Abstract
Expression of HXK2, a gene encoding a Saccharomyces cerevisiae bifunctional protein with catalytic and regulatory functions, is controlled by glucose availability, being activated in the presence of glucose and inhibited when the levels of the sugar are low. In the present study, we identified Rgt1 as a transcription factor that, together with the Med8 protein, is essential for repression of the HXK2 gene in the absence of glucose. Rgt1 represses HXK2 expression by binding specifically to the motif (CGGAAAA) located at −395 bp relative to the ATG translation start codon in the HXK2 promoter. Disruption of the RGT1 gene causes an 18-fold increase in the level of HXK2 transcript in the absence of glucose. Rgt1 binds to the RGT1 element of HXK2 promoter in a glucose-dependent manner, and the repression of target gene depends on binding of Rgt1 to DNA. The physiological significance of the connection between two glucose-signalling pathways, the Snf3/Rgt2 that causes glucose induction and the Mig1/Hxk2 that causes glucose repression, was also analysed.
Keywords: glucose repression, glucose signalling, HXK2 gene, Med8, Rgt1, Saccharomyces cerevisiae
Abbreviations: ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamidino-2-phenylindole; DRS, downstream repressing sequence; EMSA, electrophoretic mobility-shift assay; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; Hxk2, hexokinase 2
INTRODUCTION
Glucose is known to affect a variety of processes in Saccharomyces cerevisiae. These include glucose repression of genes for growth on alternative carbon sources and the induction of genes necessary for glucose transport [1–3]. Thus, depending on the availability of glucose, cells significantly change expression of many genes through mechanisms by which yeast cells sense the level of extracellular glucose [4]. Indeed, several signalling pathways governing these processes have been identified in yeast. In the Snf3/Rgt2 pathway, these two proteins act as glucose receptors. The Rgt2 and Snf3 proteins resemble hexose transporters in structure but have long cytoplasmic tails required for signal transduction [5]. Glucose binding to these transmembrane proteins initiates signals that activate the pathway and allow hexose transporter gene expression by repressing Rgt1 function [6]. Thus glucose in yeast acts similarly to a growth hormone in mammalian systems.
Glucose also activates another pathway involved in gene repression, which is not needed during the growth on glucose. In this pathway, both the Mig1 and Hxk2 (hexokinase 2) proteins are necessary to generate the glucose repression signal. The mechanism by which Mig1, a transcriptional repressor responsible for glucose repression of many genes, operates has been extensively investigated [7–9]. On the other hand, the mechanism by which Hxk2, in addition to its classical metabolic role, plays an important function in glucose signalling is only beginning to be understood [10,11]. Recently, it was described that, in S. cerevisiae, Hxk2 partially localizes to the nucleus in a glucose-dependent manner. Mig1 is required to sequester Hxk2 into the nucleus to generate a repressor complex during its growth in glucose medium [12].
So far, experimental evidence for a connection between the two glucose-sensing pathways is scarce. Only two main connections have been established [13]. These include, first, the effect of the Snf3/Rgt2 induction pathway on the glucose-repression pathway by the induction of the transcription of MIG2, which encodes a secondary repressor of the Mig1 repressor pathway [14]. Secondly, the Mig1/Hxk2 repression pathway contributes to glucose induction by repressing the expression of SNF3 and MTH1 genes [15–17] that encode a regulator of the Rgt1 transcription factor.
In the present study, we characterize the factors involved in the repression of HXK2 gene in the absence of glucose. We present evidence that repression of HXK2 requires a functional Rgt1 protein together with the product of the essential gene MED8. The Med8 protein specifically binds to a DRS (downstream repressing sequence) of the HXK2 gene [18,19] and constitutes a component of the large multiprotein complex called Srb-mediator complex. The latter enhances basal and facilitates activated transcription by interacting with the C-terminal domain of RNA polymerase II [20,21]. Transcription cofactors, such as Med8, mediate access to genes in chromatin and they help to establish, maintain or activate regulatory networks. Thus a possible role of Med8 could be to affect the formation and activity of basal initiation complexes by linking the specific DNA–protein regulatory complexes to the RNA polymerase II holoenzyme transcriptional machinery. Since the Hxk2 protein is also necessary to generate the glucose-repression signal through the Mig1/Hxk2-mediated glucose-signalling pathway, a new connection between both the pathways could be deduced.
MATERIALS AND METHODS
Strains and growth media
S. cerevisiae strain DBY1315 (MATα ura3-52 leu2-3,2-112 lys2-801 gal2) was donated by D. Botstein (Stanford University, Stanford, CA, U.S.A.) and was used as a wild-type strain and recipient in the transformation experiments. An RGTI-deficient strain was obtained by using a loxP-KlURA3-LoxP disruption cassette as described in [22]. Bacterial transformation and preparation of recombinant plasmid DNA were performed in Escherichia coli MC1061 [hsdR mcrB araD139Δ(araABC-leu)7679Δlacx74 galU galK rpsL thi]. Expressions of fusion proteins were obtained in E. coli BL21(DE3)pLysS (Promega, Madison, WI, U.S.A.).
Yeast cells were grown in the following media: rich media based on 1% (w/v) yeast extract and 2% (w/v) peptone (YEP), 2% (w/v) glucose (YEPD) or 3% (v/v) ethanol (YEPE) were added as carbon sources. Synthetic media consisted of 0.67% yeast nitrogen base without amino acids, supplemented with amino acids as required and 2% glucose or 3% ethanol. This medium was utilized to select for transformants when plasmids carrying URA3 or LEU2 were used. Solid media contained 2% (w/v) agar in addition to the components described above.
General DNA techniques
Restriction enzymes and T4 DNA ligase were from Roche (Indianapolis, IN, U.S.A.), and radioactive isotopes were from Amersham International (Arlington, IL, U.S.A.). DNA manipulations were performed as described previously [23].
Plasmid constructions
Plasmid pGEX/RGT1 was constructed by subcloning a BamHI fragment from plasmid pGBKT7/RGT1 in frame with pGEX-4T (Amersham Biosciences, Piscataway, NJ, U.S.A.). Plasmid pGBKT7/RGT1 carried a 3512 bp BamHI fragment with the complete coding region of the RGT1 gene in pGBKT7 (ClonTech Laboratories, Basingstoke, U.K.). The RGT1 insert was synthesized by PCR using genomic DNA as a template with the primer pair OL1+OL2 (OL1: AAGGATCCATGAACGAGCTGAACACTGT and OL2: ATGGATCCTCAATACCAGCCTAACTCGG). DNA sequencing verified this PCR-generated construct.
The yeast expression vector YEp352/MIG1::gfp was constructed as follows: a 969 bp BamHI–BglII fragment containing the GFP (green fluorescent protein) gene was subcloned into a YEp352/MIG1 plasmid cleaved with BamHI. YEp352/MIG1 plasmid contains a 2.77 kb SacI–BamHI fragment with the complete MIG1 gene under the control of its own promoter. The yeast expression vector YEp352/HXK2::gfp was constructed as indicated in [12]. All the clones used were verified by sequencing the fusion points.
The HXK2 reporter plasmid YIp-HXK2+404 was constructed by placing sequences from −838 to +404 bp relative to the HXK2 translation start codon, upstream of a lacZ reporter gene on the integrative yeast vector YIp357. An HXK2 reporter plasmid lacking the DRS elements was constructed by placing sequences spanning from −838 to +39 relative to the HXK2 ATG, upstream of a lacZ reporter gene on an integrative yeast vector to create the yeast reporter plasmid YIp-HXK2+39 [24].
Plasmid YIp and YEp are yeast-E. coli shuttle vectors suitable for use respectively as integrative or episomal vectors. These vectors have an URA3 yeast selectable marker [25].
RNA preparation and Northern blotting
Total yeast RNA was isolated from cells grown in the indicated medium until the absorbance (A) at 600 nm was 1.0 as described previously [26]. The RNA samples were separated by formaldehyde gel electrophoresis and transferred on to a Hybond-N membrane (Amersham Biosciences). To ensure uniform loading and transfer of RNA, ethidium bromide was added to the samples, and the stained rRNA was visualized on the blots under UV illumination. Northern blots were probed with a 1.5 kb PstI fragment from HXK2. Blots were also probed with a radiolabelled 1.4 kb BamHI–HindIII fragment from the ACT1 gene to confirm uniform loading. All probes were radiolabelled by using [α-32P]dCTP (3000 μCi/nmol) and random priming. Blots were analysed by using a Packard Instantimager and the Instantimager software.
Preparation of crude protein extracts
Yeast protein extracts were prepared as follows: yeasts were grown in 10–20 ml of rich medium (YEPD) at 28 °C to A600 1.0. Cells were collected, washed twice with 1 ml of 1 M sorbitol and suspended in 100 μl of 50 mM Tris/HCl (pH 7.5) buffer containing 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM PMSF, 0.42 M NaCl and 1.5 mM MgCl2. The cells were broken by vortex-mixing (6×20 s) in the presence of glass beads (0.5 g), and 400 μl of the same buffer was added to the suspension. After centrifugation at 19000 g (14000 rev./min) for 15 min at 4 °C, the supernatant was used as crude protein extract.
Enzyme assays
Invertase activity was assayed in whole cells as described previously [27] and expressed as (μmol of glucose released)·min−1·[100 mg of cells (dry weight)]−1. For β-galactosidase activity determinations, crude extracts were prepared with glass beads as described above and 2 mg/ml o-nitrophenol β-D-galactopyranoside was used as a substrate [23]. Specific activity was calculated in relation to total protein in the crude extract, using BSA as the standard.
Preparation of yeast nuclear extracts
Nuclear extracts were prepared by a method described in [28] with the modifications indicated in [29].
Heterologous expression of S. cerevisiae RGT1 gene in E. coli and purification of Rgt1 protein
E. coli cells containing the expression plasmid pGEX/RGT1 were used to produce the S. cerevisiae protein Rgt1 as a fusion protein with the Schistosoma japonicum GST (glutathione S-transferase) using the procedure described previously [18]. Rgt1 was obtained from GST–Rgt1 fusion protein coupled with glutathione–Sepharose beads by site-specific separation of the GST affinity tag using 2.5 units of thrombin.
DNA probes and EMSA (electrophoretic mobility-shift assay)
To investigate the interaction of Rgt1 with the sequence carrying the RGT1 element of the HXK2 promoter, we reconstituted the fragment from two complementary oligonucleotides (RGT1HXK2 sense, 5′-tcgaGCAGTTTTTCCGGTCGAT-3′; and RGT1HXK2 antisense, 5′-tcgaATCGACCGGAAAAACTGC-3′). The complementary strands were annealed and either end was labelled with [α-32P]dCTP by fill-in, using the Klenow fragment of DNA polymerase I. The labelled double-stranded DNA was used as a probe and the unlabelled DNA was used as a specific competitor in gel-retardation assays. Calf-thymus DNA was used as a non-specific competitor.
Binding reaction mixtures contained 10 mM Hepes (pH 7.5), 1 mM dithiothreitol, 1–5 μg of poly(dI-dC)·(dI-dC) and 0.5 ng of end-labelled DNA in a volume of 25 μl. The amount of unlabelled competitor DNA added is indicated in the Figure legends. The binding reaction mixtures included 12 μg (6 μl) of protein from a nuclear extract, and after 30 min of incubation at room temperature (20 °C), they were loaded on to a 4% (w/v) non-denaturing polyacrylamide gel. Electrophoresis was performed at 10 V/cm of gel for 45 min to 1 h in 0.5×TBE buffer (45 mM Tris/borate and 1 mM EDTA). Gels were dried and autoradiographed at −70 °C with an intensifying screen.
ChIP (chromatin immunoprecipitation) assays
Rgt1 and Med8 binding to the HXK2 promoter in vivo was assayed by ChIP as described previously [30,31]. Genomic DNA fragments cross-linked to HA–Rgt1 (where HA stands for haemagglutinin) or HA–Med8 were immunoprecipitated with anti-HA antibody and Protein A–Sepharose beads (Amersham Biosciences). The DNA sequences upstream (containing the RGT1 element) and downstream (containing the MED8 element) of the HXK2 translation start codon were amplified by PCR using respectively the primer pairs: OL3+OL4 (OL3, ACTACGAGTTTTCTGAACCTCC; and OL4, TAATTTCGTGGATCTCGAATC) and OL5+OL6 (OL5, GGAATTGATGCAACAAATTGAG; and OL6, GATTGAGTGGTGTCAAAGGTAC).
Fluorescence microscopy
An rgt1 mutant strain expressing the Hxk2–GFP or Mig1–GFP fusion proteins was grown to early-log phase (A600<0.7) in a synthetic medium with the carbon sources indicated and with selection for maintenance of plasmids. The cells (25 μl) were loaded on to poly(L-lysine)-coated slides and the remaining suspension was immediately withdrawn by aspiration. An aliquot (2 μl) of DAPI (4,6-diamidino-2-phenylindole; 2.5 μg/ml in 80% glycerol) was added and a covert slide was placed over the microscope slide. GFP and DAPI localization in live cultures was monitored by direct fluorescence using a Leica DMR-XA fluorescent microscope. Images were taken with a Leica Q550 camera using Leica QWin software and processed in Adobe Photoshop 6.0.
RESULTS
Rgt1 negatively regulates HXK2 gene expression in response to extracellular glucose
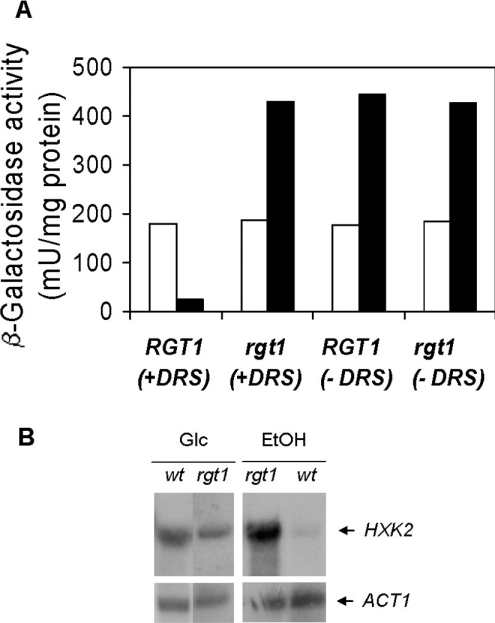
To gain more insight into the mode of HXK2 gene expression, we have analysed the upstream sequence of the HXK2 promoter using the yeast reporter plasmid YIp-HXK2+404. Since this promoter drives a glucose-regulated expression and its in silico analysis revealed the presence of a putative RGT1 element located at −395 bp, we measured reporter gene expression in an rgt1 null mutant. We found that in ethanol-grown cells, lacZ expression increased 18-fold, whereas in glucose-grown cells, Rgt1 did not affect β-galactosidase activity (Figure 1A).
Figure 1. Rgt1 and Med8 proteins repress HXK2 expression in the absence of glucose.
(A) Hxk2 expression was measured by using the lacZ expression as reporter gene. One copy of the HXK2+404::lacZ, containing the Med8 binding downstream regulatory sequence (+DRS), or the HXK2+39::lacZ, lacking the Med8 binding downstream regulatory sequence (−DRS), constructs were integrated in the chromosome at URA3 locus of the wild-type strain DBY1315 or the mutant strain rgt1. β-Galactosidase activities are averages of the results obtained for four to five independent experiments. Average values have standard errors of 10% or less. Yeasts were grown on YEPD medium (open bars) or YEPE medium (solid bars) until the A600 reached 1.0 [3.0 mg (wet weight)/ml]. β-Galactosidase activity was assayed in crude extracts. (B) Effect of RGT1 gene disruption on the expression of HXK2. A wild-type yeast strain (DBY1315) and the isogenic rgt1 mutant strain were grown using glucose (YEPD) or ethanol (YEPE) as carbon source as described above each lane. Total RNA was isolated, size-separated using a horizontal agarose gel and transferred on to a nylon membrane that was then hybridized to probes derived from HXK2 as described in the Materials and methods section. The probe used is indicated at the side of each panel.
To confirm these results, we also determined the transcription levels of HXK2. For this purpose, we isolated RNA from wild-type and rgt1 mutant cells grown with glucose or ethanol as carbon sources. As shown in Figure 1(B), strong signals were obtained in a Northern blot when wild-type or rgt1 mutant cells were grown on glucose at a size of approx. 1.45 kb and a weak signal was detected with RNA extracted from wild-type cells grown on ethanol medium. These results indicate, in accordance with previous results [32–34], that the expression of the HXK2 gene is regulated by the carbon source present in the culture medium. However, a high amount of mRNA from HXK2 was detected in the rgt1 strain grown on ethanol medium, confirming the essential role played by Rgt1 in the repression of the HXK2 gene.
In previous works, the Med8 protein was identified as a transcription factor that binds to DRS within the HXK2 gene and participates in the regulation of HXK2 expression [18,35]. A typical result of the HXK2 gene expression in the absence of Med8 binding sites in the HXK2 gene regulatory region is shown in Figure 1(A). To assay the contribution of Med8 to HXK2 gene expression, both in the presence and in the absence of the Rgt1 protein, we transformed the wild-type and the rgt1 mutant strains with the YIp-HXK2+39 plasmid and then measured β-galactosidase activity. We found that Med8 repressed lacZ expression 18 times in a wild-type strain containing the HXK2+39 reporter gene in response to growth in ethanol, whereas in glucose-grown cells, Med8 did not affect HXK2 transcription (Figure 1A). Moreover, a double defect in DRS function and Rgt1 protein also increases lacZ expression 18 times in cells growing in ethanol (Figure 1A).
Rgt1 binds to a specific site within the HXK2 promoter
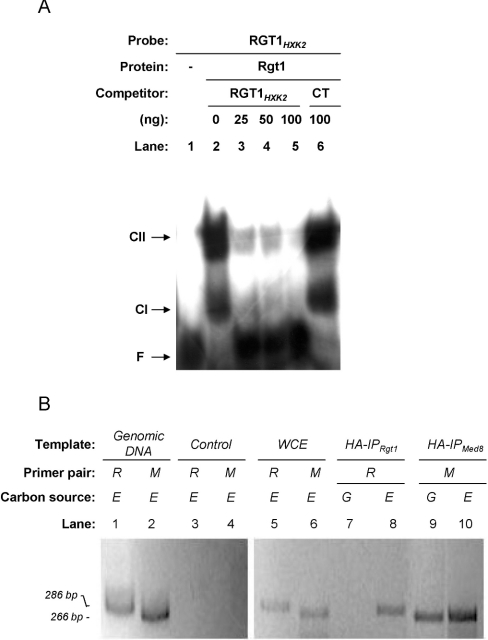
To test whether the Rgt1 protein binds specifically to the putative RGT1 element located at −395 bp in the HXK2 promoter, one synthetic oligonucleotide from −390 to −410 bp of the HXK2 promoter, containing the RGT1 consensus sequence, was used as a probe for gel EMSA. First, we tested the ability of a recombinant Rgt1 protein from E. coli cells to bind to the DNA probe. Two significantly shifted bands are produced when Rgt1 was added, indicating that the protein indeed binds to the RGT1HXK2 element (Figure 2A). The fact that two complexes, CI and CII, are observed in band-shift experiments with purified Rgt1 protein suggests that Rgt1 can bind to the RGT1HXK2 element both as a monomer and as a dimer.
Figure 2. Rgt1 protein binds to the RGT1 element of the HXK2 promoter in vitro and in vivo.
(A) Gel mobility-shift analysis of Rgt1 binding to the RGT1 regulatory element of the HXK2 promoter. The specific competitor for binding was the unlabelled RGT1-annealed oligonucleotides. The non-specific competitor for binding was 100 ng of calf-thymus DNA (CT). Nucleoprotein complexes were resolved from free DNA by non-denaturing PAGE. For the control (lane 1), the radiolabelled DNA fragment was added alone. F, unbound fragment; CI and CII, positions of shifted bands observed with purified Rgt1 protein. (B) ChIP analysis of Rgt1 binding to the RGT1 regulatory element of the HXK2 promoter. Chromatin was prepared from wild-type yeast cells containing HA-tagged Rgt1 or HA-tagged Med8 proteins. The cells were grown on YEPG (G) or YEPE (E) media as indicated above each lane, and their chromatin was immunoprecipitated using anti-HA monoclonal antibodies. The HXK2 regulatory regions in the immunoprecipitated DNA (IP) was detected by ethidium bromide staining after amplifying it in a PCR by using the primer pair OL3+OL4 (R) or OL5+OL6 (M) and resolving it by electrophoresis on a 2% agarose gel. Lanes show DNA amplified from genomic DNA, whole-cell extract before immunoprecipitation or extracts from cells without tagged protein (Control).
To confirm that Rgt1 and Med8 control HXK2 transcription by direct binding to the RGT1 and DRS elements of the gene regulatory region, we also used ChIP analyses. Our results showed that Rgt1 was recruited to HXK2 promoter in the absence of glucose (Figure 2B, lane 8). However, binding of Rgt1 to the HXK2 promoter was abolished in the presence of glucose (Figure 2B, lane 7). To test whether Med8 was also present at the DRS regulatory regions of the HXK2 gene, we also utilized ChIP analyses. As shown in Figure 2 (B), lanes 9 and 10, Med8 was present at the DRS in cells growing both in the presence and in the absence of glucose.
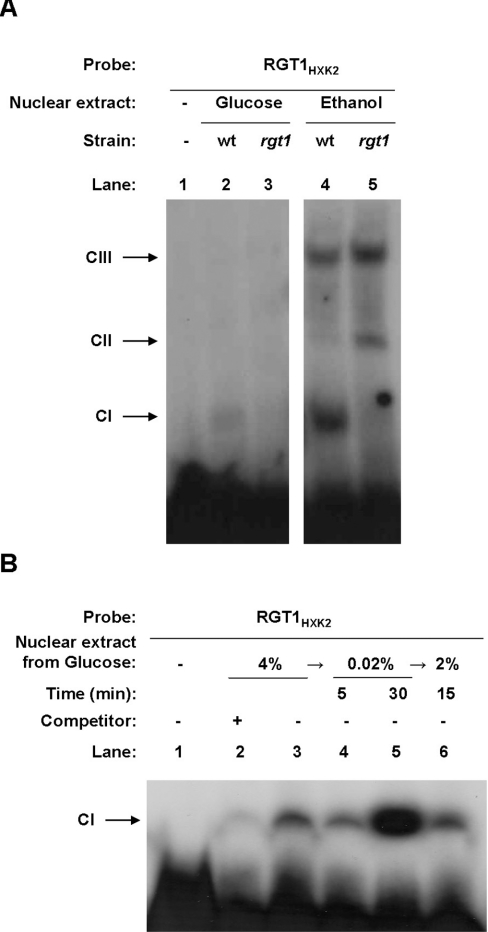
Thus our results indicate that, in the absence of glucose, Rgt1 and Med8 are present at the HXK2 promoter and co-repress gene transcription. Moreover, when glucose is present in the culture medium, Rgt1 has no regulatory function in the HXK2 gene expression. Previous studies showed that in glucose-rich media, Rgt1 becomes phosphorylated and dissociates from the repressor complex involved in the expression of HXT genes [36,37]. To examine whether the presence or absence of glucose could determine a differential pattern of band shifting with nuclear extracts prepared from wild-type and rgt1 mutant strains, we use the RGT1 element of the HXK2 promoter in EMSA assays. As shown in Figure 3(A), we observed three main DNA–protein complexes with nuclear extracts from cells grown in ethanol medium (Figure 3A, lanes 4 and 5). Among these complexes, the CI complex was formed with extracts from wild-type cells grown in the presence or absence of glucose. In extracts from cells grown in ethanol medium, comparatively strong signals appeared. In contrast, signal strength is greatly decreased by utilizing nuclear extracts from cells grown in high levels of glucose. The CI complex was not detected in nuclear extracts from rgt1 mutant cells grown in the presence or absence of glucose. Moreover, CI complex formation is reversible in response to glucose. If glucose was added back to cells after a shift to low glucose, the CI complex rapidly decreased (Figure 3B). The specificity of the binding was demonstrated by competition assays with the non-labelled oligonucleotide (Figure 3B, lane 2). Together, these results indicate that the Rgt1 protein is required in the CI complex formation and that, in cells growing in glucose, the Rgt1 protein dissociates from the repressor complex involved in the HXK2 gene expression.
Figure 3. The Rgt1 protein binds to the RGT1 element of the HXK2 promoter in a glucose-dependent manner.
(A) Yeast cells were grown in YEPD-2% glucose medium or in YEPE-3% ethanol to an A600 1.0. Nuclear extracts were prepared as indicated in the Materials and methods section and used for EMSA analysis. (B) Yeast cells were grown in YEPD-4% glucose medium to an A600 of 1.0 and shifted to YEPD-0.02% glucose medium for 5 or 30 min; then 2% glucose was added to the medium and cells were collected after 15 min of incubation. Nuclear extracts were prepared as described in the Materials and methods section and used for EMSA analysis. Unlabelled RGT1HXK2 probe (25 ng) was used as a specific competitor in lane 2.
Rgt1 is not involved in the glucose-repression-signalling pathway controlling SUC2 repression
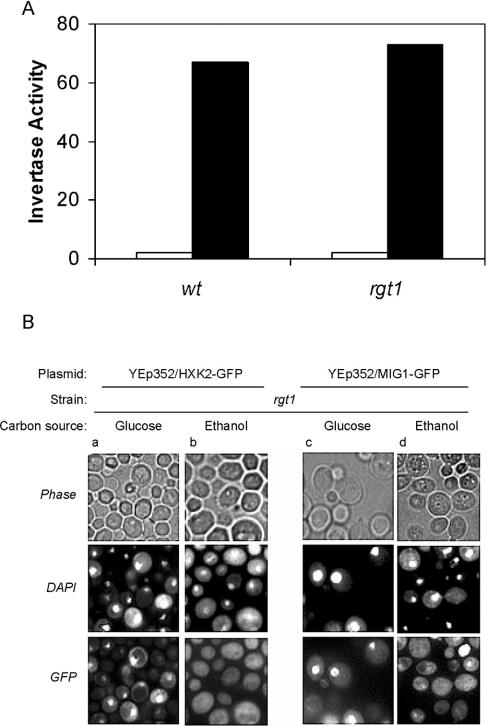
The Hxk2 protein phosphorylates glucose and also regulates glucose repression [12]. Since the rgt1 mutant cells growing in a non-fermentable carbon source have high levels of Hxk2 protein, it was interesting to investigate whether under these growth conditions, the rgt1 mutant cells still respond to glucose repression. To address this issue, we tested the extracellular invertase activity in rgt1 mutants under repressing (high glucose) and inducing (low glucose) conditions. As can be seen in Figure 4(A), invertase activity was not affected by the RGT1 gene deletion either under repressing or inducing growth conditions. This indicates that a high level of Hxk2 under low-glucose growing conditions is not sufficient for repression signalling in yeast.
Figure 4. Effect of RGT1 gene disruption on the expression of SUC2.
(A) The wild-type yeast strain (DBY1315) and its isogenic rgt1 null mutant were grown using 2% glucose (open bars) or 3% ethanol plus 0.05% glucose (solid bars) as carbon sources, until the A600 reached 1.0 [3.0 mg (wet weight)/ml]. Invertase activity was assayed in whole cells. The average values of the results obtained for four independent experiments have standard errors ≤10%. (B) Yeast rgt1 mutant strain expressing Hxk2–GFP (a and b) from the plasmid YEp352/HXK2::gfp and Mig1–GFP (c and d) from the plasmid YEp352/MIG1::gfp were grown on SD-Ura− medium supplemented with glucose or ethanol as carbon source and fluorescence was imaged. The cells were stained with DAPI for DNA and then imaged for GFP fluorescence and for DAPI fluorescence by phase-contrast optics.
To investigate why a high level of Hxk2 protein is not sufficient for repression signalling under low-glucose growing conditions, we studied the intracellular localization of Hxk2 and Mig1 in rgt1 mutants by using Hxk2–GFP and Mig1–GFP fusion proteins expressed from multicopy plasmids. As shown in Figure 4(B), in rgt1 mutant cells, grown in the presence of glucose, Hxk2–GFP is predominantly localized in the cytoplasm although a fraction is located in the nucleus (Figure 4B, column a). In contrast, Mig1–GFP is mainly located in the nucleus (Figure 4B, column c). In rgt1 mutant cells grown overnight on 3% ethanol, condition under which repression does not occur, both Hxk2 and Mig1 proteins are found in the cytoplasm, apparently excluded from the nucleus (Figure 4B, columns b and d).
DISCUSSION
Several lines of evidence provided in the present study suggest that Rgt1 and Med8 are repressors regulating transcription of the HXK2 gene in the absence of glucose. First, rgt1 null mutants, when grown in ethanol medium, exhibit intensely increased β-galactosidase activities from a lacZ gene used as a reporter for HXK2 gene expression. Identical results were obtained for a promoter lacking the Med8 binding sites to control the transcriptional expression of the lacZ reporter gene. However, a strain lacking both the Rgt1 protein and the Med8 binding sites in the HXK2 promoter does not show any synergistic effect on β-galactosidase activity. Thus the results presented in this paper support the conclusion that both Rgt1 and Med8 proteins repress HXK2 expression probably by interacting directly with repressing elements located upstream and downstream respectively in the HXK2 gene in the absence of glucose. Apparently, both factors are essential to repress HXK2 expression, because deletion of either the DRS element of the HXK2 gene or the RGT1 gene abolished to the same extent HXK2 gene repression in the absence of glucose. Secondly, Northern-blot analyses show an increased HXK2 transcription in rgt1 mutants growing in ethanol medium. These experiments confirm the results obtained by using lacZ as a fusion reporter gene. Thirdly, EMSA experiments demonstrated that Rgt1 binds specifically to a motif of the HXK2 gene promoter located at −395 bp (relative to the ATG translation start codon) containing the sequence CGGAAAA in accordance with the consensus sequence recently proposed for Rgt1 binding to DNA [37]. Finally, both ChIP and EMSA experiments demonstrated that the binding of Rgt1 to the HXK2 gene promoter element is regulated by external glucose concentrations. Rgt1 binds to the HXK2 gene promoter in the absence of glucose and has low affinity for its binding element at high levels of glucose. This result could be explained if glucose regulates the transcriptional activity of Rgt1 towards HXK2 expression by inducing changes in the phosphorylation state of the protein in a similar manner as reported for the transcriptional control of the HXT1 gene [36,37].
These observations and previous results [13,38] suggest that the Rgt1 repressor together with the classical function dedicated to regulating the expression of HXT1 genes [5] is also involved in the repression of the HXK2 gene, connecting two glucose signalling pathways that cause glucose induction and glucose repression of gene expression. Earlier results demonstrated that the MIG2 gene, encoding a repressor that collaborates with Mig1 in some target promoters, is induced by glucose through the Rgt2/Snf3-Rgt1 signalling pathway [13]. On the basis of our observations that the transcription of HXK2 is regulated by Rgt1, we suggest a simplified model (Figure 5) of how the glucose signal reaches Rgt1 through the Snf3/Rgt2 pathway and could then be transferred to the Mig1/Hxk2 repression pathway. Rgt1, in the absence of glucose, binds to the upstream sequence of HXK2 gene to repress its expression. Since Hxk2 levels are regulated by Rgt1, new possibilities of cross-talk between these two glucose-signalling pathways could be opened. To test these possibilities, we examined whether Rgt1 is involved in the glucose-repression-signalling pathway controlling SUC2 repression. Our results show that rgt1 mutant cells respond to glucose repression in a similar manner as was reported for wild-type cells. This observation could be explained by the fact that the intracellular distribution of both Hxk2 and Mig1, two essential factors of the glucose repression signalling pathway, in rgt1 mutant cells is identical as in a wild-type strain [12]. These results suggest that the increase in Hxk2 levels in the rgt1 mutant strain in the absence of glucose is not sufficient to establish a complete glucose repression, because Mig1 localization was not affected by RGT1 gene deletion and, in the absence of glucose, is localized in the cytoplasm [39]. This observation, in accordance with previous ones, suggests that the cell modulates the Hxk2 regulatory function by controlling the amount of protein localized in the nucleus and that Hxk2 is sequestered in the nucleus through its interaction with Mig1 [12]. Thus Rgt1 does not contribute to glucose repression of the SUC2 gene because the Hxk2 protein is excluded from the nucleus in the absence of high levels of glucose in the culture medium. Thus, in accordance with the results we obtained for SUC2 expression, the physiological significance of the proposed cross-communication between the Snf3–Rgt2 induction pathway and the Mig1–Hxk2 repression pathway remains to be clarified.
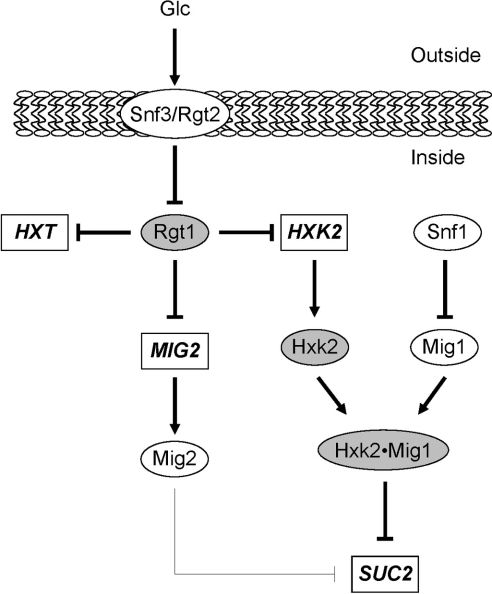
Figure 5. Model for the integration of the glucose signal through distinct signalling pathways.
Glucose signals could take several possible routes, namely Snf3/Rgt2 and Mig1/Hxk2 pathways. Binding of glucose to the transmembrane proteins Snf3/Rgt2 inactivates the Rgt1 repressor function, leading to expression of several genes (HXTs, MIG2 and HXK2). Rgt1 is the main downstream repressor of the Snf3/Rgt2 pathway controlling expression in the absence of glucose of several targets genes. Two of the targets, Hxk2 and Mig2, are directly involved, together with Mig1, in glucose signalling through the Mig1/Hxk2 pathway. Genes are depicted by rectangles, proteins by ovals and shaded proteins are discussed in the present work.
Acknowledgments
We are grateful to J. Heinisch (Universität Osnabrück, Fachbereich Biologie/Chemie, AG Genetik, Osnabrück, Germany) for fruitful discussions and a critical reading of this paper. The present study was supported by grants PB97-1213-C02-02 and BFU2004-02855-C02-02 from the DGESIC (Spain). A.P. was supported by Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología (FICYT; Oviedo, Spain).
References
- 1.Gancedo J. M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 3.Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 4.Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J. M. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 2004;29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Özcan S., Johnston M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Özcan S., Leong T., Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundin M., Nehlin J. O., Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treitel M. A., Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno F., Herrero P. The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2002;26:83–90. doi: 10.1111/j.1574-6976.2002.tb00600.x. [DOI] [PubMed] [Google Scholar]
- 11.Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., Hwang I., Jones T., Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 12.Ahuatzi D., Herrero P., de la Cera T., Moreno F. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 2004;279:14440–14446. doi: 10.1074/jbc.M313431200. [DOI] [PubMed] [Google Scholar]
- 13.Kaniak A., Xue Z., Macool D., Kim J. H., Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutfiyya L. L., Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt M. C., McCartney R. R., Zhang X., Tillman T. S., Solimeo H., Wolfl S., Almonte C., Watkins S. C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafuente M. J., Gancedo C., Jauniaux J. C., Gancedo J. M. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- 17.Lakshmanan J., Mosley A. L., Özcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- 18.Chaves R. S., Herrero P., Moreno F. Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun. 1999;254:345–350. doi: 10.1006/bbrc.1998.9954. [DOI] [PubMed] [Google Scholar]
- 19.Moreno-Herrero F., Herrero P., Colchero J., Baro A. M., Moreno F. Analysis by atomic force microscopy of Med8 binding to cis-acting regulatory elements of the SUC2 and HXK2 genes of Saccharomyces cerevisiae. FEBS Lett. 1999;459:427–432. doi: 10.1016/s0014-5793(99)01289-2. [DOI] [PubMed] [Google Scholar]
- 20.Myers L. C., Kornberg R. D. Mediator of transcriptional regulation. Annu. Rev. Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 21.Yudkovsky N., Ranish J. A., Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature (London) 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 22.Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 24.Herrero P., Ramírez M., Martínez-Campa C., Moreno F. Identification and characterisation of two transcriptional repressor elements within the coding sequence of the Saccharomyces cerevisiae HXK2 gene. Nucleic Acids Res. 1996;24:1822–1828. doi: 10.1093/nar/24.10.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 26.Song W., Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gascón S., Lampen J. O. Purification of the internal invertase of yeast. J. Biol. Chem. 1968;243:1567–1572. [PubMed] [Google Scholar]
- 28.Allen J. L., Douglas M. G. Organization of the nuclear pore complex in Saccharomyces cerevisiae. J. Ultrastruc. Mol. Struct. Res. 1989;102:95–108. doi: 10.1016/0889-1605(89)90047-5. [DOI] [PubMed] [Google Scholar]
- 29.Zachariae W., Kuger P., Breunig K. D. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9. Nucleic Acids Res. 1993;21:69–77. doi: 10.1093/nar/21.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alepuz P. M., Jovanovic A., Reiser V., Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 31.Tomas-Cobos L., Casadome L., Mas G., Sanz P., Posas F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J. Biol. Chem. 2004;279:22010–22019. doi: 10.1074/jbc.M400609200. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez A., De La Cera T., Herrero P., Moreno F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 2001;355:625–631. doi: 10.1042/bj3550625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrero P., Galíndez J., Ruiz N., Martínez-Campa C., Moreno F. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast. 1995;11:137–144. doi: 10.1002/yea.320110205. [DOI] [PubMed] [Google Scholar]
- 34.DeRisi J. L., Iyer V. R., Brown P. O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 35.de la Cera T., Herrero P., Moreno-Herrero F., Chaves R. S., Moreno F. Mediator factor Med8p interacts with the hexokinase 2: implication in the glucose signalling pathway of Saccharomyces cerevisiae. J. Mol. Biol. 2002;319:703–714. doi: 10.1016/S0022-2836(02)00377-7. [DOI] [PubMed] [Google Scholar]
- 36.Mosley A. L., Lakshmanan J., Aryal B. K., Özcan S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 2003;278:10322–10327. doi: 10.1074/jbc.M212802200. [DOI] [PubMed] [Google Scholar]
- 37.Kim J. H., Polish J., Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol. Cell. Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomas-Cobos L., Sanz P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 2002;368:657–663. doi: 10.1042/BJ20020984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vit M. J., Waddle J. A., Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]