Abstract
Sex chromosomes have evolved in many plant species with separate sexes. Current plant research is shifting from examining the structure of sex chromosomes to exploring their functional aspects. New studies are progressively unveiling the specific genetic and epigenetic mechanisms responsible for shaping distinct sexes in plants. While the fundamental methods of molecular biology and genomics are generally employed for the analysis of sex chromosomes, it is often necessary to modify classical procedures not only to simplify and expedite analyses but sometimes to make them possible at all. In this review, we demonstrate how, at the level of structural and functional genetics, cytogenetics, and bioinformatics, it is essential to adapt established procedures for sex chromosome analysis.
Keywords: Bioinformatics, chromosome dissection, cytogenetics, dioecious plants, epigenetics, functional genetics, sex chromosomes, tandem repeats, transposable elements
This review explores adapted or newly developed methods customized for comprehensive analysis of sex chromosomes in dioecious plants. We emphasize methodological approaches for understanding XY chromosome structure and function.
Introduction
Dioecy represents an extreme strategy of sexual reproduction where sex-specific structures emerge on distinct plants. The existence of different sexes frequently gives rise to what are known as sex chromosomes (typically X and Y or Z and W). The widespread occurrence of recombination suppression within sex chromosomes is a common evolutionary trend, typically accompanied by degeneration and the loss of genes in the non-recombining region of the sex-limited chromosome (Y or W) (Fig. 1). The evolution of the Y(W) chromosome, or Yh in papaya (VanBuren et al., 2015; Yue et al., 2022), involves key stages such as the establishment of the sex-determining region, suppression of recombination, accumulation of repeats, gene degeneration, and reduction through deletions. Expansion and shrinkage are frequently concurrent processes that shape the Y chromosome structure, exerting varying impacts on Y chromosome dynamics throughout different stages of sex chromosome evolution (Vyskot and Hobza, 2015).
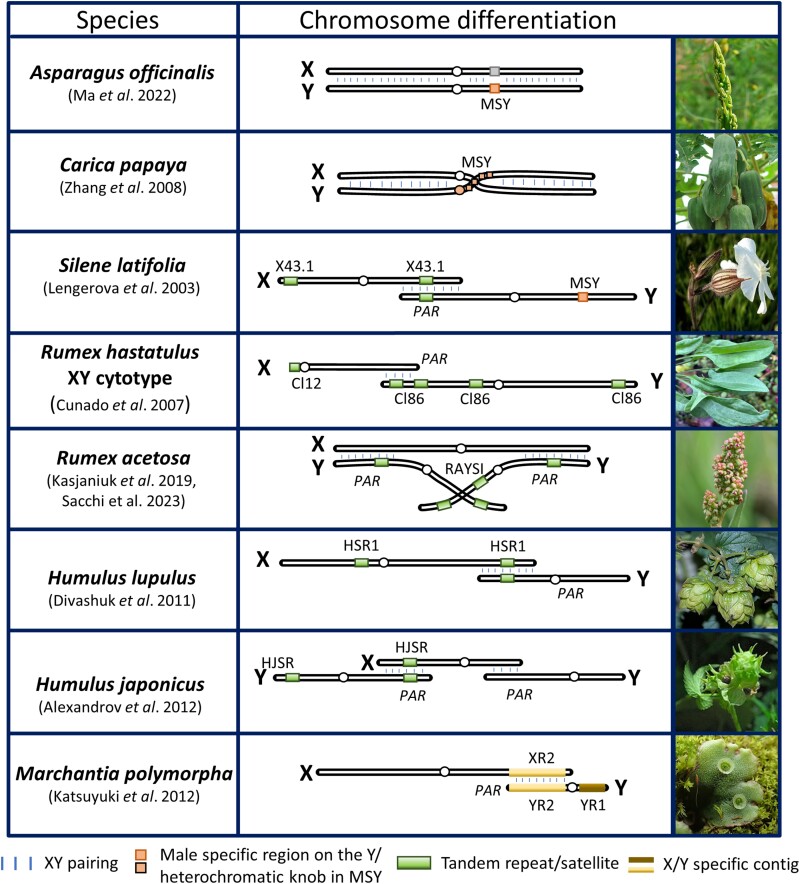
Fig. 1.
Schematic diagram of sex chromosome evolution in dioecious plants. Species are shown according to their level of sex chromosome differentiation and Y chromosome asynapsis. In S. oleracea, A. officinalis, and C. papaya, the sex chromosomes are mostly homomorphic with recently formed non-recombining regions (region with suppressed recombination). The non-recombining region is largely extended almost to the entire chromosomal length in species with heteromorphic sex chromosomes, namely in S. latifolia, R. hastatulus (XY cytotype), R. acetosa, H. lupulus, H. japonicus, and M. polymorpha. The position of the centromere, the PAR length, and the ratio between X and Y is illustrative.
The nature and complexity of sex chromosomes often demands cutting-edge technologies for comprehensive analyses of their evolution and correct assembly of non-recombining regions (Fig. 1). Consequently, classical methods in genetics that utilize genetic maps for comparative analyses, genome rearrangement analysis, and gene identification are limited due to the repetitive fraction within the sex chromosomes and suppressed recombination. Even genetic mapping based on deletion mutant lines can be challenging if those deletions are lethal during gametogenesis. The approach used to avoid this limiting factor was for a long time the use of radiation hybrid (RH) or HAPPY mapping approaches (for a review, see Riera-Lizarazu et al., 2008). However, the application of these methods in plants is still in the experimental phase and quite challenging. Currently, the huge progress in methods improving genome assemblies, such as optical genome mapping, makes the study of long non-recombining regions more feasible. This progress opens avenues for deeper exploration, potentially uncovering novel insights into sex chromosome evolution and facilitating more accurate assembly of non-recombining regions. The integration of third-generation sequencing techniques, supported by functional analysis and cytogenetics, not only will enhance our current understanding of sex chromosome origin and the role of chromosomal rearrangements during sex chromosome formation, but also paves the way for future discoveries regarding the non-recombining region and evolutionary strata.
In this review, we aim to highlight some peculiarities of sex chromosome analysis resulting from the aforementioned aspects. The purpose of the review is not to enumerate successful applications of individual methods across all plant species with sex chromosomes but to demonstrate their suitability and utility through specific examples.
Dissecting sex chromosomes: a swift transition from disorder to understanding
Laser microdissection and chromosome sorting of plant chromosomes represent distinct technology designed to simplify the analysis of large plant genomes by physical separation of their specific parts (Fig. 2). Flow sorting is a method of choice when a large volume of high molecular weight DNA suitable for further detailed analysis is required. Flow sorting is relatively easy to perform and, once adapted (e.g. time or strength of fixative), it usually takes from several hours up to several days. In contrast, chromosome microdissection typically ensures higher purity of isolated chromosomes (almost 100%) and it might be usable for a wider range of applications (Hobza and Vyskot, 2007; Soares et al., 2020). Nevertheless, both manual and laser beam-based microdissection methods provide significantly smaller amounts of material compared with flow sorting technology. Moreover, microdissection-based methods rely heavily on the expertise of the personnel involved, and the collection of plant material may take from several days up to weeks.
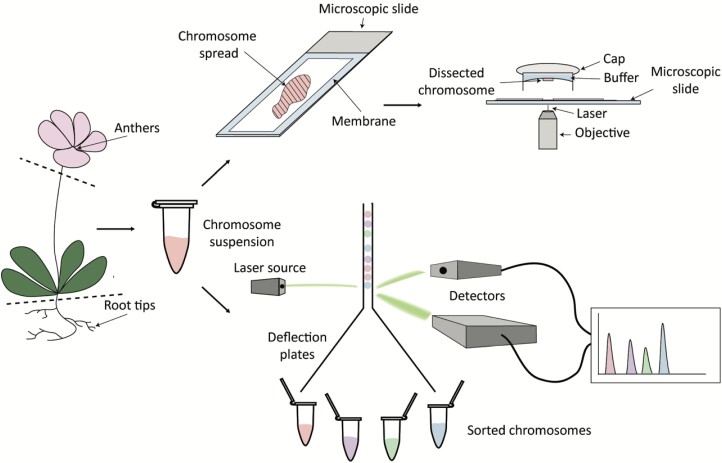
Fig. 2.
Laser microdissection as a tool to reduce genome complexity. Sex chromosomes in metaphase are isolated from plant cells (mostly pollen mother cells or root tips) and subsequently spread on a special microscopic slide covered with a membrane. After microdissection, chromosomes are transferred into a tube and processed by other applications. In the case of chromosome sorting, the chromosome suspension is stained with a DNA-specific dye and introduced into a flow chamber. Within this chamber, individual chromosomes interact with a laser beam, and the scattered light and emitted fluorescence are measured. Through this process, a histogram of fluorescence intensity (known as a flow karyotype) is generated. Sorting is accomplished by breaking the liquid stream into droplets and electrically charging the droplets containing the chromosomes of interest.
In plants, chromosome sorting and microdissection techniques have been extensively applied, particularly in the analysis of sex chromosomes. Indeed, the dissection of the largest chromosome in spinach (Spinacia oleracea) and its amplification by degenerate oligonucleotide-primed (DOP)-PCR helped to identify a male-specific marker (T11A) that was isolated from amplified DNA (Onodera et al., 2008). This led to the direct evidence of the Y chromosome. The X/Y chromosomes in spinach were recently assembled using single-chromosome sequencing and the advantage of manual microdissection (Li et al., 2023). In addition to identification of sex-specific markers and sequencing projects, the microdissection of single chromosomes further helped to develop complex chromosome painting probes as in the case of white campion (Silene latifolia) (Hobza et al., 2004) and Japanese hop (Humulus japonicus) (Yakovin et al., 2014). Alternative applications fulfil diverse objectives, including the targeted development of DNA markers and the construction of DNA libraries (Požárková et al., 2002; Hobza et al., 2006), physical mapping of individual markers and genes (Cegan et al., 2010), gene cloning (Thind et al., 2017), identification of horizontal gene transfer (Talianová et al., 2012), PCR-based mapping of markers on individual chromosomal arms, genome sequencing (Martis et al., 2013), and validation of whole-genome shotgun sequence assemblies (Kreplak et al., 2019).
While the popularity of sex chromosome (laser) microdissection seems to have dwindled nowadays, it is still a powerful tool to address many questions. The efficacy of laser microdissection extends seamlessly to other fields such as transcriptomics and proteomics, where precise tissue separation based on cellular anatomy or morphology is indispensable (reviewed in Misra et al., 2014; Yin et al., 2023). Overall, this method continues to be utilized in genomic analysis and is likely to remain a cost-effective choice for various genomic analysis, especially in non-model organisms with large genomes.
Cytogenetic tools to study sex-specific traits
Recent developments in cytogenetic techniques and significant advances in spatial resolution allowed researchers to study various aspects of plant genome architecture. Since Winge’s identification of basic chromosome number in hop (Humulus lupulus; Winge, 1923) and Blackburn’s detailed description of sex chromosome in S. latifolia (Blackburn, 1923), cytogenetic applications have been fundamental methods for rapid chromosome identification and sex chromosome characterization (Hobza et al., 2018; Charlesworth and Charlesworth, 2020; Muyle et al., 2022). With the combination of high-quality chromosome preparations from a single root tip of a small seedlings or hairy root cell lines (for more details, see Hobza and Vyskot, 2007; Bačovský et al., 2018) and single leaves of living plants (Janousek et al., 2022), it is possible to analyse the karyotype of single plants. In this section, we describe the most used techniques of fluorescent in situ hybridization (FISH) and discuss the need for correlation between DNA sequence and molecular data with the structure and organization of plant nuclei (Hobza et al., 2015; Vyskot and Hobza, 2015). FISH is particularly suited to the study of single markers, low copy bacterial artificial chromosomes (BACs), and repetitive sequences, namely (i) transposable elements (TEs) with dispersed genomic distribution and (ii) tandem repeats (satellites) usually occupying isolated genomic loci (Fig. 3).
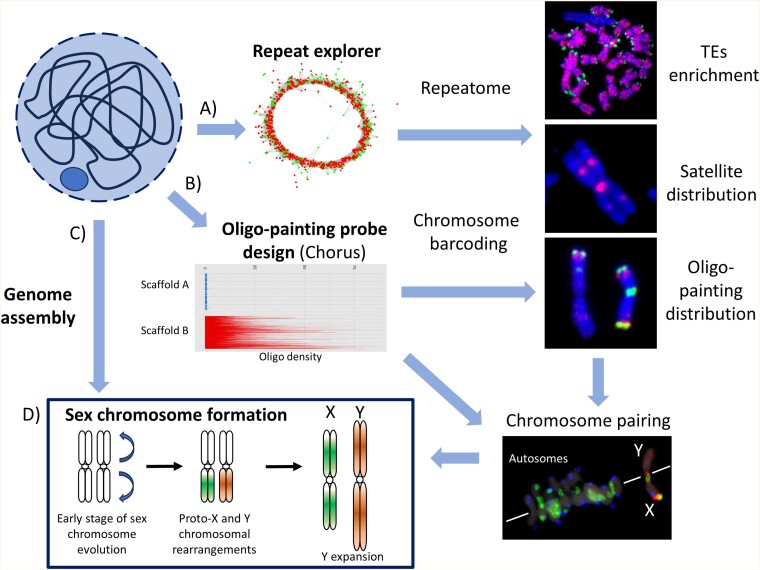
Fig. 3.
Cytogenetic tools to study sex chromosome origin and evolution. Cytogenetics nowadays combines genomic tools to study repeat fractions including transposable elements (TEs) and satellites (A), to design unique barcodes to distinguish particular chromosome or chromosomal domains using chromosome oligo-painting probe design (B), and bioinformatic tools to dissect single chromosomes or genome parts (C). The combination of the above methods helps to understand sex chromosome evolution regarding their autosomal origin, chromosomal rearrangements, and Y(W) chromosome differentiation. Arrows represent evolutionary steps during sex chromosome divergence (D). The sex chromosome barcoding allows understanding of meiotic pairing which in turn supports chromosomal fusions and inversion/translocations. Chromosomes belong to species with references, from top to bottom, as follows: S. latifolia Ogre retroelement (Kubat et al., 2014), R. hastatulus XY cytotype satellite Cl135 (Sacchi et al., 2023, Preprint), S. latifolia PAR oligo-painting probe with the subtelomeric satellite X43.1 and centromeric satellite STAR-C (Bačovský et al., 2020), and the same DNA probes on chromosomes in metaphase I in S. latifolia (Bernasconi et al., 2009; Bačovský et al., 2022).
The use of transposable elements for monitoring sex chromosome history
The key feature allowing the use of TEs in cytogenetic studies is their uneven distribution on sex chromosomes compared with autosomes (Cermak et al., 2008; Filatov et al., 2009; Steflova et al., 2013; Kralova et al., 2014; Kubat et al., 2014), contrasting with the uniformity in genomes of hermaphroditic species (Cegan et al., 2012; Wicker et al., 2018; Flasch et al., 2019). The likely cause is that TEs are preferentially active in either the male or female lineage as discussed elsewhere (Hobza et al., 2017). The male-active TEs are therefore accumulating on the Y chromosome while simultaneously being under-represented on the X chromosome, and vice versa for the female-active TEs. This allows TEs to be used (i) to estimate the size of the pseudoautosomal region (PAR), (ii) to determine the boundaries, and (iii) to determine the ages of evolutionary strata arising from the spread of the non-recombining region (Hobza et al., 2015; Vyskot and Hobza, 2015). Early studies based on this principle were limited to a single TE (Filatov et al., 2009) but, by including diverse TE lineages, a more detailed view can be obtained (Cermak et al., 2008; Puterova et al., 2018). This is because TE lineages active at different stages of sex chromosome evolution leave fingerprints (relative insertion densities) from which the history of non-recombining region expansion can be inferred. The conventional approach used to investigate this phenomenon is multicolour FISH simultaneously analysing multiple TE-derived probes. With the increasing availability of new cytological techniques and whole-genome assemblies, the precision of this approach can be expected to increase through a combination of super-resolution microscopy, such as structured illumination microscopy (SIM) and in silico analysis. In silico analysis requires precise annotation of individual TE lineages and must include assessment of their past transposition activity based on the determination of TE insertion ages (see below). When these conditions are met, TEs, alongside sex-linked genes, can become a powerful tool to study evolution of non-recombining sex chromosomes and to identify cryptic sex-linked regions in homomorphic sex chromosomes.
Satellite analysis in the context of sex chromosome biology
Repetitive sequences in non-recombining regions of sex chromosomes undergo rapid evolution, diversification, and expansion (Lengerova and Vyskot, 2001; Mariotti et al., 2008; Navajas-Pérez et al., 2009), followed by chromatin changes (Kubat et al., 2008; Steflova et al., 2013; Sacchi et al., 2023, Preprint) and the formation of inactive chromatin regions visible as DAPI banding on the metaphase Y chromosomes (Jesionek et al., 2021). The rapid expansion of repetitive sequences leads to genetic divergence and has far-reaching biological consequences, including the formation of reproductive barriers that further fix genetic differences (Kirkpatrick, 2017; O’Neill and O’Neill, 2018; Hooper et al., 2019). It also allows the use of repeats to reconstruct the evolution of sex chromosomes at the interspecific level and within a species. The established starting point is the identification and characterization of repeats from short genomic reads by specialized bioinformatics tools (e.g. RepeatExplorer, see below) followed by physical mapping using multicolour FISH.
The most suitable candidates for these analyses are satellites creating large arrays in lengths of tens to thousands of kilobases. Among satellites, we also count robust cytogenetic markers of rDNA clusters (45S, 5S). The cytological mapping of 5S rDNA in Rumex hastatulus XY and XYY cytotypes helped to identify the autosomal pair that fused to sex chromosomes resulting in the formation of the neo-XYY sex chromosome system (Grabowska-Joachimiak et al., 2015). Additionally, phased assemblies of both R. hastatulus cytotypes revealed that neo-sex chromosomes in younger cytotypes (XYY) were formed by two events: an X–autosome fusion and a reciprocal translocation between homologous autosomes and the Y chromosome. These rearrangements were supported by physical localization of eight satellites indicating that the formation of the new cytotype was accompanied by chromosome shattering (Sacchi et al., 2023, Preprint). Hence, both classes of DNA repeats (TEs and satellite clusters) provide fast information about genome reorganization and are valuable in the identification of chromosomal rearrangements during sex chromosome evolution (Fig. 3A).
Advances of chromosome-specific labeling
The most significant improvement in FISH application is the development of synthetic oligonucleotide probes (Fig. 3B–D) that are specific for chromosomal region(s), chromosomal arm(s), or whole chromosomes (Jiang and Gill, 2006; Jiang, 2019). With the increasing number of high-quality genome assemblies, such oligo painting probes are expected to become an important and essential tool to study genome evolution between related species as shown for S. latifolia and identification of XY-orthologous regions in S. vulgaris (Bačovský et al., 2020). Recently, the oligo painting probes helped to anchor Y-specific contigs in the genomic context of S. latifolia (Akagi et al., 2023, Preprint).
Epigenomic landscape of sex chromatin
While most epigenetic methods used in the field of sex chromosome biology are adopted from model organisms, the application of these methods often brings surprising and pivotal conclusions regarding the divergence of XY chromosomes. In this regard, the question of whether epigenetic changes are the cause or consequence of the evolution of sex chromosomes and related phenomena, such as dosage compensation, is still unresolved (reviewed in Muyle et al., 2017, 2022).
The term epigenetics represents all heritable and stable changes in gene expression that occur through alterations in chromatin structure and DNA methylation. These alterations are profoundly influenced by various developmental and environmental factors, driving spatio-temporal chromatin dynamics and the overall structure of the epigenomic landscape. We describe recent methodological improvements that increased our knowledge of sex chromosome epigenomics and discuss the possible use of techniques that are being adopted now or will be in the near future in plant research.
Chromatin structure of plant sex chromosomes
Before the development of advanced next-generation sequencing (NGS) techniques, such as ChIP sequencing (ChIP-seq), methylated DNA immunoprecipitation sequencing (MeDIP-seq), and other NGS-based methods, plant cytogeneticists were among the first researchers who studied sex chromosomes at the chromatin level. Indeed, early cytogenetic findings in Silene and Rumex revealed phenomena related to late replication of X chromosomes (Siroky et al., 1994, 1999) or formation of Y-chromosome bodies at the cell nucleus periphery in males (Lengerova and Vyskot, 2001; Vyskot and Hobza, 2015). A fresh perspective on immunolocalization involves the use of super-resolution microscopy techniques such as SIM and stimulated emission depletion microscopy (STED), which enable researchers to visualize objects beyond the diffraction limit of light. This innovative approach offers unprecedented clarity and detail, allowing for a deeper understanding of cellular structures and molecular interactions, as was demonstrated for pericentromeric histone modifications and their Y chromosome localization in Coccinia grandis (Sousa et al., 2016, 2017). Recent interest in the development of artificial intelligence- (AI) assisted image analysis together with high-content imaging technology further opens up new avenues in the research of various biological related phenomena, for example in meiotic stability of sex chromosomes or tissue sectioning and plant development (Pegoraro and Misteli, 2017; Bitton et al., 2021). Utilization of immunolabelling with high-content imaging remains to be adopted in plant sex chromosome research.
A complex screening of active and repressive histone marks can provide a missing link between early cytogenetic findings (Siroky et al., 1999; Bačovský et al., 2019) and RNA-seq studies (Muyle et al., 2018). Such an approach can be later complemented by the already mentioned NGS technique(s) and its modification(s). Combining sequencing with ChIP-seq with appropriate antibodies against, for example, active histone modifications, allows the deciphering of the evolutionary state of sex-linked genes and their level of epidegeneration. ChIP is a robust method and can be considered an enrichment-based technique like DNA immunoprecipitation (DIP). Following the ChIP protocol, DNA-bound protein is immunoprecipitated using a specific antibody, and the bound DNA is then co-precipitated for further analysis. Additionally, subsequent DNA purification allows either the study of selected genes through quantitative reverse transcription–PCR (qRT–PCR), or an analysis of precipitated DNA using whole-genome sequencing (Rodríguez Lorenzo et al., 2020). Low input and single-cell methods are sometimes required due to limited plant material, for example endosperm studies or single meristematic tissues. A new and versatile method named cleavage under targets and release using nuclease (CUT&RUN) utilizes a new strategy apart from ChIP-seq. CUT&RUN targets micrococcal nuclease (MNase) to binding sites of the protein of interest through specific interactions, allowing it to have a higher signal-to-background noise ratio and analysis of only thousands of cells per sample. The CUT&RUN approach was successfully utilized in Arabidopsis as an alternative and efficient strategy for plant epigenomic studies but remains to be adopted for dioecious plant research (Zheng and Gehring, 2019).
Recently, the entire Y chromosome assembly complemented by bisulfite whole-genome sequencing in S. latifolia helped to uncover the DNA methylation levels within the non-recombining region (Akagi et al., 2023, Preprint; Moraga et al., 2023, Preprint). The sodium bisulfite protocol has been widely used as a method for DNA methylation analysis for decades. This chemical deaminates non-methylated cytosines to uracil and leaves methylated cytosines unchanged. Compared with MeDIP, it allows a more accurate mapping and identification of methylation at single-base resolution, as well as determination of the average methylation level in the CG, CHG, and CHH context and the identification of differentially methylated regions (DMRs). The main weakness of bisulfite modification is that it is impossible to discriminate between methyl cytosine and other enzymatic oxidation derivatives (oxi-mCs). Oxi-mCs are present in significant amounts in plants, and specific DNA modification protocols are available for each modification (Plongthongkum et al., 2014; Wang et al., 2015; Kalinka et al., 2023).
The cutting-edge sequencing technology of Oxford Nanopore Technology (ONT), Illumina short reads, and Hi-C led to the fine-tuned genome assembly in a female willow tree (Salix dunnii) (He et al., 2021). Third-generation sequencing techniques represented by single molecule real-time sequencing (SMRT) from Pacific BioSciences and nanopore sequencing from ONT provide longer reads than conventional methods, starting from an average length of 10 kb up to ultra-long reads >100 kb. Regardless of the detection specificity, both methods can identify epigenetic changes in the nucleotides without previous enrichment or chemical modification (Chen et al., 2023). Combined with the length of the reads, these methods are becoming a promising tool in sex chromosome epigenetics, including, among others, oxi-mC detection. Nevertheless, the functional role of these derivatives in the context of sex chromosome evolution remains to be elucidated.
Increasing knowledge of epigenetic mechanisms related to sex determination in poplar (Bräutigam et al., 2017), melon (Latrasse et al., 2017), and Japanese persimmon (Akagi et al., 2016) shows the importance of chromatin analysis in studies focused on regulation of female and male development. Although the regulatory hierarchy of histone marks and DNA methylation is still elusive, with the substantial improvements in genomics and cytogenetics it is now possible to assess complex regulatory networks and to study remarkable evolutionary convergence of sex chromosomes.
Bioinformatics of sex chromosomes: unique tools and approaches
Due to the complexity and unique biology of sex chromosomes, it is necessary to develop and modify traditional bioinformatic tools to account for biological phenomena associated with sex chromosomes, specifically considering the segregation of X and Y (Z and W) chromosomes and the presence of a large region with suppressed recombination. The sequencing of complex regions of the Y(W) chromosome is challenging for assembly and subsequent analyses. This is illustrated by the history of assembling the human Y chromosome (the complete sequence was published recently, Rhie et al., 2023). Indeed, reports on dioecious plant genome assemblies with repeat annotation of sex chromosomes are rather sparse so far. It is becoming increasingly apparent that overcoming these difficulties is a priority task, because detailed annotation of repeats can contribute significantly to understanding the evolution of sex chromosomes, for example the expansion of the Y non-recombining region as discussed above. In addition, sex chromosomes appear to be an excellent model system to study the biology of repeat accumulation and evolution, such as the causes of sex-biased TE activity (Kubat et al., 2014; Hobza et al., 2017) and the evolution of satellites located in different genomic contexts in terms of selection and recombination frequency (Jesionek et al., 2021).
Detection of repeats in full-length sex chromosome assemblies
The list of approaches utilized for repetitive DNA determination in assemblies of dioecious plant genomes is summarized in Supplementary Table S1. As can be seen for R. hastatulus and Silene spp., the most recent approaches for repeat detection in full-length plant sex chromosomes are based in paticular on employing the Extensive de novo TE Annotator (EDTA; Ou et al., 2019). This tool has the capacity to reveal most of the transposons and their (super)families [long terminal repeat (LTR) retrotransposons; terminal inverted repeats (TIRs); miniature inverted transposable elements (MITEs); and Helitrons]. Nevertheless, when applied for de novo identification of TEs without the availability of a species-specific TE library, RepeatModeler2 (wrapped within EDTA) is needed for generation of the corresponding LTR retrotransposon sequence library. The TEs are coarsely designed as Ty1/Copia, Ty3/Gypsy, and Unknown, requiring manual annotation. Beside the complex EDTA pipeline, the RepeatMasker (Smit et al., 2015) is employed using a TE species-specific library generated either de novo with RepeatModeler2 (Cannabis sativa, Hippophae rhamnoides, Salix viminalis, S. cheanomeloides, and S. arbutifolia; Supplementary Table S1) or using available repeat elements from databases (TIGR Plant Repeat Databases, Ouyang and Buell, 2004; and/or RepBase, Bao et al., 2015; e.g. Carica papaya, Supplementary Table S1).
Regardless of the presence of sex chromosomes, annotation of the dominant component of repeats in plant genomes, the LTR retrotransposons, suffers from (i) poor lineage-level annotation (superfamily level only) and their (ii) full-length reconstruction from fragments due to multiple mutual nesting (e.g. Jedlicka et al., 2019). A rather general affiliation into superfamilies and/or unknown category can be fine-tuned using tools for annotation of LTR retrotransposon protein domains as well as Domain based annotation of transposable elements (DANTE; https://github.com/kavonrtep/dante) and/or an LTR retrotransposon classification tool such as Tesorter (Zhang et al., 2022). Even though most studies avoid nested TE analysis due to its complexity, there are some tools utilized for successful assembly. One of them is the TE-greedy-nester (Lexa et al., 2020), which employs an iterative greedy algorithm for reconstruction of full-length TEs. This tool provides the most reliable results in combination with Tesorter and REXdb (Neumann et al., 2019) as presented in TE annotation of Syzygium tree genomes (Ouadi et al., 2023).
Identification of repeats using short reads only: RepeatExplorer employment
Due to the above-mentioned obstacles with sex chromosome assemblies, so far most conducted approaches have started with RepeatExplorer (Novák et al., 2010, 2013) run on low coverage short read sequences (e.g. Puterova et al., 2017, 2018; Jesionek et al., 2021; Sousa et al., 2021). These studies used the convenience of RepeatExplorer for producing consensus sequences for (usually) full satellite monomers, which were in turn used for sex chromosome-specific probe design and subsequent visualization with FISH. Beside the satellites, the clusters of LTR retrotransposon fragments were manually curated and used for reconstruction of partial- or full-length Ty3/Gypsy and Ty1/Copia retrotransposons (e.g. Puterova et al., 2017, 2018). In summary, the most reliable approach used for repetitive DNA detection is a combination of reference repeat detection and annotation from the assembled genomic and/or short read sequences using a combination of RepeatExplorer, RepeatModeler2, DANTE, and Tesorter with subsequent run of robust pipelines as well as EDTA or RepeatMasker exploiting the convenience of the obtained annotated references libraries.
Identification of sex-linked genes
Identification of sex-linked genes, sex-determining regions, and sex chromosome-specific sequences can be done using different approaches (Supplementary Table S2) which can be divided into three main groups based on the input data [comparison of the coverage in male and female genomic data, transcriptomic or genomic sequences from defined crosses, and association and single nucleotide polymorphism (SNP)-based methods applied to natural populations]. All the presented tools and their corresponding links, including their pros and cons, are listed in Supplementary Table S2.
Genomic approaches
One of the first described approaches used to systematically discover Y chromosome genes was the chromosome quotient (CQ) (Hall et al., 2013). In the CQ method, genomic DNA from males and females is sequenced independently and aligned to candidate reference sequences. The female to male ratio of the number of alignments to a reference sequence is used to determine whether the sequence is Y-linked. Another option can be a k-mer-based approach which was used by Akagi et al. (2014, 2018) for identification of the sex-determining region in Diospyros kaki (Akagi et al., 2014) and in kiwifruit (Akagi et al., 2018). Briefly, reads from samples of the same gender were pooled and searched for the presence of gender-specific 35 bp k-mers. Reads including male-specific k-mers were assembled to generate Y-linked genomic contigs. Rangavittal et al. (2019) introduced a k-mer-based method called DiscoverY, which combines proportion sharing with female reads with depth of coverage from male reads to classify contigs as Y chromosomal. DiscoverY is an effective method to isolate Y-specific contigs from a whole-genome assembly. However, regions homologous to the X chromosome remain difficult to detect. Another recently developed tool is FindZX (Sigeman et al., 2022), an automated Snakemake-based computational pipeline for sex chromosome identification and visualization through differences in genome coverage and heterozygosity between males and females.
Transcriptomic/expression-based approaches in controlled crosses
Transcriptomic approaches represent relatively cheap and very efficient tools for the study of sex-determining systems in non-model organisms. Several tools were utilized to identify sex-linked genes and have been adopted for species without a reference genome assembly.
The LINKYX pipeline is based on the utilization of data obtained by transcriptome sequencing of parents and separately pooled male and female progeny (Michalovova et al., 2015). The main aim of this pipeline is to identify putatively sex-linked markers that should be further experimentally tested. In addition to the X- (or Z-linked) linked SNPs that are identified with the LINKYX_X variant of the pipeline, LinkYX enables identification of putative sex-specific genes (Y specific or W specific) based on the quantitative study of the transcription level in parents and in the dataset of pooled male and female progeny (LINKYX_Y). LINKYX_X and LinkYX have been successfully applied to the study of sex determination in S. otites, S. borysthenica, and S. colpophylla (Balounova et al., 2019). SEX-DETector and SEX-DETector++ (Muyle et al., 2016) are, similarly to LINKYX, based on the study of the transcriptomes in the population obtained by a controlled cross. This method has been shown to work well with as few as five offspring of each sex and has been used successfully in several dioecious species (Muyle et al., 2018; Martin et al., 2019; Veltsos, 2019; Badouin et al., 2020; Fruchard et al., 2020; Prentout et al., 2020). SEX-DETector and its updated version can in fact be used for multiple purposes: identification of sex-linked genes and sex chromosomes in the studied organism (XY or ZW), haplotype reconstruction of the gametologue copies, and estimation of allelic expression of each of the copies. However, because of its requirement for a controlled cross, the use of this method is limited to species that can be easily bred or cultivated in controlled conditions. Therefore, this hinders its application to species with a long generation time.
Association and SNP-based methods in wild populations
To characterize the sex determination system, genome-wide association studies (GWASs) were used in several species (e.g. Salix nigra, Sanderson et al., 2021; Dioscorea alata, Mondo et al., 2021; P. euphratica and P. alba, Yang et al., 2020). As input, the genomic sequences, DarTSeq reads (Diversity array technology), and capture array were used for mapping to reference sequences. For use of restriction site-associated DNA sequencing data to study sex determination, a computational workflow RADSex (Feron et al., 2021) was developed. This tool was developed for Japanese medaka (Oryza latipes) (Feron et al., 2021), but it can be adopted in other species, including plants. Sex-specific markers can be further identified with Double digest restriction-site associated DNA sequencing (DdRADseq), a method that was used in Nepenthes (Scharmann et al., 2019) and Amaranthus (Montgomery et al., 2019). To improve the robustness and transparency in sex-linked sequences identification, Grayson et al. (2022, Preprint) prepared a comprehensive workflow called SexFindR. This workflow combines coverage-based analysis and a variety of population genomic analyses such as the reference-based SNP density, GWAS, and FST, as well as the reference-free k-mers GWAS to screen for common candidate sex-linked regions. Sdpop (Käfer et al., 2021) has similar goals to SEX-DETector and SEX-DETector++ but is based on different models and so it enables identification of sex-linked transcripts in natural populations. This approach has been used to study sex chromosomes in Amborella trichopoda (Käfer et al., 2022).
New bioinformatics tools and approaches substantially increased the number of dioecious plants identified to date These tools helped to characterize the biological nature of sex chromosomes, leading to important discoveries in epigenetics and functional genomics, enabling the study of sex-linked genes through genome editing. In the last section, we summarize the historical and the most used techniques in the field of functional genetics and discuss future directions.
Functional genetics of plant sex chromosomes
Sex chromosomes carry the decisive information as to whether the individual will become male or female; however, functional studies of plant sex chromosomes and sex-linked genes are generally not straightforward. While classical genetics rely on recombination mapping for identification of causal genes, this approach is largely unfeasible as sex-linked genes are located within non-recombining regions. Moreover, extensive accumulation of TEs and/or tandem repeats makes genome assembly difficult, which is especially the case for Y/W chromosomes (for details see previous sections). On the other hand, the scientific community developed and applied diverse strategies to overcome the issues related to studying functional aspects of plant sex chromosomes (Fig. 4A–E).
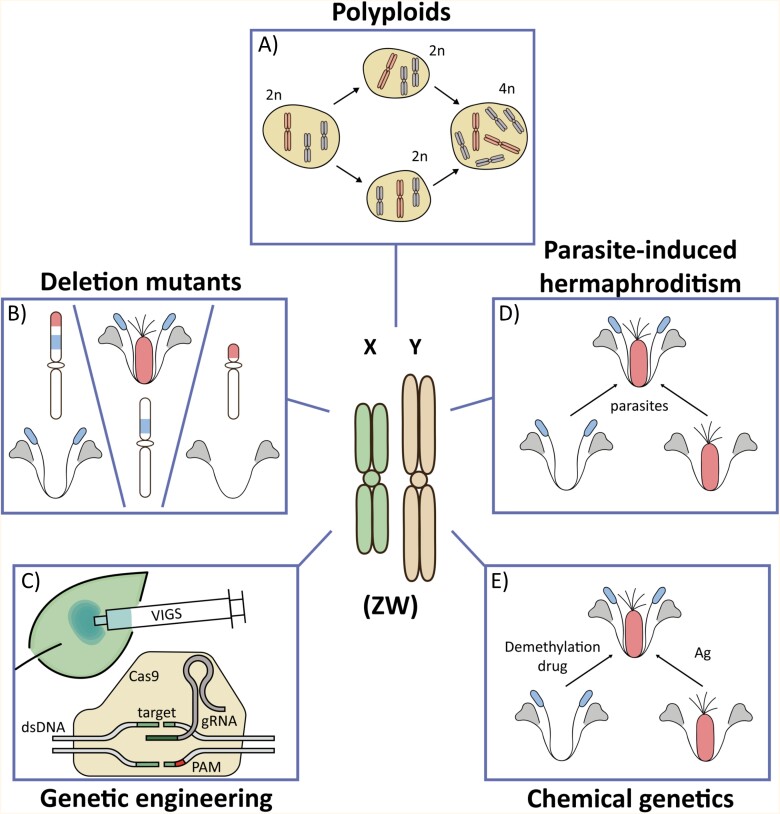
Fig. 4.
Strategies to assess the function of sex chromosomes in plants. Experimental assays with polyploids (alternatively aneuploids) represent the classical way to determine the role of individual sex chromosomes (A). These assays with plants of various ploidy levels are usually supported by analyses of deletion lines (plants carrying short chromosomal deletions or microdeletions) (B) that allowed researchers to identify sex-linked regions involved in sex determination and floral development. Modern assays using reverse genetics, such as CRISPR/Cas9, virus-induced gene silencing (VIGS), or peptide treatment of shoot apical meristems (C), provide direct evidence of the gene function and its contribution to the development of reproductive organs. Parasite-infected (D) or chemically induced (E) hermaphrodites from either female or male individuals, such as Silene or kaki, led to the identification of key mechanisms and genes that regulate sexual phenotypes, and to understanding of the regulatory networks leading to separate sexes.
Classical strategies to study functional aspects of plant sex chromosomes
Although cytogenetic techniques allowed the identification of heteromorphic sex chromosomes in numerous plant species (for a review, see Ming et al., 2011), their role in the sex determination mechanism was not immediately clear. Initially, phenotypic analyses of individuals with numerical chromosomal abnormalities were essential for deciphering the contribution of particular chromosomes to sex determination (Parker and Clark, 1991). Polyploid and aneuploid plants are key materials to elucidate whether sex is determined by an active Y(W) chromosome (as in humans) or by the X:autosome ratio (the system known from Drosophila). The active Y(W) chromosome has been observed in the majority of dioecious plants, such as Carica papaya (Hofmeyr and van Elden, 1942; Hofmeyr, 1944), Coccinia grandis (Kumar and Viseveshwaraiah, 1952), Silene latifolia (Ono, 1939; Warmke and Blakeslee, 1939; Westergaard, 1940), Silene otites (Warmke, 1942), and Populus tremula (Johnsson, 1940, 1942, 1945). Conversely, the X:autosome type that is characterized by no effect of Y(W) on sex determination seems to be infrequent in plants, as it was confirmed only in hop (genus Humulus; Neve, 1961; Shephard et al., 2000) and several sorrel species such as Rumex acetosa or Rumex hastatulus (Ono, 1928; Smith, 1963). Despite the fact that the studies of polyploids and/or aneuploids mostly explored the sex determination systems solely on the broad level of whole chromosomes, this classical methodological approach undoubtedly laid the cornerstone of plant sex chromosome research (Fig. 4A).
Deletion mutants represent another significant methodological step toward understanding the function of plant sex chromosomes (Fig. 4B). In his seminal experiments, Mogens Westergaard obtained S. latifolia individuals harbouring Y chromosome deletions (Westergaard, 1946a, b, 1948), some of which resulted in remarkable sexual phenotypes. Plants missing the distal part of the Y p-arm developed hermaphroditic flowers, whereas the absence of the proximal segment of the same arm led to the formation of asexual flowers. Based on these observations, Westergaard defined the gynoecium suppression factor (GSF) and the stamen-promoting factor (SPF). Westergaard’s results provided the first mechanistic evidence that two separate loci are involved in the establishment of dioecy. As such, the so-called ‘two-gene model’ for the evolution of dioecy was proposed based on these findings (Westergaard, 1953; Charlesworth and Charlesworth, 1978), and S. latifolia became a textbook example for explaining sex determination in plants.
The analyses of Y-linked deletions became a fundamental approach to studying the sex determination in S. latifolia (Donnison et al., 1996; Lardon et al., 1999; Lebel-Hardenack et al., 2002; Zluvova et al., 2007; Fujita et al., 2012; Kazama et al., 2016). The mutations were induced by X-irradiation, γ-irradiation, or heavy ion beam irradiation, all of which create deletions spanning relatively short genomic regions, allowing more precise mapping and marker identification (Fig. 4B). In addition, a thorough characterization of these deletion mutants was carried out including extensive cytogenetic and histological analyses, spatiotemporal gene expression profiling, and electron microscopy (Zluvova et al., 2006; Koizumi et al., 2010). By combining comprehensive phenotyping with physical mapping techniques, the detailed map of the S. latifolia Y chromosome was constructed (Kazama et al., 2016) and, recently, comparative genomics using wild-type plants and deletion mutants led to the identification of candidate genes for sex determination (Kazama et al., 2022; Akagi et al., 2023, Preprint; Moraga et al., 2023, Preprint). Deletion mutants are a powerful tool for the identification of sex-determining genes especially in species with small non-recombining regions as described in Asparagus (Harkess et al., 2017, 2020).
Interestingly, in some cases it is possible to obtain Y chromosome-linked deletions caused by storage of pollen in inappropriate conditions. This phenomenon has been described in S. latifolia as one of the causes of hermaphroditism (gerontogony) (van Nigtevecht, 1966). It is possible to hypothesize that the non-recombining regions can be prone to various kinds of genetic damage. This phenomenon has not yet been studied on a detailed level. There are even no proper data from irradiation experiments as the analyses were always focused on the plants showing aberrant phenotypes in the first generation which leads to a high prevalence of Y deletions in the further studied material, and the frequency of autosomal deletions with recessive phenotype remained unknown.
Some level of phenotypic instability is present in most plant sex-determining systems (Delph and Meagher, 1995). Hermaphrodites originating spontaneously enabled ascertainment of the type of heterogamety in some species (e.g. in S. dioica), already at the beginnings of genetics (Shull, 1911). In S. latifolia, androhermaphrodites were successfully induced by global genome demethylation and/or inhibiting histone deacetylation (Janousek et al., 1996; Bačovský et al., 2022). Likewise, (imperfect) stamen development can be activated in female plants using silver thiosulfate (Law et al., 2002) and Microbotryum infection (Strassburger, 1900; Uchida et al., 2003). A putative female suppressor gene was identified in dioecious buffalo grass (Bouteloua dactyloides) using pistil smut- (Salmacisia buchloëana) induced androhermaphrodites (Chandra and Huff, 2010). Both andro- and gynohermaphrodites represent remarkable tools for studying sex determination and sex-specific development, because, apart from deletion mutants, they contain complete genomes (Fig. 4D, E). For example, a process of stamen development can be studied in the XX background as well as gynoecium formation in individuals harbouring a Y chromosome (Law et al., 2002; Uchida et al., 2003; Kazama et al., 2005; Zemp et al., 2015; Bačovský et al., 2022) (Fig. 4).
A strategy so far not yet widely exploited for exploring the evolution of sex-determining systems from a functional point of view is the study of interspecific hybrids. The sex-linked genes are often subject to adaptive evolution (Zemp et al., 2018; Muyle et al., 2021) and their change can probably influence the rest of the genome (Zluvova et al., 2021). These changes can be visualized in interspecific hybrids between dioecious species and their hermaphrodite relatives; for example, similar histological phenotypes were observed in some deletion mutants in S. latifolia and in the interspecific hybrid between an S. latifolia female and S. viscosa male (Zluvova et al., 2005). The divergent evolution of the genes related to sexual dimorphism can be observed even in closely related species (Demuth et al., 2014; Baena-Díaz et al., 2019; Liu et al., 2020). The divergent gene evolution in very closely related dioecious species can be revealed on a phenotypic level in subsequent generations of brother×sister mating after interspecific crosses (Winge, 1931). This process overcomes functional redundancy widely present in plant genomes and reveals complex interactions between genes in pathways involved in sex determination and sexual dimorphism. Combined phenotypical, genetic, and genomic analyses of recombinant inbred lines (for a review, see Pollard, 2012) of related dioecious species which so far have not been undertaken could shed new light on the complex evolution of sex chromosomes and the rest of the genomes of dioecious species from both a qualitative and quantitative point of view.
Reverse genetics tools for investigating the function of sex-linked genes
Identification of sex-linked genes with the aforementioned bioinformatic and NGS methods has opened the door to reverse genetics studies (Fig. 4C). However, none of the dioecious plants with sex chromosomes has become a broadly used model system in molecular biology. Therefore, easily accessible tools for solving complex questions related to sex chromosome function are still lacking. Candidate genes for sex determination have been described in still increasing number of dioecious plants including Diospyrus (Akagi et al., 2014, 2016), Asparagus (Harkess et al., 2017, 2020), date palm (Torres et al., 2018), poplar (Müller et al., 2020; Xue et al., 2020), willow (Sanderson et al., 2021; Hu et al., 2023), kiwifruit (Akagi et al., 2018, 2019), Silene (Kazama et al., 2022; Akagi et al., 2023, Preprint; Moraga et al., 2023, Preprint), and many others. Although CRISPR/Cas9 [clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 9] gene editing represents a very powerful tool in model organisms, the functional evaluation of putative plant sex determination genes has been accomplished so far only in poplar (Müller et al., 2020). Most candidate genes were evaluated by heterologous expression in either Arabidopsis or tobacco (Akagi et al., 2014, 2018, 2019; Kazama et al., 2022). In S. latifolia, a combination of virus-induced gene silencing (Akagi et al., 2023, Preprint) and shoot apical meristem treatment with synthetic peptides. Kazama et al. (2022) suggested the role of the CLAVATA3 gene in gynoecium suppression. Interestingly, these studies showed that the divergence of sex chromosomes led to the loss of function in the X-linked copy whereas the Y-copy of CLAVATA3 remained conserved and fully functional. Though the treatment with synthetic peptides did not lead to complete organ suppression in females, it is a suitable tool for models in which genome engineering is not possible, inefficient, or time-consuming, such as S. latifolia (Hudzieczek et al., 2019). With the increasing number of new tools in functional genetics, more biological phenomena associated with plant sex chromosomes such as sexual antagonism, dosage compensation, or sexual dimorphism will be investigated from a functional perspective. Such areas can be examined from a new angle, offering valuable insights into their complex mechanisms and evolution.
Conclusion
This review highlights methodological approaches that are adjusted, utilized, or entirely developed de novo for the purpose of sex chromosome analyses in plants. Investigation of plant sex chromosomes often requires adapting the current methods and optimizing their use for dioecious species as in the case of CRISPR/Cas9 technology mentioned above (Fig. 4C). As more sophisticated genetic engineering tools are still emerging (Capdeville et al., 2023) and are being adapted to non-model species (Lee and Wang, 2023; Yan et al., 2023), we will probably witness extensive functional genetic studies of plant sex chromosomes in the near future. Newly developed NGS techniques and bioinformatic workflows are invaluable for genome comparative analysis, transcription profiling, and epigenomic studies regarding sex chromosome evolution. It can be anticipated that the integration of diverse methods from various disciplines, as recently elucidated in the genus Cucumis (Pichot et al., 2022), will provide a more comprehensive understanding of chromatin regulation and sex chromosome compartmentalization within the genome of studied organisms, thereby allowing for a careful rediscovery and revision of important biological questions regarding the origin of sex chromosomes.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Repeat DNA detection approaches used for dioecious plant genome assemblies.
Table S2. Tools for sex chromosome and sex determination analysis.
Contributor Information
Roman Hobza, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Václav Bačovský, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Radim Čegan, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Lucie Horáková, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic; Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.
Marcel Hubinský, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic; Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.
Tomáš Janíček, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic; Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.
Bohuslav Janoušek, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Pavel Jedlička, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Jana Kružlicová, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic; Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.
Zdeněk Kubát, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
José Luis Rodríguez Lorenzo, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Pavla Novotná, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic; Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.
Vojtěch Hudzieczek, Department of Plant Developmental Genetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Kralovopolska 135, 612 00 Brno, Czech Republic.
Martin Janda, University of South Bohemia in České Budějovice, Czech Republic.
Author contributions
RH: creating and structured the main concept and writing the first draft of the manuscript. All authors contributed equally to manuscript writing and revision, and read and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This research was funded by the Czech Science Foundation (grant awards 22-00301S, 22-00364S). The work was further supported from the project TowArds Next GENeration Crops, reg. no. CZ.02.01.01/00/22_008/0004581 of the ERDF Programme Johannes Amos Comenius.
References
- Akagi T, Fujita N, Masuda K, et al. 2023. Rapid and dynamic evolution of a giant Y chromosome in Silene latifolia. BioRxiv. doi: 10.1101/2023.09.21.558759. [Preprint]. [DOI] [Google Scholar]
- Akagi T, Henry IM, Kawai T, Comai L, Tao R.. 2016. Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. The Plant Cell 28, 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R.. 2018. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. The Plant Cell 30, 780–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L.. 2014. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. [DOI] [PubMed] [Google Scholar]
- Akagi T, Pilkington SM, Varkonyi-Gasic E, et al. 2019. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nature Plants 5, 801–809. [DOI] [PubMed] [Google Scholar]
- Bačovský V, Čegan R, Šimoníková D, Hřibová E, Hobza R.. 2020. The formation of sex chromosomes in Silene latifolia and S. dioica was accompanied by multiple chromosomal rearrangements. Frontiers in Plant Science 11, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bačovský V, Hobza R, Vyskot B.. 2018. Technical review: cytogenetic tools for studying mitotic chromosomes. Methods in Molecular Bology 1675, 509–535. [DOI] [PubMed] [Google Scholar]
- Bačovský V, Houben A, Kumke K, Hobza R.. 2019. The distribution of epigenetic histone marks differs between the X and Y chromosomes in Silene latifolia. Planta 250, 487–494. [DOI] [PubMed] [Google Scholar]
- Bačovský V, Janíček T, Hobza R.. 2022. The sister chromatid division of the heteromorphic sex chromosomes in Silene species and their transmissibility towards the mitosis. International Journal of Molecular Sciences 23, 2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouin H, Velt A, Gindraud F, et al. 2020. The wild grape genome sequence provides insights into the transition from dioecy to hermaphroditism during grape domestication. Genome Biology 21, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Díaz F, Zemp N, Widmer A.. 2019. Insights into the genetic architecture of sexual dimorphism from an interspecific cross between two diverging Silene (Caryophyllaceae) species. Molecular Ecology 28, 5052–5067. [DOI] [PubMed] [Google Scholar]
- Balounova V, Gogela R, Cegan R, et al. 2019. Evolution of sex determination and heterogamety changes in section Otites of the genus Silene. Scientific Reports 9, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi G, Antonovics J, Biere A, et al. 2009. Silene as a model system in ecology and evolution. Heredity 103, 5–14. [DOI] [PubMed] [Google Scholar]
- Bitton A, Sambrano J, Valentino S, Houston JP.. 2021. A review of new high-throughput methods designed for fluorescence lifetime sensing from cells and tissues. Frontiers in Physics 9, 648553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn KB. 1923. Sex chromosomes in plants. Nature 112, 687–688. [Google Scholar]
- Bräutigam K, Soolanayakanahally R, Champigny M, Mansfield S, Douglas C, Campbell MM, Cronk Q.. 2017. Sexual epigenetics: gender-specific methylation of a gene in the sex determining region of Populus balsamifera. Scientific Reports 7, 45388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdeville N, Schindele P, Puchta H.. 2023. Getting better all the time—recent progress in the development of CRISPR/Cas-based tools for plant genome engineering. Current Opinion in Biotechnology 79, 102854. [DOI] [PubMed] [Google Scholar]
- Cegan R, Marais GA, Kubekova H, Blavet N, Widmer A, Vyskot B, Dolezel J, Safár J, Hobza R.. 2010. Structure and evolution of Apetala3, a sex-linked gene in Silene latifolia. BMC Plant Biology 10, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegan R, Vyskot B, Kejnovsky E, Kubat Z, Blavet H, Šafář J, Doležel J, Blavet N, Hobza R.. 2012. Genomic diversity in two related plant species with and without sex chromosomes—Silene latifolia and S. vulgaris. PLoS One 7, e31898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Kubat Z, Hobza R, Koblizkova A, Widmer A, Macas J, Vyskot B, Kejnovsky E.. 2008. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Research 16, 961–976. [DOI] [PubMed] [Google Scholar]
- Chandra A, Huff DR.. 2010. A fungal parasite regulates a putative female-suppressor gene homologous to maize tasselseed2 and causes induced hermaphroditism in male buffalograss. Molecular Plant-Microbe Interactions 23, 239–250. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1978. A model for the evolution of dioecy and gynodioecy. The American Naturalist 112, 975–997. [Google Scholar]
- Charlesworth B, Charlesworth D.. 2020. Evolution: a new idea about the degeneration of Y and W chromosomes. Current Biology 30, R871–R873. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Shu X, Song C-X.. 2023. Mapping epigenetic modifications by sequencing technologies. Cell Death & Differentiation 10.1038/s41418-023-01213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph LF, Meagher TR.. 1995. Sexual dimorphism masks life history trade-offs in the dioecious plant Silene latifolia. Ecology 76, 775–785. [Google Scholar]
- Demuth JP, Flanagan RJ, Delph LF.. 2014. Genetic architecture of isolation between two species of Silene with sex chromosomes and Haldane’s rule. Evolution 68, 332–342. [DOI] [PubMed] [Google Scholar]
- Donnison IS, Siroky J, Vyskot B, Saedler H, Grant SR.. 1996. Isolation of Y chromosome-specific sequences from Silene latifolia and mapping of male sex-determining genes using representational difference analysis. Genetics 144, 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron R, Pan Q, Wen M, et al. 2021. RADSex: a computational workflow to study sex determination using restriction site-associated DNA sequencing data. Molecular Ecology Resources 21, 1715–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Howell EC, Groutides C, Armstrong SJ.. 2009. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 181, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flasch DA, Macia A, Sánchez L, Ljungman M, Heras SR, García-Pérez JL, Wilson TE, Moran JV.. 2019. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177, 837–851.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchard C, Badouin H, Latrasse D, Devani RS, Muyle A, Rhoné B, Renner SS, Banerjee AK, Bendahmane A, Marais GAB.. 2020. Evidence for dosage compensation in Coccinia grandis, a plant with a highly heteromorphic XY system. Genes 11, 787–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Torii C, Ishii K, Aonuma W, Shimizu Y, Kazama Y, Abe T, Kawano S.. 2012. Narrowing down the mapping of plant sex-determination regions using new Y-chromosome-specific markers and heavy-ion beam irradiation-induced Y-deletion mutants in Silene latifolia. G3 2, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska-Joachimiak A, Kula A, Książczyk T, Chojnicka J, Sliwinska E, Joachimiak AJ.. 2015. Chromosome landmarks and autosome–sex chromosome translocations in Rumex hastatulus, a plant with XX/XY1Y2 sex chromosome system. Chromosome Research 23, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Wright A, Garroway CJ, Docker MF.. 2022. SexFindR: a computational workflow to identify young and old sex chromosomes. BioRxiv doi: 10.1101/2022.02.21.481346. [Preprint]. [DOI] [Google Scholar]
- Hall AB, Qi Y, Timoshevskiy V, Sharakhova MV, Sharakhov IV, Tu Z.. 2013. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics 14, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Huang K, van der Hulst R, Tissen B, Caplan JL, Koppula A, Batish M, Meyers BC, Leebens-Mack J.. 2020. Sex determination by two Y-linked genes in garden asparagus. The Plant Cell 32, 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, et al. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nature Communications 8, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Jia KH, Zhang RG, et al. 2021. Chromosome-scale assembly of the genome of Salix dunnii reveals a male-heterogametic sex determination system on chromosome 7. Molecular Ecology Resources 21, 1966–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Cegan R, Jesionek W, Kejnovsky E, Vyskot B, Kubat Z.. 2017. Impact of repetitive elements on the Y chromosome formation in plants. Genes 8, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Hrusakova P, Safar J, Bartos J, Janousek B, Zluvova J, Michu E, Dolezel J, Vyskot B.. 2006. MK17, a specific marker closely linked to the gynoecium suppression region on the Y chromosome in Silene latifolia. Theoretical and Applied Genetics 113, 280–287. [DOI] [PubMed] [Google Scholar]
- Hobza R, Hudzieczek V, Kubat Z, Cegan R, Vyskot B, Kejnovsky E, Janousek B.. 2018. Sex and the flower—developmental aspects of sex chromosome evolution. Annals of Botany 122, 1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E.. 2015. Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Research 23, 561–570. [DOI] [PubMed] [Google Scholar]
- Hobza R, Lengerova M, Cernohorska H, Rubes J, Vyskot B.. 2004. FAST-FISH with laser beam microdissected DOP-PCR probe distinguishes the sex chromosomes of Silene latifolia. Chromosome Research 12, 245–250. [DOI] [PubMed] [Google Scholar]
- Hobza R, Vyskot B.. 2007. Laser microdissection-based analysis of plant sex chromosomes. Methods in Cell Biology 82, 433–453. [DOI] [PubMed] [Google Scholar]
- Hofmeyr JDJ. 1944. Further studies of tetraploidy in Carica papaya, L. South African Journal of Science 41, 225–230. [Google Scholar]
- Hofmeyr L, van Elden. 1942. Tetraploidy in Carica papaya L. induced by colchicine. South African Journal of Science 38, 181–185. [Google Scholar]
- Hooper DM, Griffith SC, Price TD.. 2019. Sex chromosome inversions enforce reproductive isolation across an avian hybrid zone. Molecular Ecology 28, 1246–1262. [DOI] [PubMed] [Google Scholar]
- Hu N, Sanderson BJ, Guo M, et al. 2023. Evolution of a ZW sex chromosome system in willows. Nature Communications 14, 7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudzieczek V, Cegan R, Cermak T, Bacovska N, Machalkova Z, Dolezal K, Plihalova L, Voytas D, Hobza R, Vyskot B.. 2019. Agrobacterium rhizogenes-mediated transformation of a dioecious plant model Silene latifolia. New Biotechnology 48, 20–28. [DOI] [PubMed] [Google Scholar]
- Janousek B, Gogela R, Bacovsky V, Renner SS.. 2022. The evolution of huge Y chromosomes in Coccinia grandis and its sister, Coccinia schimperi. Philosophical Transactions of the Royal Society B: Biological Sciences 377, 20210294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janousek B, Siroký J, Vyskot B.. 1996. Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Molecular & General Genetics 250, 483–490. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Lexa M, Vanat I, Hobza R, Kejnovsky E.. 2019. Nested plant LTR retrotransposons target specific regions of other elements, while all LTR retrotransposons often target palindromes and nucleosome-occupied regions: in silico study. Mobile DNA 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesionek W, Bodláková M, Kubát Z, Čegan R, Vyskot B, Vrána J, Šafář J, Puterova J, Hobza R.. 2021. Fundamentally different repetitive element composition of sex chromosomes in Rumex acetosa. Annals of Botany 127, 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. 2019. Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Research 27, 153–165. [DOI] [PubMed] [Google Scholar]
- Jiang J, Gill BS.. 2006. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Johnsson H. 1940. Cytological studies of diploid and triploid Populus tremula and of crosses between them. Hereditas 26, 321–352. [Google Scholar]
- Johnsson H. 1942. Cytological studies of triploid progenies of Populus tremula. Hereditas 28, 306–312. [Google Scholar]
- Johnsson H. 1945. Chromosome numbers of the progeny from the cross triploid × tetraploid Populus tremula. Hereditas 31, 500. [PubMed] [Google Scholar]
- Käfer J, Bewick A, Andres-Robin A, et al. 2022. A derived ZW chromosome system in Amborella trichopoda, representing the sister lineage to all other extant flowering plants. New Phytologist 233, 1636–1642. [DOI] [PubMed] [Google Scholar]
- Käfer J, Lartillot N, Marais GAB, Picard F.. 2021. Detecting sex-linked genes using genotyped individuals sampled in natural populations. Genetics 218, iyab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka A, Starczak M, Gackowski D, Stępień E, Achrem M.. 2023. Global DNA 5-hydroxymethylcytosine level and its chromosomal distribution in four rye species. Journal of Experimental Botany 74, 3488–3502. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Ishii K, Aonuma W, et al. 2016. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Scientific Reports 6, 18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y, Kitoh M, Kobayashi T, Ishii K, Krasovec M, Yasui Y, Abe T, Kawano S, Filatov DA.. 2022. A CLAVATA3-like gene acts as a gynoecium suppression function in white campion. Molecular Biology and Evolution 39, msac195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y, Koizumi A, Uchida W, Ageez A, Kawano S.. 2005. Expression of the floral B-function gene SLM2 in female flowers of Silene latifolia infected with the smut fungus Microbotryum violaceum. Plant and Cell Physiology 46, 806–811. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. 2017. The evolution of genome structure by natural and sexual selection. Journal of Heredity 108, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi A, Yamanaka K, Nishihara K, Kazama Y, Abe T, Kawano S.. 2010. Two separate pathways including SlCLV1, SlSTM and SlCUC that control carpel development in a bisexual mutant of Silene latifolia. Plant and Cell Physiology 51, 282–293. [DOI] [PubMed] [Google Scholar]
- Kralova T, Cegan R, Kubat Z, Vrana J, Vyskot B, Vogel I, Kejnovsky E, Hobza R.. 2014. Identification of a novel retrotransposon with sex chromosome-specific distribution in Silene latifolia. Cytogenetic and Genome Research 143, 87–95. [DOI] [PubMed] [Google Scholar]
- Kreplak J, Madoui M-A, Cápal P, et al. 2019. A reference genome for pea provides insight into legume genome evolution. Nature Genetics 51, 1411–1422. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Hobza R, Vyskot B, Kejnovsky E.. 2008. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 51, 350–356. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Zluvova J, Vogel I, Kovacova V, Cermak T, Cegan R, Hobza R, Vyskot B, Kejnovsky E.. 2014. Possible mechanisms responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytologist 202, 662–678. [DOI] [PubMed] [Google Scholar]
- Kumar LSS, Viseveshwaraiah S.. 1952. Sex mechanism in Coccinia indica Wight and Arn. Nature 170, 330–331. [DOI] [PubMed] [Google Scholar]
- Lardon A, Georgiev S, Aghmir A, Le Merrer G, Negrutiu I.. 1999. Sexual dimorphism in white campion: complex control of carpel number is revealed by Y chromosome deletions. Genetics 151, 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Rodriguez-Granados NY, Veluchamy A, et al. 2017. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics & Chromatin 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law TF, Lebel-Hardenack S, Grant SR.. 2002. Silver enhances stamen development in female white campion (Silene latifolia [Caryophyllaceae]). American Journal of Botany 89, 1014–1020. [DOI] [PubMed] [Google Scholar]
- Lebel-Hardenack S, Hauser E, Law TF, Schmid J, Grant SR.. 2002. Mapping of sex determination loci on the white campion (Silene latifolia) Y chromosome using amplified fragment length polymorphism. Genetics 160, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Wang K.. 2023. Strategies for genotype-flexible plant transformation. Current Opinion in Biotechnology 79, 102848. [DOI] [PubMed] [Google Scholar]
- Lengerova M, Vyskot B.. 2001. Sex chromatin and nucleolar analyses in Rumex acetosa L. Protoplasma 217, 147–153. [DOI] [PubMed] [Google Scholar]
- Lexa M, Jedlicka P, Vanat I, Cervenansky M, Kejnovsky E.. 2020. TE-greedy-nester: structure-based detection of LTR retrotransposons and their nesting. Bioinformatics 36, 4991–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhou J, Zhang W, et al. 2023. A rapid method for assembly of single chromosome and identification of sex determination region based on single-chromosome sequencing. New Phytologist 240, 892–903. [DOI] [PubMed] [Google Scholar]
- Liu X, Glémin S, Karrenberg S.. 2020. Evolution of putative barrier loci at an intermediate stage of speciation with gene flow in campions (Silene). Molecular Ecology 29, 3511–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti B, Manzano S, Kejnovský E, Vyskot B, Jamilena M.. 2008. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Molecular Genetics and Genomics 281, 249–259. [DOI] [PubMed] [Google Scholar]
- Martin H, Carpentier F, Gallina S, Gode C, Schmitt E, Muyle A, Marais GAB, Touzet P.. 2019. Evolution of young sex chromosomes in two dioecious sister plant species with distinct sex determination systems. Genome Biology and Evolution 11, 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martis MM, Zhou R, Haseneyer G, et al. 2013. Reticulate evolution of the rye genome. The Plant Cell 25, 3685–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalovova M, Kubat Z, Hobza R, Vyskot B, Kejnovsky E.. 2015. Fully automated pipeline for detection of sex linked genes using RNA-Seq data. BMC Bioinformatics 16, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner SS.. 2011. Sex chromosomes in land plants. Annual Review of Plant Biology 62, 485–514. [DOI] [PubMed] [Google Scholar]
- Misra BB, Assmann SM, Chen S.. 2014. Plant single-cell and single-cell-type metabolomics. Trends in Plant Science 19, 637–646. [DOI] [PubMed] [Google Scholar]
- Mondo JM, Agre PA, Asiedu R, Akoroda MO, Asfaw A.. 2021. Genome-wide association studies for sex determination and cross-compatibility in water yam (Dioscorea alata L.). Plants 10, 1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JS, Sadeque A, Giacomini DA, Brown PJ, Tranel PJ.. 2019. Sex-specific markers for waterhemp (Amaranthus tuberculatus) and Palmer amaranth (Amaranthus palmeri). Weed Science 67, 412–418. [Google Scholar]
- Moraga C, Branco C, Rougemont Q, et al. 2023. The Silene latifolia genome and its giant Y chromosome. BioRxiv doi: 10.1101/2023.09.21.558754. [Preprint]. [DOI] [Google Scholar]
- Müller NA, Kersten B, Leite Montalvão AP, et al. 2020. A single gene underlies the dynamic evolution of poplar sex determination. Nature Plants 6, 630–637. [DOI] [PubMed] [Google Scholar]
- Muyle A, Käfer J, Zemp N, Mousset S, Picard F, Marais GA.. 2016. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biology and Evolution 8, 2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Marais GAB, Bačovský V, Hobza R, Lenormand T.. 2022. Dosage compensation evolution in plants: theories, controversies and mechanisms. Philosophical Transactions of the Royal Society B: Biological Sciences 377, 20210222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Martin H, Zemp N, et al. 2021. Dioecy is associated with high genetic diversity and adaptation rates in the plant genus Silene. Molecular Biology and Evolution 38, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Shearn R, Marais GA.. 2017. The evolution of sex chromosomes and dosage compensation in plants. Genome Biology and Evolution 9, 627–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Zemp N, Fruchard C, et al. 2018. Genomic imprinting mediates dosage compensation in a young plant XY system. Nature Plants 4, 677–680. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Quesada del Bosque ME, Garrido-Ramos MA.. 2009. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Molecular Genetics and Genomics 282, 395–406. [DOI] [PubMed] [Google Scholar]
- Neumann P, Novák P, Hoštáková N, Macas J.. 2019. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R. 1961. Sex determination in the cultivated hop, Humulus lupulus. PhD thesis, University of London (Wye College).
- Novák P, Neumann P, Macas J.. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J.. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29, 792–793. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, O’Neill RJ.. 2018. Sex chromosome repeats tip the balance towards speciation. Molecular Ecology 27, 3783–3798. [DOI] [PubMed] [Google Scholar]
- Ono T. 1928. Further investigations on the cytology of Rumex. IV. Botanical Magazine Tokyo 42, 524–533. [Google Scholar]
- Ono T. 1939. Polyploidy and sex determination in Melandrium. Shokubutsugaku Zasshi 53, 549–556. [Google Scholar]
- Onodera Y, Yonaha I, Niikura S, Yamazaki S, Mikami T.. 2008. Monoecy and gynomonoecy in Spinacia oleracea L.: morphological and genetic analyses. Scientia Horticulturae 118, 266–269. [Google Scholar]
- Ou S, Su W, Liao Y, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biology 20, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouadi S, Sierro N, Kessler F, Ivanov NV.. 2023. Chromosome-scale assemblies of S. malaccense, S. aqueum, S. jambos, and S. syzygioides provide insights into the evolution of Syzygium genomes. Frontiers in Plant Science 14, 1248780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Buell CR.. 2004. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Research 32, D360–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Clark MS.. 1991. Dosage sex-chromosome systems in plants. Plant Science 80, 79–92. [Google Scholar]
- Pegoraro G, Misteli T.. 2017. High-throughput imaging for the discovery of cellular mechanisms of disease. Trends in Genetics 33, 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichot C, Djari A, Tran J, et al. 2022. Cantaloupe melon genome reveals 3D chromatin features and structural relationship with the ancestral cucurbitaceae karyotype. iScience 25, 103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plongthongkum N, Diep DH, Zhang K.. 2014. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nature Reviews. Genetics 15, 647–661. [DOI] [PubMed] [Google Scholar]
- Pollard DA. 2012. Design and construction of recombinant inbred lines. Methods in Molecular Biology 871, 31–39. [DOI] [PubMed] [Google Scholar]
- Požárková D, Koblížková A, Román B, Torres AM, Lucretti S, Lysák M, Doležel J, Macas J.. 2002. Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biologia Plantarum 45, 337–345. [Google Scholar]
- Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, Käfer J, Karlov G, Marais GAB.. 2020. An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Research 30, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Kubat Z, Kejnovsky E, Jesionek W, Cizkova J, Vyskot B, Hobza R.. 2018. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Razumova O, Martinek T, Alexandrov O, Divashuk M, Kubat Z, Hobza R, Karlov G, Kejnovsky E.. 2017. Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biology and Evolution 9, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangavittal S, Stopa N, Tomaszkiewicz M, Sahlin K, Makova KD, Medvedev P.. 2019. DiscoverY: a classifier for identifying Y chromosome sequences in male assemblies. BMC Genomics 20, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Nurk S, Cechova M, et al. 2023. The complete sequence of a human Y chromosome. Nature 621, 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Vales MI, Kianian SF.. 2008. Radiation hybrid (RH) and HAPPY mapping in plants. Cytogenetic and Genome Research 120, 233–240. [DOI] [PubMed] [Google Scholar]
- Rodríguez Lorenzo JL, Hubinský M, Vyskot B, Hobza R.. 2020. Histone post-translational modifications in Silene latifolia X and Y chromosomes suggest a mammal-like dosage compensation system. Plant Science 299, 110528. [DOI] [PubMed] [Google Scholar]
- Sacchi B, Humphries Z, Kružlicová J, et al. 2023. Phased assembly of neo-sex chromosomes reveals extensive Y degeneration and rapid genome evolution in Rumex hastatulus. BioRxiv doi: 10.1101/2023.09.26.559509. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson BJ, Feng G, Hu N, et al. 2021. Sex determination through X–Y heterogamety in Salix nigra. Heredity 126, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharmann M, Grafe TU, Metali F, Widmer A.. 2019. Sex is determined by XY chromosomes across the radiation of dioecious Nepenthes pitcher plants. Evolution Letters 3, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shephard HL, Parker JS, Darby P, Ainsworth CC.. 2000. Sexual development and sex chromosomes in hop. New Phytologist 148, 397–411. [DOI] [PubMed] [Google Scholar]
- Shull GH. 1911. Reversible sex-mutants in Lychnis dioica. Botanical Gazette 52, 329–368. [Google Scholar]
- Sigeman H, Sinclair B, Hansson B.. 2022. Findzx: an automated pipeline for detecting and visualising sex chromosomes using whole-genome sequencing data. BMC Genomics 23, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroky J, Hodurkova J, Negrutiu I, Vyskot B.. 1999. Functional and structural chromosome analyses in autotetraploid Silene latifolia. Annals of Botany 84, 633–638. [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. 2013–2015. www.repeatmasker.org. [Google Scholar]
- Smith BW. 1963. The mechanism of sex determination in Rumex hastatulus. Genetics 48, 1265–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares FAF, Carvalho CR, Sattler MC, Silva JC, Pinto DEE, Passamani PZ, Silva AJ, Clarindo WR.. 2020. Plant chromosome-specific probes by microdissection of a single chromosome: is that a reality? Frontiers in Plant Science 11, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Bellot S, Fuchs J, Houben A, Renner SS.. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. The Plant Journal 88, 387–396. [DOI] [PubMed] [Google Scholar]
- Sousa A, Fuchs J, Renner SS.. 2017. Cytogenetic comparison of heteromorphic and homomorphic sex chromosomes in Coccinia (Cucurbitaceae) points to sex chromosome turnover. Chromosome Research 25, 191–200. [DOI] [PubMed] [Google Scholar]
- Sousa A, Schubert V, Renner SS.. 2021. Centromere organization and UU/V sex chromosome behavior in a liverwort. The Plant Journal 106, 133–141. [DOI] [PubMed] [Google Scholar]
- Steflova P, Tokan V, Vogel I, Lexa M, Macas J, Novak P, Hobza R, Vyskot B, Kejnovsky E.. 2013. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biology and Evolution 5, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburger E. 1900. Versuche mit diöcischen Pflanzen in Rücksicht auf Geschlechtsverteilung. Biologische Zentralblatt 20, 721–731. [Google Scholar]
- Talianová M, Žlůvová J, Hobza R, Vyskot B, Janoušek B.. 2012. Identification and characterization of a bacteria-like sequence in the genome of some Silene species. Biologia Plantarum 56, 247–253. [Google Scholar]
- Thind AK, Wicker T, Šimková H, Fossati D, Moullet O, Brabant C, Vrána J, Doležel J, Krattinger SG.. 2017. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nature Biotechnology 35, 793–796. [DOI] [PubMed] [Google Scholar]
- Torres MF, Mathew LS, Ahmed I, Al-Azwani IK, Krueger R, Rivera-Nuñez D, Mohamoud YA, Clark AG, Suhre K, Malek JA.. 2018. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nature Communications 9, 3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida W, Matsunaga S, Sugiyama R, Kazama Y, Kawano S.. 2003. Morphological development of anthers induced by the dimorphic smut fungus Microbotryum violaceum in female flowers of the dioecious plant Silene latifolia. Planta 218, 240–248. [DOI] [PubMed] [Google Scholar]
- VanBuren R, Zeng F, Chen C, et al. 2015. Origin and domestication of papaya Y(h) chromosome. Genome Research 25, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nigtevecht G. 1966. Genetic studies in dioecious Melandrium. II. Genetica 37, 281–306. [Google Scholar]
- Veltsos P. 2019. Not all sex-biased genes are the same. New Phytologist 221, 10–11. [DOI] [PubMed] [Google Scholar]
- Vyskot B, Hobza R.. 2015. The genomics of plant sex chromosomes. Plant Science 236, 126–135. [DOI] [PubMed] [Google Scholar]
- Wang X, Song S, Wu Y-S, et al. 2015. Genome-wide mapping of 5-hydroxymethylcytosine in three rice cultivars reveals its preferential localization in transcriptionally silent transposable element genes. Journal of Experimental Botany 66, 6651–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke HE. 1942. A new method for determining the sex heterozygote in species with morphologically undifferentiated sex chromosomes, and its application to Silene otites. Genetics 27, 174. [Google Scholar]
- Warmke HE, Blakeslee AF.. 1939. Sex mechanism in polyploids of Melandrium. Science 89, 391–392. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1940. Studies on cytology and sex determination in polyploid forms of Melandrium album. Dansk Botanisk Arkiv 10, 1–131. [Google Scholar]
- Westergaard M. 1946a. Structural changes of the Y chromosome in the offspring of polyploid Melandrium. Hereditas 32, 60–64. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1946b. Aberrant Y chromosomes and sex expression in Melandrium album. Hereditas 32, 419–443. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1948. The relation between chromosome constitution and sex in the off-spring of triploid Melandrium. Hereditas 34, 257–279. [Google Scholar]
- Westergaard M. 1953. Über den Mechanismus der Geschlechtsbestimmung bei Melandrium album. Naturwissenschaften 40, 253–260. [Google Scholar]
- Wicker T, Gundlach H, Spannagl M, et al. 2018. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biology 19, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge O. 1923. On sex chromosomes, sex determination and preponderance of females in some dioecious plants. Comptes-rendus des travaux du Laboratoire de Carlsberg 15, 1–16. [Google Scholar]
- Winge O. 1931. X- and Y-linked inheritance in Melandrium. Hereditas 15, 127–165. [Google Scholar]
- Xue L, Wu H, Chen Y, et al. 2020. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nature Communications 11, 5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovin NA, Divashuk MG, Razumova OV, Soloviev AA, Karlov GI.. 2014. Use of laser microdissection for the construction of Humulus japonicus Siebold et Zuccarini, 1846 (Cannabaceae) sex chromosome-specific DNA library and cytogenetics analysis. Comparative Cytogenetics 8, 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Hou Q, Wei X, Qi Y, Pu A, Wu S, An X, Wan X.. 2023. Promoting genotype-independent plant transformation by manipulating developmental regulatory genes and/or using nanoparticles. Plant Cell Reports 42, 1395–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Wang D, Li Y, et al. 2020. A general model to explain repeated turnovers of sex determination in the salicaceae. Molecular Biology and Evolution 38, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Xia K, Xu X.. 2023. Spatial transcriptomics drives a new era in plant research. The Plant Journal 116, 1571–1581. [DOI] [PubMed] [Google Scholar]
- Yue J, VanBuren R, Liu J, et al. 2022. SunUp and Sunset genomes revealed impact of particle bombardment mediated transformation and domestication history in papaya. Nature Genetics 54, 715–724. [DOI] [PubMed] [Google Scholar]
- Zemp N, Tavares R, Widmer A.. 2015. Fungal infection induces sex-specific transcriptional changes and alters sexual dimorphism in the dioecious plant Silene latifolia. PLoS Genetics 11, e1005536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp N, Widmer A, Charlesworth D.. 2018. Has adaptation occurred in males and females since separate sexes evolved in the plant Silene latifolia? Proceedings of the Royal Society B: Biological Sciences 285, 20172824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RG, Li GY, Wang XL, Dainat J, Wang ZX, Ou S, Ma Y.. 2022. TEsorter: an accurate and fast method to classify LTR-retrotransposons in plant genomes. Horticulture Research 9, uhac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Gehring M.. 2019. Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reproduction 32, 63–75. [DOI] [PubMed] [Google Scholar]
- Zluvova J, Georgiev S, Janousek B, Charlesworth D, Vyskot B, Negrutiu I.. 2007. Early events in the evolution of the Silene latifolia Y chromosome: male specialization and recombination arrest. Genetics 177, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J, Lengerova M, Markova M, Hobza R, Nicolas M, Vyskot B, Charlesworth D, Negrutiu I, Janousek B.. 2005. The inter-specific hybrid Silene latifolia × S. viscosa reveals early events of sex chromosome evolution. Evolution and Development 7, 327–336. [DOI] [PubMed] [Google Scholar]
- Zluvova J, Hobza R, Janoušek B.. 2021. Adaptation of the autosomal part of the genome on the presence of dioecy. Philosophical Transactions of the Royal Society B: Biological Sciences 377, 20210228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J, Nicolas M, Berger A, Negrutiu I, Monéger F.. 2006. Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Silene latifolia. Proceedings of the National Academy of Sciences, USA 103, 18854–18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.