The crystal structure of the compound described here features both C—H⋯O and N—H⋯O intramolecular hydrogen bonds, as well as a myriad of intermolecular C—H⋯O hydrogen-bonding interactions.
Keywords: crystal structure, carbamoylmethylphosphine oxide, intramolecular hydrogen bond, intermolecular hydrogen bond
Abstract
The molecular structure of the tripodal carbamoylmethylphosphine oxide compound diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate, C25H52N3O12P3, features six intramolecular hydrogen-bonding interactions. The phosphonate groups have key bond lengths ranging from 1.4696 (12) to 1.4729 (12) Å (P=O), 1.5681 (11) to 1.5811 (12) Å (P—O) and 1.7881 (16) to 1.7936 (16) Å (P—C). Each amide group adopts a nearly perfect trans geometry, and the geometry around each phophorus atom resembles a slightly distorted tetrahedron.
1. Chemical context
The carbamoylmethylphosphine oxide (CMPO) group (Fig. 1 ▸) has been utilized by researchers in the area of f-element coordination chemistry to prepare compounds with an affinity for lanthanide and actinide metals. Perhaps the most well known use of this metal chelator is as part of the TRUEX (transuranium extraction) process for the remediation of spent nuclear fuel (Horwitz et al., 1985 ▸). Various research groups have studied the coordination complexes of CMPO-containing compounds with f-elements and found that, depending on the identity of the metal, two to three CMPO groups are able to coordinate to the metal center simultaneously (Horwitz et al., 1987 ▸). Based on these results, research groups have used a variety of di-, tri- and tetrapodal scaffolds to tether multiple CMPO groups together with the aim of preparing chelators for f-elements that have stronger binding affinities and higher extraction selectivities than their monomeric counterparts (Dam et al., 2007 ▸; Leoncini et al., 2017 ▸; Werner & Biros, 2019 ▸). To this end, we have prepared a tripodal CMPO compound based on a tris(2-aminoethyl)methane scaffold and report here its characterization by X-ray diffraction and NMR spectroscopy.
Figure 1.
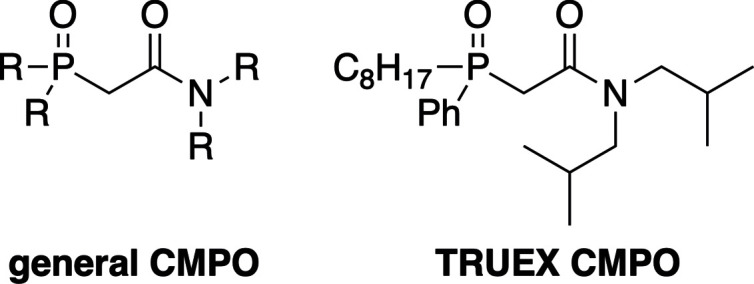
The general structure of the CMPO motif, along with the structure of the CMPO compound used in the TRUEX process.
2. Structural commentary
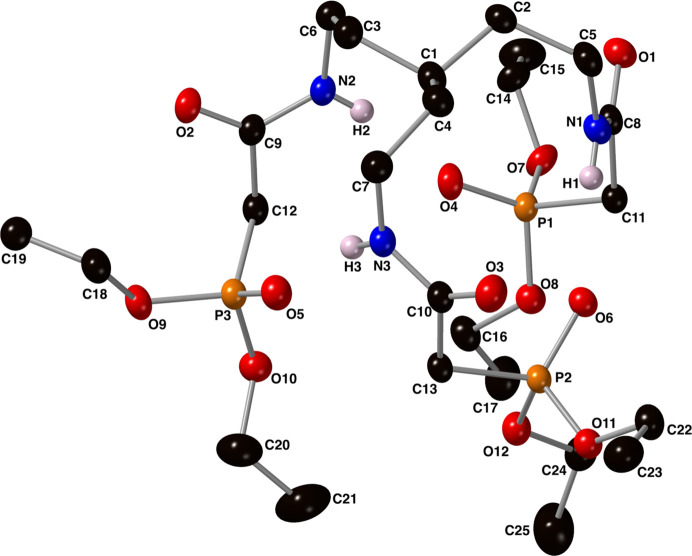
The molecular structure of compound I is shown in Fig. 2 ▸ along with the atom-numbering scheme. The electron density corresponding to the capping carbon atoms C2, C3 and C4 was disordered and was modeled over two positions with a 0.676 (3):0.324 (3) occupancy ratio (see the Refinement section for more details). The three CMPO arms are oriented on the same side of the molecule, and each phopshonate group is engaged in intramolecular hydrogen bonds with a neighboring amide group (vide infra). For the phosphonate groups, the three P=O bond lengths have values of 1.4696 (12), 1.4722 (12) and 1.4729 (12) Å. The longer P—O bond lengths range from 1.5681 (11) to 1.5811 (12) Å with P—C bond lengths ranging from 1.7881 (16) to 1.7936 (16) Å. Each phosphorus atom has a τ4 descriptor of fourfold coordination of 0.92 (where 0.00 = square planar, 0.85 = trigonal pyramidal, and 1.00 = tetrahedral; Yang et al., 2007 ▸), indicating that the geometry around these atoms resembles a slightly distorted tetrahedron. The C=O bond lengths of the amide groups are nearly identical with values of 1.231 (2), 1.231 (2) and 1.230 (2) Å. The C(O)—N bond lengths range from 1.335 (2) to 1.344 (2) Å, and each amide group adopts a nearly perfect trans geometry with H—N—C—O torsion angles of 176.9 (19), 177.9 (18) and 179.0 (16)°.
Figure 2.
The molecular structure of compound I, with the atom-labeling scheme. Displacement ellipsoids are drawn at the 50% probability level, and hydrogen atoms bonded to carbon atoms have been omitted for clarity. With regard to the disordered atoms, only the major component is shown.
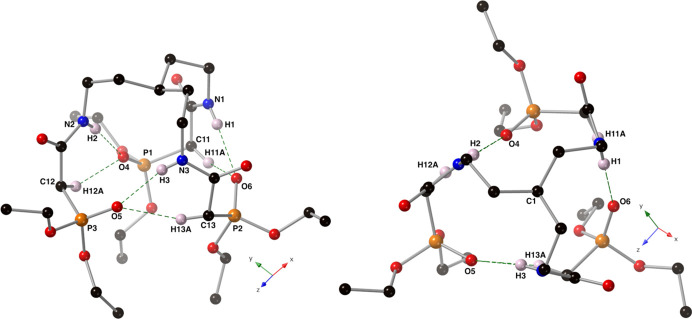
Intramolecular N—H⋯O and C—H⋯O hydrogen bonds are present in the crystal of compound I between each of the P=O oxygen atoms and a neighboring amide group (Fig. 3 ▸ and Table 1 ▸). These interactions have an average D⋯A distance of 2.886 Å and an average D—H⋯A angle of 169° for the N—H⋯O interactions, and an average D⋯A distance of 3.250 Å and an average D—H⋯A angle of 148° for the C—H⋯O interactions.
Figure 3.
Depictions of the intramolecular C—H⋯O and N—H⋯O hydrogen bonds (green, dashed lines) present in the crystal of compound I using a ball-and-stick model with standard CPK colors. With regard to the disordered atoms, only the major component is shown.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O6 | 0.86 (2) | 2.07 (2) | 2.9138 (18) | 168.8 (19) |
| N2—H2⋯O4 | 0.79 (2) | 2.06 (2) | 2.8465 (18) | 170 (2) |
| N3—H3⋯O5 | 0.82 (2) | 2.10 (2) | 2.8975 (19) | 167 (2) |
| C11—H11A⋯O6 | 0.99 | 2.36 | 3.2433 (19) | 148 |
| C11—H11B⋯O6i | 0.99 | 2.48 | 3.3235 (19) | 143 |
| C12—H12A⋯O4 | 0.99 | 2.37 | 3.2476 (19) | 148 |
| C12—H12B⋯O2ii | 0.99 | 2.35 | 3.321 (2) | 168 |
| C13—H13A⋯O5 | 0.99 | 2.37 | 3.259 (2) | 149 |
| C14—H14A⋯O1 | 0.99 | 2.56 | 3.326 (2) | 135 |
| C17—H17B⋯O3iii | 0.98 | 2.65 | 3.427 (3) | 137 |
| C18—H18A⋯O2 | 0.99 | 2.57 | 3.215 (2) | 122 |
| C22—H22B⋯O1i | 0.99 | 2.80 | 3.472 (2) | 126 |
| C23—H23C⋯O3 | 0.98 | 2.69 | 3.460 (2) | 135 |
| C24—H24A⋯O1i | 0.99 | 2.55 | 3.480 (2) | 156 |
| C24—H24B⋯O8 | 0.99 | 2.57 | 3.444 (2) | 147 |
| C4A—H4AA⋯O2iv | 0.99 | 2.39 | 3.241 (5) | 144 |
Symmetry codes: (i) 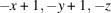 ; (ii)
; (ii) 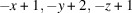 ; (iii)
; (iii) 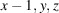 ; (iv)
; (iv) 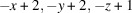 .
.
3. Supramolecular features
In the crystal, molecules of the title compound form supramolecular sheets that bisect the y- and z-axes. These sheets are held together by C—H⋯O hydrogen bonds (Table 1 ▸). Additional C—H⋯O hydrogen bonds are found between the supramolecular sheets.
4. Database survey
A search of the Cambridge Structure Database (CSD version 5.44 with updates through June 2024; Groom et al., 2016 ▸) for structures containing the general CMPO motif returned 104 hits, 63 of which were exclusively organic compounds. Of these 63 compounds, 14 structures contained the CMPO moiety tethered to a di-, tri- or tetrapodal scaffold. Structures CIWFAR (Ouizem et al., 2014 ▸) and GOGZAG (VanderWeide et al., 2019 ▸) contain aromatic rings decorated with two CMPO groups. Structures containing three CMPO groups tethered together can be found in entries IMIDEP (Coburn et al., 2016 ▸), XILJOR (Peters et al., 2002 ▸), JIVSUD and JIVTAK (Matloka et al., 2007 ▸). Lastly, a calix[4]arene scaffold was used to link four CMPO groups together in structures OLUWEX (Schmidt et al., 2003 ▸), CUVNEN and CUVNIR (Rudzevich et al., 2010 ▸).
5. Synthesis and crystallization
A 25 mL round-bottom flask was charged with 1.15 g (7.90 mmol) of freshly distilled 1,1,1-tris(2-aminoethyl)methane (Archer et al., 2004 ▸) and 1.0 mL of methanol. Under an atmosphere of nitrogen, the solution was cooled to ca. 230 K with a liquid N2/EtOAc bath. Triethylphosphonoacetate (6.50 mL, 32.8 mmol) was added slowly to the flask via syringe, and the reaction was allowed to warm to room temperature. The reaction was stirred under an inert atmosphere for 3 days, and the volatiles were removed under reduced pressure. The crude product was purified via silica gel column chromatography (5–10% MeOH/CH2Cl2 gradient) to give compound I as a slightly yellow, waxy solid (typical yield = 50–60%, Rf in 10% MeOH/CH2Cl2 = 0.4). Crystals suitable for analysis by X-ray diffraction were grown serendipitously from a concentrated solution of compound I in methanol upon standing in the refrigerator for many months. NMR data was acquired with a JEOL ECZS 400 NMR spectrometer: 1H NMR (400 MHz, CDCl3) δ 8.24 (broad, 3H), 4.10 (m, 12 H), 3.22 (m, 6H), 2.95 (d, JP–H = 21.6 Hz, 6H), 1.68 (septet, J = 6.8 Hz, 3H), 1.40 (dt, J = 6.3, 13.7 Hz, 6H), 1.29 (t, J = 7.1 Hz, 18H); 13C NMR (100 MHz, CDCl3) δ 164.8 (d, JC–P = 5.2 Hz), 62.5 (d, JC-P = 6.4 Hz), 36.3 (s), 35.0 (d, JC–P = 132 Hz), 31.8 (s), 25.2 (s), 16.4 (d, JC–P = 6.3 Hz); 31P NMR (161 MHz, CDCl3) δ 24.4.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms bonded to carbon atoms were placed in calculated positions and refined as riding: C—H = 0.95–1.00 Å with Uiso(H) = 1.2Ueq(C) for methylene and methine groups, and Uiso(H) = 1.5Ueq(C) for methyl groups. Hydrogen atoms bonded to nitrogen atoms were located using electron-density difference maps. The disordered electron density corresponding to C2/C2A, C3/C3A and C4/C4A was modeled over two positions and refined against a free variable to give a relative occupancy ratio of 0.676 (3):0.324 (3). This disorder reverberated to the nearby carbon atoms C5, C6 and C7 to give two orientations of the attached hydrogen atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C25H52N3O12P3 |
| M r | 679.60 |
| Crystal system, space group | Triclinic, P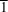
|
| Temperature (K) | 100 |
| a, b, c (Å) | 10.02487 (11), 11.92992 (15), 16.6237 (2) |
| α, β, γ (°) | 100.4792 (11), 100.124 (1), 111.1313 (11) |
| V (Å3) | 1759.25 (4) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ (mm−1) | 2.06 |
| Crystal size (mm) | 0.16 × 0.09 × 0.04 |
| Data collection | |
| Diffractometer | XtaLAB Synergy-S, Dualflex, HyPix-6000HE |
| Absorption correction | Gaussian (CrysAlis PRO; Oxford Diffraction, 2006 ▸) |
| Tmin, Tmax | 0.700, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 27502, 7504, 6481 |
| R int | 0.043 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.037, 0.099, 1.07 |
| No. of reflections | 7504 |
| No. of parameters | 438 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.61, −0.35 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024008478/vu2007sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024008478/vu2007Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989024008478/vu2007Isup3.cml
CCDC reference: 2379899
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We are grateful to GVSU (Chemistry department Weldon Fund, CSCE) for financial support of this research, as well as Dr Randy Winchester (GVSU) for helpful conversations. We also thank GVSU’s Office of Undergraduate Research for a Student Supplies Grant to B. Wackerle.
supplementary crystallographic information
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Crystal data
| C25H52N3O12P3 | Z = 2 |
| Mr = 679.60 | F(000) = 728 |
| Triclinic, P1 | Dx = 1.283 Mg m−3 |
| a = 10.02487 (11) Å | Cu Kα radiation, λ = 1.54184 Å |
| b = 11.92992 (15) Å | Cell parameters from 13811 reflections |
| c = 16.6237 (2) Å | θ = 4.1–79.9° |
| α = 100.4792 (11)° | µ = 2.06 mm−1 |
| β = 100.124 (1)° | T = 100 K |
| γ = 111.1313 (11)° | Irregular, colourless |
| V = 1759.25 (4) Å3 | 0.16 × 0.09 × 0.04 mm |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Data collection
| XtaLAB Synergy-S, Dualflex, HyPix-6000HE diffractometer | 6481 reflections with I > 2σ(I) |
| Detector resolution: 10.0000 pixels mm-1 | Rint = 0.043 |
| ω scans | θmax = 80.2°, θmin = 2.8° |
| Absorption correction: gaussian (CrysAlisPro; Oxford Diffraction, 2006) | h = −12→12 |
| Tmin = 0.700, Tmax = 1.000 | k = −15→14 |
| 27502 measured reflections | l = −20→21 |
| 7504 independent reflections |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: mixed |
| wR(F2) = 0.099 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0443P)2 + 0.5632P] where P = (Fo2 + 2Fc2)/3 |
| 7504 reflections | (Δ/σ)max = 0.001 |
| 438 parameters | Δρmax = 0.61 e Å−3 |
| 0 restraints | Δρmin = −0.34 e Å−3 |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| P1 | 0.35890 (4) | 0.68848 (4) | 0.12527 (2) | 0.02154 (10) | |
| P2 | 0.53282 (4) | 0.38100 (4) | 0.20227 (2) | 0.02071 (9) | |
| P3 | 0.47290 (4) | 0.71266 (4) | 0.45959 (2) | 0.02314 (10) | |
| O1 | 0.62953 (12) | 0.82490 (11) | 0.04850 (8) | 0.0286 (2) | |
| O2 | 0.69588 (13) | 0.99779 (11) | 0.49775 (7) | 0.0314 (3) | |
| O3 | 0.86359 (13) | 0.48663 (12) | 0.29651 (8) | 0.0336 (3) | |
| O4 | 0.45143 (13) | 0.75898 (12) | 0.21164 (7) | 0.0310 (3) | |
| O5 | 0.58783 (13) | 0.66867 (11) | 0.44337 (8) | 0.0304 (3) | |
| O6 | 0.59114 (12) | 0.46394 (10) | 0.14955 (7) | 0.0261 (2) | |
| O7 | 0.28427 (12) | 0.76460 (10) | 0.07967 (8) | 0.0283 (2) | |
| O8 | 0.22248 (12) | 0.56567 (11) | 0.11756 (8) | 0.0276 (2) | |
| O9 | 0.48562 (12) | 0.75646 (11) | 0.55729 (7) | 0.0274 (2) | |
| O10 | 0.30802 (13) | 0.61449 (11) | 0.42340 (8) | 0.0299 (3) | |
| O11 | 0.53576 (12) | 0.24858 (10) | 0.17424 (7) | 0.0249 (2) | |
| O12 | 0.36511 (12) | 0.34604 (11) | 0.20133 (7) | 0.0267 (2) | |
| N1 | 0.71852 (14) | 0.70410 (14) | 0.11270 (9) | 0.0242 (3) | |
| H1 | 0.693 (2) | 0.636 (2) | 0.1277 (13) | 0.025 (5)* | |
| N2 | 0.66717 (15) | 0.94335 (13) | 0.35606 (9) | 0.0248 (3) | |
| H2 | 0.614 (2) | 0.896 (2) | 0.3124 (15) | 0.030 (5)* | |
| N3 | 0.82763 (15) | 0.64181 (13) | 0.37830 (9) | 0.0250 (3) | |
| H3 | 0.766 (2) | 0.661 (2) | 0.3961 (14) | 0.029 (5)* | |
| C1 | 0.89282 (17) | 0.85561 (16) | 0.29672 (10) | 0.0270 (3) | |
| H1A | 0.791 (2) | 0.7973 (18) | 0.2865 (13) | 0.025 (5)* | |
| C5 | 0.87290 (17) | 0.79247 (17) | 0.13561 (11) | 0.0296 (3) | |
| H5AA | 0.893030 | 0.827828 | 0.087376 | 0.036* | 0.676 (3) |
| H5AB | 0.937439 | 0.747536 | 0.145957 | 0.036* | 0.676 (3) |
| H5BC | 0.878553 | 0.876708 | 0.134409 | 0.036* | 0.324 (3) |
| H5BD | 0.924175 | 0.767149 | 0.094618 | 0.036* | 0.324 (3) |
| C6 | 0.81521 (18) | 1.02970 (15) | 0.35924 (11) | 0.0282 (3) | |
| H6AA | 0.846342 | 1.105489 | 0.406285 | 0.034* | 0.676 (3) |
| H6AB | 0.812996 | 1.055031 | 0.305686 | 0.034* | 0.676 (3) |
| H6BC | 0.882153 | 1.049504 | 0.416062 | 0.034* | 0.324 (3) |
| H6BD | 0.811939 | 1.108309 | 0.349705 | 0.034* | 0.324 (3) |
| C7 | 0.97830 (17) | 0.73567 (16) | 0.39527 (11) | 0.0287 (3) | |
| H7AA | 1.049062 | 0.696112 | 0.404807 | 0.034* | 0.676 (3) |
| H7AB | 0.999863 | 0.800928 | 0.447785 | 0.034* | 0.676 (3) |
| H7BC | 1.030570 | 0.707191 | 0.356172 | 0.034* | 0.324 (3) |
| H7BD | 1.034145 | 0.749897 | 0.454197 | 0.034* | 0.324 (3) |
| C8 | 0.60896 (16) | 0.72964 (15) | 0.07237 (9) | 0.0223 (3) | |
| C9 | 0.62078 (18) | 0.93312 (14) | 0.42602 (10) | 0.0243 (3) | |
| C10 | 0.78257 (17) | 0.52657 (15) | 0.32878 (10) | 0.0249 (3) | |
| C11 | 0.45376 (16) | 0.63076 (14) | 0.05793 (10) | 0.0223 (3) | |
| H11A | 0.459884 | 0.554470 | 0.070441 | 0.027* | |
| H11B | 0.398214 | 0.608946 | −0.002128 | 0.027* | |
| C12 | 0.46608 (17) | 0.83407 (15) | 0.41182 (10) | 0.0249 (3) | |
| H12A | 0.419042 | 0.798180 | 0.350200 | 0.030* | |
| H12B | 0.404881 | 0.872627 | 0.436542 | 0.030* | |
| C13 | 0.61943 (17) | 0.44487 (15) | 0.31384 (10) | 0.0258 (3) | |
| H13A | 0.569838 | 0.494936 | 0.339364 | 0.031* | |
| H13B | 0.609095 | 0.376363 | 0.341386 | 0.031* | |
| C14 | 0.36017 (19) | 0.89977 (15) | 0.09918 (12) | 0.0308 (4) | |
| H14A | 0.447502 | 0.922709 | 0.075691 | 0.037* | |
| H14B | 0.394467 | 0.936564 | 0.161344 | 0.037* | |
| C15 | 0.2526 (2) | 0.94742 (19) | 0.06022 (16) | 0.0436 (5) | |
| H15A | 0.217905 | 0.909149 | −0.001106 | 0.065* | |
| H15B | 0.301497 | 1.038212 | 0.071238 | 0.065* | |
| H15C | 0.167935 | 0.926141 | 0.085007 | 0.065* | |
| C16 | 0.1174 (2) | 0.56953 (19) | 0.16770 (12) | 0.0359 (4) | |
| H16A | 0.136218 | 0.657160 | 0.193669 | 0.043* | |
| H16B | 0.129442 | 0.528868 | 0.213731 | 0.043* | |
| C17 | −0.0359 (2) | 0.5030 (2) | 0.11089 (15) | 0.0528 (6) | |
| H17A | −0.046762 | 0.543631 | 0.065403 | 0.079* | |
| H17B | −0.107391 | 0.505756 | 0.143687 | 0.079* | |
| H17C | −0.054202 | 0.416041 | 0.086221 | 0.079* | |
| C18 | 0.62943 (17) | 0.81483 (17) | 0.61951 (10) | 0.0287 (3) | |
| H18A | 0.708701 | 0.850973 | 0.591946 | 0.034* | |
| H18B | 0.650082 | 0.752204 | 0.645208 | 0.034* | |
| C19 | 0.6248 (2) | 0.91549 (17) | 0.68647 (12) | 0.0343 (4) | |
| H19A | 0.612075 | 0.980235 | 0.661174 | 0.051* | |
| H19B | 0.717792 | 0.952208 | 0.731421 | 0.051* | |
| H19C | 0.541637 | 0.879707 | 0.710529 | 0.051* | |
| C20 | 0.2548 (2) | 0.5026 (2) | 0.45184 (15) | 0.0457 (5) | |
| H20A | 0.209606 | 0.517026 | 0.498961 | 0.055* | |
| H20B | 0.338692 | 0.481182 | 0.472970 | 0.055* | |
| C21 | 0.1447 (3) | 0.3995 (2) | 0.38158 (18) | 0.0587 (7) | |
| H21A | 0.191132 | 0.382672 | 0.336272 | 0.088* | |
| H21B | 0.063215 | 0.422058 | 0.359740 | 0.088* | |
| H21C | 0.105937 | 0.324849 | 0.401657 | 0.088* | |
| C22 | 0.64205 (19) | 0.22577 (17) | 0.13194 (11) | 0.0291 (3) | |
| H22A | 0.708598 | 0.304258 | 0.122922 | 0.035* | |
| H22B | 0.589477 | 0.164576 | 0.075948 | 0.035* | |
| C23 | 0.7309 (2) | 0.17610 (18) | 0.18627 (13) | 0.0364 (4) | |
| H23A | 0.797129 | 0.153545 | 0.156173 | 0.055* | |
| H23B | 0.663732 | 0.102142 | 0.198646 | 0.055* | |
| H23C | 0.789814 | 0.240173 | 0.239481 | 0.055* | |
| C24 | 0.25179 (18) | 0.28757 (16) | 0.12104 (11) | 0.0298 (3) | |
| H24A | 0.282073 | 0.234942 | 0.081284 | 0.036* | |
| H24B | 0.237004 | 0.352005 | 0.095038 | 0.036* | |
| C25 | 0.1120 (2) | 0.2097 (3) | 0.13888 (17) | 0.0668 (8) | |
| H25A | 0.032790 | 0.170273 | 0.085977 | 0.100* | |
| H25B | 0.084140 | 0.262460 | 0.179047 | 0.100* | |
| H25C | 0.127335 | 0.145065 | 0.163357 | 0.100* | |
| C2 | 0.9138 (3) | 0.9024 (2) | 0.21687 (16) | 0.0273 (5) | 0.676 (3) |
| H2A | 1.018299 | 0.961080 | 0.227461 | 0.033* | 0.676 (3) |
| H2B | 0.850660 | 0.948454 | 0.206276 | 0.033* | 0.676 (3) |
| C3 | 0.9259 (3) | 0.9732 (2) | 0.37186 (16) | 0.0286 (6) | 0.676 (3) |
| H3A | 0.927444 | 0.947845 | 0.425396 | 0.034* | 0.676 (3) |
| H3B | 1.025648 | 1.037297 | 0.377840 | 0.034* | 0.676 (3) |
| C4 | 1.0018 (2) | 0.7972 (2) | 0.32176 (16) | 0.0276 (6) | 0.676 (3) |
| H4A | 1.103868 | 0.862754 | 0.338242 | 0.033* | 0.676 (3) |
| H4B | 0.993196 | 0.733465 | 0.271490 | 0.033* | 0.676 (3) |
| C2A | 0.9477 (5) | 0.7944 (5) | 0.2254 (3) | 0.0269 (11) | 0.324 (3) |
| H2AA | 1.056167 | 0.840578 | 0.236832 | 0.032* | 0.324 (3) |
| H2AB | 0.928389 | 0.707658 | 0.227536 | 0.032* | 0.324 (3) |
| C3A | 0.8789 (5) | 0.9696 (5) | 0.2877 (3) | 0.0263 (11) | 0.324 (3) |
| H3AA | 0.812478 | 0.952468 | 0.231216 | 0.032* | 0.324 (3) |
| H3AB | 0.977608 | 1.031831 | 0.289758 | 0.032* | 0.324 (3) |
| C4A | 0.9682 (6) | 0.8531 (5) | 0.3825 (3) | 0.0292 (12) | 0.324 (3) |
| H4AA | 1.070549 | 0.918112 | 0.398768 | 0.035* | 0.324 (3) |
| H4AB | 0.916861 | 0.878735 | 0.423347 | 0.035* | 0.324 (3) |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| P1 | 0.01846 (17) | 0.02538 (19) | 0.01788 (19) | 0.00752 (15) | 0.00438 (14) | 0.00221 (14) |
| P2 | 0.01955 (18) | 0.02467 (19) | 0.01902 (18) | 0.00989 (14) | 0.00611 (13) | 0.00554 (14) |
| P3 | 0.02313 (19) | 0.02747 (19) | 0.01929 (19) | 0.01144 (15) | 0.00539 (14) | 0.00510 (14) |
| O1 | 0.0275 (6) | 0.0315 (6) | 0.0313 (6) | 0.0127 (5) | 0.0109 (5) | 0.0142 (5) |
| O2 | 0.0338 (6) | 0.0328 (6) | 0.0204 (6) | 0.0105 (5) | 0.0035 (5) | −0.0005 (5) |
| O3 | 0.0279 (6) | 0.0382 (7) | 0.0339 (7) | 0.0157 (5) | 0.0088 (5) | 0.0023 (5) |
| O4 | 0.0263 (5) | 0.0411 (7) | 0.0188 (6) | 0.0095 (5) | 0.0052 (4) | 0.0013 (5) |
| O5 | 0.0339 (6) | 0.0344 (6) | 0.0303 (6) | 0.0201 (5) | 0.0119 (5) | 0.0095 (5) |
| O6 | 0.0278 (5) | 0.0287 (6) | 0.0236 (6) | 0.0116 (4) | 0.0083 (4) | 0.0098 (4) |
| O7 | 0.0268 (5) | 0.0222 (5) | 0.0293 (6) | 0.0099 (4) | 0.0001 (4) | −0.0014 (4) |
| O8 | 0.0227 (5) | 0.0292 (6) | 0.0307 (6) | 0.0088 (4) | 0.0128 (5) | 0.0056 (5) |
| O9 | 0.0212 (5) | 0.0406 (6) | 0.0191 (5) | 0.0114 (5) | 0.0051 (4) | 0.0074 (5) |
| O10 | 0.0274 (6) | 0.0306 (6) | 0.0272 (6) | 0.0084 (5) | 0.0021 (5) | 0.0092 (5) |
| O11 | 0.0248 (5) | 0.0267 (5) | 0.0265 (6) | 0.0124 (4) | 0.0106 (4) | 0.0072 (4) |
| O12 | 0.0210 (5) | 0.0334 (6) | 0.0258 (6) | 0.0122 (4) | 0.0069 (4) | 0.0046 (5) |
| N1 | 0.0208 (6) | 0.0309 (7) | 0.0218 (6) | 0.0101 (5) | 0.0067 (5) | 0.0090 (5) |
| N2 | 0.0257 (6) | 0.0264 (7) | 0.0185 (6) | 0.0089 (5) | 0.0036 (5) | 0.0026 (5) |
| N3 | 0.0223 (6) | 0.0299 (7) | 0.0205 (6) | 0.0094 (5) | 0.0056 (5) | 0.0043 (5) |
| C1 | 0.0221 (7) | 0.0363 (9) | 0.0229 (8) | 0.0120 (7) | 0.0064 (6) | 0.0082 (6) |
| C5 | 0.0193 (7) | 0.0441 (9) | 0.0247 (8) | 0.0098 (7) | 0.0094 (6) | 0.0102 (7) |
| C6 | 0.0287 (8) | 0.0259 (8) | 0.0253 (8) | 0.0074 (6) | 0.0054 (6) | 0.0050 (6) |
| C7 | 0.0219 (7) | 0.0327 (8) | 0.0254 (8) | 0.0080 (6) | −0.0002 (6) | 0.0063 (6) |
| C8 | 0.0217 (7) | 0.0294 (7) | 0.0168 (7) | 0.0107 (6) | 0.0076 (5) | 0.0058 (6) |
| C9 | 0.0284 (7) | 0.0246 (7) | 0.0214 (7) | 0.0140 (6) | 0.0057 (6) | 0.0040 (6) |
| C10 | 0.0250 (7) | 0.0307 (8) | 0.0188 (7) | 0.0116 (6) | 0.0046 (6) | 0.0068 (6) |
| C11 | 0.0208 (7) | 0.0262 (7) | 0.0198 (7) | 0.0097 (6) | 0.0056 (5) | 0.0050 (6) |
| C12 | 0.0273 (7) | 0.0291 (8) | 0.0202 (7) | 0.0143 (6) | 0.0066 (6) | 0.0047 (6) |
| C13 | 0.0269 (7) | 0.0293 (8) | 0.0196 (7) | 0.0090 (6) | 0.0083 (6) | 0.0058 (6) |
| C14 | 0.0298 (8) | 0.0217 (7) | 0.0369 (9) | 0.0092 (6) | 0.0091 (7) | 0.0008 (6) |
| C15 | 0.0374 (10) | 0.0335 (9) | 0.0681 (14) | 0.0189 (8) | 0.0184 (9) | 0.0191 (9) |
| C16 | 0.0316 (9) | 0.0430 (10) | 0.0327 (9) | 0.0110 (8) | 0.0190 (7) | 0.0078 (8) |
| C17 | 0.0273 (9) | 0.0609 (13) | 0.0509 (13) | 0.0022 (9) | 0.0211 (9) | −0.0080 (10) |
| C18 | 0.0213 (7) | 0.0405 (9) | 0.0211 (8) | 0.0107 (7) | 0.0019 (6) | 0.0082 (7) |
| C19 | 0.0344 (9) | 0.0334 (9) | 0.0278 (9) | 0.0085 (7) | 0.0051 (7) | 0.0051 (7) |
| C20 | 0.0384 (10) | 0.0418 (11) | 0.0491 (12) | 0.0078 (8) | −0.0002 (9) | 0.0229 (9) |
| C21 | 0.0462 (12) | 0.0369 (11) | 0.0729 (17) | 0.0088 (9) | −0.0149 (11) | 0.0139 (11) |
| C22 | 0.0301 (8) | 0.0363 (9) | 0.0292 (8) | 0.0201 (7) | 0.0137 (7) | 0.0086 (7) |
| C23 | 0.0276 (8) | 0.0367 (9) | 0.0468 (11) | 0.0175 (7) | 0.0056 (7) | 0.0101 (8) |
| C24 | 0.0239 (7) | 0.0346 (8) | 0.0293 (8) | 0.0143 (7) | 0.0017 (6) | 0.0047 (7) |
| C25 | 0.0233 (9) | 0.094 (2) | 0.0500 (14) | −0.0012 (11) | 0.0086 (9) | −0.0044 (13) |
| C2 | 0.0212 (11) | 0.0321 (12) | 0.0270 (12) | 0.0070 (9) | 0.0073 (9) | 0.0113 (10) |
| C3 | 0.0223 (11) | 0.0332 (12) | 0.0235 (12) | 0.0072 (9) | 0.0019 (9) | 0.0038 (10) |
| C4 | 0.0183 (10) | 0.0355 (13) | 0.0273 (12) | 0.0089 (9) | 0.0056 (9) | 0.0091 (10) |
| C2A | 0.019 (2) | 0.030 (2) | 0.027 (3) | 0.0078 (19) | 0.0054 (18) | 0.0021 (19) |
| C3A | 0.025 (2) | 0.025 (2) | 0.025 (2) | 0.0073 (19) | 0.0072 (19) | 0.0052 (19) |
| C4A | 0.026 (2) | 0.029 (2) | 0.024 (3) | 0.007 (2) | 0.0009 (19) | 0.0017 (19) |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Geometric parameters (Å, º)
| P1—O4 | 1.4696 (12) | C9—C12 | 1.519 (2) |
| P1—O7 | 1.5811 (12) | C10—C13 | 1.519 (2) |
| P1—O8 | 1.5681 (11) | C11—H11A | 0.9900 |
| P1—C11 | 1.7881 (16) | C11—H11B | 0.9900 |
| P2—O6 | 1.4722 (12) | C12—H12A | 0.9900 |
| P2—O11 | 1.5761 (12) | C12—H12B | 0.9900 |
| P2—O12 | 1.5759 (11) | C13—H13A | 0.9900 |
| P2—C13 | 1.7936 (16) | C13—H13B | 0.9900 |
| P3—O5 | 1.4729 (12) | C14—H14A | 0.9900 |
| P3—O9 | 1.5803 (12) | C14—H14B | 0.9900 |
| P3—O10 | 1.5697 (12) | C14—C15 | 1.495 (3) |
| P3—C12 | 1.7899 (17) | C15—H15A | 0.9800 |
| O1—C8 | 1.231 (2) | C15—H15B | 0.9800 |
| O2—C9 | 1.231 (2) | C15—H15C | 0.9800 |
| O3—C10 | 1.230 (2) | C16—H16A | 0.9900 |
| O7—C14 | 1.457 (2) | C16—H16B | 0.9900 |
| O8—C16 | 1.4612 (19) | C16—C17 | 1.490 (3) |
| O9—C18 | 1.4571 (18) | C17—H17A | 0.9800 |
| O10—C20 | 1.448 (2) | C17—H17B | 0.9800 |
| O11—C22 | 1.4515 (19) | C17—H17C | 0.9800 |
| O12—C24 | 1.455 (2) | C18—H18A | 0.9900 |
| N1—H1 | 0.86 (2) | C18—H18B | 0.9900 |
| N1—C5 | 1.457 (2) | C18—C19 | 1.502 (3) |
| N1—C8 | 1.344 (2) | C19—H19A | 0.9800 |
| N2—H2 | 0.79 (2) | C19—H19B | 0.9800 |
| N2—C6 | 1.455 (2) | C19—H19C | 0.9800 |
| N2—C9 | 1.336 (2) | C20—H20A | 0.9900 |
| N3—H3 | 0.82 (2) | C20—H20B | 0.9900 |
| N3—C7 | 1.460 (2) | C20—C21 | 1.463 (3) |
| N3—C10 | 1.335 (2) | C21—H21A | 0.9800 |
| C1—H1A | 0.97 (2) | C21—H21B | 0.9800 |
| C1—C2 | 1.550 (3) | C21—H21C | 0.9800 |
| C1—C3 | 1.587 (3) | C22—H22A | 0.9900 |
| C1—C4 | 1.532 (3) | C22—H22B | 0.9900 |
| C1—C2A | 1.564 (5) | C22—C23 | 1.501 (2) |
| C1—C3A | 1.447 (5) | C23—H23A | 0.9800 |
| C1—C4A | 1.503 (5) | C23—H23B | 0.9800 |
| C5—H5AA | 0.9900 | C23—H23C | 0.9800 |
| C5—H5AB | 0.9900 | C24—H24A | 0.9900 |
| C5—H5BC | 0.9900 | C24—H24B | 0.9900 |
| C5—H5BD | 0.9900 | C24—C25 | 1.491 (3) |
| C5—C2 | 1.577 (3) | C25—H25A | 0.9800 |
| C5—C2A | 1.540 (5) | C25—H25B | 0.9800 |
| C6—H6AA | 0.9900 | C25—H25C | 0.9800 |
| C6—H6AB | 0.9900 | C2—H2A | 0.9900 |
| C6—H6BC | 0.9900 | C2—H2B | 0.9900 |
| C6—H6BD | 0.9900 | C3—H3A | 0.9900 |
| C6—C3 | 1.497 (3) | C3—H3B | 0.9900 |
| C6—C3A | 1.619 (5) | C4—H4A | 0.9900 |
| C7—H7AA | 0.9900 | C4—H4B | 0.9900 |
| C7—H7AB | 0.9900 | C2A—H2AA | 0.9900 |
| C7—H7BC | 0.9900 | C2A—H2AB | 0.9900 |
| C7—H7BD | 0.9900 | C3A—H3AA | 0.9900 |
| C7—C4 | 1.546 (3) | C3A—H3AB | 0.9900 |
| C7—C4A | 1.490 (6) | C4A—H4AA | 0.9900 |
| C8—C11 | 1.521 (2) | C4A—H4AB | 0.9900 |
| O4—P1—O7 | 112.90 (7) | O7—C14—H14A | 110.2 |
| O4—P1—O8 | 116.05 (7) | O7—C14—H14B | 110.2 |
| O4—P1—C11 | 114.00 (7) | O7—C14—C15 | 107.68 (14) |
| O7—P1—C11 | 108.90 (7) | H14A—C14—H14B | 108.5 |
| O8—P1—O7 | 102.52 (6) | C15—C14—H14A | 110.2 |
| O8—P1—C11 | 101.26 (7) | C15—C14—H14B | 110.2 |
| O6—P2—O11 | 113.64 (6) | C14—C15—H15A | 109.5 |
| O6—P2—O12 | 115.95 (7) | C14—C15—H15B | 109.5 |
| O6—P2—C13 | 114.70 (7) | C14—C15—H15C | 109.5 |
| O11—P2—C13 | 108.50 (7) | H15A—C15—H15B | 109.5 |
| O12—P2—O11 | 101.94 (6) | H15A—C15—H15C | 109.5 |
| O12—P2—C13 | 100.62 (7) | H15B—C15—H15C | 109.5 |
| O5—P3—O9 | 112.76 (7) | O8—C16—H16A | 110.0 |
| O5—P3—O10 | 116.32 (7) | O8—C16—H16B | 110.0 |
| O5—P3—C12 | 114.34 (7) | O8—C16—C17 | 108.68 (15) |
| O9—P3—C12 | 108.77 (7) | H16A—C16—H16B | 108.3 |
| O10—P3—O9 | 102.72 (6) | C17—C16—H16A | 110.0 |
| O10—P3—C12 | 100.66 (7) | C17—C16—H16B | 110.0 |
| C14—O7—P1 | 119.89 (10) | C16—C17—H17A | 109.5 |
| C16—O8—P1 | 119.61 (11) | C16—C17—H17B | 109.5 |
| C18—O9—P3 | 120.98 (10) | C16—C17—H17C | 109.5 |
| C20—O10—P3 | 121.18 (11) | H17A—C17—H17B | 109.5 |
| C22—O11—P2 | 123.56 (10) | H17A—C17—H17C | 109.5 |
| C24—O12—P2 | 119.46 (10) | H17B—C17—H17C | 109.5 |
| C5—N1—H1 | 121.2 (14) | O9—C18—H18A | 110.2 |
| C8—N1—H1 | 117.1 (14) | O9—C18—H18B | 110.2 |
| C8—N1—C5 | 121.51 (15) | O9—C18—C19 | 107.77 (14) |
| C6—N2—H2 | 119.9 (16) | H18A—C18—H18B | 108.5 |
| C9—N2—H2 | 117.7 (16) | C19—C18—H18A | 110.2 |
| C9—N2—C6 | 122.08 (14) | C19—C18—H18B | 110.2 |
| C7—N3—H3 | 119.5 (15) | C18—C19—H19A | 109.5 |
| C10—N3—H3 | 117.9 (15) | C18—C19—H19B | 109.5 |
| C10—N3—C7 | 122.40 (15) | C18—C19—H19C | 109.5 |
| C2—C1—H1A | 109.5 (12) | H19A—C19—H19B | 109.5 |
| C2—C1—C3 | 107.39 (17) | H19A—C19—H19C | 109.5 |
| C3—C1—H1A | 108.6 (12) | H19B—C19—H19C | 109.5 |
| C4—C1—H1A | 111.6 (12) | O10—C20—H20A | 109.7 |
| C4—C1—C2 | 110.97 (17) | O10—C20—H20B | 109.7 |
| C4—C1—C3 | 108.61 (16) | O10—C20—C21 | 109.73 (18) |
| C2A—C1—H1A | 103.4 (12) | H20A—C20—H20B | 108.2 |
| C3A—C1—H1A | 103.2 (12) | C21—C20—H20A | 109.7 |
| C3A—C1—C2A | 114.5 (3) | C21—C20—H20B | 109.7 |
| C3A—C1—C4A | 118.3 (3) | C20—C21—H21A | 109.5 |
| C4A—C1—H1A | 104.2 (12) | C20—C21—H21B | 109.5 |
| C4A—C1—C2A | 111.1 (3) | C20—C21—H21C | 109.5 |
| N1—C5—H5AA | 109.0 | H21A—C21—H21B | 109.5 |
| N1—C5—H5AB | 109.0 | H21A—C21—H21C | 109.5 |
| N1—C5—H5BC | 110.0 | H21B—C21—H21C | 109.5 |
| N1—C5—H5BD | 110.0 | O11—C22—H22A | 109.9 |
| N1—C5—C2 | 113.05 (14) | O11—C22—H22B | 109.9 |
| N1—C5—C2A | 108.5 (2) | O11—C22—C23 | 108.74 (14) |
| H5AA—C5—H5AB | 107.8 | H22A—C22—H22B | 108.3 |
| H5BC—C5—H5BD | 108.4 | C23—C22—H22A | 109.9 |
| C2—C5—H5AA | 109.0 | C23—C22—H22B | 109.9 |
| C2—C5—H5AB | 109.0 | C22—C23—H23A | 109.5 |
| C2A—C5—H5BC | 110.0 | C22—C23—H23B | 109.5 |
| C2A—C5—H5BD | 110.0 | C22—C23—H23C | 109.5 |
| N2—C6—H6AA | 109.2 | H23A—C23—H23B | 109.5 |
| N2—C6—H6AB | 109.2 | H23A—C23—H23C | 109.5 |
| N2—C6—H6BC | 109.5 | H23B—C23—H23C | 109.5 |
| N2—C6—H6BD | 109.5 | O12—C24—H24A | 110.2 |
| N2—C6—C3 | 111.97 (16) | O12—C24—H24B | 110.2 |
| N2—C6—C3A | 110.5 (2) | O12—C24—C25 | 107.54 (16) |
| H6AA—C6—H6AB | 107.9 | H24A—C24—H24B | 108.5 |
| H6BC—C6—H6BD | 108.1 | C25—C24—H24A | 110.2 |
| C3—C6—H6AA | 109.2 | C25—C24—H24B | 110.2 |
| C3—C6—H6AB | 109.2 | C24—C25—H25A | 109.5 |
| C3A—C6—H6BC | 109.5 | C24—C25—H25B | 109.5 |
| C3A—C6—H6BD | 109.5 | C24—C25—H25C | 109.5 |
| N3—C7—H7AA | 109.1 | H25A—C25—H25B | 109.5 |
| N3—C7—H7AB | 109.1 | H25A—C25—H25C | 109.5 |
| N3—C7—H7BC | 110.1 | H25B—C25—H25C | 109.5 |
| N3—C7—H7BD | 110.1 | C1—C2—C5 | 112.41 (18) |
| N3—C7—C4 | 112.46 (15) | C1—C2—H2A | 109.1 |
| N3—C7—C4A | 107.8 (2) | C1—C2—H2B | 109.1 |
| H7AA—C7—H7AB | 107.8 | C5—C2—H2A | 109.1 |
| H7BC—C7—H7BD | 108.5 | C5—C2—H2B | 109.1 |
| C4—C7—H7AA | 109.1 | H2A—C2—H2B | 107.9 |
| C4—C7—H7AB | 109.1 | C1—C3—H3A | 108.8 |
| C4A—C7—H7BC | 110.1 | C1—C3—H3B | 108.8 |
| C4A—C7—H7BD | 110.1 | C6—C3—C1 | 113.63 (17) |
| O1—C8—N1 | 123.86 (14) | C6—C3—H3A | 108.8 |
| O1—C8—C11 | 121.44 (14) | C6—C3—H3B | 108.8 |
| N1—C8—C11 | 114.70 (14) | H3A—C3—H3B | 107.7 |
| O2—C9—N2 | 123.68 (15) | C1—C4—C7 | 114.59 (17) |
| O2—C9—C12 | 121.02 (15) | C1—C4—H4A | 108.6 |
| N2—C9—C12 | 115.30 (14) | C1—C4—H4B | 108.6 |
| O3—C10—N3 | 124.25 (15) | C7—C4—H4A | 108.6 |
| O3—C10—C13 | 120.97 (15) | C7—C4—H4B | 108.6 |
| N3—C10—C13 | 114.78 (14) | H4A—C4—H4B | 107.6 |
| P1—C11—H11A | 109.7 | C1—C2A—H2AA | 108.8 |
| P1—C11—H11B | 109.7 | C1—C2A—H2AB | 108.8 |
| C8—C11—P1 | 109.70 (10) | C5—C2A—C1 | 113.6 (3) |
| C8—C11—H11A | 109.7 | C5—C2A—H2AA | 108.8 |
| C8—C11—H11B | 109.7 | C5—C2A—H2AB | 108.8 |
| H11A—C11—H11B | 108.2 | H2AA—C2A—H2AB | 107.7 |
| P3—C12—H12A | 109.5 | C1—C3A—C6 | 114.5 (3) |
| P3—C12—H12B | 109.5 | C1—C3A—H3AA | 108.6 |
| C9—C12—P3 | 110.80 (11) | C1—C3A—H3AB | 108.6 |
| C9—C12—H12A | 109.5 | C6—C3A—H3AA | 108.6 |
| C9—C12—H12B | 109.5 | C6—C3A—H3AB | 108.6 |
| H12A—C12—H12B | 108.1 | H3AA—C3A—H3AB | 107.6 |
| P2—C13—H13A | 109.6 | C1—C4A—H4AA | 107.4 |
| P2—C13—H13B | 109.6 | C1—C4A—H4AB | 107.4 |
| C10—C13—P2 | 110.07 (11) | C7—C4A—C1 | 119.9 (3) |
| C10—C13—H13A | 109.6 | C7—C4A—H4AA | 107.4 |
| C10—C13—H13B | 109.6 | C7—C4A—H4AB | 107.4 |
| H13A—C13—H13B | 108.2 | H4AA—C4A—H4AB | 106.9 |
| P1—O7—C14—C15 | −169.31 (13) | N2—C9—C12—P3 | 114.58 (14) |
| P1—O8—C16—C17 | 131.70 (16) | N3—C7—C4—C1 | 55.4 (2) |
| P2—O11—C22—C23 | 119.26 (14) | N3—C7—C4A—C1 | −62.0 (4) |
| P2—O12—C24—C25 | 151.86 (17) | N3—C10—C13—P2 | 126.59 (13) |
| P3—O9—C18—C19 | 142.53 (12) | C5—N1—C8—O1 | 3.3 (2) |
| P3—O10—C20—C21 | −145.69 (17) | C5—N1—C8—C11 | −176.51 (13) |
| O1—C8—C11—P1 | −70.88 (17) | C6—N2—C9—O2 | 2.7 (3) |
| O2—C9—C12—P3 | −65.17 (18) | C6—N2—C9—C12 | −177.08 (14) |
| O3—C10—C13—P2 | −53.21 (19) | C7—N3—C10—O3 | 3.7 (3) |
| O4—P1—O7—C14 | 31.54 (14) | C7—N3—C10—C13 | −176.07 (14) |
| O4—P1—O8—C16 | 52.59 (15) | C8—N1—C5—C2 | 78.0 (2) |
| O4—P1—C11—C8 | −41.31 (13) | C8—N1—C5—C2A | 138.4 (2) |
| O5—P3—O9—C18 | 33.23 (14) | C9—N2—C6—C3 | 87.1 (2) |
| O5—P3—O10—C20 | 62.32 (17) | C9—N2—C6—C3A | 143.7 (2) |
| O5—P3—C12—C9 | −45.33 (13) | C10—N3—C7—C4 | 79.6 (2) |
| O6—P2—O11—C22 | 27.99 (14) | C10—N3—C7—C4A | 132.3 (3) |
| O6—P2—O12—C24 | 56.73 (14) | C11—P1—O7—C14 | −96.14 (13) |
| O6—P2—C13—C10 | −34.53 (14) | C11—P1—O8—C16 | 176.58 (13) |
| O7—P1—O8—C16 | −70.93 (14) | C12—P3—O9—C18 | −94.69 (13) |
| O7—P1—C11—C8 | 85.75 (12) | C12—P3—O10—C20 | −173.55 (16) |
| O8—P1—O7—C14 | 157.15 (12) | C13—P2—O11—C22 | −100.87 (13) |
| O8—P1—C11—C8 | −166.67 (11) | C13—P2—O12—C24 | −178.94 (12) |
| O9—P3—O10—C20 | −61.33 (16) | C2—C1—C3—C6 | 66.6 (2) |
| O9—P3—C12—C9 | 81.70 (12) | C2—C1—C4—C7 | −174.01 (17) |
| O10—P3—O9—C18 | 159.22 (12) | C3—C1—C2—C5 | −175.92 (16) |
| O10—P3—C12—C9 | −170.81 (11) | C3—C1—C4—C7 | 68.2 (2) |
| O11—P2—O12—C24 | −67.24 (13) | C4—C1—C2—C5 | 65.5 (2) |
| O11—P2—C13—C10 | 93.74 (12) | C4—C1—C3—C6 | −173.29 (18) |
| O12—P2—O11—C22 | 153.50 (12) | C2A—C1—C3A—C6 | 177.4 (3) |
| O12—P2—C13—C10 | −159.72 (12) | C2A—C1—C4A—C7 | −47.4 (5) |
| N1—C5—C2—C1 | 61.5 (2) | C3A—C1—C2A—C5 | −43.5 (4) |
| N1—C5—C2A—C1 | −69.9 (3) | C3A—C1—C4A—C7 | 177.1 (3) |
| N1—C8—C11—P1 | 108.97 (13) | C4A—C1—C2A—C5 | 179.2 (3) |
| N2—C6—C3—C1 | 62.7 (2) | C4A—C1—C3A—C6 | −48.6 (5) |
| N2—C6—C3A—C1 | −60.9 (4) |
Diethyl {[(5-[2-(diethoxyphosphoryl)acetamido]-3-{2-[2-(diethoxyphosphoryl)acetamido]ethyl}pentyl)carbamoyl]methyl}phosphonate. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O6 | 0.86 (2) | 2.07 (2) | 2.9138 (18) | 168.8 (19) |
| N2—H2···O4 | 0.79 (2) | 2.06 (2) | 2.8465 (18) | 170 (2) |
| N3—H3···O5 | 0.82 (2) | 2.10 (2) | 2.8975 (19) | 167 (2) |
| C11—H11A···O6 | 0.99 | 2.36 | 3.2433 (19) | 148 |
| C11—H11B···O6i | 0.99 | 2.48 | 3.3235 (19) | 143 |
| C12—H12A···O4 | 0.99 | 2.37 | 3.2476 (19) | 148 |
| C12—H12B···O2ii | 0.99 | 2.35 | 3.321 (2) | 168 |
| C13—H13A···O5 | 0.99 | 2.37 | 3.259 (2) | 149 |
| C14—H14A···O1 | 0.99 | 2.56 | 3.326 (2) | 135 |
| C17—H17B···O3iii | 0.98 | 2.65 | 3.427 (3) | 137 |
| C18—H18A···O2 | 0.99 | 2.57 | 3.215 (2) | 122 |
| C22—H22B···O1i | 0.99 | 2.80 | 3.472 (2) | 126 |
| C23—H23C···O3 | 0.98 | 2.69 | 3.460 (2) | 135 |
| C24—H24A···O1i | 0.99 | 2.55 | 3.480 (2) | 156 |
| C24—H24B···O8 | 0.99 | 2.57 | 3.444 (2) | 147 |
| C4A—H4AA···O2iv | 0.99 | 2.39 | 3.241 (5) | 144 |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+1, −y+2, −z+1; (iii) x−1, y, z; (iv) −x+2, −y+2, −z+1.
Funding Statement
This work was funded by National Science Foundation, Directorate for Mathematical and Physical Sciences grant CHE-2102576; National Science Foundation, Directorate for Mathematical and Physical Sciences grant CHE-2102381; National Science Foundation, Directorate for Mathematical and Physical Sciences grant CHE-1919565; National Science Foundation, Directorate for Mathematical and Physical Sciences grant CHE-1559886.
References
- Archer, C. M., Wadsworth, H. J. & Engell, T. (2004). US Patent Application US 2004/0258619 A1.
- Bourhis, L. J., Dolomanov, O. V., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2015). Acta Cryst. A71, 59–75. [DOI] [PMC free article] [PubMed]
- Coburn, K. M., Hardy, D. A., Patterson, M. G., McGraw, S. N., Peruzzi, M. T., Boucher, F., Beelen, B., Sartain, H. T., Neils, T., Lawrence, C. L., Staples, R. J., Werner, E. J. & Biros, S. M. (2016). Inorg. Chim. Acta, 449, 96–106.
- Dam, H. H., Reinhoudt, D. N. & Verboom, W. (2007). Chem. Soc. Rev.36, 367–377. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst.42, 339–341.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Horwitz, E. P., Diamond, H., Martin, K. A. & Chiarizia, R. (1987). Solvent Extr. Ion Exch.5, 419–446.
- Horwitz, E. P., Kalina, D. G., Diamond, H., Vandegrift, G. F. & Schulz, W. W. (1985). Solvent Extr. Ion Exch.3, 75–109.
- Leoncini, A., Huskens, J. & Verboom, W. (2017). Chem. Soc. Rev.46, 7229–7273. [DOI] [PubMed]
- Matloka, K., Sah, A. K., Peters, M. W., Srinivasan, P., Gelis, A. V., Regalbuto, M. & Scott, M. J. (2007). Inorg. Chem.46, 10549–10563. [DOI] [PubMed]
- Ouizem, S., Rosario-Amorin, D., Dickie, D. A., Paine, R. T., de Bettencourt-Dias, A., Hay, B. P., Podair, J. & Delmau, L. H. (2014). Dalton Trans.43, 8368–8386. [DOI] [PubMed]
- Oxford Diffraction (2006). CrysAlis PRO. Oxford Diffraction Ltd, Abingdon, England.
- Palmer, D. (2007). CrystalMaker. CrystalMaker Software, Bicester, England.
- Peters, M. W., Werner, E. J. & Scott, M. J. (2002). Inorg. Chem.41, 1707–1716. [DOI] [PubMed]
- Rudzevich, V., Kasyan, O., Drapailo, A., Bolte, M., Schollmeyer, D. & Böhmer, V. (2010). Chem. Asian J.5, 1347–1355. [DOI] [PubMed]
- Schmidt, C., Saadioui, M., Böhmer, V., Host, V., Spirlet, M.-R., Desreux, J. F., Brisach, F., Arnaud-Neu, F. & Dozol, J.-F. (2003). Org. Biomol. Chem.1, 4089–4096. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- VanderWeide, A. I., Staples, R. J. & Biros, S. M. (2019). Acta Cryst. E75, 991–996. [DOI] [PMC free article] [PubMed]
- Werner, E. J. & Biros, S. M. (2019). Org. Chem. Front.6, 2067–2094.
- Yang, L., Powell, D. R. & Houser, R. P. (2007). Dalton Trans. pp. 955–964. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024008478/vu2007sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024008478/vu2007Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989024008478/vu2007Isup3.cml
CCDC reference: 2379899
Additional supporting information: crystallographic information; 3D view; checkCIF report