Abstract
Background.
Each day, humans produce approximately 0.5 through 1.5 liters of saliva, a biofluid that is rich in biological omic constituents. Our lack of understanding how omic biomarkers migrate from diseased tissue to the saliva has impeded the clinical translation of saliva testing. Although such biomarkers must be conveyed via the vascular and lymphatic systems to the salivary glands, the molecular mechanisms that underlie this transport remain unclear. Although COVID-19 highlighted the need for rapid and reliable testing for infectious diseases, it represents only one of the many health conditions that potentially can be diagnosed using a saliva sample.
Types of Studies Reviewed.
The authors discuss salivaomics, saliva exosomics, and the mechanisms on which saliva diagnostics are based and introduce a novel electrochemical sensing technology that may be exploited for saliva liquid biopsy.
Results.
The utility of saliva for screening for lung cancer is under investigation. Saliva testing may be used to stratify patients, monitor treatment response, and detect disease recurrence. The authors also highlight the landscapes of saliva-based SARS-CoV-2 testing and ultrashort cell-free DNA and outline how these fields are likely to evolve in the near future.
Practical Implications.
Breakthroughs in the study of saliva research, therefore, will facilitate clinical deployment of saliva-based testing.
Keywords: Saliva diagnostics, salivaomics, saliva exosomics, liquid biopsy, biomarker
Historically, many diseases have been diagnosed via detection of biomarkers in the blood. However, drawing blood causes patient discomfort and is not feasible in all settings. Saliva diagnostics could offer a less invasive and more reproducible alternative.1 Furthermore, routine saliva testing of at-risk patients may reduce cancer-associated mortality. For example, most head and neck cancers are only diagnosed via biopsy after the appearance of symptoms or a lump. The malignancy is generally advanced at this point, and this is associated with poor prognosis. Presymptomatic saliva testing would facilitate earlier treatment for patients with head and neck cancer and thereby improve patient outcomes. In support of this approach, a saliva-based molecular test was able to detect oral cancer earlier, and this led to improved prognosis.2
However, the lack of basic mechanistic understanding of how disease-related biomarkers reach the saliva still limits the translation of saliva diagnostics from bench to bedside. However, concerted efforts from the academic community and from the National Institutes of Health (NIH) are beginning to provide mechanistic insight into the systemic transport of biomarkers from peripheral tissues to the oral compartment. For example, a US-based consortium has generated a comprehensive catalog of the salivary proteome of healthy people, revealing that 20% through 30% of the salivary proteome overlaps with the plasma proteome.3–5 This observation, together with the close physical proximity of saliva and blood, suggests that many salivary constituents are derived from the blood. Furthermore, the NIH-led Rapid Accelerated Diagnostic (RADx) initiative has accelerated basic research into saliva diagnostics to provide rapid, accurate, and reproducible COVID testing via this method.6 This has shone the spotlight on this novel area of diagnostics and may help pave the way for similar tests to detect diseases driven by other pathogens or etiologies. Spurred on by such investments, the academic and industry sectors are working on 3 core elements of saliva-based diagnostics: salivaomics, saliva exosomics, and saliva liquid biopsy.
SALIVAOMICS
Salivaomics is defined as the integrative study of saliva and its constituents and functions, using omics technologies.7,8 High throughput methodologies enable researchers to identify DNA-, RNA-, protein-, and metabolite-based biomarkers via omic studies; they also facilitate the study of salivary microbiomes. In a field only a decade old, researchers have identified and are working through the various technical difficulties that have become apparent. For example, it is challenging to identify subpopulations of clinically relevant biomarkers from the analysis of bulk saliva samples. In addition, other elements in the oral cavity can add to the biological noise in saliva samples. These include food and oral bacteria, as well as whether the sample is taken during rest or shortly after eating, when the process of mastication can alter the composition of saliva.9 Even after all the above variables are controlled for, it is critical to prevent degradation of molecules in the saliva to maximize the opportunity for biomarker discovery.10 Protease inhibitors are required to block amylase-dependent degradation of protein biomarkers, whereas ribonuclease inhibitors must be added to ensure transcriptomic analyses are not compromised.11,12
With regard to this latter point, it is encouraging to note the progress of the Extracellular RNA Communication Consortium (ERCC), another NIH initiative, designed to profile the extracellular RNA (exRNA) from all bodily fluids, including saliva.13 Figure 1 shows that saliva exRNAs account for 2.5% (256/9,892) of all the RNA-seq data deposited in the exRNA Atlas (https://exrna-atlas.org).14 These data provide further proof of concept that saliva biopsies can be exploited for biomarker analysis.
Figure 1.
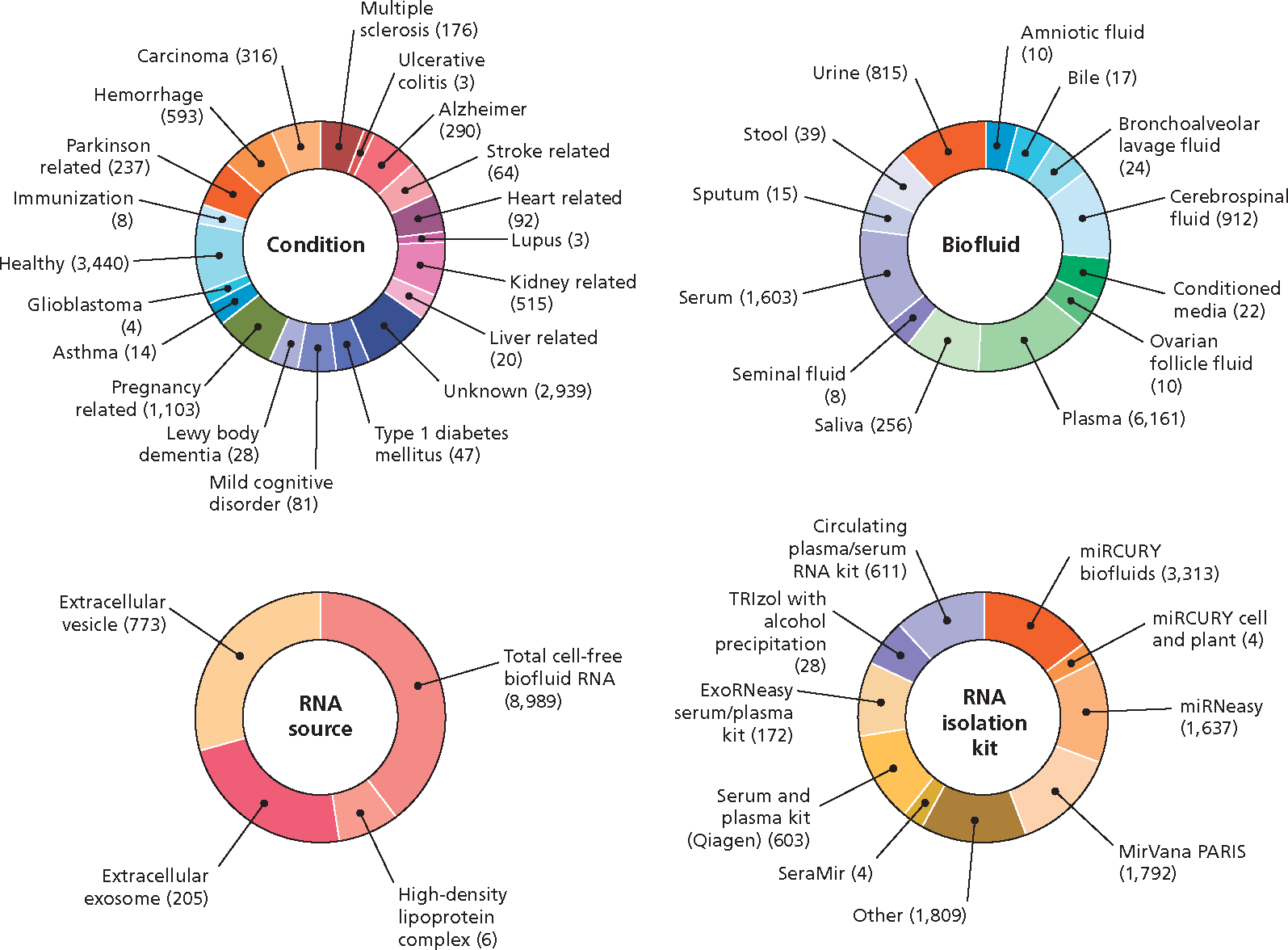
Extracellular RNA (exRNA) Atlas. exRNA profiles are derived from various biofluids, including saliva. Small RNA sequencing assays are used to profile various disease conditions from diverse RNA sources and via different isolation methods. All the RNA-seq data, including saliva exRNAs, were extracted from the results deposited in the exRNA Atlas (https://exrna-atlas.org). Manufacturer information was not available in the database except for the serum and plasma kit.
SALIVA EXOSOMICS
Saliva exosomics is an emerging subfield of salivaomics research; its focus is the analysis of the molecular cargos carried by small (30–100 nm diameter) membrane-enclosed extracellular structures called exosomes.15 Originating in the endosomal pathway and secreted by almost all cell types, exosomes move through the vasculature to distal sites, including the salivary glands. In addition, exosomes are found in other body fluids such as blood, urine, breast milk, and cerebrospinal fluids.16 Exosomes can modulate intercellular signaling and cellular homeostasis via transporting information-rich molecular cargos between cells of origin and other tissues.17 As they constitute a specific subpopulation of salivary fluid, they are essentially naturally enriched sources of biomarker information that are free from many of the other salivary contaminants listed above (Figure 2).18 In addition, the lipid bilayer of exosomes acts as a natural barrier to proteases and ribonucleases that would otherwise degrade their contents. This makes exosomes an attractive source of material for diagnostic procedures.19 Exosomes have a well-documented role in cancer biology, and we therefore look forward to future studies that explore the link between tumor cells and tumor cell–derived exosomes that appear in the saliva. We infer that molecular profiling of both compartments will lead to the identification of biomarkers of tumor initiation, progression, and response to treatment.
Figure 2.
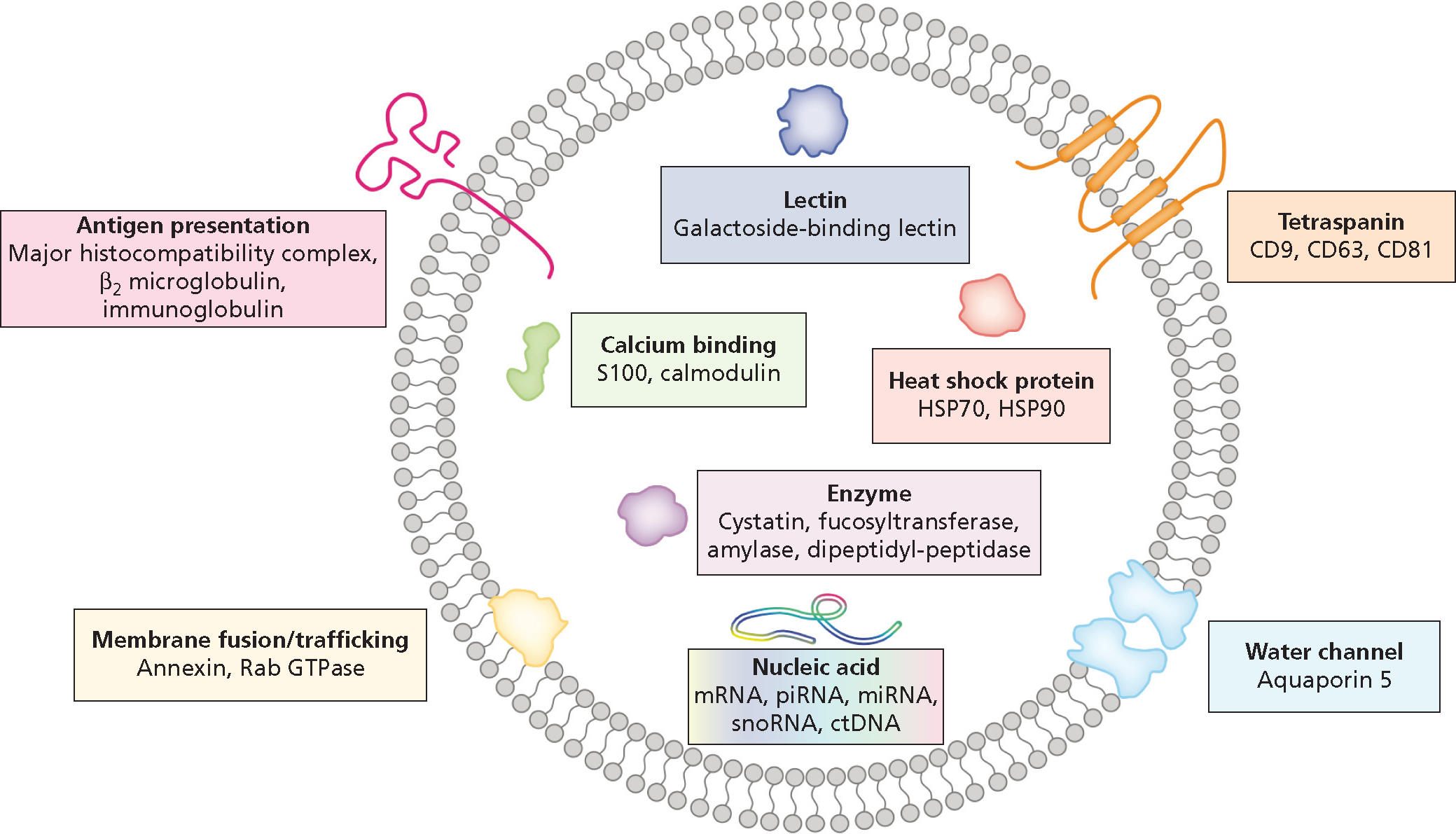
Structure and contents of typical salivary exosomes. The exosome is surrounded by a phospholipid bilayer, carrying many cell type–specific cargos. CD: Cluster of differentiation. ctDNA: Circulating tumor DNA. mRNA: Messenger RNA. miRNA: MicroRNA. piRNA: Piwi-interacting RNA. snoRNA: Small nucleolar RNA. Adapted with permission of the publisher from Nonaka and Wong.1
MECHANISTIC LINK BETWEEN SALIVARY EXOSOMES AND SYSTEMIC DISEASES
Cancer-derived exosomes now are known to be a rich source of information that reflects the genetic composition and status of their parent tumors. A murine orthotopic pancreatic cancer xenograft model has been used to provide proof of concept that salivary exosomes can be exploited for cancer detection.20 The introduction of a dominant negative Rab11 GTPase in the tumor cells prevented exosome biogenesis and removed tumor-derived messenger RNA signals from salivary samples. This clearly shows that cancer-derived messenger RNAs are the cargo of exosomes and likely reach the salivary gland via the circulation (Figure 3). In another study, human CD63-GFP-positive vesicles and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were isolated from the saliva of mice harboring human lung cancer xenografts.21 This supports the results from the orthotopic pancreatic cancer model and confirms the movement of vesicles from cancer cells to distal sites via the circulation.
Figure 3.
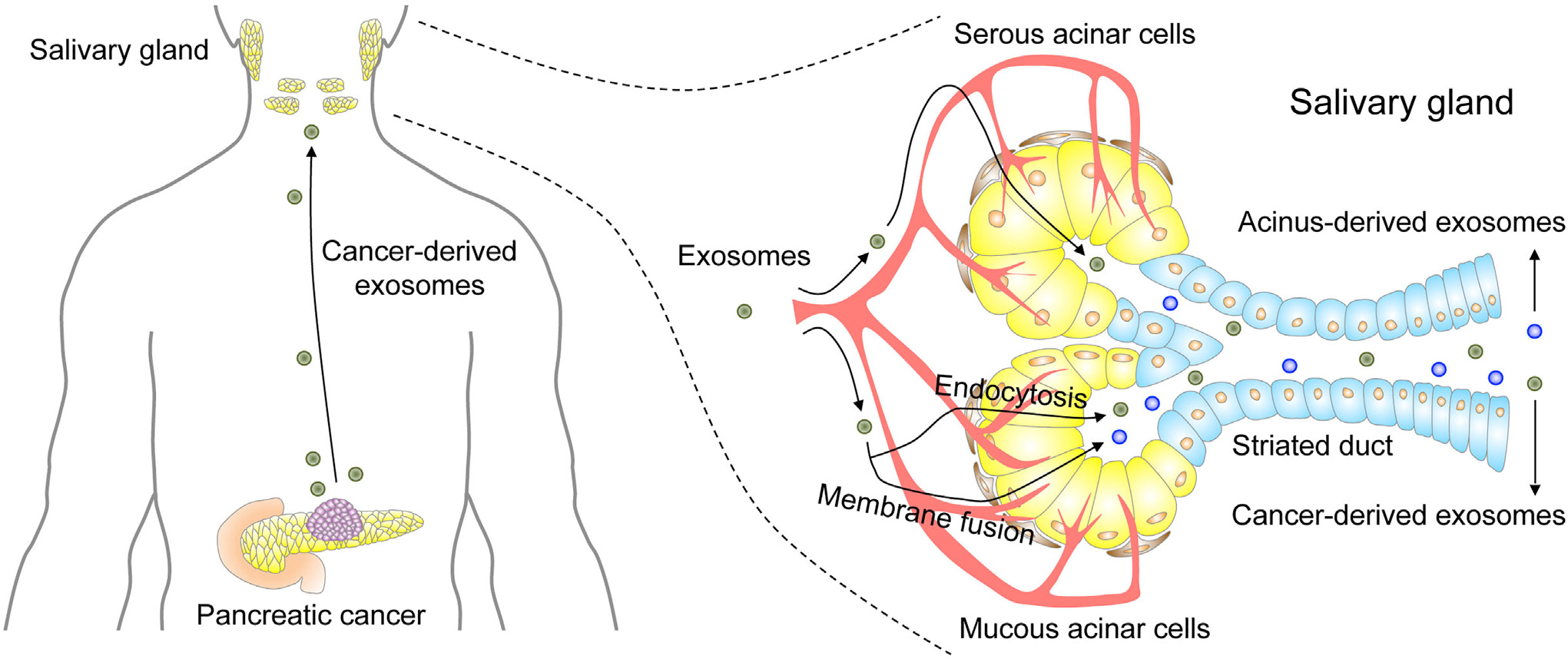
Mechanistic link between cancer and saliva. Cancer-derived exosomes enter the circulation and reach the salivary glands. Exosome uptake at salivary gland acinar cells occurs via endocytosis or membrane fusion. Two types of salivary exosomes are released into the saliva. Cancer-derived exosomes are released through exocytosis, and acinus-derived exosomes are released through fusion of multivesicular bodies with the plasma membrane. Both types of salivary exosomes carry cargos that include cancer-derived products. Adapted with permission of Elsevier from Nonaka and Wong.15
The pancreatic cancer model also has been used to show that the cancer cell–derived exosomes are biologically active. Transfer of saliva from tumor-bearing mice to control animals suppressed the expression of the activated natural killer cell markers CD69 and NKG2D, and dominant negative Rab11 GTPase reversed these effects.22
SALIVA LIQUID BIOPSY
Early detection of cancer can improve prognosis for patients significantly. Key to early detection is the identification of biomarkers, and this is facilitated via obtaining liquid biopsy samples that contain tumor-derived materials such as circulating tumor cell (CTC), circulating tumor DNA (ctDNA), and circulating extracellular vesicle (EV). ctDNA harbors somatic mutations that are pathognomonic of certain human tumors23; the identities of these ctDNAs can be used to guide the selection of targeted therapy, whereas the amount of ctDNA is proportional to disease burden. In the latter case, a major goal is to lower the limit of detection via improvements in technology, so that disease can be picked up as early as possible. In addition, the development of multicancer early detection (MCED) tests, which contain probes for drivers of diverse cancer types, will enhance the ability of clinicians to screen individual patients for more than 1 cancer simultaneously.24
Electric field–induced release and measurement (EFIRM) is an electrochemical-based platform for rapid, noninvasive detection of molecular targets in body fluids.25,26 At its heart lie cutting edge technologies including the design of nucleic acid probes that allow the detection of low-abundance targets (< 100 molecules) without prior extraction or amplification, specially modified electrodes that increase probe surface density and improve biocompatibility, and the use of an electric waveform to enhance the incubation process.27 These combined elements make EFIRM a powerful platform for the detection of multiomic biomarkers from bodily fluids.28,29 In terms of oncology applications, EFIRM has been used to detect oncogenic mutations in the biofluids of patients with cancer,30,31 and below, we use mutant EGFR as an example of how the technology works (Figure 4).
Figure 4.
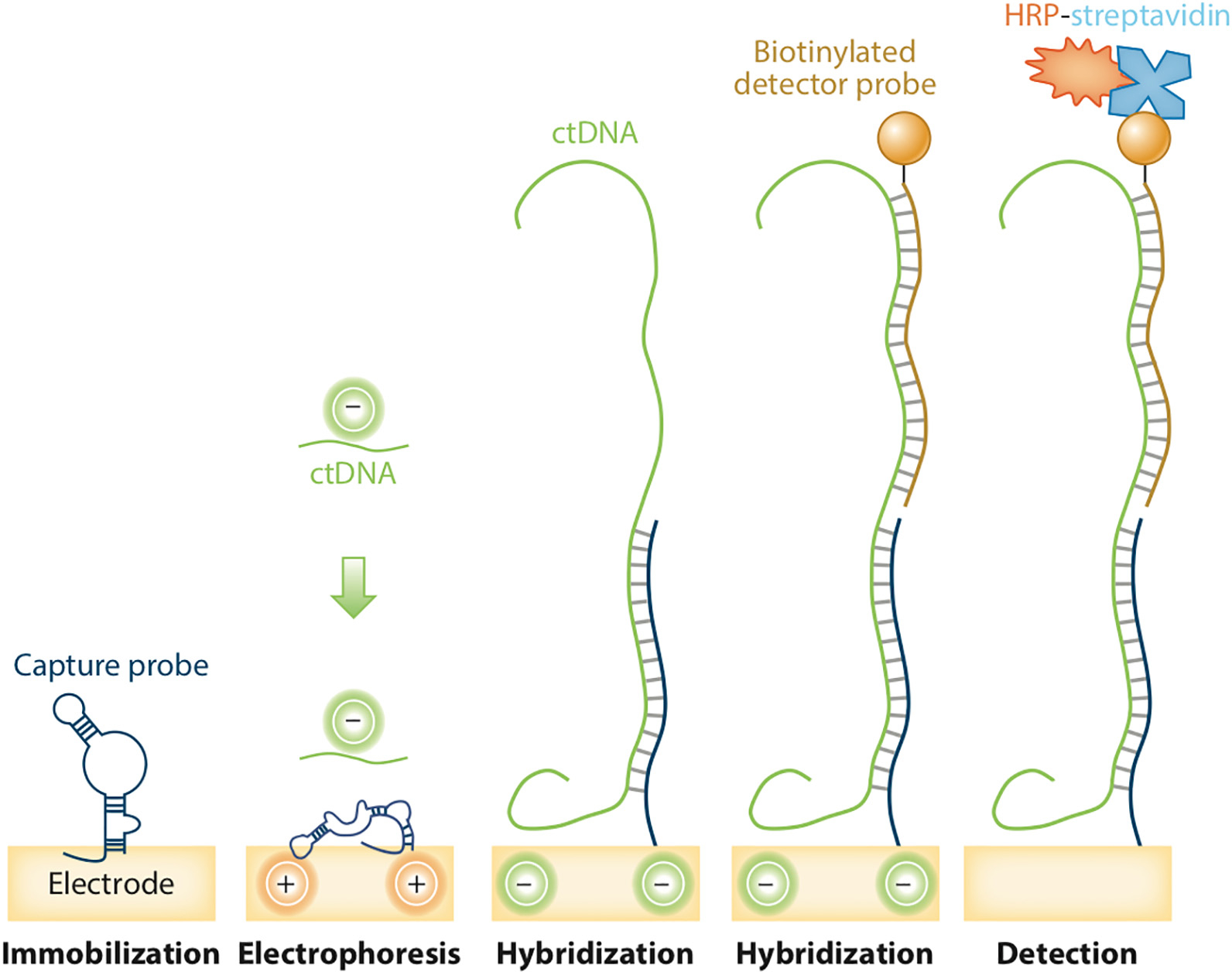
Schematic of electric field–induced release and measurement assay. The cyclic-square wave of the electrical field is applied to release and detect the mutations. Hybridized sequences are measured on the electrochemical sensor with a capture probe precoated in conducting polymer. The horseradish peroxidase (HRP)-labeled reporter probe generates amperometric signals when there is a reaction with tetramethylbenzidine substrate under a −200 mV electrical field. ctDNA: Circulating tumor DNA. Adapted with permission of the publisher from Nonaka and Wong.1
First, paired capture and detector probes specific for each EGFR mutant are designed. The capture probe is immobilized on the gold electrode via copolymerization with pyrrole in the presence of an electric field. Then, a mixture of the clinical sample (for example, plasma or saliva) and the detector probe is transferred to the electrode. Next, a special cyclic square-wave electric field is applied to create a nonuniform electric field that will allow ctDNA to be rapidly detected. The high sensitivity and specificity of EFIRM is due to 2 main properties of electric field–enhanced hybridization. First, the positive potential created by the cyclic square-wave electric field ensures that the gene target accumulates on the working electrode. Concomitantly, the negative potential ensures that weak, nonspecific binders are removed. Another factor that increases signal to noise ratio in this assay is the continuous switching of the electrical field during EFIRM; this generates near field solution mixing and accumulation, which results in hybridization between perfectly matched sequences only. Because of its high sensitivity, EFIRM can detect single nucleotide variants in complex biofluids rapidly.27 Combined with the rapid detection time (≈ 30 minutes) and low-volume (40 μL) requirements, this positions EFIRM at the leading edge in terms of biomarker detection from biofluids.
The clinical utility of saliva was shown 10 years ago when ctDNAs coding for actionable sensitizing EGFR mutations were detected in salivary samples from patients with lung cancer, using EFIRM.30 Building on this seminal study, Kim and colleagues31 showed that quantification of salivary ctDNA could be used for treatment monitoring and prediction of recurrence in lung cancer. Saliva-based EFIRM liquid biopsy was able to detect an increase in recurrence-related T790M ctDNA in 2 of 26 patients, whereas this signal was missed using digital droplet polymerase chain reaction (PCR) and next-generation sequencing. Together, these studies provide a solid scientific basis for the clinical use of saliva diagnostics and treatment monitoring (Figure 5). Concerted efforts involving collaborations between clinicians, commercial entities, and regulatory bodies are now required to harness the full power of saliva testing in next-generation clinical practice.
Figure 5.
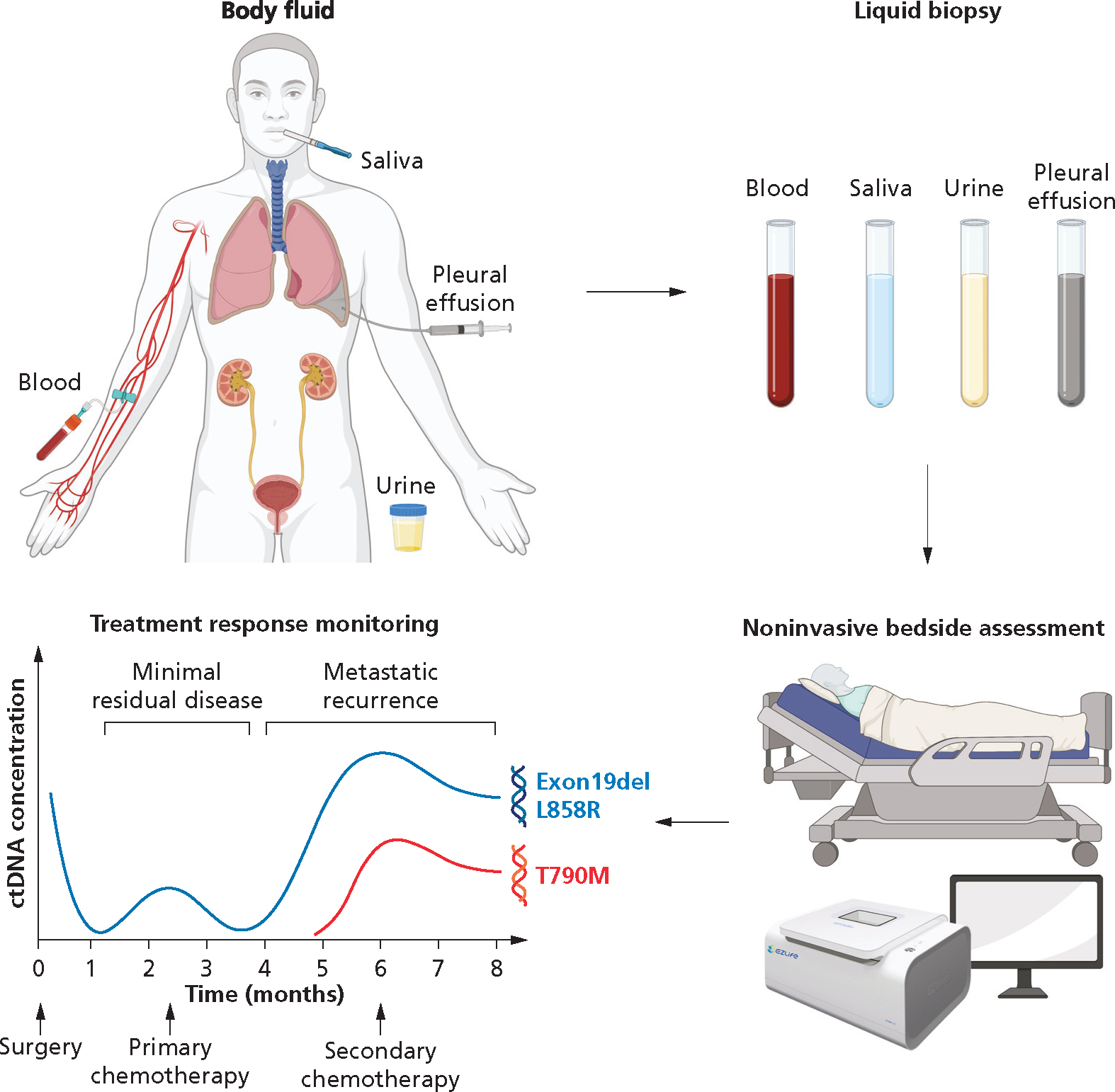
Liquid biopsy for noninvasive bedside assessment and treatment response monitoring. T790M is a recurrent missense mutation within the tyrosine kinase domain of the epidermal growth factor receptor gene. Exon 19del (Exon 19 deletion) and L858R (amino acid substitution from leucine [L] to arginine [R] at codon 858) are mutations that occur in the epidermal growth factor receptor gene. ctDNA: Circulating tumor DNA.
SALIVA-BASED SARS-CoV-2 TESTING
At the beginning of the COVID-19 pandemic, nasal swab testing rapidly became the standard method of viral detection owing to the availability of appropriate reagents and intense pressure to screen at scale. Although clearly a gold standard, this procedure does have some drawbacks. For example, irritation during nasal swabbing often induces sneezing, which can contribute to disease spreading to nearby patients and health care workers. In contrast, saliva screening is much less invasive and does not provoke a strong patient reflex. An increasing number of studies has been showing the sensitivity of saliva at detecting SARS-CoV-2 compared with PCR-based nasal swabs.32 Where it has been adopted in schools and workplaces, saliva self-testing has contributed to improved screening rates and earlier detection of infection.33–35 These findings have created a favorable public view of saliva testing for COVID-19, and both the SalivaDirect test36,37 and the Spectrum Solutions SDNA-1000 test38 have been submitted to the US Food and Drug Administration for Emergency Use Authorization (FDA EUA). Further highlighting the utility of saliva testing, University of California, Los Angeles created and deployed SwabSeq kits campuswide.39 All these initiatives have placed saliva diagnostics in a favorable light.
Although testing for SARS-CoV-2 genomic RNA from saliva is now well established, we envisioned that saliva also could be used to provide information regarding infectivity and the immune response to this pathogen.40–42 We therefore designed EFIRM tests capable of detecting either viral antigen and host-neutralizing antibodies, as well as viral genomic RNA (Table). This triplex test (which requires only 50 μL of sample) therefore provides a 360° view of a person’s infectious state and immune response. Because this EFIRM multiplex diagnostics outperforms all EUA-approved tests for performance for all 3 biomarkers (Table), it is poised to become a leading technology for COVID surveillance in the postpandemic era.
Table.
EFIRM*-based SARS-CoV-2 assay.
| EFIRM SARS- COV-2 ASSAY | SENSITIVITY; SPECIFICITY, % | LIMIT OF DETECTION, EFIRM | LIMIT OF DETECTION, FDA EUA* | PERFORMANCE VS FDA EUA† | SAMPLE VOLUME, μL | RUN TIME, MIN | COST PER REACTION, $ | MULTIPLEX |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Genomic RNA | 95; 100 | 6.25 copies per reaction | 6–12 copies per reaction | × 16 | 3 | 60 | 5.30 | Yes |
| Antigen | 95; 100 | 3.5 TCID50‡ per mL | 22.5–140 TCID50 per mL | × 6.4 | 3 | 55 | 6.46 | Yes |
| Antibody | 93; 97 | 24 pg per mL | Not applicable | Not applicable | 3 | 45 | 9.42 | Yes |
EFIRM: Electrical field-induced release and measurement.
FDA EUA: US Food and Drug Administration Emergency Use Authorization.
TCID50: 50% tissue culture infectious dose.
ULTRASHORT CELL-FREE DNA: A NEW CELL-FREE DNA SUBTYPE
The revelation that there are 2 distinct populations of cell-free DNA (cfDNA) has opened up several new avenues for scientific exploration.43 Broad range cfDNA sequencing is a novel next generation sequencing pipeline that has uncovered a distinct population of ultrashort cell-free DNA (uscfDNA) in plasma (Figure 6A).43 The length of uscfDNA ranges from 25 through 75 nucleotides, with a modal peak of 50 nucleotides and is distinct from the typically described 167 base pair mononucleosomal cell-free DNA (mncfDNA). The ability of this pipeline to discover uscfDNA is due to the pairing of a DNA extraction method optimized for short DNA fragments with single-stranded library preparation; such fragments are missed by conventional sequencing. uscfDNA abundance is generally higher in healthy people than in patients with cancer and is thought to reflect sequences that are present in open chromatin regions. The relative proportions of functional peaks (related to introns, intergenic regions, and promotors) were significantly different between uscfDNA and mncfDNA samples (Figure 6B). The uscfDNA fragments are optimal targets for detection by means of EFIRM. Combining uscfDNA biopsy with EFIRM may lead to significant enhancements in clinical performance of liquid biopsies owing to the ability to directly detect biomarkers using minimal sample volume. In the context of saliva diagnostics, it is encouraging that uscfDNA pools have been detected in plasma.44–46 Although the precise biological explanation for the generation of uscfDNA remains unclear, it is hoped that the study of differences in uscfDNA levels and sequence in plasma and saliva between healthy people and those with disease will increase diagnostic power in the clinic.
Figure 6.

Molecular profile of ultrashort cell-free DNA (uscfDNA). A. Broad range cfDNA sequencing reveals a distinct population of ultrashort low-molecular-weight cell-free DNA in plasma. B. Functional peak analysis of plasma fragments reveals an enhanced proportion of regulatory elements in the fragments between 40 base pairs (bp) and 70 bp. mncfDNA: Mononucleosomal cell-free DNA. ncRNA: Noncoding RNA. rRNA: Ribosomal RNA. TTS: Transcription termination site. UTR: Untranslated region. Adapted under a Creative Commons Attribution 4.0 International License from Cheng and colleagues.43
FUTURE PERSPECTIVES OF SALIVA DIAGNOSTICS
We highlighted how fundamental biological discoveries have accelerated the application of salivaomics to the clinical environment. The remarkable redistribution of exosomes from diseased tissue to the saliva via the circulatory system has provided “canary in the coal mine” that can be exploited for diagnostics, patient stratification, and treatment monitoring. New facets of salivaomics are continually opening up. For example, additional interactions of salivary exosomes with the gut microbiome represent a new frontier for salivaomics and will be studied in the near future.
Three EFIRM-based assays have obtained Clinical Laboratory Improvement Amendments (CLIA) certification: EGFR L858R, exon 19del, and T790M in plasma; EGFR L858R, exon 19del, and T790M in saliva; and SARS-CoV-2 immunoglobulin G S1 in saliva. Buoyed by the CLIA certification of the mutant EGFR saliva tests for lung cancer, we are hopeful that the multiplex EFIRM test for SARS-CoV-2 that we described above also will receive an Emergency Use Authorization approval. However, commercial partnerships are required to take the EFIRM tests to the next stage. Indeed, the synergy between basic and translational research is a key driver of tests moving toward regulatory approval. We look forward to additional breakthroughs from other groups in this fast-moving field, and we see a bright future ahead for saliva diagnostics.
Acknowledgments
This work was supported by grants UG3/UH3 TR002978, UH2/UH3 CA206126, U01 CA233370, and U01 DE017790 from the National Institutes of Health to Dr. Wong; and R03 DE027759, R03 DE29272, Feist-Weiller Cancer Center Foundation Legacy Fund, and Louisiana State University Collaborative Cancer Research Initiative Fund to Dr. Nonaka.
ABBREVIATION KEY
- bp
Base pair
- CD
Cluster of differentiation
- cfDNA
Cell-free DNA
- CLIA
Clinical Laboratory Improvement Amendments
- ctDNA
Circulating tumor DNA
- EFIRM
Electric field–induced release and measurement
- exRNA
Extracellular RNA
- FDA EUA
US Food and Drug Administration Emergency Use Authorization
- HRP
Horseradish peroxidase
- mRNA
Messenger RNA
- miRNA
MicroRNA
- mncfDNA
Mononucleosomal cell-free DNA
- ncRNA
Noncoding RNA
- NIH
National Institutes of Health
- PCR
Polymerase chain reaction
- piRNA
Piwi-interacting RNA
- rRNA
Ribosomal RNA
- snoRNA
Small nucleolar RNA
- TTS
Transcription termination sites
- uscfDNA
Ultrashort cell-free DNA
- UTR
Untranslated region
- TCID50
50% tissue culture infectious dose
Footnotes
Disclosures. Dr. Wong holds equity in Liquid Diagnostics and RNAmeTRIX and is a consultant to GlaxoSmithKline, PeriRx, Wrigley, Colgate-Palmolive, and Spectrum Solutions. Dr. Nonaka did not report any disclosures.
Contributor Information
Taichiro Nonaka, Department of Cellular Biology and Anatomy and Feist-Weiller Cancer Center, Louisiana State University Health Sciences Center, Shreveport, LA..
David T.W. Wong, Division of Oral and Systemic Health Sciences and Center for Oral/Head and Neck Oncology Research, School of Dentistry, University of California Los Angeles, Los Angeles, CA..
References
- 1.Nonaka T, Wong DTW. Saliva diagnostics. Annu Rev Anal Chem. 2022;15(1):107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nonaka T, Wong DTW. Liquid biopsy in head and neck cancer: promises and challenges. J Dent Res. 2018;97(6):701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8(12):5590–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan W, Apweiler R, Balgley BM, et al. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3(1):116–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny P, Hagen FK, Hardt M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7(5):1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapid acceleration of diagnostics (RADx). National Institutes of Health (NIH). Accessed January 30, 2023. https://www.nih.gov/research-training/medical-research-initiatives/radx [Google Scholar]
- 7.Ai J, Smith B, Wong DT. Saliva ontology: an ontology-based framework for a salivaomics knowledge base. BMC Bioinformatics. 2010;11:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong DT. Salivaomics. JADA. 2012;143(suppl 10):19S–24S. [DOI] [PubMed] [Google Scholar]
- 9.Mackie DA, Pangborn RM. Mastication and its influence on human salivary flow and alpha-amylase secretion. Physiol Behav. 1990;47(3):593–595. [DOI] [PubMed] [Google Scholar]
- 10.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86(8):680–693. [DOI] [PubMed] [Google Scholar]
- 11.Henson BS, Wong DT. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol Biol. 2010;666:21–30. [DOI] [PubMed] [Google Scholar]
- 12.Chiang SH, Thomas GA, Liao W, et al. RNA-Pro•SAL: a device for rapid and standardized collection of saliva RNA and proteins. Biotechniques. 2015;58(2):69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, The Extracellular RNA Communication Consortium; Ansel KM, et al. The Extracellular RNA Communication Consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177(2):231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murillo OD, Thistlethwaite W, Rozowsky J, et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019;177(2):463–477, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nonaka T, Wong DTW. Saliva-exosomics in cancer: molecular characterization of cancer-derived exosomes in saliva. Enzymes. 2017;42:125–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lässer C, Alikhani VS, Ekström K, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andaloussi SEL, Mager I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. [DOI] [PubMed] [Google Scholar]
- 18.Al-Tarawneh SK, Border MB, Dibble CF, Bencharit S. Defining salivary biomarkers using mass spectrometry-based proteomics: a systematic review. OMICS. 2011;15(6):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, Nonaka T, Wong DTW. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials (Basel). 2019;12(4):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau C, Kim Y, Chia D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288(37):26888–26897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Wei F, Schafer C, Wong DT. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One. 2014;9(11):e110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsiougiannis S, Chia D, Kim Y, Singh RP, Wong DT. Saliva exosomes from pancreatic tumor-bearing mice modulate NK cell phenotype and antitumor cytotoxicity. FASEB J. 2017;31(3):998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomans-Kropp HA, Umar A, Minasian LM, Pinsky PF. Multi-cancer early detection tests: current progress and future perspectives. Cancer Epidemiol Biomarkers Prev. 2022;31(3):512–514. [DOI] [PubMed] [Google Scholar]
- 25.Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho C-M. Bio/abiotic interface constructed from nanoscale DNA dendrimer and conducting polymer for ultrasensitive biomolecular diagnosis. Small. 2009;5(15):1784–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei F, Wang J, Liao W, Zimmermann BG, Wong DT, Ho C-M. Electrochemical detection of low-copy number salivary RNA based on specific signal amplification with a hairpin probe. Nucleic Acids Res. 2008;36(11):e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu M, Cheng J, Chen YL, et al. Electric field-induced release and measurement (EFIRM): characterization and technical validation of a novel liquid biopsy platform in plasma and saliva. J Mol Diagn. 2020;22(8):1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei F, Patel P, Liao W, et al. Electrochemical sensor for multiplex biomarkers detection. Clin Cancer Res. 2009;15(13):4446–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei F, Yang J, Wong DT. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron. 2013;44:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei F, Lin CC, Joon A, et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med. 2014;190(10):1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C, Xi L, Cultraro CM, et al. Longitudinal circulating tumor DNA analysis in blood and saliva for prediction of response to osimertinib and disease progression in EGFR-mutant lung adenocarcinoma. Cancers (Basel). 2021;13(13):3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandes LL, Pacheco VB, Borges L, et al. Saliva in the diagnosis of COVID-19: a review and new research directions. J Dent Res. 2020;99(13):1435–1443. [DOI] [PubMed] [Google Scholar]
- 33.Lai J, German J, Hong F, et al. Comparison of saliva and midturbinate swabs for detection of SARS-CoV-2. Microbiol Spectr. 2022;10(2):e0012822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol. 2021;59(5):e02881–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan C, Ditelberg S, Dutta S, et al. Saliva is comparable to nasopharyngeal swabs for molecular detection of SARS-CoV-2. Microbiol Spectr. 2021;9(1):e0016221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogels CBF, Watkins AE, Harden CA, et al. Saliva-Direct: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. 2021;2(3):263–280, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emergency Use Authorization (EUA) Review Memorandum SDNA-1000 Saliva Collection Device. US Food and Drug Administration. Updated August 4, 2021. Accessed January 30, 2023. https://www.fda.gov/media/142907/download [Google Scholar]
- 39.Bloom JS, Sathe L, Munugala C, et al. Massively scaled-up testing for SARS-CoV-2 RNA via next-generation sequencing of pooled and barcoded nasal and saliva samples. Nat Biomed Eng. 2021;5(7):657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang SH, Tu M, Cheng J, et al. Development and validation of a quantitative, non-invasive, highly sensitive and specific, electrochemical assay for anti-SARS-CoV-2 IgG antibodies in saliva. PLoS One. 2021;16(7):e0251342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu MK, Chiang SH, Bender RA, Wong DTW, Strom CM. The kinetics of COVID-19 vaccine response in a community-vaccinated population. J Immunol. 2022;208(4):819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu MK, Chiang SH, Wong DTW, Strom CM. Variability in severe acute respiratory syndrome coronavirus 2 IgG antibody affinity to Omicron and Delta variants in convalescent and community mRNA-vaccinated individuals. Immunohorizons. 2022;6(5):307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng J, Morselli M, Huang WL, et al. Plasma contains ultrashort single-stranded DNA in addition to nucleosomal cell-free DNA. iScience. 2022;25(7):104554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng LY, Dai P, Wu LR, Patel AA, Zhang DY. Direct capture and sequencing reveal ultra-short single-stranded DNA in biofluids. iScience. 2022;25(10):105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hisano O, Ito T, Miura F. Short single-stranded DNAs with putative non-canonical structures comprise a new class of plasma cell-free DNA. BMC Biol. 2021;19(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudecova I, Smith CG, Hansel-Hertsch R, et al. Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res. 2022;32(2):215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]


