML1 and pRML2. We also thank Xiaodong Wu and Danielle Foster for technical assistance and Juan Carlos de la Torre for helpful comments on the manuscript.
This work was supported by grants from the National Institutes of Health to P.G. (AI-30627) and from The R. W. Johnson Pharmaceutical Research Institute. A.A. is a fellow of the Universitywide AIDS Research Program.
Footnotes
This is publication no. 13364-IMM from the Scripps Research Institute.
REFERENCES
- 1.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Kerry D, Huang H, Borst E M, Razinsky A, Wu J, Hobom U, Messerle M, Ghazal P. Identification of a boundary domain adjacent to the potent human cytomegalovirus enhancer that represses transcription of the divergent UL127 promoter. J Virol. 2000;74:2826–2839. doi: 10.1128/jvi.74.6.2826-2839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan J S, Bittner A, Frueh K, Jackson M R, Peterson P A, Erlander M G, Ghazal P. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMarchi J M. Nature of the block in the expression of some early virus genes in cells abortively infected with human cytomegalovirus. Virology. 1983;129:287–297. doi: 10.1016/0042-6822(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 6.DeMarchi J M, Schmidt C A, Kaplan A S. Patterns of transcription of human cytomegalovirus in permissively infected cells. J Virol. 1980;35:277–286. doi: 10.1128/jvi.35.2.277-286.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galang C K, Der C J, Hauser C A. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- 8.Grzimek N K, Podlech J, Steffens H P, Holtappels R, Schmalz S, Reddehase M J. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J Virol. 1999;73:5043–5055. doi: 10.1128/jvi.73.6.5043-5055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 10.Jeang K T, Chin G, Hayward G S. Characterization of cytomegalovirus immediate-early genes. I. Nonpermissive rodent cells overproduce the IE94K protein form CMV (Colburn) Virology. 1982;121:393–403. doi: 10.1016/0042-6822(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 11.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julicher K, Vieten L, Brocker F, Bardenheuer W, Schutte J, Opalka B. Yeast artificial chromosome transfer into human renal carcinoma cells by spheroplast fusion. Genomics. 1997;43:95–98. doi: 10.1006/geno.1997.4787. [DOI] [PubMed] [Google Scholar]
- 13.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskenderian A C, Anders D G, Stenberg R M. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurz S, Steffens H P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lafemina R L, Hayward G S. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 18.Luu P, Flores O. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGeoch D J, Cook S, Dolan A, Jamieson F E, Telford E A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 20.Mocarski E S. Cytomegalovirus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. p. 2447. [Google Scholar]
- 21.Nowlin D M, Cooper N R, Compton T. Expression of a human cytomegalovirus receptor correlates with infectibility of cells. J Virol. 1991;65:3114–3121. doi: 10.1128/jvi.65.6.3114-3121.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer, Connelly C, Hieter P. Targeted recombination-based cloning and manipulation of large DNA segments in yeast. Methods. 1993;5:161–175. [Google Scholar]
- 24.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, O'Neill J, Barbosa M S. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J Virol. 1998;72:236–244. doi: 10.1128/jvi.72.1.236-244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]