Abstract
RNAi (RNA interference) and ASO (antisense oligonucleotide) technologies are the most commonly used approaches for silencing gene expression. However, the specificity of such powerful tools is an important factor to correctly interpret the biological consequences of gene silencing. In the present study, we examined the effects of acute loss of Ser/Thr kinase PDK1 (3-phosphoinositide-dependent kinase 1) expression using ASO and RNAi, and compared, for the first time, these two techniques using Affymetrix microarrays. We show that both ASO- and siRNA (small interfering RNA)-mediated knock-down of PDK1 expression strongly inhibited cell proliferation, although by different mechanisms, thereby questioning the specificity of these reagents. Using microarray analysis, we characterized the specificity of the ASO- and siRNA-mediated gene silencing of PDK1 by examining expression profiles 48 and 72 h following oligonucleotide transfection. At 48 h, a PDK1-dependent pattern of gene alterations was detectable, despite a large number of non-specific changes due to transfection of control nucleic acids. These non-specific alterations became more apparent at the 72 h time point, and obscured any PDK1-specific pattern. This study underscores the importance of defining appropriate control ASOs and siRNAs, using multiple oligonucleotides for each target and preferably short time points following transfection to avoid misinterpretation of the phenotype observed.
Keywords: antisense oligonucleotide (ASO), gene silencing, microarray analysis, 3-phosphoinositide-dependent kinase 1 (PDK1), RNA interference (RNAi), small interfering RNA (siRNA)
Abbreviations: ARF6, ADP-ribosylation factor 6; ASO, antisense oligonucleotide(s); CDK2, cyclin-dependent kinase 2; dsRNA, double-stranded RNA; eIF2α, eukaryotic translation initiation factor 2, subunit 1α; FDR, false discovery rate; HAT-1, histone acetyltransferase 1; HMOX-1, haem oxygenase (decycling) 1; LF, Lipofectin®; MM, mismatch control; OLF, Oligofectamine®; p70S6K, p70 ribosomal S6 kinase; p90RSK, p90 ribosomal S6 kinase; PDK1, 3-phosphoinositide-dependent kinase 1; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PTEN, phosphatase and tensin homologue deleted on chromosome 10; RNAi, RNA interference; SAM, significance analysis of microarrays; siRNA, small interfering RNA; SOD2, superoxide dismutase 2
INTRODUCTION
Recently, methods have been developed to study the function of a gene using acute loss of function, in addition to genetic deletion approaches. A commonly used approach has been the use of ASOs (antisense oligonucleotides), which provides a rapid and high-throughput method for silencing gene expression [1,2]. The specificity of the antisense approach is based on Watson–Crick base-pairing interactions, and the most effective knock-down using this technique is often observed with a RNase H-dependent cleavage mechanism of mRNA targets. However, over the last decade, controversy has emerged over the specificity of the biological effects observed by ASOs [3–8]. To minimize toxic and non-specific effects, modifications of the oligonucleotide backbone have been made that prevent oligonucleotide degradation (from endogenous nucleases) and improve its specificity and its affinity. For instance, it has been shown that chimeras containing a phosphorothioate backbone throughout, with 2′-methoxyethyl-modified nucleotide regions at the 5′ and 3′ ends, are nuclease resistant and exhibit increased stability and increased affinity to the target mRNA, yet still allow RNase-mediated degradation [9,10]. Antisense technology has been recently complemented by the use of RNAi (RNA interference), which represents a powerful tool for inhibiting gene expression in cultured cells, as well as in animal models [11–13]. First discovered in plants and Caenorhabditis elegans, RNAi is a highly conserved cellular mechanism of defence, involving a sequence-specific post-transcriptional process mediated by dsRNA (double-stranded RNA). Once delivered into cells, long dsRNA is cleaved by an endogenous RNase III enzyme called Dicer, generating siRNAs (small interfering RNAs) of 21–23 nt. These siRNAs are then recognized by a protein complex called RISC (RNA-induced silencing complex), which unwinds the two strands of siRNAs and targets one of the two strands to the homologous mRNA by Watson–Crick base-pairing interactions. This interaction leads to the final endonucleolytic cleavage of mRNA at a single site in the centre of the duplex region [11]. The use of RNAi in mammals has been limited, as non-specific effects induced by dsRNA longer than 35 nt have been reported. These non-specific effects are caused by either activation of the PKR (dsRNA-dependent protein kinase) pathway, resulting in inhibition of protein translation [14], or by activation of the 2′–5′-oligoadenylate synthase pathway, triggering mRNA degradation via RNase L [15]. To circumvent this problem, synthetic siRNAs shorter than 30 nt (approx. 21 nt) have instead been used in several models with success [16–18]. However, it has been shown recently that even siRNAs may induce an interferon response and/or off-target effects in some cell systems and under certain conditions [19–23]. Because it is crucial to demonstrate biological consequences beyond protein level reduction, we evaluate, in the present study, these two knock-down technologies at the global transcriptional level and compare the biological responses. As a model system, we targeted the Ser/Thr kinase PDK1 (3-phosphoinositide-dependent kinase 1), a critical regulator of many protein kinases in the AGC superfamily (initially categorized as kinases showing strong homology to protein kinases A, G and C).
PDK1 was initially identified by its ability to phosphorylate the activating loop phosphorylation site of PKB (protein kinase B)/Akt in the presence of lipid products of PI3K (phosphoinositide 3-kinase) [24–27]. Subsequently, PDK1 has been shown to phosphorylate the analogous phosphorylation sites on a number of additional protein kinases in vitro, including p70S6K (p70 ribosomal S6 kinase), p90RSK (p90 ribosomal S6 kinase), different isoforms of PKC (protein kinase C), PKA (protein kinase A) and SGK (serum- and glucocorticoid-stimulated protein kinase) [28–32].
The role of PDK1 in vivo has been addressed in different organisms by genetic deletion of PDK1 homologues in several models, such as Schizosaccharomyces pombe [33], Saccharomyces cerevisiae [34,35], Caenorhabditis elegans [36], Drosophila [37] and mice [38]. These knock-out studies reveal an essential role of PDK1 for the viability and normal development in these models. Furthermore, a study performed in PDK1-null mouse embryonic stem cells by Williams et al. [39] showed that these cells were viable and failed to activate PKB, p70S6K and p90RSK, providing the first evidence of PDK1 substrates in vivo. Interestingly, the same group generated PDK1 knock-out mice and showed that mice lacking PDK1 die at embryonic day 9.5 from multiple developmental defects [38]. In contrast, PDK1-hypomorphic mutant mice, expressing lower levels (approx. 10%) of PDK1 in all tissues tested, were viable and exhibited a small size phenotype (40–50% smaller than wild-type). This was due to a reduction of cell size, rather than cell number, suggesting a role for PDK1 in regulating cell growth [38].
In the present study we have demonstrated that siRNA knocked down PDK1 expression to a similar extent as using previously characterized ASOs. However, some biological differences were seen between these different approaches, leading us to re-examine the specificity of these reagents. Therefore we used Affymetrix microarrays to explore the transcriptional profile of a glioblastoma cell line subjected to inhibition of PDK1 at two different time points by the two different strategies, ASOs and RNAi. Interestingly, this global view displayed many alterations dependent on the chemistry of the nucleic acid transfected and not the sequence. Nevertheless, on top of this non-specific signature, a specific pattern of PDK1-dependent gene alterations was apparent at shorter, but not at longer times, following transfection. The present study is the first report examining simultaneously ASO and siRNA against the same target (PDK1). Altogether, these results document the differences between these two reagents at the genomic level and demonstrate the biological consequences beyond protein level reduction.
EXPERIMENTAL
Cell culture and transfections
Human glioblastoma U87-MG cells were grown in a humidifier incubator with 8% CO2 at 37°C in DMEM (Dulbecco's modified Eagle's medium) containing 4.5 g/l glucose (Life Technologies) supplemented with 10% fetal bovine serum. Cells were regularly passaged to maintain exponential growth and were plated at 30% confluence the day before transfection. ASOs (300 nM) or siRNAs (at indicated concentrations) were routinely transfected twice at 24 h intervals, using either Oligofectamine® (OLF) or Lipofectin® (LF) as indicated, and cells harvested 48 and 72 h following the second transfection.
Oligonucleotide and siRNA design
For PDK1 antisense (ASO) and 8 bp mismatch control (MM) oligonucleotides, the same sequences described previously (71, 5′-TGAATGATGCCCTTGCCGTG-3′, and 98, 5′-TTACTGTTCCCGTAGCAGGG-3′) were used [40]. siRNA duplexes (21 nt) were synthesized by Qiagen and annealed according to the manufacturer's instructions. The design of each siRNA was carried out as described by Elbashir et al. [17] and Qiagen. PDK1-siRNA#1 was raised against the PDK1 coding region, corresponding to nt 301 to 322 (5′-AACTGGCAACCTCCAGAGAAT-3′), PDK1-siRNA#2 to nt 926 to 947 (5′-AAGAGACCTCGTGGAGAAACT-3′) and PTEN-siRNA raised against PTEN (phosphatase and tensin homologue deleted on chromosome 10) coding region from nt 1046 to 1067 (5′-AACAGTAGAGGAGCCGTCAAA-3′). Several siRNAs were targeted against human PDK1 sequence at least 100 nt downstream of the start codon. The GC content of the duplexes was kept within the 40–60% range. All siRNA and oligonucleotides sequences were compared against all human sequences deposited in GenBank® databases with BLAST searches using the NCBI and Smith–Waterman tools.
Immunoblotting and kinase assays
Cells were washed twice in PBS at 2 days or 3 days post-transfection and harvested in protein lysis buffer [1% Nonidet P40, 20 mM Tris/HCl, pH 7.5, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 150 mM NaCl, 1 mM dithiothreitol, 1 mM NaVO4, 1 μM microcystin and 1 pill of protease inhibitor cocktail (Boehringer Mannheim) in 10 ml of lysis buffer]. Cell lysates were prepared as described previously [40]. Equal amounts of total protein (30 μg) were separated on 4–20% gradient polyacrylamide gels and transferred on to PVDF membranes. Immunostaining was performed using antibodies generated against PDK1 (BD Biosciences), p70S6K, phospho-Thr389 p70S6K, phospho-Ser473 PKB, phospho-Thr308 PKB (Cell Signaling, Berveley, MA, U.S.A.), CDK2 (cyclin-dependent kinase 2; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), c-MET (Zymed, Victoria, BC, Canada), cyclin D1 (Santa Cruz Biotechnology), β-actin (Sigma–Aldrich), PTEN (Cascade Bioscience, Winchester, MA, U.S.A.), HAT-1 (histone acetyltransferase 1; Sigma–Aldrich), ARF6 (ADP-ribosylation factor 6; Santa Cruz Biotechnology), HMOX-1 [haem oxygenase (decycling) 1], eIF2α (eukaryotic translation initiation factor 2, subunit 1α; Cell Signaling), SOD2 (superoxide dismutase 2; StressGen Biotechnologies, Victoria, BC, Canada) and p21/Cip1 (BD Pharmingen). For the kinase assays, equal amounts of total protein were subjected to immunoprecipitation with antibodies raised against recombinant full-length PKB, PDK1, p70S6K (Santa Cruz Biotechnology) or CDK2 (Santa Cruz Biotechnology) and Protein A–Sepharose beads. Immunoprecipitates were washed twice and assayed for PKB activity using 50 μM Crosstide (GRPRTSSFAEG), for PDK1 activity using 2 μg of a recombinant PH (pleckstrin homology domain)-deleted form of PKB, for p70S6K activity using 50 μM S6 peptide, (C)FPMISKRPEHLRMNL (Calbiochem), and for CDK2 activity using 5 μg of histone H1 (Upstate Biotechnology, Lake Placid, NY, U.S.A.). All assays were performed with 5 μCi of [γ-32P]ATP and 50 μM ATP. The phosphorylated peptides were separated by SDS/PAGE (16 or 12% gel) and quantified using STORM phosphorimager (Amersham Biosciences).
Cell cycle analysis and apoptosis assays
Cells were harvested for FACS analysis as described previously [40]. Apoptosis assays were performed using annexin V-PE apoptosis detection kit (BD Biosciences). Briefly, cells were harvested by pooling the medium containing non-adherent cells with adherent cells that had undergone trypsin treatment, washed in PBS, resuspended in 1×binding buffer, and 100000 cells were double stained with annexin V-PE and 7-AAD according to the manufacturer's instructions. Cells (20000) were analysed on a FACScan (Becton Dickinson), and the results interpreted using CellQuest software. Time-lapse video microscopy was performed as described previously [40]. Measurement of caspase 3/8 activity was assayed using ApoAlert Caspase Fluorescent Assay Kits (Clontech Laboratories) according to the manufacturer's instructions.
RNA isolation and microarray analysis
Total RNA was pooled from 5 independent transfections and extracted using TRIzol reagent (Invitrogen Life Technology, Carlsbad, CA, U.S.A.) and purified on RNeasy columns (Qiagen). Biotin-labelled cRNA was prepared in triplicates according to the manufacturer's instructions and hybridized to human genome U95Av2 chips (Affymetrix, Santa Clara, CA, U.S.A.), containing approx. 12000 genes and expressed sequence tags. The quality and purity of the samples were analysed by Bioanalyzer (Agilent Technologies). Microarray data were analysed using SAM (significance analysis of microarrays) software (http://rana.lbl.gov/EisenSoftware.htm). After a multiclass comparison or pair-wise comparison, all genes regulated with a P value of <0.05% were retained for clustering analysis. Two-dimensional hierarchical clustering was performed by using Cluster/Tree view software (http://rana.lbl.gov/EisenSoftware.htm) and by using average-linkage clustering algorithm. Venn Diagrams were constructed with all overlapping SAM genes retained with P value <0.05% using pair-wise comparison method.
RESULTS
PDK1-siRNA design, and effects of ASOs and PDK1-siRNA on PDK1 and PKB activities
We reported previously that the introduction of PDK1 ASOs into a PTEN-deficient human glioblastoma cell line (U87-MG) dramatically inhibited cell proliferation, whereas 8 bp mismatch control oligonucleotides (PDK1-MM) had a weaker effect. This seemed to be mostly due to cell death (as measured using TUNEL assay), with only a minor effect on cell cycle progression [40]. This is in contrast with direct inhibition of PI3K by the inhibitor LY294002 [41], which results in G1 arrest with no effect on apoptosis. In order to investigate in more detail the potential mechanism of PDK1 knock-down-induced cell death we used additional cell death assays, such as time-lapse video microscopy, measurement of caspase 3/8 activity and annexin V staining.
Using LF as the transfection reagent, ASO-transfected cells exhibited increased annexin V staining (from 5 to 15% of annexin V-positive cells) 2 days post-transfection (Supplementary Figure 1A, left-hand panel; http://www.BiochemJ.org/bj/388/bj3880573add.htm), which is consistent with previous TUNEL staining results. The PDK1-MM-transfected population, however, displayed an intermediate level of annexin V-positive cells (approx. 10% of annexin V-positive cells). Similar effects were also observed using time-lapse video microscopy or caspase 3/8 activity quantification (results not shown), suggesting a possible toxic effect of the ASO/MM oligonucleotides on U87-MG cells. Similar effects were also seen in other glioma cell lines, expressing either wild-type or mutant PTEN, and in primary human astrocytes (results not shown). Surprisingly, during optimization experiments, we found that the cell death observed was dependent on the type of the transfection reagents used. Using OLF as another lipid-mediated transfection reagent, we observed a similar reduction of PDK1 expression following ASO transfection (Supplementary Figure 1B, right-hand panel; http://www.BiochemJ.org/bj/388/bj3880573add.htm). However, cell death was not seen in the ASO- and MM-transfected cells (Supplementary Figure 1B, left-hand panel; http://www.BiochemJ.org/bj/388/bj3880573add.htm). Taken together, these data questioned our previous conclusions concerning apoptosis induced by ASO. Therefore to address whether the ASO effects were specific or not, we decided to use RNAi as an additional powerful silencing technique to knock-down PDK1 levels.
The siRNA was designed on the basis of the method described by Elbashir et al. [17], with some revisions according to Lassus et al. [42]. Out of three different siRNAs originally designed according to the Qiagen algorithm (http://www1.qiagen.com/Products/GeneSilencing/CustomSiRna/SiRnaDesigner.aspx), only one (named siRNA#1) was retained due to its ability to efficiently reduce PDK1 expression (Figure 1A). The siRNA#1 sequence exhibits the features of siRNA candidate criteria, including AA(N)19 nucleotides with 47% GC content and no G/C stretches ≥4 bases long, satisfying the suggested rules. Using BLAST searches and the Smith–Waterman method to find potential 21 nt matches of the ASO and PDK-siRNA#1 sequences, no hits were found other than PDK1 sequences in the human genome, confirming the specificity of the ASO/siRNA#1 sequences used in the present study.
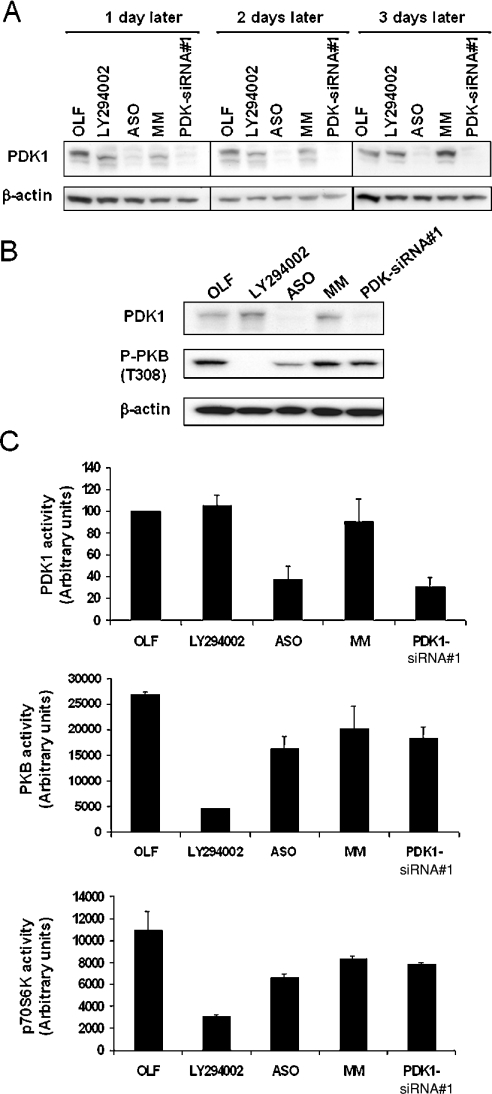
Figure 1. siRNA directed against PDK1 efficiently reduces PDK1 expression in U87-MG cells.
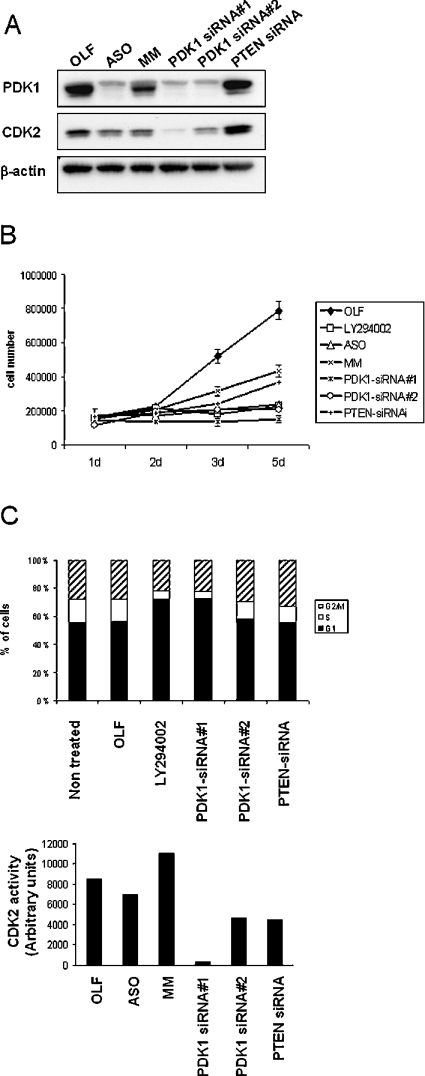
(A) Western blot analysis of PDK1 expression at 1, 2 and 3 days post-transfection. Cells were transfected with OLF alone, OLF in the presence of 300 nM PDK1 ASOs, MM oligonucleotides, PDK-siRNA#1 or treated with the PI3K inhibitor LY294002 (20 μM) as a positive control. Total cell lysates (30 μg) was subjected to immunoblot analysis using anti-PDK1 and anti-β-actin antibodies. (B) Western blot analysis of proteins (30 μg) from total cell lysates was performed 3 days post-transfection with the indicated reagents, using anti-PDK1, anti-phospho(Thr308)-PKB and anti-β-actin antibodies. (C) PDK1 (upper panel), PKB (middle panel) and p70S6K (lower panel) were individually immunoprecipitated from cells treated with the indicated reagents for 3 days, and assayed for in vitro kinase activity as described in the Experimental section. The results are expressed as the means±S.E.M. for three independent experiments. Similar results were obtained at two days post-transfection. As a positive control, U87-MG cells were incubated with 20 μM LY294002 for 20 min in presence of serum.
A titration of siRNA concentration and kinetic analysis of PDK1 expression was first performed to determine the optimal conditions for siRNA#1-mediated silencing of PDK1. The titration results indicated that a knock-down was apparent at 1 nM, but was only fully effective above 200 nM (more than 80% knock-down) using LF as a primary transfection reagent (Supplementary Figure 2, http://www.BiochemJ.org/bj/388/bj3880573add.htm). We performed a kinetic analysis using OLF reagent because of its decreased toxicity, which showed that PDK1-siRNA#1 at 300 nM, efficiently reduced PDK1 protein expression 2 days and 3 days post-transfection (Figure 1A). We next assayed ASO/MM-and PDK1-siRNA#1-transfected cells for PDK1 activity and for its downstream targets, such as PKB and p70S6K. As we expected, PDK1 activity was reduced in ASO- and siRNA#1-treated cells 2 days (results not shown) and 3 days (Figure 1C, right-hand panel) after transfection. In these assays, we used as a positive control the PI3K inhibitor LY294002, which strongly inhibited PKB and p70S6K activity. However, PKB activity and phosphorylation on Thr308 were less dramatically and less reproducibly reduced by ASO and PDK1-siRNA#1 transfections (Figures 1B and 1C, middle panels). p70S6K activity showed a similar effect observed with PKB activity (Figure 1C, lower panel). Altogether, these data are consistent with the observations reported by Lawlor et al. [38] who demonstrated that a 90% decrease of PDK1 expression did not significantly affect PKB and p70S6K upon insulin stimulation. Despite the small effect observed on PKB and p70S6K activity upon ASO or PDK1-siRNA#1 treatments, the dramatic decrease in PDK1 expression and activity lead us to explore the consequences of PDK1 knock-down in U87-MG cells.
ASO and siRNA#1 raised against PDK1 inhibit cell proliferation
It has been previously shown that ASO caused a dramatic inhibition of cell proliferation to a similar degree as LY294002-treated cells [40]. PDK1-siRNA#1 also strongly reduced cell proliferation over 5 days post-transfection (Figure 2A). We then investigated whether the mechanism involved in the inhibition of cell proliferation might be due to apoptosis or cell cycle regulation. Using annexin V staining to quantify apoptosis induction, we showed that PDK1-siRNA#1 did not induce apoptosis in U87-MG cells, similar to the ASO treatment with OLF reagent (Figure 2C and Supplementary Figure 1B, http://www.BiochemJ.org/bj/388/bj3880573add.htm). In this assay we used TNFα (tumour necrosis factor α) in the presence of cycloheximide as a positive control.
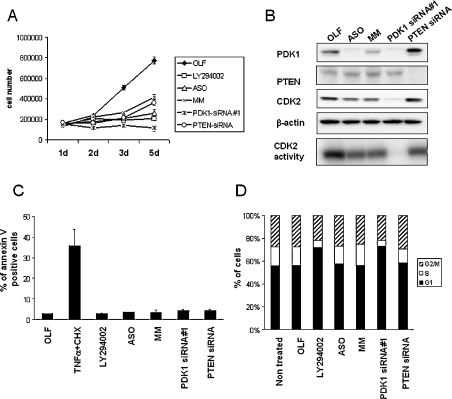
Figure 2. Inhibition of cell proliferation caused by PDK1 knock-down.
(A) Proliferation curve over 5 days (5d) performed on ASO-, MM-, PDK1-siRNA#1- and OLF-transfected cells. For LY294002 treatment, U87-MG cells were incubated with 20 μM of LY294002 for 5 days (replaced in fresh media every 24 h). U87-MG cells at 30% confluency were transfected and harvested by treatment with trypsin after transfection at indicated time points, and viable cells were counted using a haemocytometer. The results are expressed as the means±S.E.M. for three independent experiments. (B) Western blot analysis of proteins (30 μg) prepared from total cell lysates was performed 3 days post-transfection with the indicated reagents, using anti-PDK1, anti-PTEN, anti-CDK2 and anti-β-actin antibodies. CDK2-associated histone H1 kinase assay is shown on the lower panel. (C) U87-MG cells were transfected with the indicated oligonucleotides, and cells harvested 3 days later for FACS analysis of propidium iodide and annexin V staining. TNFα+CHX, tumour necrosis factor α, in the presence of cycloheximide. The number of annexin V staining cells are shown, and the results are expressed as the means±S.E.M. for four separate experiments. (D) Cell cycle distribution at 3 days post-transfection using propidium iodide staining. The number of cells containing 2n, 2n–4n and 4n DNA content, corresponding to the G1 phase, S phase and the G2/M phase respectively, were quantified using Cellquest. The results are representative of three independent experiments. Similar results were obtained at 2 days posttransfection.
As no apoptosis was found in ASO- and PDK1-siRNA#1-treated cells, we next examined cell cycle regulation. Following incubation with PDK1-siRNA#1 for 2 days U87-MG cells were arrested in G1 phase (71% of cells in G1), whereas ASO and MM had no effect on cell cycle distribution (Figure 2D). The PDK1-siRNA#1-induced G1 arrest was observed to a similar extent as reported previously for LY294002-induced G1 arrest (75% of cells in G1) [43]. This suggests that the inhibition of cell proliferation observed following PDK1-siRNA#1 and LY294002 treatment was mainly due through a G1 arrest mechanism.
To determine whether the ASO- and PDK1-siRNA#1-mediated effects were due to differences in the chemical structure, we used a control siRNA. U87-MG cells express a mutant non-functional PTEN [44], making PTEN an ideal target for monitoring potential non-specific effects mediated by siRNA in these cells. PTEN expression is severely reduced after PTEN-siRNA treatment 2 days and 3 days post-transfection without affecting PDK1 expression (Figure 2B, and results not shown). PTEN-siRNA caused a similar effect as MM oligonucleotides on cell proliferation, but the effect was not as dramatic as the PDK1-siRNA- or ASO-mediated inhibition (Figure 2A). Furthermore, as we expected, the knock-down of mutant PTEN did not induce a G1 arrest (Figure 2D). Therefore the effect mediated by PDK1-siRNA#1 on G1 arrest is unlikely to be due to its chemical backbone alone. To investigate in more details the effects seen on cell cycle following siRNA treatment, we examined the expression and activity of one of the key cell cycle regulatory proteins, CDK2. PDK1-siRNA#1 strongly reduced CDK2 expression level and dramatically abolished CDK2 activity, whereas ASO/MM and PTEN-siRNA only showed a minor effect (Figure 2B). The discrepancy in the effects of ASO and siRNA#1 on cell cycle, as well as the partial inhibitory effects of control oligonucleotides, led us to examine and compare the global effects of these reagents using oligonucleotide microarrays.
A genome-wide view of ASO and siRNA effects
Global gene expression patterns were determined using human Affymetrix microarrays (human genome U95Av2 GeneChip), representing approx. 12000 genes and expressed tag sequences. These experiments were performed at two different time points after transfection, according to the kinetic analysis shown in Figure 1(A). The time point 2 days post-transfection, which shows the earliest time point of PDK1 silencing (Figure 1A and Figure 3A), was chosen to analyse in triplicate the OLF-, MM-, ASO- and PDK1-siRNA#1-treated cells. A longer incubation period (72 h post-transfection), showing similar reduction of PDK1 expression (Figure 1A and Figure 4A) was also analysed in triplicate with the same treatment, but including PTEN-siRNA as an appropriate siRNA control (Figure 4A).
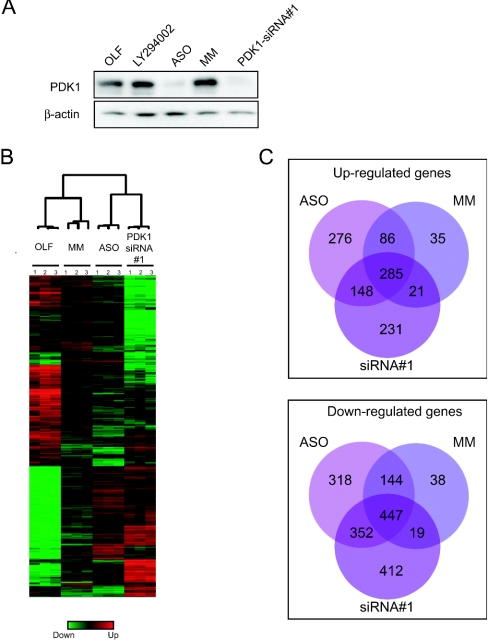
Figure 3. Alterations of gene expression induced by ASO, MM, PDK1-siRNA#1 and OLF treatments at 2 days post-transfection.
(A) PDK1 and β-actin expression levels were assayed by immunoblot 2 days post-transfection. (B) Cluster analysis of 1795 genes designated significantly affected across all four treatments by a multi-class response using the microarray analysis program SAM (see the text for details). The genes are represented by each row and the experimental samples are represented on each column (in triplicates). (C) Venn diagrams representing the number of genes significantly up-regulated or down-regulated by each reagent compared with OLF transfection (OLF versus ASO, MM or PDK1-siRNA#1), and their overlap between each other. Upper panel, up-regulated genes; lower panel, down-regulated genes. Each circle represents one treatment as indicated.
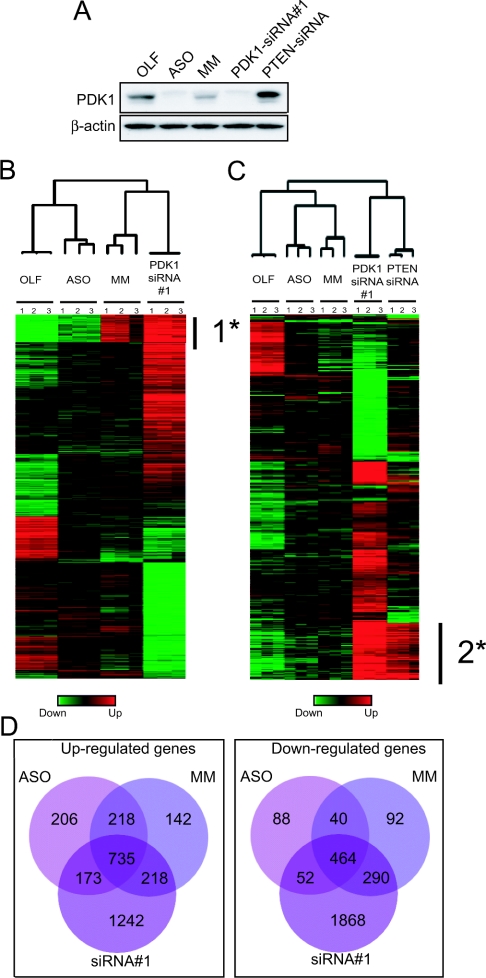
Figure 4. Alterations of gene expression induced by ASO, MM, PDK1-siRNA#1 and OLF treatments at 3 days post-transfection.
(A) PDK1 expression level in U87-MG cells 3 days post-transfection. (B) Cluster analysis of 2056 genes designated significantly affected across all four treatments by a multiclass response using the microarray analysis program SAM (see the text for details). 1* mark represents genes up-regulated in MM- and PDK1-siRNA#1-transfected cells (see the text for details). (C) Cluster analysis of 2200 genes designated significantly affected across all four treatments by a multiclass response using the microarray analysis program SAM, but including the regulation of genes affected by PTEN siRNA. (D) Venn diagram representing the number of genes affected by OLF versus PDK1-ASO, -MM or PDK1-siRNA#1, as indicated next to each circle.
We first performed a statistical test analysis (SAM) to identify significantly expressed genes that were differentially regulated across the different conditions. Using a multiclass response method with a FDR (false discovery rate)<0.03% we found that there were 1795 genes differentially regulated (Figure 3B) across the different conditions. We then applied a hierarchical two-dimensional clustering algorithm to these 1795 genes (represented on each row) and arrays or experimental samples (represented on each column) on the basis of similarity in their expression patterns (Figure 3B). As expected, each triplicate was clustered together within each experimental groups, indicating a high level of reproducibility between the biological replicates. Interestingly, the samples derived from ASO- and PDK1-siRNA#1-transfected cells were clustered together, as were the OLF and MM transfected cells (Figure 3B). The similarity between the ASO and the PDK1-siRNA#1 gene expression patterns suggested a target-specific profile, rather than an ‘off-target’-specific signature. Both increases and decreases in gene expression were observed with each treatment (Figure 3B). This gene expression profiling also revealed genes regulated uniquely by ASO, MM or PDK1-siRNA#1. To identify those genes, we determined next the transcripts whose expression level changed significantly from ‘OLF control’ expression levels in response to one of the three experimental treatments. We used a pair-wise comparison (SAM statistical test, two classes, unpaired data) with an FDR<0.05% (P<0.0005) threshold for each comparison. On the basis of these criteria we identified individual genes specifically affected by ASO (2052 genes), genes specifically regulated by MM control (1077 genes) and genes specifically affected by PDK1-siRNA#1 (1923 genes), and determined the overlap between these sets of genes. Up-regulated genes and down-regulated genes were separately analysed, and we represented the number of genes altered in the three sets of expression profiles using a Venn diagram (Figure 3C, upper and lower panels). Analysis of these Venn diagrams indicated that there are more genes down-regulated than up-regulated. A surprisingly large number of genes were affected by all three treatments (285 genes up-regulated and 447 genes down-regulated). This indicates that approx. 6% of the genes represented on the arrays were affected in a sequence-independent manner by the introduction of oligonucleotides into cells. Despite the fact we found more than 700 genes affected by all three treatments, we observed that only few genes were affected by the MM alone (35 and 38). More importantly, this analysis allowed the identification of PDK1-specific genes (represented in the set of genes co-regulated by PDK1-ASO and PDK1-siRNA#1 without being affected by the other treatments). Therefore these data revealed that at an early time point (48 h post-transfection) a PDK1-specific transcriptional signature is apparent, in addition to an oligonucleotide/siRNA response signature.
A longer incubation time reveals predominantly an ‘off-target’ signature
We then analysed the 72 h gene expression patterns. These displayed a different pattern from the 48 h time-point profile, even though PDK1 expression was reduced to a similar extent (Figure 4A, and results not shown). Using the same type of analysis described above, we generated clustering analysis on 2056 genes, which were assigned (according to SAM analysis) ‘significant’ with a FDR of <0.03%, similar to the above time point (multi-class response). In contrast with the 48 h time point, the analysis was no longer able to cluster ASO- and PDK1-siRNA#1-treated samples together, but instead grouped MM samples with PDK1-siRNA#1 samples and OLF samples with ASO samples (Figure 4B). As indicated by 1* on Figure 4(B), the most up-regulated genes induced by PDK1-siRNA#1 and MM were involved in proteasome degradation, and response to stress or oxidative stress (e.g. ubiquitin-activating enzyme E1-like or macropain). This decreased similarity between ASO and siRNA#1 samples suggested a greater degree of off-target gene regulation at longer incubation times, reflecting a possible non-specific signature.
More interestingly, if we included PTEN-siRNA samples to the analysis we observed three major clusters within the experimental samples. On the basis of the similarity in their gene expression profiles, PDK1-siRNA#1 samples were clustered with PTEN-siRNA samples, whereas ASO and MM samples were clustered together and OLF samples were separated from these two clusters (Figure 4C). Most of the upregulated genes illustrated by 2* on Figure 4(C) were involved in cytokine pathways or in inflammatory response [for example, chemokine (C-X-C motif) ligand 2 and 3, interleukin-8, interleukin-6 receptor, interleukin-7 receptor, interferon receptor-2], or more surprisingly in RNAi mechanisms, such as Dicer-1 (3.5-fold change). Therefore, bringing a siRNA control sequence into the analysis revealed a siRNAs-specific signature and an oligonucleotide-specific signature (Figure 4C), indicating that a longer incubation period dramatically affects the global gene expression profile independently of the loss of PDK1 function. Moreover, using the same criteria as the 48 h time point analysis (FDR<0.05%) a SAM pair-wise comparison identified a larger number of genes which were assigned as ‘significantly different’ between OLF-treatment and ASO treatment, MM treatment or siRNA#1 treatment (1923 genes, 2198 genes or 5032 genes respectively). The largest difference between the 48 and 72 h time points was in the total number of genes regulated by the PDK1-siRNA#1 transfection alone (600 to 2940 genes), suggesting a stronger effect of siRNA treatment on gene expression profile than the ASO/MM treatments at longer incubation time (Figure 4D). The genes affected by all three treatments (735 up-regulated and 464 down-regulated) were greatly reduced after subtraction of the PTEN-siRNA regulated genes from the Venn Diagrams shown in Figure 4(D) (and Supplementary Figure 3, http://www.BiochemJ.org/bj/388/bj3880573add.htm), indicating that PTEN siRNA in U87-MG expressing a mutant PTEN affects the expression of a large number of genes. Most of these genes commonly regulated by four different treatments encoded proteins involved in response to stress [MAPK9 (mitogen-activated protein kinase 9), Hsp70 (heat-shock protein 70)/Hsp90-organizing protein)], interferon response [interleukin-6, STAT3 (signal transduction and activators of transcription 3), CSF3 (colony-stimulating factor 3)], apoptosis (BAX, nuclear factor κB, caspase 8) and metabolism (hydrolase-activity-related genes).
PDK1 target genes defined by the 48 h time-point profiling
As we demonstrated above that the common feature of PDK1 knock-down in U87-MG cells using ASO and siRNA was an inhibition of cell proliferation, we next investigated possible mechanism(s) responsible for this by identifying the PDK1 transcriptional targets. We used the 48 h time point to define PDK1-specific gene alterations, as both the clustering analysis and the Venn diagrams showed greater specificity at this time point.
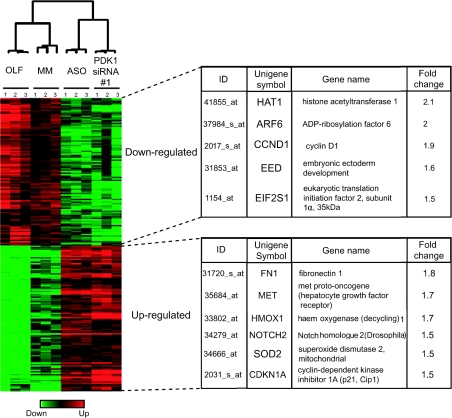
Using an FDR<0.03% and sequential SAM analysis we identified approx. 180 genes (representing 1.5% of probe sets on the array) for which the expression levels were altered (1.4-fold or greater up and down regulated) by PDK1 knockdown (Figure 5). Importantly, we found a set of genes known to be regulated by FOXO transcription factors, such as SOD2 or cyclin D1, demonstrating that some of the PDK1-regulated genes were specific for the PKB pathway. We also found that many genes regulated by PDK1 (>1.4-fold change) have been not reported as PKB targets, such as EIF2S1, Met, HAT-1, fibronectin-1, ARF6, EED (embryonic ectoderm development), HMOX-1 and NOTCH2 (Figure 5).
Figure 5. PDK1 target genes identified at 2 days post-transfection.
Up-regulated and down-regulated genes (>1.4-fold change) were selected from the overlapping ASO–PDK1-siRNA#1 cluster represented on the Venn diagrams. The 180 corresponding genes are represented using TreeView software (left-hand panel). Right panel described some of the target genes isolated. ID represents the Probe Set ID present on the chips.
A second independent siRNA targeting a distinct region of PDK1 (PDK1-siRNA#2) inhibits cell proliferation
The current siRNA protocols recommend the use of at least a second siRNA duplex to confirm the specificity of the biological effects observed. Therefore we designed and tested a second siRNA (PDK1-siRNA#2), which was as effective as siRNA#1 and ASO for its ability to knock-down PDK1 expression at the protein level (Figure 6A, upper panel). Similar to PDK1-siRNA#1 and ASO, PDK1-siRNA#2 strongly inhibited cell proliferation (Figure 6B). However, no apoptosis induction, no G1 arrest and little effect on the CDK2 protein and activity were observed in PDK1-siRNA#2-transfected cells (results not shown, and Figures 6A and 6C). Although both our microarray data, and our biochemical and biological endpoints, highlight the difficulties in interpreting specific effects, a common feature of PDK1 silencing in U87-MG cells was an inhibition of cell proliferation. This effect was also confirmed using another ASO (results not shown, and AS66 as reported previously in [40]).
Figure 6. PDK1-siRNA#2 reduced PDK1 expression and inhibits cell proliferation.
(A) PDK1 and CDK2 expression were analysed by Western blotting from ASO-, MM-, PDK1-siRNA#1- and PDK1-siRNA#2-transfected cells performed at three days post-transfection. (B) Proliferation curves over 5-day period. The results are expressed as the means±S.E.M. for two independent experiments. (C) Upper panel, cell cycle distribution of transfected cells 3 days post-transfection using propidium iodide staining. Lower panel, quantification of CDK2 activity using a STORM phosphorimager.
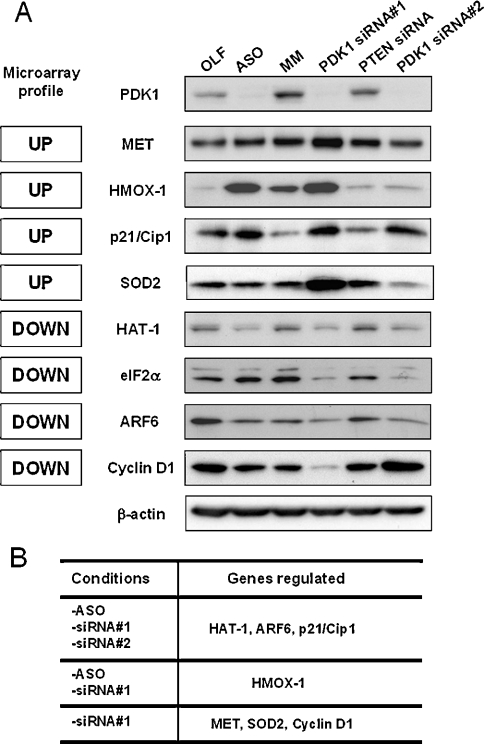
We next tested some of the candidate PDK1-specific genes by Western blotting (Figure 7). We expected that a PDK1 target gene would be regulated in the same manner in the following conditions: ASO/PDK1-siRNA#1/PDK1-siRNA#2-treated cells, without being regulated by MM, or PTEN siRNA. Out of eight candidates examined, three followed these stringent criteria (ARF6, p21/Cip1, HAT-1), strongly suggesting that these genes are PDK1 targets. However, all of the candidates were regulated by at least one of the conditions, indicating a general correlation between microarray data and protein expression in U87-MG cells (Figures 7A and 7B).
Figure 7. Validation of the PDK1-specific gene alterations by Western blot analysis.
(A) Immunoblot assay performed on nine different proteins as indicated. The nature of the regulation observed on the oligonucleotide microarray is indicated at the left of each blot. (B) Summary of the Western blot analysis in (A), describing the identity of the gene regulated in each condition.
DISCUSSION
In the present study we have documented for the first time the global effect on gene expression of ASO and siRNA against the same target (PDK1). We further examine the biological consequences of these reagents and report numerous differences, raising the danger of misinterpretation in the use of these approaches.
In recent years, PDK1 has attracted attention, since this protein kinase plays a central role in regulating a large number of other protein kinases, as well as a large number of cell responses. Several knock-out studies have underlined the importance of this kinase in regulating cell proliferation and cell growth, but the underlying mechanism(s) remains unclear [37,38,45,46]. Therefore acute inhibition of PDK1 activity would provide an ideal method to explore its role in cells.
We have previously shown in U87-MG cells that PDK1 inhibition results in decreased cell proliferation and cell survival [40]. In the present, we have demonstrated that the cell death observed was likely contributed by toxicity of the transfection reagent used (LF), as the use of OLF to introduce ASO does not cause cell death, using annexin V staining, caspase 3 activity and time-lapse video microscopy. As for all gene silencing methods, the critical step is the delivery of molecules into the cells. There has been some debate regarding the optimal transfection reagent to maximize the delivery and to avoid cytotoxicity mediated by delivery reagents. Although LF has been successfully used in some systems [9], other studies [47,48] strongly suggested that LF was toxic in other models in agreement with our present study. The intermediate inhibitory effects of MM oligonucleotides on both apoptosis in the presence of LF (Supplementary Figure 1, http://www.BiochemJ.org/bj/388/bj3880573add.htm), and proliferation in the presence of OLF (Figure 2A) led us to re-examine the specificity of the effects observed with PDK1-ASO. To address this question we compared the specificity of the ASO effects with another highly efficient silencing method, RNAi, to knockdown PDK1 gene expression. Using several guidelines, criteria and algorithms based on recent reviews [13,16] to design effective siRNAs, we analysed five different siRNA duplexes from three different companies. Our first concern was to determine the optimal concentration of siRNA capable of silencing PDK1 expression to a similar degree as ASO in order to closely compare these two techniques. Although many publications and siRNA-related guidelines recommend to use lower concentrations of siRNAs, we were not be able to detect an effective silencing of PDK1 using these siRNA duplexes with LF reagent at doses <300 nM. Other reports have also shown that lowering the concentration of siRNA to 100 nM or below can reduce silencing efficiency for some genes [21,49,50]. Importantly, using a genome-wide analysis and multiple siRNAs against the same target, Jackson et al. [51] reported that the ‘off-target’ effects observed on global gene expression profile could not be eliminated by decreasing the concentration of siRNA. One plausible explanation would be that the siRNA molecules used in our study were not optimally designed at that time we performed our experiments. Designing the optimal siRNA seems to be an empirical exercise, although new criteria have recently improved the guidelines for siRNA design [16,52–58].
As the degree of PDK1 protein knock-down was extensive (approx. 90% reduction), we were expecting to see a strong inhibition of PKB and p70S6K activities, but this was not observed. Instead, only a very weak inhibition of PKB activity and p70S6K activity was observed following ASO and siRNA treatments (Figure 1C). Similar results were observed by Lawlor et al. [38] in hypomorphic mice PDK1−/fl expressing a low level of PDK1 where normal activation of PKB, p90RSK and p70S6K upon insulin stimulation still occurred in several tissues. It is therefore likely that low levels of PDK1 in most cells and tissues are sufficient to phosphorylate and maintain high activity of these three substrates. In some experiments, PKB and p70S6K activities were more substantially reduced (more than 40%) and this was probably due to an occasional knockdown of PDK1 expression of more than 95% (results not shown).
Although we demonstrated that both ASO- and siRNA-mediated silencing of PDK1 dramatically inhibited cell proliferation, we found that the mechanism involved was totally different. A G1 arrest was apparent following transfection of PDK1-siRNA#1, which was not seen using ASO (Figure 2D). In an attempt to understand this discrepancy we decided to use microarray analysis to determine the global gene expression consequences of different oligonucleotide treatments, to clearly define a PDK1 specific signature from an ‘off-target’ signature.
Several groups have reported non-specific effects of these reagents, mainly due to toxicity, poor level of inhibition and numerous ‘off-target’ effects, but to our knowledge no genome-wide comparison between ASO and siRNA raised against the same target has been documented. Seven reports have separately addressed the specificity of ASO or RNAi using microarray analysis and led to contradictory conclusions [20,22,23,51,59–61]. Two studies reported that siRNA-mediated gene silencing exhibited a high degree of specificity, whereby the gene expression pattern observed was highly dependent on the gene being targeted [20,22]. The opposite conclusion was reached by two groups using either synthetic siRNAs or DNA vectors encoding small RNA hairpins to knock-down endogenous target genes, who observed a gene expression signature induced by all siRNAs, rather than a target-specific signature and reported the induction of an interferon response [23,59]. An additonal study examining expression profiles of cells transfected with 24 different siRNAs targeting two different genes demonstrated that each individual siRNA hallmarked its own expression profile, suggesting an individual effect of each siRNA sequence on the global gene expression pattern [51]. Similarly, a recent report showed a sequence-dependent non-specific effect of siRNA when examining p53 and p21 protein levels [21]. The reason for these discrepancies may be that in the former studies the siRNAs used were either designed against an exogenous target (green fluorescent protein) or employed at very low concentrations, which could overcome the non-specific effects observed in both the latter studies and our present experiments. We also cannot exclude that the design of some of the siRNAs used in the latter studies were not optimal and therefore more prone to trigger non-specific effects. In addition, it is important to mention that this ‘off-target’ profile has also been described using ASO approach. For instance, Fisher et al. [7] indicated that 2% of the tested genes were unrelated to the targeted gene, suggesting non-specific effects induced by ASO. Moreover, it has also been indicated that an ASO targeted to one gene (PKA subunit R1α) altered, in a sequence-specific manner, the expression of different set of genes, but a careful analysis of the microarray data revealed that many of the regulated genes were not related to the targeted gene, suggesting an ‘off-target’ profile [5,62]. Although improvements of ASO chemistry have been achieved in terms of reducing cytotoxicity, possible residual effects can persist and this may cause non-specific biological effects. One possible cause of non-specific toxic effects has been attributed to the degradation of the liberated bases of oligonucleotides [1,63,64].
In our present study, non-specific effects were detectable at both 48 h and 72 h post-transfection. However, a PDK1-specific signature was discernable at the earlier time point (48 h post-transfection), whereas the longer incubation time (72 h post-transfection) displayed a more predominant ‘off-target’ signature (Figures 3 and 4). We conclude the predominance of this non-specific signature on the basis of two criteria: (i) the clustering analysis exhibiting an oligonucleotide-specific profile, rather than a sequence-specific profile (Figures 4B and 4C), and (ii) the Venn diagram exhibiting an increased number of genes regulated by all three treatments (ASO–MM–PDK1-siRNA#1, Figure 4D). The ‘off-target’ genes are numerous, including those involved in interferon response, proteasome degradation and oxidative stress. Moreover, few reports suggested that siRNAs can tolerate one to several mismatches to the mRNA targets and that only a partial homology is sufficient for an effective silencing of untargeted protein [51,65]. A recent study [66] reported that if two mismatches are allowed, 20% of 359 siRNAs published in the literature are not specific. On the other hand, if three mismatches are tolerated, 75% of the 359 siRNAs collected have a risk of ‘off-target’ activity. Therefore we do not find it surprising that the two PDK1-siRNA duplexes (siRNA#1 and #2) used in our present study behaved differently, either phenotypically (G1 arrest) or by their ability to affect changes in protein expression. The use in our study of a second siRNA (#2) raised against a different region of PDK1 did not help our final conclusions regarding the mechanism by which PDK1 inhibits cell proliferation in U87-MG cells. As BLAST search may miss important alignments for such short oligonucleotides, we used a method with higher sensitivity, such as Smith–Waterman algorithm, and were not able to detect an unintended target with a mismatch tolerancy higher than 3.
Even though PDK1 primarily regulates its substrates via post-transcriptional mechanisms (mainly through phosphorylation), it is likely that PDK1 activity will lead to downstream transcriptional responses, which may contribute to the understanding of its cellular functions. Therefore the early time-point clustering analysis allowed us to define a PDK1-specific profile and to identify putative PDK1 transcriptional target genes. Three out of nine candidate proteins selected for biochemical analysis showed strong indication of a relevant PDK1 target gene (HAT-1, p21/Cip1 and ARF6), because their expression changes showed a strong correlation with PDK1 depletion (ASO, siRNA#1 and siRNA#2 transfection; Figures 7A and 7B). In some cases, the validation experiment became difficult to interpret, as the PDK1-siRNA#2 transfected cells did not exhibit a similar pattern of change confirming that several siRNAs need to be used in order to confirm any biological consequences of the knock-down.
Nevertheless, the common feature of ASO- or siRNA-mediated silencing of PDK1 in U87-MG cells is an inhibition of cell proliferation, which is observed with two siRNA duplexes and two ASOs (Figure 6, and results not shown). This is consistent with a recent study in vivo suggesting that PDK1 is required for T cell proliferation, as hypomorphic PDK1 mice display a severely reduced number of T-cells [45]. Although the mechanism remains unclear, we identified some of the PDK1 target genes (such as p21/Cip1) which may explain the effect on proliferation in our model. Further investigations to address this question remains to be determined.
In summary, in agreement with previous reports this study demonstrates that the use of ASO and siRNA needs to be carefully employed. This is so far the first study documenting a comparison of ASO and RNAi approaches against a single target using a genome-wide analysis. The degree of specificity for PDK1 silencing reduces from 48 h to 72 h, but the mechanism for this is unclear and we do not rule out that it can vary from one protein to another. Altogether, these findings underscore the importance of developing powerful design strategies to predict highly effective and specific siRNAs, using multiple oligonucleotide duplexes and multiple time points to correctly interpret the results. The lowest possible concentration of the most active siRNAs should be used. Moreover, we provide in the present study the use of an appropriate control for silencing: targeting a mutant and non-functional expressed gene. This study confirms and extends previous strategies for an optimal interpretation of RNAi and/or ASO approach. It is therefore crucial to examine with caution the possible side-effects of such powerful techniques in order to study gene function, or to use ASO and siRNA as potential therapeutic agents.
Multimedia adjunct
Acknowledgments
We thank Pablo Rodriguez-Viciana, Neil Torbett, Tanja Meyer, Ruth Zeidman and Mike Fried for critical and helpful discussions during preparation of the manuscript. This work was supported by National Institutes of Health grants RO1CA79548, P50 CA97257, P01 CA64602 from the NCI, and Daiichi Pharmaceuticals (D.S.), and l'Association pour la Recherche contre le Cancer (ARC) (B.B.).
References
- 1.Lebedeva I., Stein C. A. Antisense oligonucleotides: promise and reality. Annu. Rev. Pharmacol. Toxicol. 2001;41:403–419. doi: 10.1146/annurev.pharmtox.41.1.403. [DOI] [PubMed] [Google Scholar]
- 2.Scherer L. J., Rossi J. J. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 3.Bock L. C., Griffin L. C., Latham J. A., Vermaas E. H., Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature (London) 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 4.Burgess T. L., Fisher E. F., Ross S. L., Bready J. V., Qian Y. X., Bayewitch L. A., Cohen A. M., Herrera C. J., Hu S. S., Kramer T. B., et al. The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4051–4055. doi: 10.1073/pnas.92.9.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho Y. S., Kim M. K., Cheadle C., Neary C., Becker K. G., Cho-Chung Y. S. Antisense DNAs as multisite genomic modulators identified by DNA microarray. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9819–9823. doi: 10.1073/pnas.171314398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fennewald S. M., Rando R. F. Inhibition of high affinity basic fibroblast growth factor binding by oligonucleotides. J. Biol. Chem. 1995;270:21718–21721. doi: 10.1074/jbc.270.37.21718. [DOI] [PubMed] [Google Scholar]
- 7.Fisher A. A., Ye D., Sergueev D. S., Fisher M. H., Shaw B. R., Juliano R. L. Evaluating the specificity of antisense oligonucleotide conjugates. A DNA array analysis. J. Biol. Chem. 2002;277:22980–22984. doi: 10.1074/jbc.M203347200. [DOI] [PubMed] [Google Scholar]
- 8.Guvakova M. A., Yakubov L. A., Vlodavsky I., Tonkinson J. L., Stein C. A. Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J. Biol. Chem. 1995;270:2620–2627. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- 9.McKay R. A., Miraglia L. J., Cummins L. L., Owens S. R., Sasmor H., Dean N. M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-α expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 10.Monia B. P., Lesnik E. A., Gonzalez C., Lima W. F., McGee D., Guinosso C. J., Kawasaki A. M., Cook P. D., Freier S. M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 11.Hannon G. J. RNA interference. Nature (London) 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 12.Hemann M. T., Fridman J. S., Zilfou J. T., Hernando E., Paddison P. J., Cordon-Cardo C., Hannon G. J., Lowe S. W. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 13.McManus M. T., Sharp P. A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 14.Williams B. R. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem. Soc. Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 15.Player M. R., Torrence P. F. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dykxhoorn D. M., Novina C. D., Sharp P. A. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir S. M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caplen N. J., Parrish S., Imani F., Fire A., Morgan R. A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi J. T., Chang H. Y., Wang N. N., Chang D. S., Dunphy N., Brown P. O. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scacheri P. C., Rozenblatt-Rosen O., Caplen N. J., Wolfsberg T. G., Umayam L., Lee J. C., Hughes C. M., Shanmugam K. S., Bhattacharjee A., Meyerson M., Collins F. S. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semizarov D., Frost L., Sarthy A., Kroeger P., Halbert D. N., Fesik S. W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sledz C. A., Holko M., de Veer M. J., Silverman R. H., Williams B. R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 24.Alessi D. R., Deak M., Casamayor A., Caudwell F. B., Morrice N., Norman D. G., Gaffney P., Reese C. B., MacDougall C. N., Harbison D., et al. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 25.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 26.Stephens L., Anderson K., Stokoe D., Erdjument-Bromage H., Painter G. F., Holmes A. B., Gaffney P. R., Reese C. B., McCormick F., Tempst P., et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science (Washington, D.C.) 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 27.Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science (Washington, D.C.) 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X., Ma Y., Moore M., Hemmings B. A., Taylor S. S. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alessi D. R. Discovery of PDK1, one of the missing links in insulin signal transduction. Colworth Medal Lecture. Biochem. Soc. Trans. 2001;29:1–14. doi: 10.1042/0300-5127:0290001. [DOI] [PubMed] [Google Scholar]
- 30.Le Good J. A., Ziegler W. H., Parekh D. B., Alessi D. R., Cohen P., Parker P. J. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science (Washington, D.C.) 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 31.Vanhaesebroeck B., Alessi D. R. The PI3K–PDK1 connection: more than just a road to PKB. Biochem. J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- 32.Frodin M., Antal T. L., Dummler B. A., Jensen C. J., Deak M., Gammeltoft S., Biondi R. M. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 2002;21:5396–5407. doi: 10.1093/emboj/cdf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niederberger C., Schweingruber M. E. A Schizosaccharomyces pombe gene, ksg1, that shows structural homology to the human phosphoinositide-dependent protein kinase PDK1, is essential for growth, mating and sporulation. Mol. Gen. Genet. 1999;261:177–183. doi: 10.1007/s004380050955. [DOI] [PubMed] [Google Scholar]
- 34.Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 35.Inagaki M., Schmelzle T., Yamaguchi K., Irie K., Hall M. N., Matsumoto K. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol. Cell. Biol. 1999;19:8344–8352. doi: 10.1128/mcb.19.12.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paradis S., Ailion M., Toker A., Thomas J. H., Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rintelen F., Stocker H., Thomas G., Hafen E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15020–15025. doi: 10.1073/pnas.011318098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawlor M. A., Mora A., Ashby P. R., Williams M. R., Murray-Tait V., Malone L., Prescott A. R., Lucocq J. M., Alessi D. R. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams M. R., Arthur J. S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D. R. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 40.Flynn P., Wong M., Zavar M., Dean N. M., Stokoe D. Inhibition of PDK-1 activity causes a reduction in cell proliferation and survival. Curr. Biol. 2000;10:1439–1442. doi: 10.1016/s0960-9822(00)00801-0. [DOI] [PubMed] [Google Scholar]
- 41.Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J. Biol. Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 42.Lassus P., Rodriguez J., Lazebnik Y. Confirming specificity of RNAi in mammalian cells. Sci. STKE. 2002;2002:PL13. doi: 10.1126/stke.2002.147.pl13. [DOI] [PubMed] [Google Scholar]
- 43.Gottschalk A. R., Basila D., Wong M., Dean N. M., Brandts C. H., Stokoe D., Haas-Kogan D. A. p27Kip1 is required for PTEN-induced G1 growth arrest. Cancer Res. 2001;61:2105–2111. [PubMed] [Google Scholar]
- 44.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science (Washington, D.C.) 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 45.Hinton H. J., Alessi D. R., Cantrell D. A. The serine kinase phosphoinositide-dependent kinase 1 (PDK1) regulates T cell development. Nat. Immunol. 2004;5:539–545. doi: 10.1038/ni1062. [DOI] [PubMed] [Google Scholar]
- 46.Mora A., Davies A. M., Bertrand L., Sharif I., Budas G. R., Jovanovic S., Mouton V., Kahn C. R., Lucocq J. M., Gray G. A., et al. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J. 2003;22:4666–4676. doi: 10.1093/emboj/cdg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Axel D. I., Spyridopoulos I., Riessen R., Runge H., Viebahn R., Karsch K. R. Toxicity, uptake kinetics and efficacy of new transfection reagents: increase of oligonucleotide uptake. J. Vasc. Res. 2000;37:221–234. doi: 10.1159/000025737. [DOI] [PubMed] [Google Scholar]
- 48.Miyagishi M., Hayashi M., Taira K. Comparison of the suppressive effects of antisense oligonucleotides and siRNAs directed against the same targets in mammalian cells. Antisense Nucleic Acid Drug Dev. 2003;13:1–7. doi: 10.1089/108729003764097296. [DOI] [PubMed] [Google Scholar]
- 49.Harborth J., Elbashir S. M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 50.Vickers T. A., Koo S., Bennett C. F., Crooke S. T., Dean N. M., Baker B. F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 51.Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 52.Dudek P., Picard D. TROD: T7 RNAi Oligo Designer. Nucleic Acids Res. 2004;32:W121–W123. doi: 10.1093/nar/gkh360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henschel A., Buchholz F., Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–W120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naito Y., Yamada T., Ui-Tei K., Morishita S., Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–W129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 56.Schwarz D. S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P. D. Asymmetry in the assembly of the RNAi enzyme complex. Cell (Cambridge, Mass.) 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 57.Khvorova A., Reynolds A., Jayasena S. D. Functional siRNAs and miRNAs exhibit strand bias. Cell (Cambridge, Mass.) 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 58.Medema R. H. Optimizing RNA interference for application in mammalian cells. Biochem. J. 2004;380:593–603. doi: 10.1042/BJ20040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bridge A. J., Pebernard S., Ducraux A., Nicoulaz A. L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 60.Persengiev S. P., Zhu X., Green M. R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pebernard S., Iggo R. D. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation. 2004;72:103–111. doi: 10.1111/j.1432-0436.2004.07202001.x. [DOI] [PubMed] [Google Scholar]
- 62.Cho-Chung Y. S., Becker K. G. A genome-wide view of antisense. Nat. Biotechnol. 2003;21:492. doi: 10.1038/nbt0503-492a. [DOI] [PubMed] [Google Scholar]
- 63.Bennett M. R., Schwartz S. M. Antisense therapy for angioplasty restenosis: some critical considerations. Circulation. 1995;92:1981–1993. doi: 10.1161/01.cir.92.7.1981. [DOI] [PubMed] [Google Scholar]
- 64.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 65.Saxena S., Jonsson Z. O., Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 66.Snove O., Jr, Holen T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.