Abstract
Pathogens are ubiquitous and a constant threat to their hosts, which has led to the evolution of sophisticated immune systems in bacteria, archaea and eukaryotes. Bacterial immune systems encode an astoundingly large array of antiviral (antiphage) systems, and recent investigations have identified unexpected similarities between the immune systems of bacteria and animals. In this Review, we discuss advances in our understanding of the bacterial innate immune system and highlight the components, strategies and pathogen restriction mechanisms conserved between bacteria and eukaryotes. We summarize evidence for the hypothesis that components of the human immune system originated in bacteria, where they first evolved to defend against phages. Further, we discuss shared mechanisms that pathogens use to overcome host immune pathways and unexpected similarities between bacterial immune systems and interbacterial antagonism. Understanding the shared evolutionary path of immune components across domains of life and the successful strategies that organisms have arrived at to restrict their pathogens will enable future development of therapeutics that activate the human immune system for the precise treatment of disease.
Table of content:
In this Review, Ledvina and Whiteley highlight the key similarities between eukaryotic and bacterial innate immune systems, exploring conserved immune components and signaling strategies, as well as conserved mechanisms for pathogen restriction.
Introduction
Pathogens are a ubiquitous threat faced by all domains of life. To defend themselves against pathogens, host organisms deploy sophisticated, multicomponent immune systems. There are two main categories of immune systems: innate and adaptive. Innate immune systems are largely invariant and recognize conserved signatures or patterns of pathogenesis to elicit rapid immune responses1. By contrast, adaptive immune systems vary across a population and are targeted to specific pathogens based on previous exposures. Bacteria and mammals have both innate and adaptive immune systems. Although the adaptive immune system of bacteria (CRISPR–Cas) and mammals (for example, T cells, B cells and antibodies) share many parallels, in this Review we focus on the commonalities of their innate immune systems.
Historically, our understanding of innate immune signaling has been led by investigation of animals. One of the best examples is detection of bacterial lipopolysaccharides (LPS, also known as endotoxin) by Toll-like receptor 4 (TLR4)2. Almost all Gram-negative bacteria produce LPS as a critical component of their outer membrane. Mammalian cells exploit this aspect of bacterial physiology by detecting LPS as a hallmark of infection via the transmembrane receptor TLR4. The extracellular domain of TLR4 binds LPS, becomes activated and relays that signal by indirectly activating transcription of inflammatory genes. TLR4 does not require previous exposure to LPS and the genes encoding TLR4 are conserved across a population. LPS is an ideal ligand for innate immune detection because it is conspicuous (that is; not produced by other organisms or under other circumstances) and crucial (that is; it cannot be mutated or altered by the bacterium without substantial fitness cost).
Although it was previously thought that bacterial innate immune pathways were limited to restriction–modification systems, we now appreciate this was a vast underestimate. Over the past several years an expansive array of bacterial innate immune pathways has been discovered that defend against phage infection. The rapidly growing list of candidate antiphage pathways led to a surprising finding: some proteins within these pathways are structurally homologous to human innate immune proteins (Box 1). In this Review, we aim to highlight the key similarities between eukaryotic and bacterial innate immune systems.
Box 1: Identifying conservation through discovery of antiphage pathways.
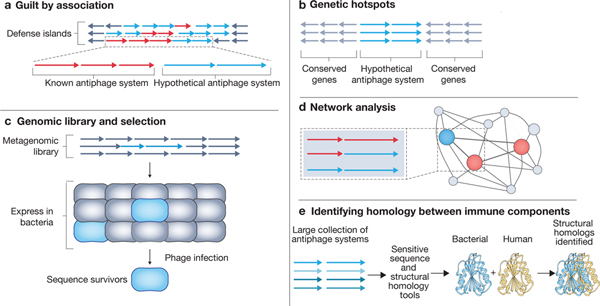
Identifying conserved immune components, strategies and signaling outcomes was made possible by the discovery of antiphage systems (see the figure). These efforts have been fueled by the considerable increase in the number of sequenced bacterial genomes, the development of sensitive bioinformatic tools and the increased interest in bacterium–phage interactions resulting from our improved understanding of CRISPR–Cas systems. The first method for identifying new antiphage operons capitalized on the tendency of these systems to cluster in microbial genomes into ‘defense islands’149, an observation that remained under-appreciated until a seminal paper identified antiphage systems and validated their function150. There have since been numerous new systems identified through this ‘guilt by association’ model35,68 (see the figure, part a). A second approach found that antiphage genes can be found in genomic hotspots, which are identified based on surrounding conserved genes and may only contain a single antiphage system (see the figure, part b). The study found that prophages often have genomic hotspots that provide a reservoir of antiphage systems36. By surveying all the genes found in hotspot locations the authors were able to identify and characterize novel systems. A conceptually similar approach was used in the genomes of Vibrio cholerae isolates151. Spurred on by the shear abundance of antiphage genes, a third approach has been identify antiphage genes from unbiased genomic libraries152 (see the figure, part c). Finally, the last approach takes advantage of the constant reshuffling and re-use of related components to identify defense genes using network analyses12,21,146 (see the figure, part d). This approach was prophetically used in various forms long before the function of many of these proteins or pathways were observed in the laboratory146.
The discovery of uncharacterized antiphage pathways was essential to identifying conserved immune components. Simply comparing the sequences of eukaryotic genes of interest to all bacterial genomes (or vice-versa) using tools like BLAST is rarely successful due to low amino acid sequence conservation. Instead, more sensitive tools that require smaller search-sets of candidate genes or curated databases that account for predicted protein structure must be used (see the figure, part e).
For the purposes of this Review, we define an immune system as a collection of multiple immune pathways that combat a wide range of threats. In turn, each immune pathway (synonymous with antiphage systems) is composed of individual immune components. We highlight three different areas of similarity between bacteria and eukaryotic innate immune systems. These similarities encompass evolutionary relationships demonstrated by homologous proteins, and thematic similarities that unify otherwise unrelated innate immune pathways.
First, we discuss conserved immune components. Second, we highlight conserved signaling strategies that extend beyond clear evolutionary relationships and illustrate shared overarching themes and mechanisms of signaling. Third, we look at conserved mechanisms used by hosts for pathogen restriction (Fig. 1).
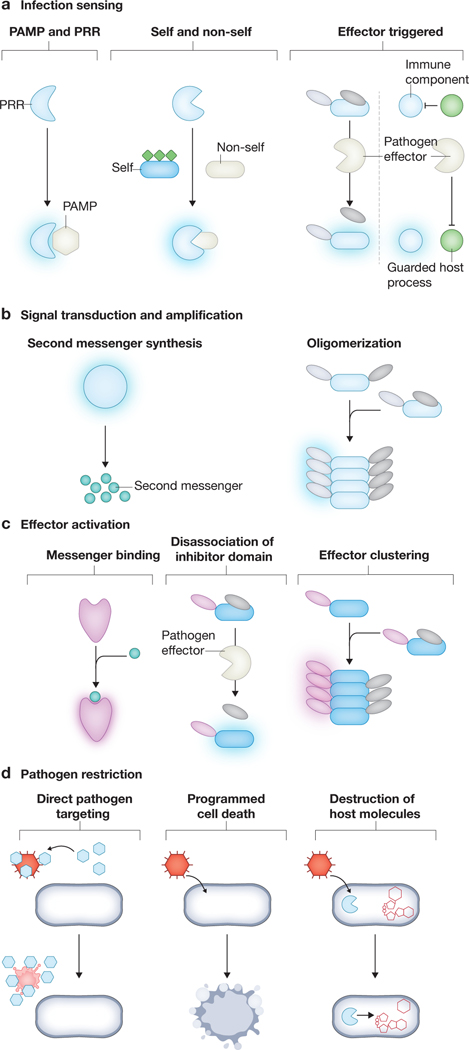
Figure 1: General steps of innate immune signalling pathways.
Innate immune signalling conforms to four general steps: an infection is detected (part a); that signal must be transduced and amplified to alert the cell (part b); the signal must activate an effector (part c); and finally the effector should directly or indirectly restrict pathogen replication (part d).
(a) Infections are detected through three mechanisms. Direct sensing can be mediated by host-encoded pathogen recognition receptors (PRRs) which bind to a specific pathogen associated molecular (PAMP). Binding activates the PRR, triggering the appropriate downstream immune response. Hosts also rely on target modifications to distinguish self and non-self thereby sensing foreign invaders. In this case, an immune component specifically recognizes the non-self version of the target (in the example shown this is the unmodified target) activating the immune response. The final form of sensing, effector triggered, relies on the activity of the pathogen The activity of a pathogen-encoded effector either inadvertently targets an immune component (left) or targets a protein/process that normally inhibits an immune component (right). In either case, pathogen activity results in activation of the appropriate immune pathway.
(b) Upon activation there are two primary mechanisms used by sensors to alert the cell. The sensor can produce a nucleotide second messenger (typically a linear or cyclic oligonucleotide) or a sensor can use oligomerization to trigger activation of effector domains.
(c) One mechanism of effector activation is through direct binding of a messenger (often a nucleotide second messenger). Activation can also occur via disassociation of an inhibitor domain, for example activity of a pathogen-encoded effector can cleave off the inhibitory domain. Clustering of effector domains can also lead to activation, often through completion of active sites.
(d) There are three primary methods of pathogen restriction. The first relies of direct targeting of a pathogen, either through extracellular (as shown in figure) or intracellular mechanisms (not depicted in figure). Infected cells can also undergo programmed cell death in response to infection to limit the pathogen’s replicative niche. Finally, a host can destroy specific host molecules that are required for the pathogen to replicate.
Conserved immune components
This section highlights the components (proteins and small molecules) of bacterial and human innate immune systems that share homology (Fig. 2). These components are the building blocks of immune pathways. Assembling a functional pathway requires a sensor that recognizes that a pathogen is present, a signal transducer or amplifier that relays the signal, and an effector that executes the immune response (Fig. 1). Each component can be a separate protein or molecule, or a single protein can facilitate multiple processes.
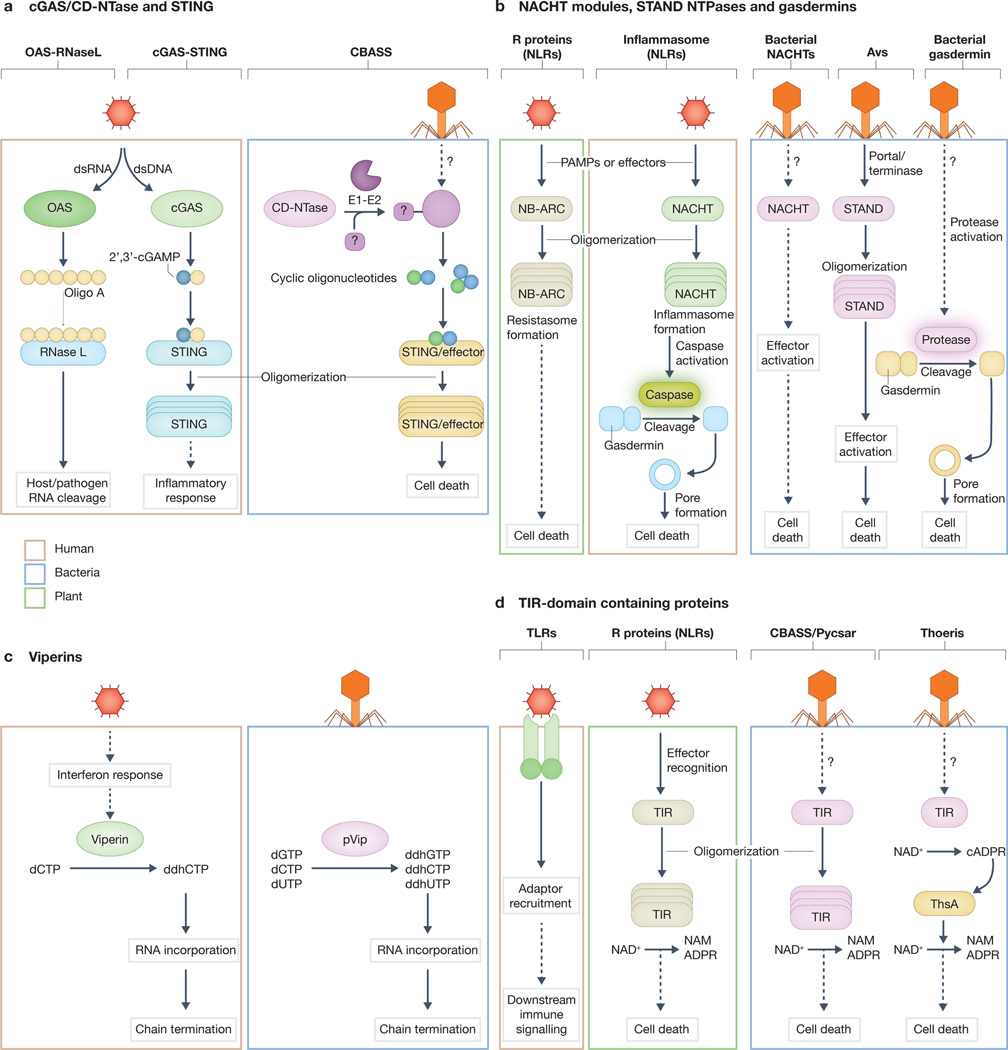
Figure 2: Components shared between bacteria, human and plant innate immune pathways.
Human (orange), plant (green) and bacterial (blue) innate immune pathways are compared to highlight their homologous components.
(a) Cyclic GMP–AMP synthase (cGAS)-like enzymes, including cGAS, 2’–5’-oligoadenylate synthase (OAS) and cGAS/dinucleotide cyclase in Vibrio (DncV)-like nucelotidyltransferase (CD-NTases), are activated by diverse stimuli to synthesize nucleotide second messengers. Human OAS and cGAS are actuvated through direct binding of pathogen-associated molecular pattern (PAMP) double-strainded RNA (dsRNA) or dsDNA to synthesize 2’–5’-oligoadenylate (oligo A) or 2’,3’-cyclic GMP–AMP (2’,3-cGAMP), respectively. These second messengers go on to bind to and activate their associated effectors with oligo A binding RNase L, leading to RNA cleavage, and cGAMP-binding stimulator of interferon genes (STING) leading to oligomerization and downstream inflammatory responses. Within bacterial type II cyclic oligonucleotide based antiphage signalling systems (CBASS), the CD-NTase is primed by an E2-E1 fusion protein Cap2 through conjugation to an unknown target (indicated by the question mark). Across other types of CBASS, the CD-NTase is activated through an unknown mechanism (indicated by the question mark) to generate one of a variety of cyclic oligonucleotide second messengers. Nucleotides in turn bind to the receptor domain (for example STING or SAVED) of an effector protein, leading to oligomerization and cell death.
(b) Signal transduction ATPases with numerous domains (STAND) NTPase modules link C-terminal sensor domains to N-terminal effector domains to condense multiple innate immune pathway components into a single open reading frame (not indicated). When the C-terminal sensor domain is a leucine-rich repeat, the proteins is called a nucleotide-binding domain and leucine-rich-repeat-containing gene family (NLR) protein. Plants use the NB-ARC subtype of STAND NTPases to sense various PAMPs or pathogen effectors. Activation leads to NB-ARC oligomerization and formation of a complex termed the resistosome, which leads to cell death. Humans use the NACHT subtype of STAND NTPases for immune signalling. Upon PAMP or effector sensing, these proteins oligomerize forming an inflammasome which, through several mechanisms, cause activation of an associated effector. A subset of mammalian inflammasomes activate a caspase-family proteins (for example, caspase1), which in turn cleaves gasdermin D, removing an inhibitory domain from the protein and initiating cell death through pore formation. Bacteria use NACHT and other antiviral STAND (Avs) NTPase subtypes. Bacterial NACHTs are activated upon phage infection through an unknown mechanism (indicated by the question mark), triggering effector activation and cell death. Avs proteins are activated by direct binding to the phage portal or terminase proteins. Binding of these PAMPs induces oligomerization, effector activation and cell death. Bacteria further use protease-activated gasdermins to initiate cell death during infection; however, these systems are activated by an unknown mechanism (indicated by the question mark).
(c) Viperin proteins generate 3′-deoxy-3′,4′-didehydro (ddh)-nucleotide triphosphates. Human viperin and Bacterial viperin (pVip) have different preferences for nucleotide substrates (for example, CTP, GTP or UTP). Human viperin is controlled transcriptionally in response to type I interferon (IFN).
(d) Toll/interleukin-1 receptor (TIR) domains can be non-enzymatically active and function to recruit adapter proteins or enzymatically active and metabolize NAD+ upon activation. Human Toll-like receptors (TLRs) become active upon binding to PAMPs, which leads to TIR-domain-mediated recruitment of adapter proteins and downstream signalling. Plant resistosome proteins (R proteins or NLRs) that use TIR domains are activated by PAMPS and pathogen effectors, which triggers oligomerization and activation of their associated TIR domains to metabolize NAD+ to nicotinamide (NAM), and ADP-ribose (ADPR) or cyclic ADP-ribose (cADPR). ADPR and cADPR variants then act as second messengers to activate cell death. Bacterial CBASS and pyrimidine cyclase system for antiphage resistance (Pycsar) activate TIR-domain effectors that oligomerize for activation and degrade the cellular pool of NAD+, resulting in cell death. The bacterial Thoeris system also encodes TIR domains that are activated as a result of phage infection but these proteins metabolise NAD+ into NAM and variants of cADPR that act as a second messengers to activate ThsB, an effector protein that also degrades NAD+, resulting in cell death.
The sensor component of innate immune systems must detect a stimulus that is specific to infection but distinct from other general signs of cellular stress. Further, the stimulus must be crucial to pathogen fitness to prevent pathogen immune evasion through altered stimulus production. Innate immune sensors have overcome these challenges by recognizing pathogen amino acid motifs that are essential to protein function, pathogen genomes or nucleic acids, and by monitoring host pathways for hijacking by pathogens. The signal transducer component has two important roles: amplification and threshold setting. By constructing a signaling pathway with an intermediary between sensor and effector, a few activated sensors can result in stoichiometrically more active effectors. However, by tuning the amount of signal transducer required for effector activation, a minimum threshold for signaling can be set. Finally, the effector component must limit pathogen replication, typically by directly targeting the pathogen or by removing an essential resource or growth niche.
This section is organized by the homologous immune components found in human (eukaryotic) antimicrobial signaling pathways and bacterial phage-defense systems. We will first discuss general features of the component, then specific details and examples found in the human and bacterial immune pathways, and finally contrast how components have diverged. A portion of these components are also found in plants and fungi. Homology between immune components is rarely detectable in the amino acid sequence and instead is found in shared structural features. The unambiguous conservation of protein structure establishes that these components evolved from a common ancestor, yet their species distribution is atypical for highly conserved proteins. In the section Horizontal gene transfer of immune components, we discuss what structural homology and species distribution mean for the evolutionary origin of immune components.
cGAS and CD-NTases
Metazoan cyclic GMP–AMP synthase (cGAS) and bacterial CD-NTase family enzymes are sensors that generate nucleotide second messengers in response to infection. These second messengers interact with and activate downstream effector proteins that mediate an appropriate immune response (Fig. 2a).
Mammalian cGAS is activated by binding double-stranded DNA in the cytoplasm3. DNA binding induces a conformational change that activates the production of the cyclic dinucleotide second messenger 2′,3′ cyclic GMP–AMP (2′,3′-cGAMP)3,4 (Fig. 2a). Eukaryotes encode a large variety of cGAS-like enzymes5. In insects, cGAS-like receptors (cGLRs) have diversified substantially with one member producing 3′,2′-cGAMP (instead of 2′,3′-cGAMP) in response to double-stranded RNA6,7. The human genome encodes many proteins that are structurally homologous to human cGAS8. The best-characterized belong to the 2′−5′-oligoadenylate synthetase (OAS) family, which sense double-stranded RNA to produce linear 2′–5′ oligoadenylate, which, in turn, activates RNase L to destroy invading RNA viruses and host RNAs9 (Fig. 2b). Some metazoan cGAS-like enzymes are paralogs of cGAS, such as MB21D2 (Ref. 5); however, it is unclear whether other cGAS structural homologs, such as OAS, evolved as a radiation of cGAS in metazoans or by distinct horizontal gene transfer events from bacterial CD-NTases.
Structure determination of a bacterial 3′,3′-cGAMP synthase, dinucleotide cyclase in Vibrio (DncV), led to the astonishing finding that cGAS and DncV adopt homologous DNA polymerase β-like nucleotidyltransferase folds10. Computational, biochemical and structural studies showed that cGAS and DncV structural homologs are encoded by >10% of bacteria11,12 and a portion of archaea13. Moreover, it was shown that subtle changes in their active sites enable the synthesis of various cyclic di- and even trinucleotides11,14. These proteins were named cGAS/DncV-like nucleotidyltransferases (CD-NTases), and they contain a second messenger oligonucleotide or dinucleotide synthetase (SMODS) protein domain, and each is encoded alongside an effector protein (also referred to as a receptor protein)11,12,15. CD-NTase operons are conserved12 and can be classified into four different subtypes13. Two subsequent studies demonstrated that these operons are antiphage, functionally linking them to human cGAS. It was shown that CD-NTase operons are enriched within defense islands and that they produce cGAMP in response to phage infection, and they were thus named cyclic oligonucleotide-based antiphage signaling systems (CBASS)15. Another study characterized a different subtype of these operons and demonstrated that CD-NTase activation was both positively and negatively regulated by other CBASS operon-encoded genes16. Both groups showed that second messengers that are synthesized by CD-NTase activate effector proteins to initiate programmed cell death15–17, so called ‘abortive infection’, which altruistically halts the viral lifecycle through suicide of the infected cell (Fig. 2a).
cGAS and CD-NTases both couple pathogen stimuli to nucleotide second messenger production, but the phage stimuli for CD-NTase activation remains poorly understood. A CD-NTase from Staphylococcus schleiferi is activated by a structured phage RNA, however it is unclear how widespread this mode is across CBASS systems18. The zinc ribbon used by mammalian cGAS to bind DNA is not present in bacterial proteins and emerged recently in animals10,19. It is possible, but unknown, if the mechanism for bacterial CD-NTase phage-sensing is conserved in some eukaryotic cGAS homologs.
STING
The stimulator of interferon genes (STING) proteins are effector proteins that are activated by cyclic dinucleotides; binding induces a conformational change in the effector that leads to the initiation of immune signaling20. STING proteins typically function downstream of cGAS and CD-NTase enzymes, which produce cyclic dinucleotides. STING proteins can have multiple domains. The conserved domain that defines STING-like proteins is the C-terminal cyclic dinucleotide-binding domain, which can be fused to various N-terminal transmembrane or Toll/interleukin-1 receptor (TIR) domains21–24.
STING was discovered in mammals where it was shown to be essential for the induction of type I interferon in response to cytoplasmic DNA4. It was later shown that STING was activated by binding 2′,3′-cGAMP, which was produced by cGAS following sensing of foreign or mislocalized DNA in the cytoplasm. STING can also bind other cyclic dinucleotides that are produced by bacteria. Nucleotide-binding induces a conformational change in STING that results in filamentation and clustering, which leads to interferon and NF-κB signaling. cGAS–STING-dependent signaling is crucial for defense against certain viral and bacterial infections, and effectiveness of some cancer immunotherapies in humans4,25 (Fig. 2a). Animal STING is predominantly transmembrane but, in rare instances, is fused to TIR domains instead21–24.
STING homologs are found in bacteria (bSTING) encoded in operons with CD-NTases15. These proteins are activated by binding their cognate CD-NTase-derived cyclic dinucleotide22 (Fig. 2a) and, excitingly, activation results in filamenting and clustering, the same conformations adopted by activated animal STING23. The activated conformation is the result of structural changes in the C-terminal STING cyclic dinucleotide-binding domain, which is the core region shared between bacterial and animal homologs. bSTING N-terminal domains vary, but each are expected to coordinate cell death when activated by conformational changes or oligomerization induced upon cyclic dinucleotide binding21,22. The STING cyclic dinucleotide-binding domain functions analogously to another cyclic oligonucleotide-binding domain encoded adjacently to CD-NTases, called SAVED, which also filaments and clusters in response to ligand binding but is confined to bacteria and archaea26 (Fig. 2a). CD-NTase–bSTING operons have not been shown to defend bacteria against phage possibly because only a few instances of these operons have been identified to date and because the organisms in which they are encoded are incompatible with heterologous experimental systems22. Despite these technical limitations, CD-NTase–bSTING operons are a type I CBASS systems and this family of genes is uniformly antiphage, which suggesting that CD-NTase–bSTING operons might have a similar role.
Like cGAS and CD-NTases, STING is hypothesized to have originated in bacteria prior to being assimilated by the animal germline through horizontal gene transfer20,21,24. Of note, the STING domain is markedly absent from archaea and plants. Mammalian STING has four transmembrane domains, a soluble cyclic dinucleotide-binding domain, and a C-terminal tail that indirectly activates interferon production to limit viral replication27. Bacteria do not have interferon; however, many animals also do not encode interferon pathways. The Cnidaria Nematostella vectensis lacks interferon but uses a STING homolog that does not encode a C-terminal tail to initiate antimicrobial signaling28. This suggests that interferon signaling was acquired after horizontal gene transfer to animals. These observations and others imply that STING signaling in animals extends beyond interferon induction, perhaps resembling an undescribed mechanism found in bacteria.
NLRs and STAND NTPases
Nucleotide-binding domain and leucine-rich repeat containing gene family (NLR) proteins are innate immune components containing a core NTPase domain that is conserved in animals29, fungi30,31, plants32 and bacteria33–36(Fig. 2b). These NTPase domains are members of the signal transduction ATPases with numerous domains (STAND) class of P-loop NTPases37. The function of the NTPase domain in NLRs and NLR-related proteins is to link a C-terminal sensor with an N-terminal effector region, mediating oligomerization and conformational changes to sensitively and specifically activate effector functions in response to infection29,37,38.
STAND NTPases are divided into families based on signature motifs37. Within eukaryotic immune signaling, animals use STAND NTPases39 and NLRs with NACHT (NAIP, CIITA, HET-E and TP1 proteins) modules, plants use NLRs that encode a subdivision of AP-ATPases called NB-ARC modules37,40, and fungi encode NACHT and AP-ATPases41 (Fig. 2b). NACHT and AP-ATPase NTPases function in a similar manner but have divergent evolutionary histories. Human NLRs such as the NAIP–NLRC4 inflammasome sense conserved pathogen-associated molecular patterns and activate inflammation and cell death42. Similarly, the plant NLR (also called R-proteins that form the resistosome) ZAR1 senses effectors secreted by diverse pathogens and activates cell death via pore formation43 (Fig. 2b). Although not thought of as ‘classic’ innate immunity, NACHT modules in fungi function in self–non-self discrimination during heterokaryon incompatibility, which limits viral transmission30,31.
Within bacteria, proteins that encode a NACHT modules or additional clades of STAND NTPases are antiphage33–36 (Fig. 2b). NACHT module proteins can be considered NLR-related because they are evolutionary ancestors of human NLRs and conceptually similar to NLRs, but do not uniformly encode the leucine-rich repeats required to be designated a NLR34. Phylogenetic analysis suggests that NACHT domains diversified in bacteria prior to being acquired horizontally by animals and fungi34. NACHT and AP-ATPase domains can also be found in archaea but they have not been investigated for antiviral signaling34,37.
For NLR and NLR-related proteins infection sensing, signal amplification and effector activation are encoded in one open reading frame. A shared mechanism of sensing for these proteins is to recognize fundamental molecular features of their stimuli (for example, conserved amino acid residues). The bacterial Avs proteins even detect conserved structural motifs of stimulating proteins33. However, there are also many variations on sensing as some NLRs detect the activity of pathogen effectors, such as plant NLRs RGA5 and Pik-1 (Ref. 44), rather than the proteins themselves. Finally, effector regions of NLR and NLR-related proteins can either indirectly initiate signaling by recruiting additional proteins, such as caspases recruited by mammalian NLRs42, or have a direct enzymatic activities, such as TIR domains33,34 (Fig. 2b).
Gasdermins
Gasdermins are effector proteins that, following activation, initiate cell death by forming pores in the cell membrane. Gasdermin proteins are held in an inactive, soluble confirmation by their C-terminus. Proteolytic cleavage releases the N-terminal fragment, which inserts into the membrane, oligomerizes to form a large pore, and results in unrestricted flux through the pore in addition to destabilizing membrane integrity45–47.
Gasdermin D was first discovered in mammals as an effector of pyroptosis, which is an inflammatory form of cell death that results from activation of inflammasomes47 (Fig. 2b). In that pathway, NLR proteins sense infection and initiate signaling through recruitment of specific caspase proteases. Caspases are signal transducers and amplifiers that cleave multiple targets, including Gasdermin D, which releases the membrane-disrupting N-terminal fragment47. In bacteria, Gasdermins are effectors in antiphage pathways that are activated by a cognate peptidase encoded in the same operon45 (Fig. 2d). The peptidase belongs to one of multiple protease families, is essential for Gasdermin activation, and is hypothesized to be the sensor of phage infection45. Gasdermins are also found in fungi where they initiate cell death in response to heterokaryon incompatibililty46,48. Interestingly, Gasdermins are not encoded by plants or archaea, which suggests horizontal gene transfer has an important role in Gasdermin evolution21 (see Horizontal gene transfer of immune components).
Viperin
Viperins are a family of potently antiviral enzymes that act as effectors by modifying ribonucleotide triphosphates to produce molecules that are unusable for viral genome replication and/or transcription49. As such, upon incorporation of these molecules into a the genome or transcripts of the pathogen, replication is limited and pathogen growth is restricted.
Viperin was first identified in mammals where the gene is substantially upregulated by type I interferon signaling50. Upon synthesis of the protein, human viperin converts cytidine triphosphate (CTP) to 3′-deoxy-3′,4′-didehydro (ddh)-cytidine triphosphate (ddhCTP), causing chain termination of elongating RNA strands and thereby limiting RNA virus genome replication and transcription of viral genes51 (Fig. 2c). Bacteria and archaea encode viperin homologs that inhibit phage replication by a similar mechanism; however these proteins modify a wide-array of nucleotide triphosphates (NTPs) and produce ddhNTP nucleotide derivatives from CTP, guanosine triphosphate (GTP) and uridine triphosphate (UTP)52 (Fig. 2c).
TIR domain-containing proteins
Toll/interleukin-1 receptor (TIR) domains are central components in numerous immune pathways and adopt a core structure with a catalytic activity separating two distinct subtypes of TIR: non-enzymatically active and enzymatically active53. Non-enzymatically active TIR domains are predominantly found in the intracellular domains of the membrane-spanning Toll-like receptors (TLRs) where they coordinate signaling via an adapter protein54. Enzymatically active TIR domains catalyze reactions that use NAD+, either by degrading NAD+ en mass to alter crucial cellular processes (for example, human SARM1 (Ref. 55), bacterial effector proteins in CBASS22, STAND NTPases33,34, Pycsar56 antiphage systems and plant RBA1 (Ref. 57)) or by converting NAD+ into a second messenger (for example, Thoeris antiphage system58 and flax plant TNL L7 (Ref. 59))(Fig. 2d). It has been hypothesized that the massive disruption in NAD+ and NADH levels results in cell death, which indirectly limits pathogen replication. TIR domains have therefore been implicated in two steps of the immune response: signal amplification (NAD+-derived second messengers) and pathogen restriction (NAD+ and NADH depletion).
TIR domains, which were originally identified in eukaryotes, were the first immune component recognized to be shared in bacterial and human innate immune pathways. Enzymatically active TIR domains are broadly distributed in bacteria, archaea and eukaryotes56,60 (Fig. 2d). Interestingly, non-enzymatic TIR domains are confined to animal and plant cells, which suggests this modification evolved later. It is also of note that synthesis of a NAD+-derived second messenger and depletion of NAD+ levels are not mutually exclusive as conversion to a second messenger may also deplete the usable pool of NAD+.
E1 and E2 domain-containing proteins
E1 and E2 domain-containing proteins are modulators of signal amplification in innate immune pathways. In general, these proteins mediate post-translation modifications to targets that alter their fate and function. Within immune signaling E1- and E2-mediated modifications have been demonstrated to potentiate, license and limit immune responses.
E1 and E2 proteins and their respective protein domains are most notorious for their role in eukaryotes where they mediate ubiquitin and ubiquitin-like protein conjugation to targets (typically proteins but also other molecules)61. Conjugation to ubiquitin results in target protein degradation, enzyme activation, changes in protein localization and numerous other cellular processes. In eukaryotes, substrates (ubiquitin-like protein) of E1 and E2 proteins are restricted to β-grasp proteins, including SUMO and the immune modulator ISG15 (Ref. 62).
More recently, E1 and E2 domains have also been identified within a subset of bacterial antiphage pathways12,63,64. In type II CBASS, E1 and E2 domains are found in a fusion protein named CD-NTase associated protein 2 (Cap2), which catalyzes ligation of the C-terminus of a substrate protein to a target molecule63,64 (Fig. 2a). However, Cap2 does not modify the C-terminus of a β-grasp family protein like ubiquitin, instead, Cap2 modifies the C-terminus of a CD-NTase, ligating the CD-NTase to proteins in vivo. Ligation of the CD-NTase to a target increases that activity of the enzyme, thereby priming the CD-NTase, a process required for phage defense63,64. The physiological target of CD-NTase ligation and the mechanism for increased enzyme activity are not known.
Cap2-mediated regulation of CD-NTases is similar to E1- and E2-mediated regulation of the human immune sensor RIG-I, which is ubiquitinated upon pathogen sensing to alleviate an autoinhibitory domain and enable robust signaling65,66. In this way, E1 and E2 domains are important regulators of immune signaling in bacteria and mammals. In the future, we expect additional examples of bacterial E1 and E2 domains that regulate immune signaling will be discovered, as there are additional candidate and verified antiphage systems encoding these domains, Cap2 homologs, and even β-grasp proteins62–64,67,68. An exciting avenue of future investigation will be understanding specific and general mechanisms that β-grasp proteins from bacteria and humans (for example, ISG15) use to protect against viral infection69,70.
HORMA and Trip13
HORMA proteins dynamically regulate various cellular processes through a conserved mechanism of protein–protein interaction. HORMA proteins bind ‘closure motifs’ on their protein partners to adopt a ‘closed’ state, and thyroid receptor-interacting protein 13 (Trip13; also known as Pch2) AAA+ ATPases disassemble HORMA–partner complexes, converting HORMA proteins back to an ‘open’ state71. In eukaryotes, HORMA proteins are involved in recombination during meiosis, the mitotic spindle assembly checkpoint and DNA repair71. Bacteria encode homologs of HORMA proteins in type III CBASS systems, sometimes in two copies named Cap7 and Cap8, and Trip13-like proteins named Cap6. HORMA proteins are required for CD-NTase activation in these systems whereas Trip13-like proteins are negative regulators of CBASS signaling16,17. Although HORMA and Trip13 proteins are not involved in the eukaryotic immune response, their evolutionary connection to antiphage components is striking, which suggests that more than just immune signaling and modulation evolved from antiphage signaling.
Argonautes
Argonautes are guide-directed nucleic acid-binding proteins encoded by animals, fungi, plants, bacteria and archaea72. Central to Argonautes are the middle (MID), P-element-induced wimpy testis (PIWI), and PIWI–Argonaute–Zwille (PAZ) domains which together coordinate loading of a guide RNA (or rarely DNA) to direct the protein to a homologous RNA or DNA target molecule. Binding to a target typically activates the PIWI domain, which results in target degradation and/or silencing72,73.
Argonautes were first characterized in eukaryotes where they are involved in numerous processes including gene regulation, generation of regulatory RNAs, and defense against invading nucleic acids or transposons. Their primary role in animals and humans is not innate immunity, but instead gene regulation72,73. Bacteria and archaea encode two distinct families of Argonautes, long pArgos and short pArgos, both of which defend against invading nucleic acids such as phages and plasmids. Long pArgos are more closely related to those found in eukaryotes and possess a true PAZ domain and a catalytically active PIWI domain. Short pArgos lack a canonical PAZ, instead encoding for an analog of PAZ (APAZ domain) and have catalytically inactive PIWI domains. Short pArgos do not directly degrade target molecules, but instead recruit accessory proteins which trigger abortive infection72. In bacteria, the precise mechanism of guide generation remains unclear; however, unlike CRISPR, preexposure to the threat is not required and therefore Argonautes seem to be a component of the bacterial innate immune system.
Conserved immune signaling strategies
This section highlights the signaling strategies that are similar between bacterial and eukaryotic immune systems. Although the individual components may not share a common ancestor, innate immune pathways often converge on shared concepts of sensing and signal transduction. These observations suggest there are ‘optimal strategies’ that enable cells to sensitively detect infection and amplify signaling, but only initiate an effector response when a threshold has been met. Common strategies are used to organize this section. Although they are presented separately, they are not mutually exclusive, and a given pathway can use more than one strategy.
PAMP–PRR
Pathogen-associated molecular patterns (PAMPs) are highly-conserved molecules that are detected by pattern recognition receptors (PRRs)1,74 (Fig. 1a). This simple ligand–receptor signaling strategy relies on PRRs evolving to recognize PAMPs that are crucial to the physiology or lifestyle of a pathogen. In this way, pathogens cannot easily evade agonizing PRRs because stopping to produce a PAMP would come at a high fitness cost. The PAMP–PRR immune strategy was first described in eukaryotes and a classic example is the PAMP bacterial flagellin binding to and activating the PRR complex NAIP5–NLRC4 (Ref. 75). Bacteria cannot simply mutate to evade detection because the flagellin motifs detected by NAIP–NLRC4 are essential to flagellin function75,76.
Although one might have predicted that the broad mutational landscape available to phage (owing to their short generation time and high replication rate) eliminated this strategy for bacterial immune systems, there are three clear examples of PAMP–PRR signaling in bacterial phage defense systems. The CapRelSJ46 system binds to and is activated by phage capsid77, the DSR system binds to and is activated by phage tail tube78, and the AVAST STAND NTPases Avs3 and Avs4 bind to and are activated by phage terminase and portal, respectively33. Bacterial PRRs recognize phage PAMPs that fulfill specialized roles in virion morphogenesis, thus limiting the availability of immune evading mutations to alleles that still produce functional proteins. For example, the capsid motif detected by CapRelSJ46 is essential for capsid assembly and cannot be mutated77. However, a pitfall of this strategy is that the CapRelSJ46 interaction with capsid is specific to the amino acid sequence of a single phage species77. Surprisingly, this is not a universal limitation. AVAST STAND NTPases can detect discrete structures of phage PAMPS. Avs 1, 2 or 3 detected terminase and Avs4 detected portal from a wide range of phages that share ≤5 % sequence identity33. Recognizing protein structure may limit the availability of escape mutations to phage. It is possible PRR detection of PAMP structures may also be found in eukaryotes or more broadly in phage defense, an exciting area for future investigation.
Self and non-self discrimination
Host organisms can modify their cellular components with chemical modifications to mark them as self thereby enabling unmodified components, such as those from a pathogen, to be distinguished as non-self and stimulate an immune response (Fig. 1a). This signaling strategy is highly effective as the chemical marker for self can vary between hosts. A limitation is that the target cellular component must be sufficiently tolerant of modifications and modified quickly enough to avoid autoimmunity. Perhaps the best example is restriction–modification (RM) in bacteria79. Conventionally, these pathways methylate host DNA (for example, the chromosome) to designated it as self. Upon infection, a phage genome without the proper methylation pattern is recognized as non-self and cut, halting infection80. Humans use a conceptually similar strategy of self–non-self discrimination for RNA using a 5′ cap structure81–83. In this case, host transcripts and other RNAs are appropriately modified with a chemical cap on their 5′ end to designated them as self. This cap not only targets the RNA for translation but also protects against exonucleases83. During an infection, viral (or other pathogenic) RNA is often not capped and is destroyed by host cells, limiting infection84. Further, uncapped RNAs are more stimulatory to the human innate immune sensors RIG-I and MDA5, which induce type I interferon in response to double-stranded RNA81,82. Viruses counter with multiple evasion mechanisms, including cap-snatching, whereby a virus will hijack the 5′ end of the host RNA in order to protect its own transcripts85.
Effector-triggered immunity
Effector-triggered immunity was first discovered in plants and describes a common immune signaling strategy that recognizes infection through pathogen-specific activities86. The term ‘effector triggered immunity’ is often used interchangeably with ‘guard-based immunity’ and ‘decoy-based immunity’. Combined, these terms refer to an immune pathway that is activated through the action of pathogen-encoded effectors, as opposed to molecules (for example, PAMPs)86 (Fig. 1a). Bacteria and eukaryotes both use effector-triggered immunity to detect pathogens. An example in humans is NLRP1b, which is natively held in an inactive confirmation until the activity of a bacterial effector or toxin inadvertently releases an active NLRP1b fragment that is capable of signaling87–89 (Fig. 1c). An example in bacteria is the retron system Ec48, which monitors the RecBCD nuclease for perturbations by phage inhibitors90,91. Core processes such as transcription and translation are also frequently monitored for sensing infection. Mammalian cells link pathogen detection to inflammation by monitoring translation using constitutively produced NF-κB inhibitors that are depleted when bacteria like Legionella pneumophila inhibit translation using secreted effectors92. Bacteria detect phage by monitoring transcription using a constitutively produced antitoxin ToxI, which is depleted when phage inhibit transcription, releasing the RNase ToxN that aborts phage infection93,94.
Another variation on effector-mediated immunity found in eukaryotes and bacteria is immune proteins monitoring themselves or other immune pathways. In humans, interferon transcription is silenced by MORC3 and MORC3 is itself also a restriction factor for viruses95,96. When pathogens such as herpes simplex virus type 1 inhibit or degrade MORC3 to evade MORC3-mediated restriction, this alleviates silencing of type I interferon and results in alternative antiviral signaling96. In bacteria, PrrC is normally silenced by a linked restriction–modification system. When the restriction–modification system is inhibited by phages, PrrC is activated and cleaves tRNAs97. The effector-mediated immunity strategy enables organisms to discriminate between threats, detect various pathogens, and activate alternative immune pathways to counter immune evasion.
Nucleotide second messenger-based signal amplification
A key step in every immune pathway is signal amplification because detecting an invading pathogen may be a rare (or rarely successful) event. A common mechanism for signal amplification found in innate immunity is production of a nucleotide second messenger (Fig. 1b). In these pathways, a sensor synthase becomes activated either through direct binding of a PAMP or through an indirect mechanism, which leads to enzymatic production of the signal transducing and amplifying nucleotide second messenger5,98. These molecules go on to allosterically activate effector proteins, which results in an immune response (Fig. 1c). Nucleotide derivatives make excellent second messengers because they are synthesized from abundant reactants (nucleoside triphosphates), highly soluble, able to diffuse throughout cells and easily recycled when degraded99. Nucleotide second messengers come in many forms. Cyclic mono-, di- and trinucleotides are used in CBASS11,15,100 and Pycsar56 antiphage pathways as well as the cGAS–STING pathway of metazoans101. Linear and cyclic oligoadenylate are used in type III CRISPR102,103 and the animal OAS-RNase L98 immune pathways. Cyclic ADP-ribose second messengers derived from NAD+ are used by the Thoeris antiphage pathway104 and the RPP1 protein in plants105,106. Nucleotide second messengers are produced by enzymes with a wide variety of protein folds, some are conserved across domains of life and others unrelated. A distinct benefit of these pathways is that the genes encoding effector proteins that bind second messengers can be interchanged in a modular fashion so long as they recognize the same second messenger. However, a weakness of these systems is their susceptibility to pathogen-encoded phophodiesterases that degrade signaling molecules (Box 2).
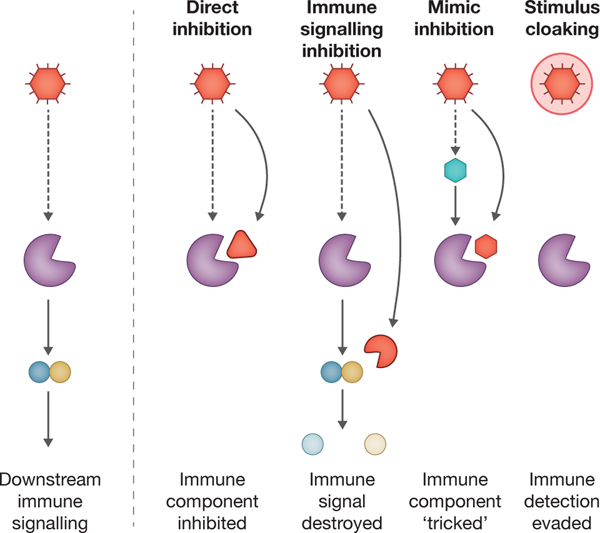
Box 2: Common strategies of pathogen immune evasion.
For every intricate pathway used by the host immune system, it seems there is an immune evasion mechanism used by pathogens. These mechanisms have converged on at least four broad strategies: direct inhibition, immune signaling inhibition, mimic inhibition and stimulus cloaking (see the figure).
Direct inhibitors bind immune components at high affinity and disable their function. In humans, Yersinia pestis YopM binds to and inactivates caspase 1 (Ref. 153) and in bacteria, phage encoded DSAD1 binds to and inactivates defense-associated sirtuins78. The disadvantage for pathogens is that direct inhibitors must be made 1:1 with their targets and are susceptible to mutations at the inhibitor–target interface. The resulting evolutionary ‘arms race’ of mutations provides an important clue when identifying sites of conflict154.
Immune signaling inhibitors are proteins that stop signal amplification. The best examples are viral phosphodiesterases that destroy nucleotide second messengers. In humans, poxvirus poxin degrades 2′,3′-cGAMP155 and coronaviruses degrade 2′–5′ oligoadenylate156. In bacteria, T4 phage Acb1 degrades cyclic dinucleotides and SBSphiJ phage Apyc1 degrades cyclic mononucleotides157. The destruction of signaling molecules has the advantage of one inhibitor silencing sensing events from across the cell. A variation on immune signaling inhibitors are Acb2 from phage T4 and PaMx33 (Ref.64,158,159) and Tad1 from phage SBSphiJ7 (Ref. 160), which neutralize immune signaling within bacterial hosts by acting as ‘sponges’ for nucleotide second messengers.
Mimic inhibitors antagonize productive immune pathway signaling by mimicking either the stimuli or a natural pathway component to trick the host pathway into binding a non-productive ligand. An example in mammals is poxvirus K3L, which mimics mammalian eIF2α161. An example in bacteria is T7 phage Ocr, which mimics the structure of DNA to inhibit restriction–modification systems162. Mimic inhibitors are particularly effective because evolving specificity for the natural component is challenging for hosts.
Finally, stimulus cloaking is the physical separation of stimulatory ligands from immune pathway components. In eukaryotes, the virus Oryctes rhinoceros nudivirus uses a membrane to pass through the cytoplasm and avoid cGAS-STING and OAS-RNase L163. In bacteria, the jumbo phage PhiKZ uses a protein shield resembling a nucleus to separate its DNA from CRISPR targeting164,165.
Oligomerization-based signal amplification
Perhaps the most pervasive mechanism of signal amplification is protein oligomerization. In this strategy, an immune protein is in an inactive confirmation until pathogen sensing, which triggers a confirmational change that leads to recruitment and oligomerization of additional monomers (Fig. 1b). Oligomerization can cluster activated monomers of the same protein (for example, gasdermins45–47, enzymatic TIR domains 23,26,107, NLR108 and NLR-related proteins33) or cluster different subunits of a signaling complex (also known as nucleated oligomerization; for example, human RIG-I oligomerized MAVS109). The resulting oligomer repositions effector domains. For enzymatic effectors, this can introduce a conformational change to activate individual effectors (for example, Cap4 (Ref. 110)) or complete an active site through interactions between two effectors (for example, TIR-STING (Ref. 23,26)). For non-enzymatic effectors, clustering effector domains appropriately recruits adapter proteins (for example, non-enzymatic TIR clustering in TLRs recruit the adaptor MyD88 which in turn leads to immune signaling via kinase recruitment54) (Fig. 1c).
Conserved pathogen restriction mechanisms
This section highlights pathogen restriction mechanisms that are similar between bacteria and eukaryotes. The ultimate signaling outcome of all immune pathways must somehow limit pathogen replication to successfully fight infection. Even when immune components and strategies differ, the mechanisms for limiting pathogen replication converge on a small number of themes. These themes are used to organize this section.
Pathogen targeting and chemical defenses
The most intuitive mechanism for immune pathways to restrict pathogen replication is through direct inactivation or destruction of the invader (Fig. 1d). A key aspect of this theme is evolving effector components that target pathogens while limiting collateral damage to the host. Both vertebrates and bacteria secrete molecules that directly neutralize or destroy pathogens prior to host cell invasion. Animal cells generate and release antimicrobial peptides that can directly disrupt the membranes of invading pathogens111. Vertebrates further use the alternative complement system to deposit pro-inflammatory and pore-forming proteins on membranes for innate defense112. Similarly, bacteria such as the Streptomyces peutetius, release numerous secondary metabolites that intercalate into DNA molecules thereby inhibiting phage infection113. Eukaryotes and bacteria also directly target pathogens after host cell invasion. Animals destroy pathogen genomes using the OAS-RNase L pathway98 and guanylate binding-proteins (GBPs) to target the membranes of bacteria, parasites and viruses114. Bacterial innate immune systems primarily destroy invading genomes using restriction modification80, BREX115,116, and DISARM104 antiphage systems. Unfortunately for the host, there appear to be relatively few targets available for direct attack on pathogens (for example, the pathogen’s genome and membrane).
Programmed cell death
A highly effective mechanism for pathogen restriction is for host cells to limit cellular resources by initiating programmed cell death in response to infection (expertly reviewed in Refs. 117,118) (Fig. 1d). When infected cells kill themselves, they create a ‘dead-end’ for their intracellular pathogens. Programmed cell death is an altruistic behavior: the cell is sacrificed to protect the population of neighboring cells. This makes immediate sense for a multicellular eukaryote but is less obviously beneficial for unicellular bacteria. However, a single T7 phage virion might produce over 300 progeny in a susceptible bacterium, and rapid programmed cell death neutralizes that virion, thus protecting kin cells in close proximity. Protected kin now survive and pass-on the protective immune pathway, which leads to population-level success. Evidence suggests that many components of eukaryotic programmed cell death, including components of apoptosis pathways, first evolved in bacteria119. Bacterial mechanisms of programmed cell death vary (Box 3) but the general signaling outcome of premature cell death during infection is called abortive infection, which is frequent and widespread15,17,33,45,58,90,117.
Box 3: Shared and distinct arsenals of cell-killing effectors.
Bacteria initiating premature cell death in response to phage infection, so called abortive infection, is an effective immune strategy to limit pathogen replication. Interestingly, the effector mechanisms and components of abortive infection are similar to the effector proteins found in kin recognition and interbacterial competition systems, including bacteriocins166, contact-dependent inhibition systems167, and type VI168 and type VII169 secretion systems. It seems that these cell-killing toxic proteins have evolved to target the same essential molecules within the cell.
Cell-killing effectors are often enzymes with common targets. Effector toxins degrade DNA en mass using HNH, restriction endonuclease-like or toxin_43 domains17,33,170. Similarly, RNA is also targeted, by the RhsP2 T6SS effector 171 and by the PrrC phage defense pathway130. Both type VI secretion system effectors172 and phage defense systems131 target essential metabolites and cofactors such as ATP and NAD+. Finally, disruption of the cytoplasmic membrane integrity is perhaps the best conserved mechanism to induce cell death, exemplified by bacteriocins166 and type VI effectors168, and the wide array of phage resistance systems that encode phospholipases173 or pore forming proteins12,174,175 that cause membrane damage.
There are also notable examples of cell-killing mechanisms that are unique to interbacterial competition and not found in phage defense. A majority of these differences derive from how the effector proteins are introduced to target cells. Interbacterial effectors often arrive at susceptible cells extracellularly and thus target external molecules (for example, peptidoglycan) whereas phage defense systems do not typically encode secreted proteins. However, other targets of interbacterial competition seem to be conspicuously absent (for example, FtsZ176). Given the convergence of effector proteins on a limited set of targets, it is likely that we have not yet discovered the antiphage system effector that shares these targets.
Mammals use a form of programmed cell death called pyroptosis to limit pathogen replication and release inflammatory signaling molecules via gasdermin D membrane pores120. The result of pyroptosis is inflammation. Evidence is beginning to emerge that bacteria too may use paracrine signaling after infection. Bacillus subtilis that succumb to phage infection are sensed by neighboring cells, which leads to a stress responses that modify the cell wall to limit phage adsorption121. This ‘phage tolerance response’ results in temporary phage resistance.
In bacteria, programmed cell death is highly nuanced and it remains unclear when cell death versus cell dormancy may occur122, or if the cell death is due to the defense system or the phage. It has been suggested that programmed cell death is overestimated as a signaling outcome and experimental approaches to address these questions have proved challenging. Programmed cell death has also been hypothesized to function as a ‘last resort’, with these pathways only becoming activate when other, non-lethal, pathogen restriction mechanisms have failed122. Although there are examples of immune pathways that monitor the function of another defense pathway in bacteria, additional studies are required to establish intricacies of cell death and a hierarchy of signaling outcomes.
Targeting host molecules and pathways
Pathogens can also be restricted through specific removal or inhibition of host cellular processes that pathogens rely on for replication (Fig. 1d). This is a similar concept to programmed cell death; however in this case the host cell can survive infection (though determining the fate of host cells, and whether host or pathogen caused ensuing cell death can be experimentally challenging). Humans primarily inhibit transcription or translation to limit pathogens during infection. Both type I interferon signaling and PKR activation inhibit translation123,124. OAS-RNase L pathway activation results in both mRNA and rRNA degradation, which not only inhibits translation but can also directly degrade pathogen-derived RNA123,125. Bacterial immune pathways that target host pathways without initiating cell death primarily destroy specific nucleotide pools. This includes an array of proteins that deaminate RNA, NTPs or dNTPs35,126–128, the PrrC system which destroys tRNA129,130, and NTPases which degrade either ATP131 or GTP68. Finally, both humans and bacteria use viperin enzymes to inhibit pathogen RNA polymerization by chain termination49,52,132.
Horizontal gene transfer of immune components
Structurally homologous components of bacterial and eukaryotic immune systems must share a common ancestor. The chance that two proteins, such as STING, could convergently evolve to adopt the identical protein fold is infinitesimal133. How then did shared components become distributed in the genomes of bacteria, archaea, animals, fungi and plants?
There are two modes of inheritance to consider: vertical and horizontal. The majority of well-known genes shared between bacteria and eukaryotes are the result of parent-to-offspring inheritance, so called ‘vertical gene transfer’ from the last universal common ancestor134. These genes are broadly distributed across bacteria, archaea and eukaryotes and encode highly conserved cellular machines (for example, the ribosome). The shared immune components described in this Review are markedly different. For example, genes such as cGAS and viperin are found in bacteria, archaea and animals, but not in fungi or plants. Genes encoding STING and gasdermins are found in bacteria, fungi and animals, but missing from archaea and plants15,21,22,45. The distribution is unlikely to be the result of gene loss, although this possibility cannot be completely rule out. Instead, the shared immune components of bacteria and eukaryotes are likely to be the result of horizontal gene transfer, DNA transfer between an unrelated donor and recipient.
One of the best examples of immune component horizontal gene transfer comes from NACHT modules34,37,40. NACHT module sequences can be divided into 25 monophyletic clades34: most clades exclusively represent bacterial proteins but some include bacteria, animals, but not fungi. Instead, fungal NACHTs are encoded in different clades. This distribution is consistent with NACHT modules diversifying in bacteria and occasionally being horizontally transferred into different lineages of eukaryotes. The direction, from bacteria to animals, is supported by the genes being polyphyletic in bacteria but monophyletic in animals34. This model of inheritance is similarly observed for CD-NTases/cGAS and viperins, which are each biochemically more diverse in bacteria than in animals11,15,52.
Each immune component has its own evolutionary history, and a lack of gene sequences limits our understanding. However, it is interesting to speculate on just how these immune genes may have moved between genomes of such disparate organisms (expertly discussed in Ref. 135). It may be that the location of bacterial antiphage genes within mobile genetic elements was crucial to enabling DNA to be introduced into eukaryotes via conjugation or transduction. Undoubtably, this event was rare. However, the evolutionary pressure imposed by viruses is likely to be so strong that eukaryotic recipients of antiviral genes quickly outcompeted kin cells. Perhaps the reason why some antiphage genes were transferred to eukaryotes, while others were not, is because the transferred genes were uniquely well suited to functioning in non-bacterial cells.
Conclusions and perspectives
The near exponential increase in the number of antiphage immune pathways reported in recent years demonstrates that we are only beginning to understand the bacterial immune system (Box 1). The discovery that some bacterial immune components are conserved within eukaryotes confirms that understanding the molecular mechanism of antiphage pathways will also increase our understanding of the human immune system. In this way, we anticipate that the fresh perspective afforded by studying bacteria and phage resistance may uncover novel features of even the most well-characterized mammalian proteins. In addition, conservation of antiviral signaling will also enable the discovery of uncharacterized antiviral genes in eukaryotes136. For these reasons, studying bacterial innate immunity has the potential to unlock both basic biology and novel therapeutic targets.
Numerous questions have emerged from the rapid advances in the field of bacterial phage defense. Perhaps the largest gap in our knowledge is understanding the viral molecules and activities that are sensed by bacterial immune pathways. These ‘phage triggers’ will spotlight fundamental (and perhaps immutable) aspects of the viral lifecycle. However, identifying phage-specific PAMPs or host pathways guarded by defense systems has been challenging137. One paradox is that many antiphage pathway components are constitutively active in vitro in the absence of stimuli (for example, viperins52, CD-NTases11,100, gasdermin-activating proteases45) — are each of these constitutively inhibited in the host cytosol until phage arrive? An additional complication is that phages encode a multitude of immune evasion mechanisms; yet characterizing these too is an important frontier in understanding the bacterial immune system138.
Another unanswered question is why phage defense genes are often clustered or co-located in genomes139. Co-localized defense genes form defense islands and are often found within mobile genetic elements. Defense islands may have evolved to help preserve or select for mobile elements by providing a fitness advantage to their host and/or functioning as addiction modules140,141. In addition, there are clear evolutionary relationships between certain components of mobile genetic elements and immune pathways141. Undoubtably, distributing phage resistance outside the core genome has advantages and enables strains within the same species to maintain different arsenals of defense genes. Variability in phage defense systems between individual strains decreases the likelihood that an invading phage has already been selected to evade every immune pathway present. Further, distributing phage defense pathways shares distributes the fitness burden of systems due to off-target or unintended effects142. But why are multiple immune pathways co-localized together in defense islands? Perhaps co-localization also drives the rapid reshuffling of immune components and even enables evolution of new immune pathways. Perhaps, synergy between specific immune pathways drives co-localization143. Or perhaps, co-localization enables layers of transcriptional, translational or post-translational regulation of those immune pathways.
The evolutionary history of conserved immune components also raises the question of how antiphage genes horizontally transferred from bacteria to primordial eukaryotes. Did transfer occur between an ancient endosymbiont and its host? Or perhaps a bacteria-eating eukaryote acquired the DNA from its prey. Or maybe it was not a direct exchange event but instead a eukaryote obtained a fragment of free DNA from the environment or a transducing viral particle. We suspect there may be examples of each scenario. Such events are likely to have been rare, but the transferred genes provided a selective advantage against viruses that allowed recipients to outcompete other early eukaryotes that did not obtain the DNA and remained susceptible to pathogens. Along these lines, an interesting observation is that bacteria that exhibit multicellular behavior are often enriched in defense systems, particularly those that result in cell death144,145. This finding has led to the provocative hypothesis that defense systems may have been a substantial driver of multicellularity in eukaryotes146.
For a long time, the antiphage pathways discussed in this Review went uncharacterized in part because they were absent from the typical laboratory strains of bacteria that had been exhaustively characterized147. Instead, most antiphage pathways have a mosaic distribution across strains of a given species, often within mobile genetic elements in the pangenome142. This antiphage pan-immune system is vast, yet still is estimated to only represent ~10% of the total accessory genome148. Are the remaining genes also defense systems? If so, what threats (other than phage) are bacteria defending themselves against? The answers to these and other questions about the bacterial immune system are just a fraction of the exciting findings yet to come.
Acknowledgements
The authors thank members of the Whiteley laboratory for advice and helpful discussion. Work in the authors laboratory was funded by the National Institutes of Health through the NIH Director’s New Innovator Award DP2AT012346 (A.T.W.), a Mallinckrodt Foundation Grant (A.T.W.) the Boettcher Foundation’s Webb-Waring Biomedical Research Program (A.T.W.), and the PEW Biomedical Scholars Program (A.T.W). H.E.L. is supported as a fellow of the Jane Coffin Childs Memorial Fund for Medical Research (61–1783).
Footnotes
Competing interests
The University of Colorado Boulder has patents for CD-NTase, Cap2, and Cap3 technologies on which H.E.L. and A.T.W are listed as inventors.
References
- 1.Vance RE, Isberg RR & Portnoy DA Patterns of Pathogenesis: Discrimination of Pathogenic and Nonpathogenic Microbes by the Innate Immune System. Cell Host Microbe 6, 10–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosadini CV & Kagan JC Early innate immune responses to bacterial LPS. Curr. Opin. Immunol. 44, 14–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L, Wu J, Du F, Chen X. & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Sun L. & Chen ZJ Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kranzusch PJ cGAS and CD-NTase enzymes: structure, mechanism, and evolution. Curr. Opin. Struct. Biol. 59, 178–187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slavik KM et al. cGAS-like receptors sense RNA and control 3′2′-cGAMP signalling in Drosophila. Nature 597, 109–113 (2021). This study finds that cGLRs have diversified within eukaryotes to sense PAMPs beyond double-stranded DNA, such as double-stranded RNA.
- 7.Holleufer A. et al. Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 597, 114–118 (2021). [DOI] [PubMed] [Google Scholar]
- 8. Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L. & Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714 (2009). This in-depth analysis of nucleotidyltransferase fold proteins identifies that humans encode a large range of cGAS-like proteins, many of which are of unknown function.
- 9.Sparrer KMJ & Gack MU Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 26, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kranzusch PJ et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158, 1011–1021 (2014). This study recognized that the bacterial enzyme DncV was structurally homologous to the human enzyme cGAS, although the two share no appreciable sequence identity.
- 11. Whiteley AT et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019). This study demonstrates that bacteria encode a wide array of cGAS-like enzymes termed CD-NTases, which produce diverse cyclic di- and trinucleotide second messengers.
- 12. Burroughs AM, Zhang D, Schäffer DE, Iyer LM & Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654 (2015). This study bioinformatically identifies numerous antiphage systems (including CBASS, PYCSAR and Thoeris) using genomic network analysis and sensitive predictions of protein structure and function.
- 13.Millman A, Melamed S, Amitai G. & Sorek R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 5, 1608–1615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govande AA, Duncan-Lowey B, Eaglesham JB, Whiteley AT & Kranzusch PJ Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense. Cell Rep. 35, 109206 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Cohen D. et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019). [DOI] [PubMed] [Google Scholar]
- 16. Ye Q. et al. HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity. Mol. Cell 77, 709–722.e7 (2020). This landmark study, together with ref. 15, establishes that operons encoding cGAS-like enzymes are antiphage systems, activation of which results in abortive infection.
- 17.Lau RK et al. Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity. Mol. Cell 77, 723–733.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banh DV et al. Bacterial cGAS senses a viral RNA to initiate immunity. Nature 623, 1001–1008 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kranzusch PJ et al. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Mol. Cell 59, 891–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolis SR, Wilson SC & Vance RE Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 38, 733–743 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Burroughs AM & Aravind L. Identification of Uncharacterized Components of Prokaryotic Immune Systems and Their Diverse Eukaryotic Reformulations. J. Bacteriol. 202, e00365–20, /jb/202/24/JB.00365–20.atom (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morehouse BR et al. STING cyclic dinucleotide sensing originated in bacteria. Nature 586, 429–433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morehouse BR et al. Cryo-EM structure of an active bacterial TIR–STING filament complex. Nature 608, 803–807 (2022). This study demonstrates that bacterial STING has the same filamentation activation mechanism as human STING.
- 24.Culbertson EM & Levin TC Eukaryotic CD-NTase, STING, and viperin proteins evolved via domain shuffling, horizontal transfer, and ancient inheritance from prokaryotes. PLOS Biol. 21, e3002436 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yum S, Li M, Frankel AE & Chen ZJ Roles of the cGAS-STING Pathway in Cancer Immunosurveillance and Immunotherapy. Annu. Rev. Cancer Biol. 3, 323–344 (2019). [Google Scholar]
- 26.Hogrel G. et al. Cyclic nucleotide-induced helical structure activates a TIR immune effector. Nature 608, 808–812 (2022). [DOI] [PubMed] [Google Scholar]
- 27.de Oliveira Mann CC et al. Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-κB Signaling Adaptation. Cell Rep. 27, 1165–1175.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Margolis SR et al. The cyclic dinucleotide 2′3′-cGAMP induces a broad antibacterial and antiviral response in the sea anemone Nematostella vectensis. Proc. Natl. Acad. Sci. 118, e2109022118 (2021). This study shows that STING activates innate immune signalling in organisms that do not encode a type I interferon pathway.
- 29.Guo H, Callaway JB & Ting JP-Y Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi GH et al. Molecular Characterization of Vegetative Incompatibility Genes That Restrict Hypovirus Transmission in the Chestnut Blight Fungus Cryphonectria parasitica. Genetics 190, 113–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paoletti M. & Saupe SJ Fungal incompatibility: Evolutionary origin in pathogen defense? BioEssays 31, 1201–1210 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Saur IML, Panstruga R. & Schulze-Lefert P. NOD-like receptor-mediated plant immunity: from structure to cell death. Nat. Rev. Immunol. 21, 305–318 (2021). [DOI] [PubMed] [Google Scholar]
- 33. Gao LA et al. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096 (2022). This exceptional study identifies phage protein structures, not amino acid sequence, which are recognized by bacterial STAND NTPases.
- 34. Kibby EM et al. Bacterial NLR-related proteins protect against phage. Cell S (2023) doi: 10.1016/j.cell.2023.04.015. This study demonstrates that NACHT modules in bacteria are widespread antiphage proteins that defend against DNA and RNA phages; this work also traces the horizontal gene transfer of NACHT modules into eukaryotes, including the predecessor of human NLRs.
- 35.Gao L. et al. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rousset F. et al. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 30, 740–753.e5 (2022). This study determines that phages, including prophages, possess genomic hotspots enriched with antiphage systems and uses this observation to discover new antiphage systems.
- 37.Leipe DD, Koonin EV & Aravind L. STAND, a Class of P-Loop NTPases Including Animal and Plant Regulators of Programmed Cell Death: Multiple, Complex Domain Architectures, Unusual Phyletic Patterns, and Evolution by Horizontal Gene Transfer. J. Mol. Biol. 343, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Sandall CF, Ziehr BK & MacDonald JA ATP-Binding and Hydrolysis in Inflammasome Activation. Molecules 25, 4572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F. et al. Human SAMD9 is a poxvirus-activatable anticodon nuclease inhibiting codon-specific protein synthesis. Sci. Adv. 9, eadh8502 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koonin EV & Aravind L. The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 25, 223–224 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Daskalov A. Emergence of the fungal immune system. iScience 26, 106793 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vance RE The NAIP/NLRC4 inflammasomes. Curr. Opin. Immunol. 32, 84–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bi G. et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e12 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Cesari S, Bernoux M, Moncuquet P, Kroj T. & Dodds PN A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy†hypothesis. Front. Plant Sci. 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson AG et al. Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clavé C. et al. Fungal gasdermin-like proteins are controlled by proteolytic cleavage. Proc. Natl. Acad. Sci. U. S. A. 119, e2109418119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broz P, Pelegrín P. & Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Daskalov A, Mitchell PS, Sandstrom A, Vance RE & Glass NL Molecular characterization of a fungal gasdermin-like protein. Proc. Natl. Acad. Sci. 117, 18600–18607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lachowicz JC, Gizzi AS, Almo SC & Grove TL Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life. Biochemistry 60, 2116–2129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin KC & Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98, 15125–15130 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gizzi AS et al. Publisher Correction: A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 562, E3 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Bernheim A. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nimma S. et al. Structural Evolution of TIR-Domain Signalosomes. Front. Immunol. 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Neill LAJ & Bowie AG The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Gerdts J, Summers DW, Sasaki Y, DiAntonio A. & Milbrandt J. Sarm1-Mediated Axon Degeneration Requires Both SAM and TIR Interactions. J. Neurosci. 33, 13569–13580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tal N. et al. Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell 184, 5728–5739.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan L. et al. TIR domains of plant immune receptors are NAD + -cleaving enzymes that promote cell death. Science 365, 799–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ofir G. et al. Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature 600, 116–120 (2021). This study expands the role of TIR domains in immune signalling by finding that Thoeris TIR domains use NAD+ to generate a variant of cyclic ADP-ribose that acts as a second messenger, activating an effector protein to further deplete the cell of NAD+.
- 59.Yu D. et al. TIR domains of plant immune receptors are 2′,3′-cAMP/cGMP synthetases mediating cell death. Cell 185, 2370–2386.e18 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Ka D, Oh H, Park E, Kim J-H & Bae E. Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+ degradation. Nat. Commun. 11, 2816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh E, Akopian D. & Rape M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 34, 137–162 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Iyer LM, Burroughs AM & Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 7, R60 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ledvina HE et al. An E1–E2 fusion protein primes antiviral immune signalling in bacteria. Nature (2023) doi: 10.1038/s41586-022-05647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jenson JM, Li T, Du F, Ea C-K & Chen ZJ Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence. Nature 616, 326–331 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu H. & Sun S-C Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang W. et al. Ubiquitin ligase enzymes and de-ubiquitinating enzymes regulate innate immunity in the TLR, NLR, RLR, and cGAS-STING pathways. Immunol. Res. (2023) doi: 10.1007/s12026-023-09400-5. [DOI] [PubMed] [Google Scholar]
- 67.Burroughs AM, Iyer LM & Aravind L. Natural history of the E1-like superfamily: Implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins Struct. Funct. Bioinforma. 75, 895–910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Millman A. et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556–1569.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Hör J, Wolf SG & Sorek R. Bacteria Conjugate Ubiquitin-like Proteins to Interfere with Phage Assembly. http://biorxiv.org/lookup/doi/10.1101/2023.09.04.556158 (2023) doi: 10.1101/2023.09.04.556158. [DOI] [PubMed] [Google Scholar]
- 70.Chambers LR et al. Bacterial Antiviral Defense Pathways Encode Eukaryotic-like Ubiquitination Systems. http://biorxiv.org/lookup/doi/10.1101/2023.09.26.559546 (2023) doi: 10.1101/2023.09.26.559546. [DOI] [Google Scholar]
- 71.Gu Y, Desai A. & Corbett KD Evolutionary Dynamics and Molecular Mechanisms of HORMA Domain Protein Signaling. Annu. Rev. Biochem. 91, 541–569 (2022). [DOI] [PubMed] [Google Scholar]
- 72.Koopal B, Mutte SK & Swarts DC A long look at short prokaryotic Argonautes. Trends Cell Biol. 33, 605–618 (2023). [DOI] [PubMed] [Google Scholar]
- 73.Wu J, Yang J, Cho WC & Zheng Y. Argonaute proteins: Structural features, functions and emerging roles. J. Adv. Res. 24, 317–324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janeway CA The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992). [DOI] [PubMed] [Google Scholar]
- 75.Lightfield KL et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tenthorey JL et al. The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang T. et al. Direct activation of a bacterial innate immune system by a viral capsid protein. Nature 612, 132–140 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Garb J. et al. Multiple phage resistance systems inhibit infection via SIR2-dependent NAD+ depletion. Nat. Microbiol. 7, 1849–1856 (2022). This study identifies the phage molecule sensed by an antiphage pathway using the elegant approach of phage mating.
- 79.Murray NE Immigration control of DNA in bacteria: self versus non-self. Microbiology 148, 3–20 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Blumenthal RM & Cheng X. Restriction-Modification Systems. in Modern Microbial Genetics (eds. Streips UN & Yasbin RE) 177–225 (John Wiley & Sons, Inc., New York, USA, 2002). doi: 10.1002/047122197X.ch7. [DOI] [Google Scholar]
- 81.Schuberth-Wagner C. et al. A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1–2’O-Methylated Self RNA. Immunity 43, 41–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Züst R. et al. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramanathan A, Robb GB & Chan S-H mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jurado AR, Tan D, Jiao X, Kiledjian M. & Tong L. Structure and Function of Pre-mRNA 5′-End Capping Quality Control and 3′-End Processing. Biochemistry 53, 1882–1898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Decroly E, Ferron F, Lescar J. & Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10, 51–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopes Fischer N, Naseer N, Shin S. & Brodsky IE Effector-triggered immunity and pathogen sensing in metazoans. Nat. Microbiol. 5, 14–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hellmich KA et al. Anthrax Lethal Factor Cleaves Mouse Nlrp1b in Both Toxin-Sensitive and Toxin-Resistant Macrophages. PLoS ONE 7, e49741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chavarría-Smith J. & Vance RE Direct Proteolytic Cleavage of NLRP1B Is Necessary and Sufficient for Inflammasome Activation by Anthrax Lethal Factor. PLoS Pathog. 9, e1003452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sandstrom A. et al. Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Millman A. et al. Bacterial Retrons Function In Anti-Phage Defense. Cell 183, 1551–1561.e12 (2020). [DOI] [PubMed] [Google Scholar]
- 91.Stokar-Avihail A. et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 186, 1863–1876.e16 (2023). [DOI] [PubMed] [Google Scholar]
- 92.Fontana MF et al. Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent Legionella pneumophila. PLoS Pathog. 7, e1001289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fineran PC et al. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. 106, 894–899 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guegler CK & Laub MT Shutoff of host transcription triggers a toxin-antitoxin system to cleave phage RNA and abort infection. Mol. Cell 81, 2361–2373.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sloan E, Orr A. & Everett RD MORC3, a Component of PML Nuclear Bodies, Has a Role in Restricting Herpes Simplex Virus 1 and Human Cytomegalovirus. J. Virol. 90, 8621–8633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaidt MM et al. Self-guarding of MORC3 enables virulence factor-triggered immunity. Nature 600, 138–142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Penner M, Morad I, Snyder L. & Kaufmann G. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 249, 857–868 (1995). [DOI] [PubMed] [Google Scholar]
- 98.Hornung V, Hartmann R, Ablasser A. & Hopfner K-P OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 14, 521–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nelson JW & Breaker RR The lost language of the RNA World. Sci. Signal. 10, eaam8812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Davies BW, Bogard RW, Young TS & Mekalanos JJ Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. cholerae Virulence. Cell 149, 358–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G. & Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Niewoehner O. et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Ofir G. et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 3, 90–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bayless AM et al. Plant and Prokaryotic TIR Domains Generate Distinct Cyclic ADPR NADase Products. http://biorxiv.org/lookup/doi/10.1101/2022.09.19.508568 (2022) doi: 10.1101/2022.09.19.508568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang S. et al. Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science 377, eabq3297 (2022). [DOI] [PubMed] [Google Scholar]
- 107.Sporny M. et al. Structural Evidence for an Octameric Ring Arrangement of SARM1. J. Mol. Biol. 431, 3591–3605 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Hu M, Qi J, Bi G. & Zhou J-M Bacterial Effectors Induce Oligomerization of Immune Receptor ZAR1 In Vivo. Mol. Plant 13, 793–801 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Wu B. & Hur S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 12, 91–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lowey B. et al. CBASS Immunity Uses CARF-Related Effectors to Sense 3′–5′- and 2′– 5′-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 182, 38–49.e17 (2020). This study demonstrated CD-NTases synthesizing cyclic oligonucleotides with 2′–5′ bonds.
- 111.Bahar A. & Ren D. Antimicrobial Peptides. Pharmaceuticals 6, 1543–1575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V. & Roumenina LT Complement System Part II: Role in Immunity. Front. Immunol. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kronheim S. et al. A chemical defence against phage infection. Nature 564, 283–286 (2018). [DOI] [PubMed] [Google Scholar]
- 114.Kutsch M. & Coers J. Human guanylate binding proteins: nanomachines orchestrating host defense. FEBS J. 288, 5826–5849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goldfarb T. et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gordeeva J. et al. BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. 47, 253–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lopatina A, Tal N. & Sorek R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 7, 371–384 (2020). [DOI] [PubMed] [Google Scholar]
- 118.Johnson AG & Kranzusch PJ What bacterial cell death teaches us about life. PLOS Pathog. 18, e1010879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koonin EV & Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9, 394–404 (2002). [DOI] [PubMed] [Google Scholar]
- 120.Cookson BT & Brennan MA Pro-inflammatory programmed cell death. Trends Microbiol. 9, 113–114 (2001). [DOI] [PubMed] [Google Scholar]
- 121.Tzipilevich E, Pollak-Fiyaksel O, Shraiteh B. & Ben-Yehuda S. Bacteria elicit a phage tolerance response subsequent to infection of their neighbors. EMBO J. 41, e109247 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koonin EV & Zhang F. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. BioEssays News Rev. Mol. Cell. Dev. Biol. 39, 1–9 (2017). [DOI] [PubMed] [Google Scholar]
- 123.Hur S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 37, 349–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hsu JC-C, Laurent-Rolle M. & Cresswell P. Translational regulation of viral RNA in the type I interferon response. Curr. Res. Virol. Sci. 2, 100012 (2021). [Google Scholar]
- 125.Chakrabarti A, Jha BK & Silverman RH New Insights into the Role of RNase L in Innate Immunity. J. Interferon Cytokine Res. 31, 49–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hsueh BY et al. Phage defence by deaminase-mediated depletion of deoxynucleotides in bacteria. Nat. Microbiol. (2022) doi: 10.1038/s41564-022-01162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tal N. et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 7, 1200–1209 (2022). [DOI] [PubMed] [Google Scholar]
- 128.Duncan-Lowey B. et al. Cryo-EM structure of the RADAR supramolecular anti-phage defense complex. Cell 186, 987–998.e15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bitton L, Klaiman D. & Kaufmann G. Phage T4-induced DNA breaks activate a tRNA repair-defying anticodon nuclease. Mol. Microbiol. 97, 898–910 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Blanga-Kanfi S, Amitsur M, Azem A. & Kaufmann G. PrrC-anticodon nuclease: functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 34, 3209–3219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rousset F. et al. A Conserved Family of Immune Effectors Cleaves Cellular ATP upon Viral Infection. http://biorxiv.org/lookup/doi/10.1101/2023.01.24.525353 (2023) doi: 10.1101/2023.01.24.525353. [DOI] [PubMed] [Google Scholar]
- 132.Vonderstein K. et al. Viperin Targets Flavivirus Virulence by Inducing Assembly of Noninfectious Capsid Particles. J. Virol. 92, e01751–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gough J. Convergent evolution of domain architectures (is rare). Bioinformatics 21, 1464–1471 (2005). [DOI] [PubMed] [Google Scholar]
- 134.Koonin EV Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136 (2003). [DOI] [PubMed] [Google Scholar]
- 135.Wein T. & Sorek R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. (2022) doi: 10.1038/s41577-022-00705-4. [DOI] [PubMed] [Google Scholar]
- 136.Cury J. et al. Conservation of Antiviral Systems across Domains of Life Reveals Novel Immune Mechanisms in Humans. http://biorxiv.org/lookup/doi/10.1101/2022.12.12.520048 (2022) doi: 10.1101/2022.12.12.520048. [DOI] [PubMed] [Google Scholar]
- 137.Huiting E. & Bondy-Denomy J. Defining the expanding mechanisms of phage-mediated activation of bacterial immunity. Curr. Opin. Microbiol. 74, 102325 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Borges AL, Davidson AR & Bondy-Denomy J. The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annu. Rev. Virol. 4, 37–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Makarova KS, Wolf YI, van der Oost J. & Koonin EV Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rocha EPC & Bikard D. Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? PLOS Biol. 20, e3001514 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koonin EV, Makarova KS, Wolf YI & Krupovic M. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat. Rev. Genet. 21, 119–131 (2020). [DOI] [PubMed] [Google Scholar]
- 142. Bernheim A. & Sorek R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 18, 113–119 (2020). This perspective article introduces the idea that the bacterial immune system is distributed throughout the pangenome and antiphage systems are maintained by individuals in different combinations, which affords a benefit in the arms race against phages.
- 143.Dupuis M-È, Villion M, Magadán AH & Moineau S. CRISPR-Cas and restriction–modification systems are compatible and increase phage resistance. Nat. Commun. 4, 2087 (2013). [DOI] [PubMed] [Google Scholar]
- 144.Kaur G, Burroughs AM, Iyer LM & Aravind L. Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. eLife 9, e52696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaur G, Iyer LM, Burroughs AM & Aravind L. Bacterial death and TRADD-N domains help define novel apoptosis and immunity mechanisms shared by prokaryotes and metazoans. eLife 10, e70394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aravind L, Iyer LM & Burroughs AM Discovering Biological Conflict Systems Through Genome Analysis: Evolutionary Principles and Biochemical Novelty. Annu. Rev. Biomed. Data Sci. 5, 367–391 (2022). [DOI] [PubMed] [Google Scholar]
- 147.Hochhauser D, Millman A. & Sorek R. The defense island repertoire of the Escherichia coli pan-genome. PLOS Genet. 19, e1010694 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koonin EV, Makarova KS & Wolf YI Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 71, 233–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Makarova KS, Wolf YI, Snir S. & Koonin EV Defense Islands in Bacterial and Archaeal Genomes and Prediction of Novel Defense Systems. J. Bacteriol. 193, 6039–6056 (2011). This study hypothesized that antiphage genes co-localize into ‘defence islands’ within bacterial genomes.
- 150. Doron S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018). This seminal study demonstrates that operons of unknown function that cluster with known phage defence genes, within so called ‘defence islands’, confer phage defence, setting the stage for the rapid advances in the field of antiphage system discovery.
- 151.Jaskólska M, Adams DW & Blokesch M. Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature 604, 323–329 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Vassallo CN, Doering CR, Littlehale ML, Teodoro GIC & Laub MT A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. 7, 1568–1579 (2022). This study uses metagenomic libraries to identify new phage defence systems, establishing a high-throughput approach and demonstrating that many antiphage systems are not located in defence islands.
- 153.LaRock CN & Cookson BT The Yersinia Virulence Effector YopM Binds Caspase-1 to Arrest Inflammasome Assembly and Processing. Cell Host Microbe 12, 799–805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sawyer SL, Wu LI, Emerman M. & Malik HS Positive selection of primate TRIM5 α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. 102, 2832–2837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Eaglesham JB, Pan Y, Kupper TS & Kranzusch PJ Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 566, 259–263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao L. et al. Antagonism of the Interferon-Induced OAS-RNase L Pathway by Murine Coronavirus ns2 Protein Is Required for Virus Replication and Liver Pathology. Cell Host Microbe 11, 607–616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hobbs SJ et al. Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature 605, 522–526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cao X. et al. Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides. Mol. Cell S1097276523009711 (2023) doi: 10.1016/j.molcel.2023.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huiting E. et al. Bacteriophages inhibit and evade cGAS-like immune function in bacteria. Cell 186, 864–876.e21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Leavitt A. et al. Viruses inhibit TIR gcADPR signalling to overcome bacterial defence. Nature 611, 326–331 (2022). [DOI] [PubMed] [Google Scholar]
- 161.Elde NC, Child SJ, Geballe AP & Malik HS Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457, 485–489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kennaway CK et al. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 37, 762–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Velamoor S. et al. Visualizing Nudivirus Assembly and Egress. mBio 11, e01333–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mendoza SD et al. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Malone LM et al. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 5, 48–55 (2020). [DOI] [PubMed] [Google Scholar]
- 166.Riley MA & Wertz JE Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84, 357–364 (2002). [DOI] [PubMed] [Google Scholar]
- 167.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ & Low DA Mechanisms and Biological Roles of Contact-Dependent Growth Inhibition Systems. Cold Spring Harb. Perspect. Med. 4, a010025–a010025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hernandez RE, Gallegos-Monterrosa R. & Coulthurst SJ Type VI secretion system effector proteins: Effective weapons for bacterial competitiveness. Cell. Microbiol. 22, e13241 (2020). [DOI] [PubMed] [Google Scholar]
- 169.Spencer BL & Doran KS Evolving understanding of the type VII secretion system in Gram-positive bacteria. PLOS Pathog. 18, e1010680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Koskiniemi S. et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U. S. A. 110, 7032–7037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bullen NP et al. An ADP-ribosyltransferase toxin kills bacterial cells by modifying structured non-coding RNAs. Mol. Cell 82, 3484–3498.e11 (2022). [DOI] [PubMed] [Google Scholar]
- 172.Ahmad S. et al. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Severin GB et al. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 115, E6048–E6055 (2018). This study showed that the nucleotide second messenger produced by DncV activates an adjacently encoded effector protein; these findings helped to solidify the signalling mechanism for what would later be called CBASS systems.
- 174.Tak U, Walth P. & Whiteley AT Bacterial cGAS-like Enzymes Produce 2′,3′-cGAMP to Activate an Ion Channel That Restricts Phage Replication. http://biorxiv.org/lookup/doi/10.1101/2023.07.24.550367 (2023) doi: 10.1101/2023.07.24.550367. [DOI]
- 175.Duncan-Lowey B, McNamara-Bordewick NK, Tal N, Sorek R. & Kranzusch PJ Effector-mediated membrane disruption controls cell death in CBASS antiphage defense. Mol. Cell 81, 5039–5051.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 176.Ting S-Y et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 175, 1380–1392.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]