Abstract
The simian virus 40 capsid is composed of 72 pentamers of VP1 protein. Although the capsid is known to dissociate to pentamers in vitro following simultaneous treatment with reducing and chelating agents, the functional roles of disulfide linkage and calcium ion-mediated interactions are not clear. To elucidate the roles of these interactions, we introduced amino acid substitutions in VP1 at cysteine residues and at residues involved in calcium binding. We expressed the mutant proteins in a baculovirus system and analyzed both their assembly into virus-like particles (VLPs) in insect cells and the disassembly of those VLPs in vitro. We found that disulfide linkages at both Cys-9 and Cys-104 conferred resistance to proteinase K digestion on VLPs, although neither linkage was essential for the formation of VLPs in insect cells. In particular, reduction of the disulfide linkage at Cys-9 was found to be critical for VLP dissociation to VP1 pentamers in the absence of calcium ions, indicating that disulfide linkage at Cys-9 prevents VLP dissociation, probably by increasing the stability of calcium ion binding. We found that amino acid substitutions at carboxy-terminal calcium ion binding sites (Glu-329, Glu-330, and Asp-345) resulted in the frequent formation of unusual tubular particles as well as VLPs in insect cells, indicating that these residues affect the accuracy of capsid assembly. In addition, unexpectedly, amino acid substitutions at any of the calcium ion binding sites tested, especially at Glu-157, resulted in increased stability of VLPs in the absence of calcium ions in vitro. These results suggest that appropriate affinities of calcium ion binding are responsible for both assembly and disassembly of the capsid.
Simian virus 40 (SV40), a member of the Papovaviridae, is a small, nonenveloped tumorigenic virus. Its genome consists of double-stranded circular DNA of 5,243 bp, which encodes three structural proteins (VP1, VP2, and VP3) and two nonstructural proteins (large T antigen and small T antigen). The structural proteins are expressed in late infection, and viral capsids are formed in the nucleus of infected susceptible host cells (32). The SV40 capsid is about 45 to 50 nm in diameter, and its major component is VP1. The capsid is formed by the arrangement of 72 VP1 pentamers in a T=7d icosahedral lattice. The three-dimensional structure of the SV40 virion has been elucidated by X-ray crystallography, which showed that the capsid is formed from three nonequivalent types of interactions between pentameric capsomeres (α-α′, β-β′, and γ-γ) (20, 30).
SV40 capsids, as well as those of the closely related murine polyomavirus, dissociate to VP1 pentamers following treatment with dithiothreitol (DTT) and EGTA in vitro (2–4, 7), indicating that disulfide linkage and calcium ion-mediated interactions between pentamers are important for capsid formation in these viruses. Consistent with these observations, crystallographic analysis of SV40 virions has identified disulfide linkage between Cys-104 and Cys-104 in β-β′ interpentameric interactions and has shown that the two calcium ions which are bound per VP1 molecule are also involved in interpentameric interaction (20, 30). In spite of these observations, however, the functional roles of these interactions in association and dissociation of the capsid are largely unknown.
We have previously reported that recombinant SV40 VP1 protein expressed in a baculovirus expression system assembled into virus-like particles (VLPs) in insect cells in the absence of VP2, VP3, T antigens, and genomic DNA. The VLPs were morphologically indistinguishable from wild-type SV40 and could be dissociated into VP1 pentamers by treatment with DTT and EGTA in vitro (19). Thus, the structural properties of the recombinant VLPs were very similar to those of wild-type SV40 particles, making this system a useful tool with which to study the assembly and disassembly of the SV40 capsid.
To elucidate the functional roles of disulfide linkage and calcium ion-mediated interactions, we produced a series of VP1 proteins mutated at cysteine residues and putative calcium ion binding sites, and analyzed the assembly in insect cells and disassembly in vitro of the VLPs formed from these mutant proteins.
MATERIALS AND METHODS
Construction of recombinant baculovirus.
The construction and structure of baculovirus expressing wild-type VP1 are described elsewhere (19). Point mutations were introduced by PCR-based site-directed mutagenesis (8) using pUCVP1 (19) as the initial PCR template. The primers used were as follows (mutation sites are underlined): WT5′T (terminal) S (sense) (5′-AAG GGT CGA CAT GAA GAT GGC CCC AAC AAA AAG-3′), C9A S (5′-AAG GGT CGA CAT GAA GAT GGC CCC AAC AAA AAG AAA AGG AAG TGC TCC AGG GGC-3′), C104S S (5′-GGA CTT AAC CTC TGG AAA TAT TTT G-3′), C104S AS (anti-sense) (5′-CAA AAT ATT TCC AGA GGT TAA GTC C-3′), C207S S (5′-CCA GTG GAG TCC TGG GTT CCT G-3′), C207S AS (5′-CAG GAA CCC AGG ACT CCA CTG G-3′), C254S S (5′-GGG CCC TTG TCC AAA GCT GAC-3′), C254S AS (5′-GTC AGC TTT GGA CAA GGG CCC-3′), E46,48Q dm (double mutation) S (5′-CTT CAC TCA GGT GCA GTG CTT TTT AAA TCC-3′), E46,48Q dm AS (5′-AAA AAG CAC TGC ACC TGA GTG AAG CTG TC-3′), E157Q S (5′-GTT GGT GGG CAA CCT TTG GAG C-3′), E157Q AS (5′-GCT CCA AAG GTT GCC CAC CAA C-3′), E160Q S (5′-GGA ACC TTT GCA GCT GCA GGG-3′), E160Q AS (5′-CCC TGC AGC TGC AAA GGT TCC-3′), E157,160Q dm S (5′-GTT GGT GGG CAA CCT TTG CAG CTG CAG GG-3′), E157,160Q dm AS (5′-CCC TGC AGC TGC AAA GGT TGC CCA CCA AC-3′), S213A,K214A,E216Q tm (triple mutation) S (5′-CCT GAT CCA GCT GCA AAT CAA AAC ACT AGA TAT TTT GGA ACC TAC-3′), S213A,K214A,E216Q tm AS (5′-GTG TTT TGA TTT GCA GCT GGA TCA GGA ACC CAG CAC TCC-3′), E329,330Q dm S (5′-CCT CTC AAG TAC AGC AGG TTA GGG-3′), E329,330Q dm AS (5′-CCC TAA CCT GCT GTA CTT GAG AGG-3′), D345N S (5′-GGG GAT CCA AAC ATG ATA AGA TAC-3′), D345N AS (5′-GTA TCT TAT CAT GTT TGG ATC CCC-3′), WT3′T AS (5′-CCG GCT CGA GTC ACT GCA TTC TAG TTG TGG TTT G-3′). The WT5′T S and WT3′T AS primers were used in the second round of PCR. A SalI restriction site was introduced at the 5′ end of the mutated DNA fragments to facilitate further subcloning. The amplified products were digested with SalI and inserted into the SalI and blunt-ended HindIII sites of pBluescriptII to make pBSmt1 to pBSmt6 and pBSmtA to pBSmtH. The nucleotide sequences of the entire coding regions of all mutants were confirmed by dideoxynucleotide sequencing analysis.
For baculovirus expression, pBSmt1 to pBSmt6 and pBSmtA to pBSmtH were digested with SalI and EcoRV and the inserts were recloned into the SalI and blunt-ended HindIII sites of pFastBac1 (Gibco BRL). Recombinant baculoviruses were produced using the Bac-To-Bac system (Gibco BRL).
Expression of VP1 proteins in insect cells.
Spodoptera frugiperda (Sf9) cells were maintained in spinner culture flasks at 27°C in Sf-900II medium (Gibco BRL) supplemented with streptomycin, penicillin, and 0.3% heat-inactivated fetal bovine serum.
For protein expression, Sf9 cells were seeded onto tissue culture dishes at 2 × 105 cells/cm2 and allowed to attach for at least 1 h at room temperature. Recombinant baculoviruses encoding mutant VP1 genes were used to infect the attached Sf9 cells (5 × 107 cells at the time of infection) at a multiplicity of infection of 5 to 10. After 1 h, TC-100 medium (Gibco BRL) supplemented with streptomycin, penicillin, and 10% heat-inactivated fetal bovine serum was added and the cells were incubated at 27°C. The infected cells were harvested at 72 h postinfection (p.i.). After being washed twice with phosphate-buffered saline, the cells were sonicated in 1 ml of sonication buffer (20 mM Tris-HCl [pH 7.9], 1% sodium deoxycholate, 2 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of chymostatin per ml, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of antipain per ml, 1 μg of pepstatin per ml) and centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were collected. For cysteine mutants mt1, mt5, and mt6, nuclear extracts of the infected Sf9 cells were prepared as described elsewhere (29) with some modifications. A 1-ml volume of buffer A (10 mM HEPES-NaOH [pH 7.9], 10 mM KCl, 2 mM CaCl2, 0.5 mM PMSF, 1 μg of chymostatin per ml, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of antipain per ml, 1 μg of pepstatin per ml), 62.5 μl of 10% Nonidet P-40 (NP-40), and 250 μl of buffer C (20 mM HEPES-NaOH [pH 7.9]), 0.4 M NaCl, 2 mM CaCl2, 1 mM PMSF, 1 μg of chymostatin per ml, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of antipain per ml, 1 μg of pepstatin per ml) were used per 200 μl of packed cells.
Purification of VP1 proteins.
Whole-cell lysates or nuclear extracts (mt1, mt5, and mt6) of infected Sf9 cells (400 μl) were gently loaded onto 4 ml of preformed 20 to 50% (wt/vol) cesium chloride density gradients in 20 mM Tris-HCl (pH 7.9) and centrifuged at 35,000 rpm for 3 h at 4°C in an SW55Ti rotor (Beckman). After centrifugation, fractions were collected from the tops of the tubes and a small aliquot of each fraction was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were visualized by staining with Coomassie brilliant blue (CBB). Fractions containing VLPs were filled up with 37% (wt/vol) cesium chloride solution in 20 mM Tris-HCl (pH 7.9) and centrifuged at 50,000 rpm for 20 h at 4°C in an SW55Ti rotor. After centrifugation, fractions were collected as described above. For mt1, mt5, and mt6, 2 mM CaCl2 was added to all cesium chloride solutions. Finally, fractions containing VLPs were adjusted to a final concentration of 0.1% NP-40 and dialyzed against 20 mM Tris-HCl (pH 7.9)–0.1% NP-40 at 4°C for at least 15 h with several buffer changes. After dialysis, samples were collected and centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were stored at −80°C.
Electron microscopy.
Baculovirus-infected Sf9 cells were harvested, washed with 100 mM cacodylate (pH 7.2)–2.5 mM CaCl2, and fixed with glutaraldehyde at 8 days p.i. The cells were then fixed with osmium tetroxide, stained with uranyl acetate, and processed for thin sectioning. For negative staining of purified VLPs or pentamer, 3 μl of each sample was absorbed onto glow-discharged carbon-coated copper grids. The grids were washed with water, stained with 2% uranyl acetate, and air dried. The specimens were examined with an H-7500 electron microscope (Hitachi) at 80 kV.
Sucrose gradient centrifugation.
A 20-μl volume of each cell lysate was diluted tenfold with 20 mM Tris-HCl (pH 7.9) and loaded onto a 0.6-ml preformed 10 to 30% sucrose gradient in 20 mM Tris-HCl (pH 7.9) in a 5- by 41-mm open-top tube (Beckman). Using the appropriate adapters, the tubes were centrifuged at 45,000 rpm for 1 h at 4°C in an SW55Ti rotor. After centrifugation, 55-μl fractions were collected from the tops of the tubes and the bottom of each tube was washed with 55 μl of 2× SDS sample-loading buffer. A 7-μl volume of each fraction was separated by SDS-PAGE and immunoblotted with anti-SV40 VP1 polyclonal antibody (courtesy of M. Ikeda and I. Tamai, MBL, Nagoya, Japan). Immunoreactive bands were detected using the enhanced chemiluminescence system (Amersham).
For preparation of VP1 pentamer, purified VLP preparations were adjusted to final concentrations of 25 mM EGTA and 30 mM DTT, incubated for 1 h at 37°C, and separated on a Superdex 200 gel filtration chromatography column (Pharmacia) in 20 mM Tris-HCl (pH 7.9)–150 mM NaCl–5 mM EGTA–5 mM DTT at 4°C. The peak fractions corresponding in size to approximately 200 kDa were collected and stored at −80°C.
SDS-PAGE under nonreducing conditions.
Purified VLPs (10-ng portions) were incubated in 20 mM CHES (N-cyclohexyl-2-aminoethanesulfonic acid) (pH 9.0)–1% SDS–2 mM N-ethylmaleimide at 37°C for 10 min to block free thiols. Samples were boiled for 5 min after addition of SDS sample-loading buffer containing no β-mercaptoethanol and then boiled again in the presence or absence of 100 mM DTT. The proteins were separated by SDS-PAGE (8% polyacrylamide) and immunoblotted with anti-VP1 antibody.
Degradation by proteinase K.
Proteinase K (0.1, 0.3, or 1 ng) was added to 10 ng of purified VLPs, and the reaction mixtures were incubated at 37°C for 15 min. The reactions were stopped by addition of SDS sample-loading buffer and immediate boiling. The samples were separated by SDS-PAGE (12% polyacrylamide) followed by immunoblotting with anti-VP1 antibody.
Native agarose gel electrophoresis.
Purified VLPs (20-ng portions) were incubated for 1 h in 20 μl of 20 mM Tris-HCl (pH 7.9) with or without 30 mM DTT in combination with 2 mM CaCl2 or 25 mM BAPTA (a highly selective calcium ion-chelating agent) (33). Samples containing BAPTA or DTT were incubated at 37°C, and all other samples were incubated at 4°C. After incubation, 5 μl of sample-loading buffer (250 mM Tris-acetate [pH 8.1], 25% [vol/vol] glycerol, 0.125% bromophenol blue) was added to each sample and 7 μl of sample was loaded onto 0.8% agarose gels in 50 mM Tris-acetate (pH 8.1) with or without 2 mM CaCl2. Electrophoresis was carried out at 4°C in the same buffer as in the gel until the dye reached the bottom of the gel (approximately 2 h). After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes in 50 mM NaOH for 3 to 8 h at room temperature by a standard capillary transfer method and then immunoblotted with anti-VP1 antibody.
RESULTS
SV40 VP1 VLP formation in the nucleus of insect cells and dissociation to pentamers in vitro.
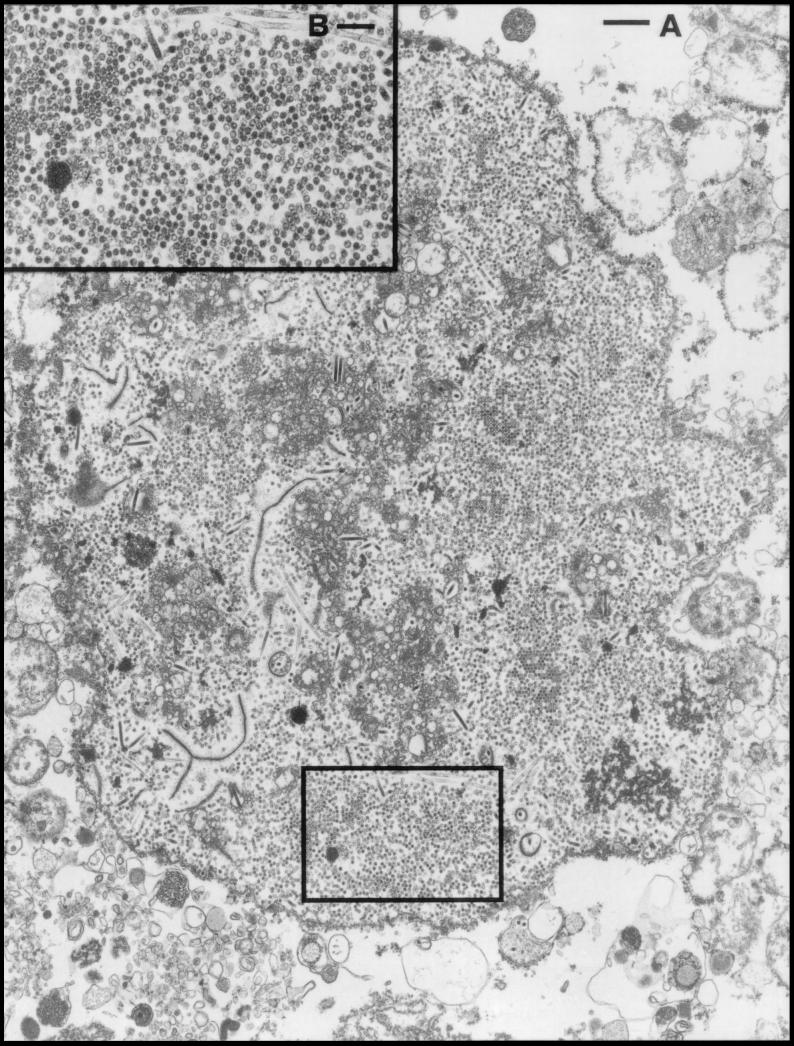
We previously constructed a recombinant baculovirus expressing VP1 and showed that VP1 was efficiently accumulated and assembled into VLPs in insect cells (19). We further assessed this system for use as a model of assembly and disassembly of the SV40 capsid. Figure 1 shows an electron microscopic view of baculovirus-infected Sf9 cells. Spherical particles approximately 45 nm in diameter accumulated abundantly in the nucleus, which is the location of wild-type SV40 particle formation in susceptible host cells. No such particles were formed in the cytoplasm or in mock-infected Sf9 cells (data not shown).
FIG. 1.
(A) Electron micrograph of an ultrathin section of an Sf9 cell 8 days after infection with baculovirus expressing VP1. (B) Higher magnification of the region indicated in panel A. Scale bars, 530 nm (A) and 220 nm (B).
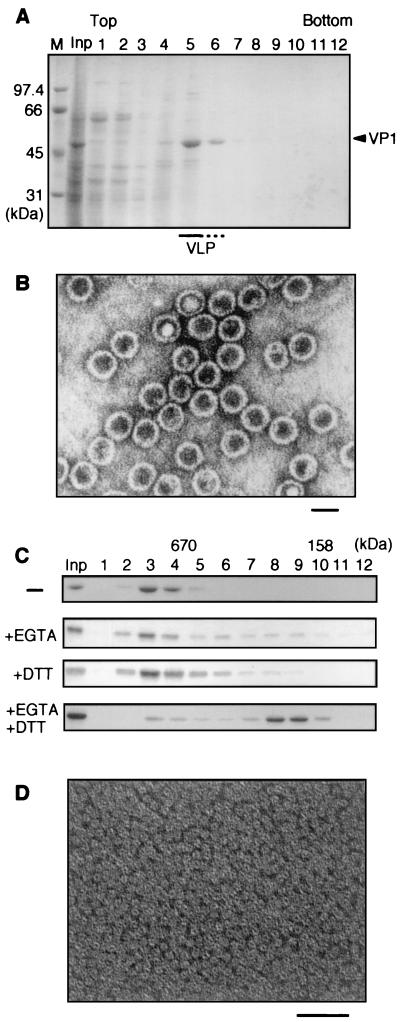
The VLPs were easily and efficiently purified by cesium chloride density gradient ultracentrifugation (Fig. 2A). We obtained 6 to 8 mg of VP1 protein at more than 90% homogeneity from 5 × 108 insect cells. Electron microscopic observation confirmed that VP1 formed VLPs which were morphologically indistinguishable from wild-type SV40 particles (Fig. 2B).
FIG. 2.
Purification of wild-type VLPs and their dissociation to VP1 pentamers in vitro. (A) Purification of VLPs by cesium chloride density gradient centrifugation. Aliquots of fractions after ultracentrifugation were separated by SDS-PAGE and stained with CBB. M, molecular mass standards; Inp, input. VP1 protein was detected in fractions 5 and 6. (B) Negatively stained electron micrograph of purified VLPs. Scale bar, 50 nm. (C) Size exclusion chromatography. The purified VLPs were incubated for 1 h at 37°C with EGTA and/or DTT as indicated and loaded on a Superdex 200 gel filtration chromatography column (Pharmacia). The minus sign indicates the VLP preparation prior to incubation. Fractions were separated by SDS-PAGE and stained with CBB. Molecular mass standards are thyroglobulin (670 kDa) and bovine gamma globulin (158 kDa). (D) Electron micrograph of the 200-kDa fraction of the bottom chromatogram in panel C. Scale bar, 50 nm.
For in vitro dissociation analysis, purified VLPs were incubated at 37°C for 1 h with EGTA and/or DTT. Size exclusion chromatography showed that VP1 protein in the form of VLPs was detected in the void volume (Fig. 2C). Following addition of both DTT and EGTA, approximately 80% of VP1 was detected in fractions corresponding to a molecular mass of approximately 200 kDa, in which VP1 pentamers would be expected, suggesting efficient dissociation of VLPs to VP1 pentamers. No VP1 proteins were detected in fractions corresponding in size to VP1 monomer (data not shown). Consistent with these observations, electron microscopic observation of the 200-kDa fraction revealed pentamer-like structures but not VLPs (Fig. 2D). Addition of either DTT or EGTA alone resulted in only partial dissociation (Fig. 2C).
These results demonstrate that the recombinant VP1 assembled into VLPs in the nucleus and dissociated to VP1 pentamers following treatment with DTT and EGTA in vitro. These characteristics are similar to those of wild-type SV40 particles, indicating that VLPs composed of recombinant VP1 can be used as a model of the association and dissociation of SV40 capsid. Using this system, we analyzed the roles of disulfide linkage and calcium ion-mediated interactions in the assembly and disassembly of VLPs by introducing amino acid substitutions at cysteine residues and putative calcium ion binding sites.
Cysteine mutants.
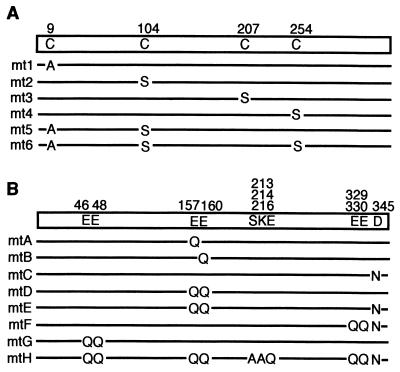
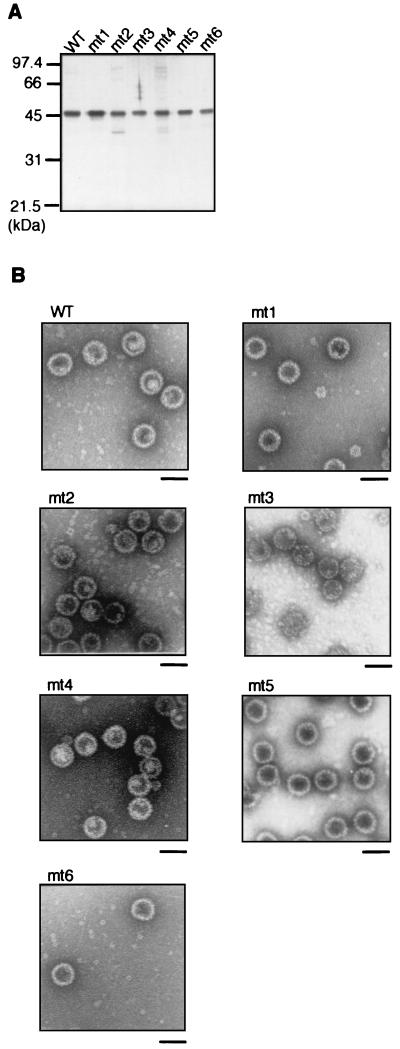
The VP1 cysteine residue mutants used in the present study are shown schematically in Fig. 3A. Of the seven cysteine residues in SV40 VP1, Cys-9, Cys-104, and Cys-254 are conserved between SV40 and murine polyomavirus (20, 31), suggesting that these residues may be of particular functional importance. These three cysteine residues were mutated singly in mt1, mt2, and mt4 and in combination in mt5 and mt6. We included a mutation at Cys-207 (mt3) as a control. The mutated VP1 constructs were introduced into baculovirus vectors, which were used to infect Sf9 cells. SDS-PAGE analysis of whole-cell extracts of mutant baculovirus-infected Sf9 cells showed that soluble proteins of about 50 kDa were specifically expressed at a level comparable to the expression of wild-type VP1 (Fig. 4A). Western blotting analysis with anti-VP1 antibody confirmed that these ∼50-kDa proteins were VP1 (Fig. 4B).
FIG. 3.
Schematic representation of SV40 VP1 mutants. (A) Cysteine mutants. (B) Calcium ion binding site mutants. Amino acid residues are represented in the one-letter code and numbered as in references 20 and 30.
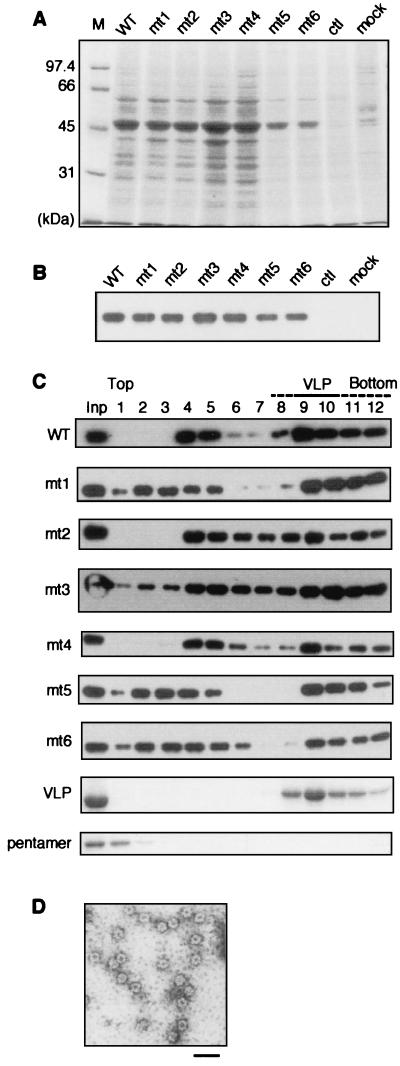
FIG. 4.
Expression and VLP formation in Sf9 cells of VP1 cysteine mutants. (A) SDS-PAGE analysis of whole-cell extracts of Sf9 cells infected with baculoviruses expressing cysteine mutants. The soluble fractions of whole-cell lysates at 3 days p.i. were separated by SDS-PAGE and stained with CBB. WT, wild type; ctl (control), lysate of cells infected with a baculovirus encoding no exogenous gene; mock, mock-infected cell lysate. (B) Detection of mutant VP1 proteins by immunoblotting with anti-VP1 antibody. (C) Analysis of VLP formation in Sf9 cells. Whole-cell lysates were separated by 10 to 30% sucrose gradient centrifugation and fractionated from the tops of the tubes. The fractions were separated by SDS-PAGE and subjected to Western blotting with anti-VP1 antibody (wild type [WT] and mt1 to mt6) or stained with CBB (VLP and pentamer). VLP and pentamer, purified wild-type VLPs and VP1 pentamer, respectively. (D) Electron micrograph of the material from the middle of the gradient. Fractions 4 and 5 of wild-type VP1 were further purified by cesium chloride density gradient ultracentrifugation, and VP1-containing fractions were negatively stained and observed by electron microscopy. Scale bar, 50 nm.
To analyze the ability of the mutant VP1 proteins to form VLPs in insect cells, the cell lysates were subjected to sucrose gradient centrifugation followed by immunoblotting with anti-VP1 antibody (Fig. 4C). Purified VLPs composed of wild-type VP1 were detected in fractions 8 to 12 (Fig. 4C, VLP), and pentamers remained at the top of the gradient (Fig. 4C, pentamer). The majority of wild-type VP1 was detected in VLP fractions but not in fractions corresponding to pentamer (or monomer, which has a smaller sedimentation coefficient and also remains at the top of the gradient), indicating that VLPs were efficiently formed in insect cells. VP1 protein was also detected in intermediate fractions between VLP and pentamer (fractions 4 and 5). Further purification of VP1 from these fractions followed by electron microscopic observation revealed that the protein was present in particles approximately 20 nm in diameter (Fig. 4D; see Fig. 2B for comparison). Particles considerably smaller than virus capsid are also formed from recombinant polyomavirus VP1 pentamers under certain in vitro conditions (28). These small particles probably form as a result of inaccurate interactions between pentamers during capsid assembly. All of the cysteine mutants were detected as a peak in fractions 8 to 12 (Fig. 4C), indicating that these mutants formed VLPs in insect cells. mt2, mt3, and mt4 also showed a peak in fractions 4 and 5, similar to wild-type VP1. However, mt1, mt5, and mt6, which all contain a mutation at Cys-9, were detected in fractions 2, 3, and 4, suggesting inefficient or unstable interpentameric assembly.
The observation that all of these mutants formed VLPs prompted us to purify the VLPs by cesium chloride density gradient ultracentrifugation. SDS-PAGE and silver staining demonstrated that all of the VLPs were purified to near homogeneity (Fig. 5A). Electron microscopic observation showed that all of the mutant VLPs were morphologically indistinguishable from wild-type VLPs (Fig. 5B). These results indicate that disulfide linkages at the mutated cysteines are not essential for the formation of VLPs in insect cells. However, the final yields of mt1, mt5, and mt6 VLPs were 1/10 to 1/40 of the yield of wild-type VLPs, because of dissociation or degradation during the purification process (data not shown and see below). Together with the results of sucrose gradient centrifugation analysis (Fig. 4C), these observations suggest that Cys-9, probably in a disulfide linkage, may be important in the stabilization of VLPs.
FIG. 5.
Purification and morphological analysis of cysteine mutant VLPs. (A) Silver staining of purified mutant VP1 proteins. Cysteine mutant VLPs were purified by cesium chloride density gradient centrifugation. VLP fractions were separated by SDS-PAGE, and proteins were visualized by silver staining. (B) Electron micrographs. Purified VLPs were negatively stained and observed by electron microscopy. Scale bar, 50 nm.
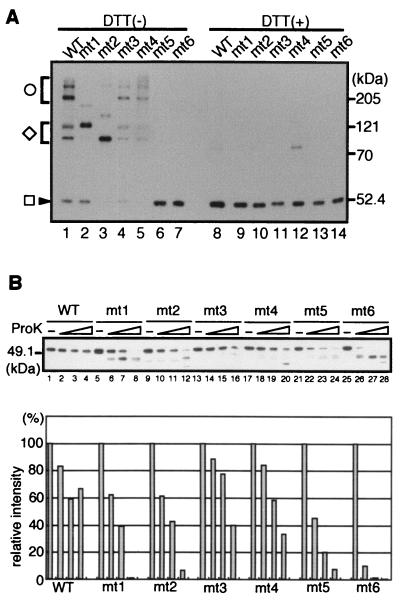
To determine which cysteines form intermolecular disulfide linkages in VLPs, cysteine mutant VLPs were analyzed by SDS-PAGE under nonreducing or reducing conditions and then immunoblotted with anti-VP1 antibody. Under nonreducing conditions, wild-type VLPs dissociated to multimers (more than 200 kDa in size) and dimers (approximately 100 kDa) as well as monomer (50 kDa) after denaturation by boiling in SDS (Fig. 6A, lane 1). These oligomers were completely dissociated to monomer by the addition of DTT (lane 8). These results showed that intermolecular disulfide linkages were formed in wild-type VLPs. mt3 and mt4 VLPs exhibited the same patterns as wild-type VLPs (lanes 4 and 5), indicating that Cys-207 and Cys-254 are not involved in intermolecular disulfide linkage. In contrast, mt1 and mt2 VLPs were dissociated to dimer and monomer but did not appear as multimers (lanes 2 and 3). The different electrophoretic mobilities of the dimers of these mutants may be due to different conformations (see Discussion). Since mt5 and mt6, which carry mutations on both Cys-9 and Cys-104, formed VLPs which completely dissociated to monomer in the absence of DTT (lanes 6 and 7), it appears that both Cys-9 and Cys-104 form intermolecular disulfide linkages and that no disulfide linkages which do not involve these cysteines are formed in VLPs. The partner cysteines forming disulfide linkages with Cys-9 and Cys-104 are discussed below. The observation that even mt5 and mt6, which no longer formed intermolecular disulfide linkages, formed VLPs (Fig. 5B) clearly demonstrates that intermolecular disulfide linkages are not essential for VLP formation.
FIG. 6.
Analysis of intermolecular disulfide linkages and sensitivity to protease digestion in cysteine mutant VLPs. (A) SDS-PAGE analysis under reducing or nonreducing conditions. Purified VLPs assembled from the indicated mutant VP1 proteins were added to SDS sample loading buffer without β-mercaptoethanol. After the mixture was boiled, DTT (lanes 8 to 14) or nothing (lanes 1 to 7) was added and the samples were boiled again. The samples were separated by SDS-PAGE (8% polyacrylamide) and immunoblotted with anti-VP1 antibody. The circle, diamond, and box indicate the VP1 multimer, dimer, and monomer, respectively. WT, wild type. (B) Analysis of VLP sensitivity to proteinase K digestion. Purified VLPs were incubated with 0.1 ng (lanes 2, 6, 10, 14, 18, 22, and 26), 0.3 ng (lanes 3, 7, 11, 15, 19, 23, and 27) or 1 ng (lanes 4, 8, 12, 16, 20, 24, and 28) of proteinase K, or without proteinase K (lanes 1, 5, 9, 13, 17, 21, and 25), at 37°C for 15 min. The samples were separated by SDS-PAGE and immunoblotted with anti-VP1 antibody. The lower panel shows the relative intensity of bands representing intact VP1 protein.
To test the possibility that disulfide linkages help to stabilize VLPs, we analyzed the sensitivity of the VLPs to protease digestion. Purified VLPs were treated with increasing amounts of proteinase K, and the remaining VP1 proteins were detected by Western blotting with anti-VP1 antibody (Fig. 6B, upper panel). The relative intensities of the bands representing intact VP1 proteins are also indicated in the figure (Fig. 6B, lower panel). mt1, mt2, mt5, and mt6 were more sensitive to protease than was wild-type VP1, while mt3 and mt4 showed wild-type protease resistance. These results indicate that intermolecular disulfide linkages at Cys-9 and Cys-104 contribute to the resistance of VLPs to protease.
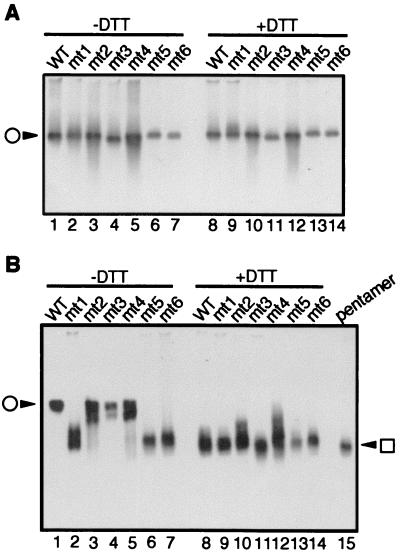
We also tested the effect of each disulfide linkage on dissociation caused by DTT and EGTA treatment in vitro. Purified VLPs were treated with DTT in either the presence (Fig. 7A) or absence (Fig. 7B) of calcium ions and were separated by native agarose gel electrophoresis. In the presence of calcium ions, all mutants remained in the VLP form (Fig. 7A, lanes 1 to 7), and DTT had no effect (lanes 8 to 14), demonstrating that the reducing agent did not affect VLP dissociation in the presence of calcium ions. In contrast, in the absence of calcium ions (note that chelating agent was not added), all VLPs dissociated to pentamers following the addition of DTT (Fig. 7B, lanes 8 to 15). When no DTT was added, mt2, mt3, mt4 and wild-type VP1 remained in VLP form (lanes 1, 3, 4, and 5). However, VLPs formed from mt1, mt5, and mt6, all of which carry a mutation at Cys-9, dissociated to pentamers (lanes 2, 6, and 7). These results indicate that in the absence of calcium ions, VLP dissociation to pentamers depends on the reduction of a disulfide linkage at Cys-9. The results also suggest that the interpentamer disulfide linkage at Cys-9 prevents VP1 protein from releasing bound calcium ions, which results in the stabilization of VLPs through calcium ion-mediated interactions.
FIG. 7.
Analysis of the dissociation of cysteine mutant VLPs to pentamers in vitro. (A) Native agarose gel electrophoresis in the presence of calcium ions. Purified VLPs assembled from the indicated mutants were incubated for 1 h with (lanes 8 to 14) or without (lanes 1 to 7) 30 mM DTT in the presence of 2 mM CaCl2 and were separated on a 0.8% agarose gel containing 2 mM CaCl2 in both gel and electrophoresis buffer. Proteins were transferred to a polyvinylidene difluoride membrane and immunoblotted with anti-VP1 antibody. The circle indicates VLPs. WT, wild type. (B) Native agarose gel electrophoresis in the absence of calcium ions. Purified VLPs were processed as in panel A except that no CaCl2 was added to the samples, gel, or electrophoresis buffer. No chelating agent was added. Purified wild-type VP1 pentamer was loaded as a control (lane 15). The circle and box indicate VLPs and VP1 pentamer, respectively.
This idea was further supported by our observation that mt1, mt5, and mt6 were more susceptible to dissociation or degradation during the purification process. Moreover, modification of the purification procedure (use of nuclear extracts instead of sonicated whole-cell lysates of infected Sf9 cells as starting material, and addition of 2 mM CaCl2 in all purification steps) resulted in an increase in the final yields of purified mt1, mt5, and mt6 VLPs to levels comparable to wild-type VLP yields (data not shown).
Calcium ion binding site mutants.
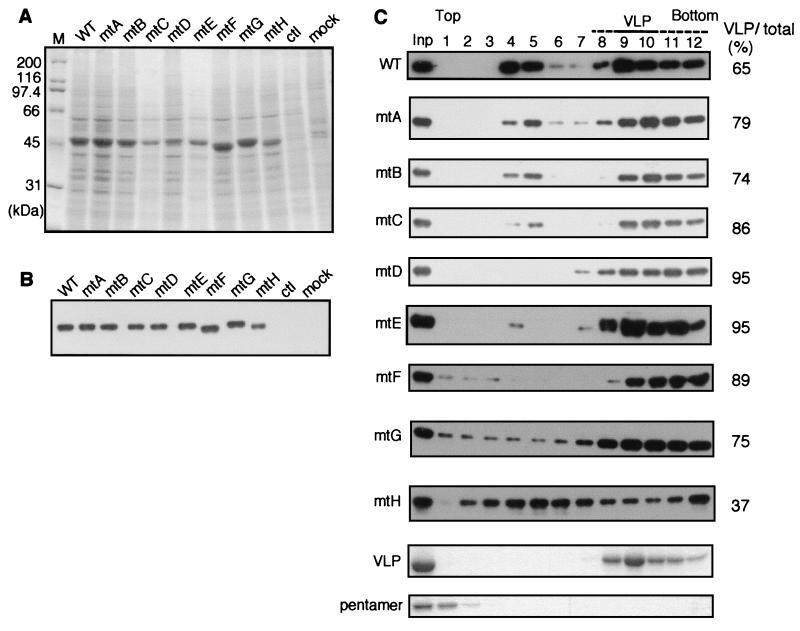
X-ray analyses of SV40 virions have revealed that two calcium ions are bound per VP1 molecule (calcium ions 1 and 2), and the amino acid residues interacting with these calcium ions have been identified. Three VP1 molecules are linked by these two calcium ions. Calcium ion 1 interacts with Ser-213 and Glu-216 of the first VP1 molecule, Glu-46 and Glu-48 of the second VP1 in the same pentamer, and Glu-330 of the third VP1 in a neighboring pentamer. Calcium ion 2 interacts with Glu-157, Glu-160, Lys-214, and Glu-216 of the first VP1 and Asp-345 of the third VP1 (20, 30). To elucidate the functional roles of these interactions, we introduced mutations at these residues, as shown schematically in Fig. 3B. Amino acids which bind to calcium ion 2 (Glu-157, Glu-160, and Asp-345) were mutated singly (mtA, mtB, and mtC) or in combination (mtD and mtE). The carboxy terminus of VP1 invades and links to VP1 in the neighboring pentamer through calcium ions 1 and 2; residues implicated in this interaction (Glu-330 and Asp-345) were substituted in mtF. In this mutant, we also substituted Glu-329 to avoid the possibility that calcium ion binding to this adjacent residue might substitute for binding to Glu-330. mtG was mutated at amino acids near the amino terminus which interact with VP1 molecules in the same pentamer (Glu-46 and Glu-48). In mtH, all of the amino acid residues involved in calcium ion binding were mutated. All of these mutants were introduced into baculovirus vectors and successfully expressed in Sf9 cells at levels comparable to wild-type VP1 expression (Fig. 8A and B). mtF and mtH showed slightly higher electrophoretic mobility (apparent molecular mass, approximately 45kDa) than did the other mutants or wild-type VP1 (approximately 50 kDa), possibly because of changes in electric charge caused by the mutations.
FIG. 8.
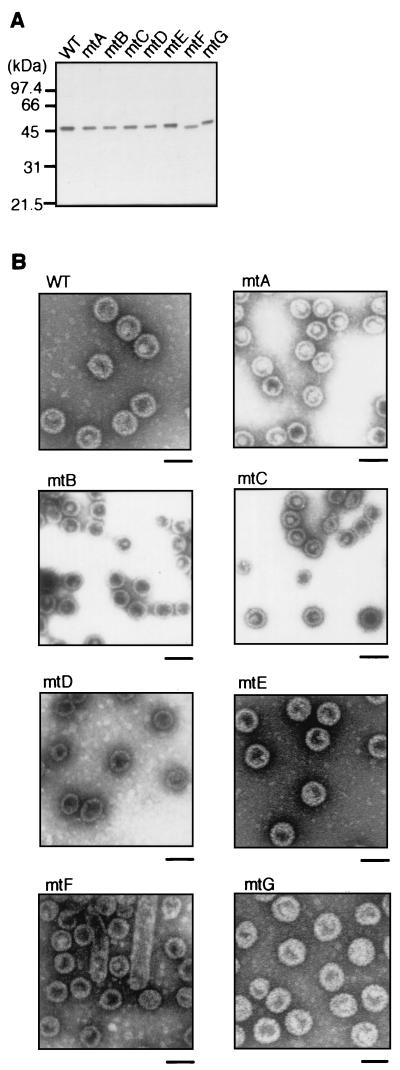
Expression and VLP formation in Sf9 cells of VP1 calcium binding site mutants. (A) Analysis of expression in insect cells. Sf9 cells infected with baculoviruses expressing calcium binding site mutants were lysed at 3 days p.i., and the soluble fractions of whole-cell lysates were separated by SDS-PAGE and stained with CBB. WT, wild type. (B) Immunoblotting with anti-VP1 antibody of an identical gel to that shown in panel A. (C) Analysis of VLP formation in insect cells. Whole-cell lysates were subjected to 10 to 30% sucrose gradient centrifugation, and fractions were separated by SDS-PAGE and immunoblotted with anti-VP1 antibody. The percentages of total VP1 protein represented by the material observed in fractions 8 to 12 are shown on the right. Inp, input.
To analyze VLP formation by these mutant proteins in insect cells, whole-cell lysates of infected cells were subjected to sucrose gradient centrifugation (Fig. 8C). Peaks of all mutants except for mtH were detected in fractions 8 to 12, indicating that they formed VLPs efficiently in insect cells, whereas mtH VLP formation was relatively inefficient. More mtF VP1 was detected in fractions 11 and 12 than in fractions 9 and 10, suggesting that this mutant formed particles larger than normal VLPs. Interestingly, all of the mutants except mtH showed a lower tendency to form the small particles detected in fractions 4 and 5 than did wild-type VP1; mtD, mtE, and mtF formed particularly low levels of the small particles. Densitometric quantification also showed that a greater proportion of total VP1 was detected in fractions 8 to 12 in the analysis of the mutants (except mtH) than in the analysis of wild-type VP1. These results indicate that the mutant VP1 proteins formed intact VLPs more efficiently than wild-type VP1 did.
All mutant VLPs except for those assembled from mtH were purified to near homogeneity by cesium chloride density gradient ultracentrifugation (Fig. 9A). mtH could not be purified in quantities sufficient for further analysis even by the modified purification procedures described above. Electron microscopic analysis showed that all of the purified mutant VLPs were morphologically indistinguishable from wild-type VLPs (Fig. 9B). mtF was unique in that it frequently formed tubular particles, which may have a larger sedimentation coefficient than VLPs, as well as intact VLPs (Fig. 9B). These results indicate that the carboxy-terminal calcium ion binding sites of VP1 protein affect the accuracy of viral assembly. No homogeneous particles, not even the small particles, were observed in mtH preparations (data not shown). The inability of this mutant to form VLPs indicates that calcium ion-mediated interactions are essential for VLP formation, although there remains the alternative possibility that mtH VP1 could no longer achieve the necessary conformation because of the (calcium-independent) effects of its many amino acid substitutions.
FIG. 9.
Purification and morphological analysis of calcium binding site mutant VLPs. (A) Purification of mutant VLPs by cesium chloride density gradient centrifugation. Fractions containing VLPs were separated by SDS-PAGE, and proteins were visualized by silver staining. WT, wild type. (B) Electron micrographs. Purified VLPs were negatively stained and observed by electron microscopy. Scale bar, 50 nm.
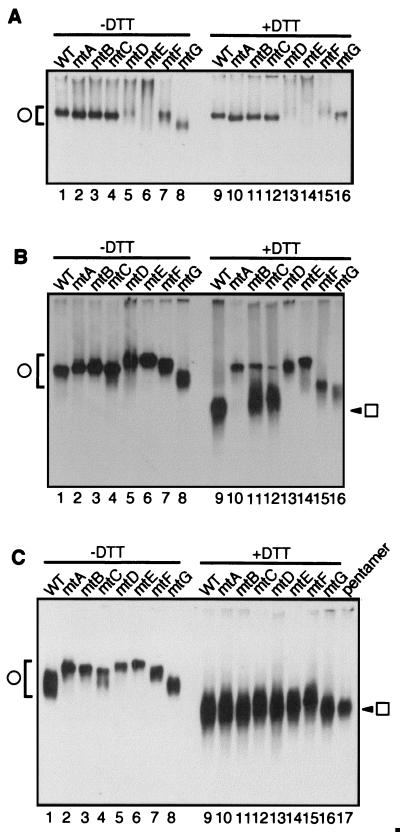
We next analyzed the dissociation properties of these mutants under various conditions by native agarose gel electrophoresis (Fig. 10). In the presence of calcium ions (Fig. 10A), mtA, mtB, mtC, mtF, mtG, and wild-type VP1 remained in VLP form in the absence or the presence of DTT (Fig. 10A, lanes 1 to 4, 7, 8, 9 to 12, 15, and 16). mtG VLPs migrated faster than the other VLPs (lane 8) despite being indistinguishable from the other VLPs in shape or size. The difference in migration rate may be due to changes in electric charge on the surface of the mtG VLP. The relative mobilities of mtD and mtE VLPs were reduced whether or not DTT was added (lanes 5, 6, 13, and 14), indicating that these mutant VLPs formed aggregates in the presence of calcium ions.
FIG. 10.
Dissociation of calcium ion binding site mutant VLPs to pentamers in vitro. Purified VLPs assembled from the indicated mutants were incubated for 1 h with (lanes 9 to 16) or without (lanes 1 to 8) 30 mM DTT in the presence of 2 mM CaCl2 (A), in the absence of either calcium ion or chelating agent (B), or in the presence of BAPTA, an efficient calcium ion-chelating agent (C). The samples were separated on 0.8% native agarose gels with (A) or without (B and C) 2 mM CaCl2 in both gel and electrophoresis buffer. VP1 proteins were detected by immunoblotting with anti-VP1 antibody. WT, wild type; pentamer, purified wild-type VP1 pentamer (panel C, lane 17). The circle and box indicate VLPs and pentamer, respectively.
In the absence of calcium ions (Fig. 10B), all mutants as well as wild-type VP1 remained in VLP form in the absence of DTT (Fig. 10B, lanes 1 to 8). Again, mtG VLPs migrated a little faster than the other VLPs did. Under these conditions, VLPs of mtD and mtE did not aggregate. All of the calcium binding site mutants behaved differently from wild-type VP1 in the presence of DTT (lanes 9 to 16). Under reducing conditions, wild-type VLPs dissociated to pentamers (Fig. 7B, lanes 8 and 15) whereas mtA, mtD, and mtE VLPs did not dissociate at all (Fig. 10B, lanes 10, 13, and 14). mtB, mtC, mtF, and mtG VLPs dissociated, but the mobility of the dissociated products was lower than that of VP1 pentamer, indicating incomplete dissociation (lanes 11, 12, 15, and 16). In addition, a fraction of mtB and mtC VLPs remained intact (lanes 11 and 12). These results were remarkable because all of the mutant VLPs were more resistant to dissociation than were wild-type VLPs, suggesting that calcium ion-interacting amino acids, especially Glu-157, contribute to the flexibility of VLPs.
We further analyzed the behavior of these mutant VLPs in the presence of BAPTA, an efficient calcium ion-chelating agent. Under these conditions, addition of DTT caused all VLPs to dissociate completely to VP1 pentamers (Fig. 10C, lanes 9 to 17). However, in the absence of DTT, none of the mutant VLPs dissociated at all, while wild-type VLPs dissociated only partially (lanes 1 to 8). This observation further indicates that the calcium binding site mutant VLPs were more rigid than the wild type and that the high stability of the mutants was due to stronger calcium ion-mediated interactions. These results, taken together, demonstrate that the conformation of calcium ion binding sites moderates the affinity of VLPs for calcium ions and may confer a greater tendency to dissociation on the SV40 capsid.
DISCUSSION
Utility of the baculovirus expression system for the study of viral capsid assembly and disassembly.
The advantages of baculovirus expression systems are that relatively large amounts of recombinant protein can be expressed in a eukaryotic environment, and folding of the protein is expected to be similar to that of native protein (24). To date, recombinant viral capsid proteins from polyomaviruses (6, 10, 11, 21, 25), papillomaviruses (17, 18, 26), and adeno-associated virus (12, 13) have been shown to assemble into VLPs in insect cells. We also utilized this system and demonstrated that VP1 of SV40 formed VLPs in the nucleus (in which location the capsid of wild-type SV40 is naturally formed) of insect cells (Fig. 1); that the VLPs were morphologically indistinguishable from wild-type SV40 particles (Fig. 2B) (19); and that, like wild-type SV40 particles (7), the VLPs dissociated to VP1 pentamers following treatment with DTT and EGTA in vitro (Fig. 2). Thus, this expression system offers a powerful and convenient system for the study of assembly and disassembly of viral capsid proteins.
Disulfide linkages between cysteine residues stabilize VLPs.
Although a study of SV40 VP1 cysteine mutants using a rabbit reticulocyte translation system has been reported (15), this system could produce only pentamers and pentamer-oligomers but not VLPs due to the low yield of VP1. Thus, it was not suitable for analysis of particle formation. In the present study, using a baculovirus expression system, we demonstrated that both Cys-9 and Cys-104 formed intermolecular disulfide linkages in VLPs, that these disulfide linkages were not essential for the formation of VLPs in vivo, and that these disulfide linkages increased the resistance of VLPs to protease. We also showed that a disulfide linkage at Cys-9 was critical for maintenance of VLPs at low calcium ion concentrations and contributed to the stabilization of VLPs against dissociation to pentamers by preventing the release of calcium ion.
Previous studies of VP1 cysteine mutants using a cell-free expression system have shown that Cys-9, Cys-104, and Cys-207 are necessary and sufficient for disulfide linkage between pentamers (15). In contrast, our present results show that Cys-207 did not form any intermolecular disulfide linkage in VLPs (Fig. 6A). This inconsistency may result from differences in the protein expression system. Proteins synthesized in a cell-free system may be exposed to air oxidization, which allows nonspecific disulfide linkages. Crystallographic analyses have shown that Cys-207 is located inside the VP1 molecule in pentamers (20, 30), so that formation of intermolecular disulfide linkages at Cys-207 seems unlikely. Thus, we propose that Cys-207 does not form intermolecular disulfide linkages in the viral capsid.
On the other hand, our observation that Cys-104 formed a disulfide linkage agrees with the results from the cell-free system (15). Crystallographic analyses of the SV40 virion have shown that Cys-104 is located in a loop of VP1 that is very close to another Cys-104 loop on a neighboring pentamer (20), and a disulfide linkage involving Cys-104 has been identified in β-β′ interpentamer interaction (30). Thus, the VP1 dimer shown in Fig. 6A, lane 2, is probably the result of a disulfide linkage between Cys-104 and Cys-104.
Our observation that Cys-9 formed a disulfide linkage is also consistent with the results from the cell-free system (15). The structure of the amino-terminal 13 amino acid residues of VP1, a region which includes Cys9, has not been crystallographically determined, probably because it is located inside the capsid (20, 30). However, since crystallographic analysis has shown that no cysteines other than Cys-9 are located inside the virion, the most likely disulfide bonding partner of Cys-9 is Cys-9. Thus, the VP1 dimer shown in Fig. 6A, lane 3, whose mobility is different from that of the dimer linked by Cys-104, is probably due to a disulfide linkage between Cys-9 and Cys-9. Although the structures of the amino termini of SV40 and polyomavirus VP1 proteins in the capsid have yet to be determined (20, 30, 31), two functional domains have been localized to this region. The first is a nonspecific DNA binding domain (5, 22; P. P. Li, C.-K. Sun, A. Miyao, and H. Kasamatsu, Abstr. 8th Annu. Meet. Am. Soc. Virol., p. 107, 1997), which supports the idea that the VP1 amino terminus extends inward to interact with viral DNA. The second is a nuclear localization signal (14, 23). In addition to these functions, we demonstrated that the amino terminus of VP1 plays a novel functional role in strengthening the calcium ion binding affinity by forming an interpentamer disulfide bond at Cys-9, which results in stabilization of VLPs.
Multimers of mt3, mt4, and wild-type VP1, formed by disulfide linkages, were observed in nonreducing SDS-PAGE (Fig. 6A). Thus, the partner VP1 molecule forming disulfide linkages with Cys-9 and Cys-104 appears to be different in some VP1 molecules, suggesting that disulfide linkages form an extensive covalent network throughout the VLP and contribute to its stability.
Calcium ion-mediated interactions of VP1 proteins affect viral assembly and stability.
Studies of the assembly and disassembly of SV40 and murine polyomavirus capsids have indicated that calcium ions are important for capsid formation (2–4, 7). In addition, the assembly into VLPs of recombinant polyomavirus VP1 expressed in Escherichia coli was driven by calcium ions at physiological pH and ionic strength in vitro (27, 28). Further evidence of the importance of calcium ions in capsid formation is provided by the observation that the increase in calcium ion concentration induced by treatment with the calcium ionophore ionomycin causes recombinant polyomavirus VP1 to assemble into VLPs not only in the nucleus but also in the cytoplasm of insect cells, at which location VLPs are not formed under normal physiological conditions (21).
Crystallographic studies of SV40 VP1 have identified three amino acid residues (Glu-157, Glu-160, and Asp-345) which bind to calcium ion 2 (20). More refined crystallographic analysis has shown that Lys-214 and Glu-216 are also involved in binding to calcium ion 2 and that calcium ion 1 interacts with Glu-46, Glu-48, Ser-213, Glu-216, and Glu-330 (30). These studies have revealed that calcium ion-mediated interactions form bridges between a carboxy-terminal arm extended from one pentamer and the internal loops of a neighboring pentamer. In the present study, we analyzed the functional roles of the amino acids involved in calcium ion binding sites.
We demonstrated that calcium ion binding sites on the carboxy-terminal arm affect the association of VP1. Unexpectedly, we found that mutations at any of the calcium ion-interacting amino acids tested, especially Glu-157, resulted in the formation of more rigid VLPs than were formed from wild-type VP1. We also showed that VP1 mutated at all of these amino acids (mtH) was deficient for VLP formation, indicating that calcium ion-mediated interactions are essential for capsid assembly, although there remains the possibility that the large number of amino acid substitutions in mtH VP1 resulted in sufficient conformational change to inhibit assembly.
The VP1 carboxy terminus plays an important role in interpentameric interactions. Crystallographic studies of SV40 showed that in the fully assembled virus shell, most pentamers contact each other only through the carboxy-terminal 49 amino acid residues (residues 313 to 361) protruding into an adjacent pentamer (20, 30). In addition, deletion of the corresponding region of recombinant polyomavirus VP1 led to deficient VLP assembly in vitro, despite the ability of the mutant VP1 to form pentamers (9). It is interesting that mtF, which has mutations in the carboxy-terminal arm, frequently formed tubular particles as well as intact VLPs in insect cells. It has been shown that wild-type polyomavirus forms such tubular particles at low frequency in vivo, a phenomenon which is considered to result from incorrect selection among three possible configurations of interpentameric interactions (1). These observations suggest that the carboxy-terminal calcium ion binding sites affect the specificity of interpentameric interactions during capsid assembly.
We found that amino acid substitutions at any of the calcium ion binding sites, especially Glu-157, resulted in the formation of VLPs which were more rigid than wild-type VLPs in the absence of calcium ions. It thus appears that these calcium ion binding sites moderate calcium ion binding affinity, which provides flexibility to the viral capsid and promotes dissociation during viral uncoating.
We also note that mtD and mtE formed VLPs more efficiently than did wild-type VP1 in insect cells and that these VLPs were the most rigid of the mutant VLPs, suggesting that these mutants might be useful as capsules to package DNA for gene therapy.
Biological significance.
Taken together, our present observations provide a preliminary model of the molecular mechanisms of SV40 capsid assembly and disassembly in vivo. The concentration of calcium ions is much higher in blood (approximately 2 mM) than in cells (approximately 0.05 to 0.3 μM). That VLPs are much more stable and resistant to reducing agents in the presence of calcium ions indicates the reason for SV40 virion survival in extracellular environments. Studies of early events in SV40 infection have indicated that for the transportation of its genomic DNA into the nucleus, where replication and transcription occur, the capsid must dissociate at least partially in the cytoplasm, because the capsid is too large to pass through the nuclear pore complex (reviewed in reference 16). Thus, it is thought that after binding to cellular receptors and endocytotic internalization, the low calcium ion concentration and reducing conditions in the cytoplasmic environment lead to viral uncoating. During this step, reduction of the Cys-9–to–Cys-9 disulfide linkage may trigger dissociation, and the calcium ion binding sites probably facilitate the reaction. In the late phase of infection, newly synthesized VP1 proteins are translocated and assembled into capsids in the nucleus. At this stage, calcium ion-mediated interactions at the VP1 carboxy terminus are important whereas intermolecular disulfide linkages between cysteines are not essential.
Studies of the behavior in mammalian cells of recombinant SV40 carrying the VP1 mutations presented here may help to further clarify the precise molecular mechanisms of capsid assembly and disassembly.
ACKNOWLEDGMENTS
This work was supported by a Research Grant from Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation (JST); a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture; and a grant for research and development projects in Cooperation with Academic Institutions from New Energy and Industrial Technology Development Organization (NEDO).
We are grateful to Peggy Li and Harumi Kasamatsu (University of California, Los Angeles, Calif.) for helpful suggestions. We thank Kenji Suzuki (National Institute of Infectious Diseases, Tokyo, Japan) for help with electron microscopy.
REFERENCES
- 1.Baker T S, Caspar D L, Murakami W T. Polyoma virus “hexamer” tubes consist of paired pentamers. Nature. 1983;303:446–448. doi: 10.1038/303446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady J N, Winston V D, Consigli R A. Dissociation of polyoma virus by the chelation of calcium ions found associated with purified virions. J Virol. 1977;23:717–724. doi: 10.1128/jvi.23.3.717-724.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady J N, Winston V D, Consigli R A. Characterization of a DNA-protein complex and capsomere subunits derived from polyoma virus by treatment with ethyleneglycol-bis-N,N′-tetraacetic acid and dithiothreitol. J Virol. 1978;27:193–204. doi: 10.1128/jvi.27.1.193-204.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady J N, Kendall J D, Consigli R A. In vitro reassembly of infectious polyoma virions. J Virol. 1979;32:640–647. doi: 10.1128/jvi.32.2.640-647.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang D, Cai X, Consigli R A. Characterization of the DNA binding properties of polyomavirus capsid protein. J Virol. 1993;67:6327–6331. doi: 10.1128/jvi.67.10.6327-6331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang D, Fung C Y, Ou W C, Chao P C, Li S Y, Wang M, Huang Y L, Tzeng T Y, Tsai R T. Self-assembly of the JC virus major capsid protein, VP1, expressed in insect cells. J Gen Virol. 1997;78:1435–1439. doi: 10.1099/0022-1317-78-6-1435. [DOI] [PubMed] [Google Scholar]
- 7.Colomar M C, Degoumois-Sahli C, Beard P. Opening and refolding of simian virus 40 and in vitro packaging of foreign DNA. J Virol. 1993;67:2779–2786. doi: 10.1128/jvi.67.5.2779-2786.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack B. Directed mutagenesis using polymerase chain reaction. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 8.5.7–8.5.9. [Google Scholar]
- 9.Garcea R L, Salunke D M, Caspar D L. Site-directed mutation affecting polyomavirus capsid self-assembly in vitro. Nature. 1987;329:86–87. doi: 10.1038/329086a0. [DOI] [PubMed] [Google Scholar]
- 10.Gillock E T, Rottinghaus S, Chang D, Cai X, Smiley S A, An K, Consigli R A. Polyomavirus major capsid protein VP1 is capable of packaging cellular DNA when expressed in the baculovirus system. J Virol. 1997;71:2857–2865. doi: 10.1128/jvi.71.4.2857-2865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldmann C, Petry H, Frye S, Ast O, Ebitsch S, Jentsch K D, Kaup F J, Weber F, Trebst C, Nisslein T, Hunsmann G, Weber T, Luke W. Molecular cloning and expression of major structural protein VP1 of the human polyomavirus JC virus: formation of virus-like particles useful for immunological and therapeutic studies. J Virol. 1999;73:4465–4469. doi: 10.1128/jvi.73.5.4465-4469.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoque M, Shimizu N, Ishizu K, Yajima H, Arisaka F, Suzuki K, Watanabe H, Handa H. Chimeric virus-like particle formation of adeno-associated virus. Biochem Biophys Res Commun. 1999;266:371–376. doi: 10.1006/bbrc.1999.1831. [DOI] [PubMed] [Google Scholar]
- 13.Hoque M, Ishizu K, Matsumoto A, Han S I, Arisaka F, Takayama M, Suzuki K, Kato K, Kanda T, Watanabe H, Handa H. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J Virol. 1999;73:7912–7915. doi: 10.1128/jvi.73.9.7912-7915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii N, Minami N, Chen E Y, Medina A L, Chico M M, Kasamatsu H. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J Virol. 1996;70:1317–1322. doi: 10.1128/jvi.70.2.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jao C C, Weidman M K, Perez A R, Gharakhanian E. Cys9, Cys104 and Cys207 of simian virus 40 Vp1 are essential for inter-pentamer disulfide-linkage and stabilization in cell-free lysates. J Gen Virol. 1999;80:2481–2489. doi: 10.1099/0022-1317-80-9-2481. [DOI] [PubMed] [Google Scholar]
- 16.Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Annu Rev Microbiol. 1998;52:627–686. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- 17.Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. In vitro construction of pseudovirions of human papillomavirus type 16: incorporation of plasmid DNA into reassembled L1/L2 capsids. J Virol. 1998;72:10298–10300. doi: 10.1128/jvi.72.12.10298-10300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosukegawa A, Arisaka F, Takayama M, Yajima H, Kaidow A, Handa H. Purification and characterization of virus-like particles and pentamers produced by the expression of SV40 capsid proteins in insect cells. Biochim Biophys Acta. 1996;1290:37–45. [PubMed] [Google Scholar]
- 20.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 21.Montross L, Watkins S, Moreland R B, Mamon H, Caspar D L, Garcea R L. Nuclear assembly of polyomavirus capsids in insect cells expressing the major capsid protein VP1. J Virol. 1991;65:4991–4998. doi: 10.1128/jvi.65.9.4991-4998.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreland R B, Montross L, Garcea R L. Characterization of the DNA-binding properties of the polyomavirus capsid protein VP1. J Virol. 1991;65:1168–1176. doi: 10.1128/jvi.65.3.1168-1176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreland R B, Garcea R L. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology. 1991;185:513–518. doi: 10.1016/0042-6822(91)90811-o. [DOI] [PubMed] [Google Scholar]
- 24.O'Reilly D, Miller L, Luckow V. Baculovirus expression vectors. Oxford, United Kingdom: Oxford University Press; 1994. [Google Scholar]
- 25.Pawlita M, Muller M, Oppenlander M, Zentgraf H, Herrmann M. DNA encapsidation by viruslike particles assembled in insect cells from the major capsid protein VP1 of B-lymphotropic papovavirus. J Virol. 1996;70:7517–7526. doi: 10.1128/jvi.70.11.7517-7526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose R C, Bonnez W, Reichman R C, Garcea R L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salunke D M, Caspar D L, Garcea R L. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 28.Salunke D M, Caspar D L, Garcea R L. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 31.Stehle T, Harrison S C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 32.Tooze J, editor. DNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 33.Tsien R Y. New calcium indicators and buffers with high selectivity against magnesium and protons: design, synthesis, and properties of prototype structures. Biochemistry. 1980;19:2396–2404. doi: 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]