Abstract
Transmission of cauliflower mosaic virus (CaMV) by aphids requires two viral nonstructural proteins, the open reading frame (ORF) II and ORF III products (P2 and P3). An interaction between a C-terminal domain of P2 and an N-terminal domain of P3 is essential for transmission. Purified particles of CaMV are efficiently transmitted only if aphids, previously fed a P2-containing solution, are allowed to acquire a preincubated mixture of P3 and virions in a second feed, thus suggesting a direct interaction between P3 and coat protein. Herein we demonstrate that P3 directly interacts with purified viral particles and unassembled coat protein without the need for any other factor and that P3 mediates the association of P2 with purified virus particles. The interaction domain of P3 is located in its C-terminal half, downstream of the P3-P2 interaction domain but overlapping a region which binds nucleic acids. Mutagenesis of P3 which interferes with the interaction between P3 and virions is correlated with the loss of transmission by aphids. Taken together, our results demonstrate that P3 plays a crucial role in the formation of the CaMV transmissible complex by serving as a bridge between P2 and virus particles.
Cauliflower mosaic virus (CaMV), the type member of the genus Caulimovirus, is transmitted between host plants by several aphid species in a noncirculative manner (for a review, see reference 18). It is characterized by icosahedral particles, 50 nm in diameter, and a genome of double-stranded circular DNA (8 kbp) which is replicated by reverse transcription of a pregenomic 35S RNA. The DNA codes for six proteins which are believed to be independently expressed from the polycistronic 35S RNA and the monocistronic 19S RNA (for a review, see reference 19). Proteins P1 (40 kDa) and P2 (18 kDa) are involved in virus cell-to-cell spread and in transmission by aphids, respectively. P4 (57 kDa) is the precursor of four N- and C-terminally truncated forms of the protein with molecular masses of 42, 39, 37, and 35 kDa, whereas protein P5 (78 kDa) has aspartate proteinase and reverse transcriptase activities at its N and C termini, respectively. P6 (62 kDa) is the major component of viroplasms. It has been reported to activate the translation of the 35S RNA (4) and to influence host specificity and symptomatology (21).
CaMV P3 protein (15 kDa), encoded by open reading frame (ORF) III, is a small basic protein (129 amino acids) organized into two functional domains located towards the N and C termini. Both domains are required for CaMV infectivity (13). The C-terminal region contains a short lysine- and proline-rich domain that binds DNA and RNA in a sequence-nonspecific manner (11, 17). The N-terminal domain can be folded into an α-helix secondary structure which can form coiled coils that allow tetramerization of P3 in vitro (14) and in vivo (23). Recently, it was demonstrated that the 30 N-terminal amino acids of P3 interact specifically with the C-terminal region of CaMV P2 protein and that P3 represents a second CaMV factor of transmission by aphids (15). This finding provides a solution to the long-standing question of why aphids previously fed a P2-containing solution (P2-loaded aphids) are able to transmit nontransmissible isolates of CaMV from infected plants, or crude extracts thereof, but not from highly purified virus preparations (2, 15). The transmission of purified CaMV by P2-loaded aphids has been shown to be efficient only when P3 is preincubated with virions. The P3-binding domain was mapped to the C-terminal 60 amino acids of P2, precisely matching the domain previously thought to interact with the virus particle (20). A possible interpretation of this finding is that P2 does not interact directly with the viral particles but instead binds to the particle via the bridging action of P3, which is thus predicted to interact directly with the CaMV coat protein (CP).
In the present study, we have investigated the capacities of P3 and P2 to interact with the viral particle and/or unassembled CP. We have demonstrated that P3 interacts specifically with both purified CaMV particles and CP. The domain of P3 involved in this interaction was mapped between amino acid residues 61 and 122, downstream of the P3-P2 interaction domain but overlapping the nucleic acid-binding domain. The ability of P3 to bind virus particles was shown to be of functional importance since its disruption correlates with the loss of P3 activity in transmission by aphids. We have also established that P2 can interact with the virus particle only in the presence of P3, thereby confirming that P3 is a pivotal transmission factor that acts as a bridge between P2 and the CaMV particle.
MATERIALS AND METHODS
Cloning of CaMV ORF III and ORF IV.
The complete ORF III and deleted versions were excised from CaMV strain Cabb-S, cloned into the SalI site of pBR322, and inserted between the NdeI and CelI sites of the procaryotic expression vector pET-3a (15). CaMV ORF IV was obtained by PCR amplification of DNA of strain Cabb-B JI cloned into the SacI site of pAT-153 (8). The oligonucleotides used as primers were the following: 5′ CGTTCGCATATGGCCGAATCAATTTTAGACAGGACC 3′ and 5′ TCGGGATCCGAGCTCTCAGTCTGAGTCTGGAGTCTTCAGAAGT 3′ as forward and reverse primers, respectively (nucleotides in bold and italics correspond to the NdeI or BamHI site and to the ORF IV sequence, respectively). PCR products were cloned between the NdeI and BamHI sites of vector pET-3a in frame with the ATG initiation codon present in the NdeI restriction site. The construct was confirmed to be error free by sequencing.
Expression of CaMV P3 and P4 proteins in Escherichia coli BL21/DE3(pLysS).
P3 and its different versions were produced and semipurified as previously described (11). Expression of the precursor of CP (preCP) in E. coli was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 2 h. Bacteria were collected by centrifugation, resuspended in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 12 mM MgCl2, and lysed using a French press. After centrifugation at 10,000 × g for 5 min, inclusion bodies were resuspended in 0.5 ml of the same buffer and frozen at −20°C. The suspension of inclusion bodies containing P4 was diluted in 1 volume of 1 M NaCl and vigorously mixed before being used as an overlay in far-Western assays. P2 was produced in sf9 insect cells via a recombinant baculovirus as described previously (2).
Western and far-Western experiments.
Different forms of the CP from purified virions (10 μg) and P3 or P2 protein (2 μg) were separated by 18% polyacrylamide gel electrophoresis under denaturing conditions (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) and transferred onto a nitrocellulose membrane (Schleicher and Schuell). Western and far-Western assays were performed as described previously (15), using rabbit antibodies raised against P3 (diluted to 1/10,000), P2 (diluted to 1/5,000), or the 37-kDa CP (diluted to 1/10,000). Secondary antibodies were goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase, diluted, and used according to the manufacturer's instructions (Sigma). For each experiment, the same blot was separated into strips in order to test the various lanes with different overlays. The immunological reactions were done under strictly the same conditions.
Virus purification.
CaMV virions (strain Cabb-S) were purified as described previously (10), with an additional step of cesium chloride gradient centrifugation at 200,000 × g for 16 h.
Transmission assays.
CaMV strain Cabb-S, used for transmission testing with Myzus persicae, was propagated in turnip plants grown under greenhouse conditions (20 to 23°C, photoperiod of 16 h). Transmission assays were performed as previously described (2). In a first feed, aphids were allowed to acquire P2 (0.5 μg/μl) through a Parafilm membrane, and in a second feed, they were allowed to acquire purified virions (0.1 μg/μl) mixed with wild-type P3 or a derivative. The latter were partially purified from recombinant bacteria (11). Aphids were then transferred onto healthy plants (10 aphids per plant), and symptom appearance was noted 3 weeks postinoculation. Transmission of wild-type and mutant P3 was tested in two or three independent experiments with 40 plants each.
RESULTS
CaMV P3 is able by itself to interact with virus particles.
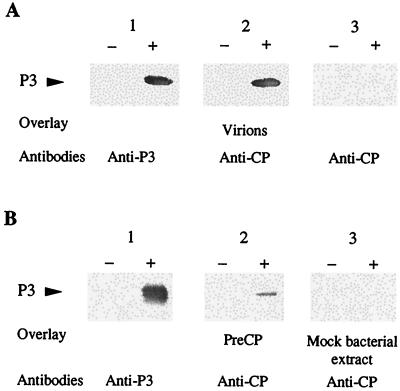
Very small amounts of P3 have previously been found to copurify with CaMV particles (7). To determine whether CaMV P3 protein interacts directly with virions, far-Western assays were performed with P3 produced in E. coli and partially purified by heat treatment of the bacterial extract. After fractionation by electrophoresis in a polyacrylamide gel under denaturing conditions (SDS-PAGE) and transfer onto a nitrocellulose membrane, P3 was tested for its capacity to bind to purified virions of CaMV strain Cabb-S, which was used as an overlay. Detection with polyclonal antibodies raised against the 37-kDa CP (anti-CP), a major capsid component, revealed a single band at the position of P3 as identified by Western blot analysis (Fig. 1A, lanes 1+ and 2+). No band was visible in the control lane, containing an extract of bacteria transformed with an empty vector (mock bacterial extract [Fig. 1A, lane 2−]), confirming that virions interact with P3 and not with a bacterial protein. The specificity of the interaction between P3 and virus particles was evidenced by the lack of a band at the position of P3 when virions were absent from the overlay solution (Fig. 1A, lane 3+). In additional far-Western assays, purified CaMV was replaced by an E. coli extract containing preCP as the overlay. A band was observed at the position of P3 (Fig. 1B, lane 2+) but not when the overlay was a mock bacterial extract (Fig. 1B, lane 3+). These results show that preCP is also able to bind to P3. The relative faintness of the band obtained with preCP, compared to that observed when virions were used as the overlay (Fig. 1A, lane 2+), might be due to the reduced amount of soluble preCP present, since this viral protein aggregates when expressed in E. coli (5). Moreover, a lower affinity of preCP for P3 cannot be totally excluded.
FIG. 1.
Detection of interactions between CaMV P3 and either purified virus particles (A) or preCP (B). E. coli extracts containing P3 (+) or not (−) were fractionated by SDS-PAGE (18% polyacrylamide) and transferred onto a nitrocellulose membrane. P3 was detected with anti-P3 antibodies (lanes 1). (A) Strips from the same blot were incubated either with purified virions (lanes 2) or with buffer (lanes 3), then washed, and treated with anti-CP antibodies raised against the 37-kDa CP form. (B) Strips were incubated with bacterial extracts containing preCP (lanes 2) or not (lanes 3), then washed, and treated with anti-CP antibodies.
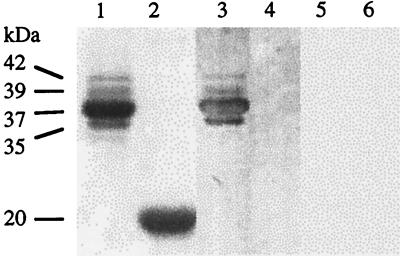
Whether P3 interacts preferentially with one or more of the different capsid protein forms (Fig. 2, lane 1) was also investigated by far-Western assays. Purified CaMV virions were fractionated by SDS-PAGE, and the proteins were revealed by Coomassie blue staining or electrotransferred onto a nitrocellulose membrane. Staining of the polyacrylamide gel displayed a major capsid protein of 37 kDa and three minor components of 42, 39, and 35 kDa but not preCP. The presence of preCP and the proportion of its processed forms depend on the CaMV preparation. When the membrane was incubated with P3 as the overlay, signals with different intensities were detected at the positions of the four capsid proteins (Fig. 2, compare lane 3 to lane 1), reflecting the relative amounts of the various CP forms (Fig. 2, lane 1) rather than a differential affinity of P3 for one of the capsid proteins. The specificity of P3-CP interaction was confirmed by the fact that under the same conditions P3 did not bind to the 20-kDa turnip yellow mosaic virus CP (Fig. 2, lane 4) or to the tobacco mosaic virus or beet necrotic yellow vein virus CP (not illustrated).
FIG. 2.
Detection of interactions between CaMV P3 and different forms of the capsid protein. Purified particles of CaMV (lanes 1, 3, and 5) and of turnip yellow mosaic virus, serving as a control (lanes 2, 4, and 6), were fractionated by SDS-PAGE (18% polyacrylamide). Proteins were stained with Coomassie blue (lanes 1 and 2) or transferred onto nitrocellulose membranes (lanes 3, 4, 5, and 6). Membranes were incubated with bacterial extracts containing (lanes 3 and 4) or not containing (lanes 5 and 6) P3 and then were incubated with anti-P3 antibodies. Molecular masses of capsid proteins are listed to the left.
As P3 and CP are able to bind RNA and DNA in a nonspecific manner (5, 13, 17), far-Western experiments were also performed using as the overlay partially purified P3 treated previously with RNase and DNase. Such treatment did not suppress the P3-CP interaction, excluding therefore the possibility that the latter is mediated by nucleic acid fragments (data not shown).
The interaction between P3 and unassembled CPs is of double significance since it demonstrates that (i) P3 interacts directly with the CaMV CP and no additional plant or viral factors are required and (ii) P3 probably recognizes a specific sequence and/or conformation of the unassembled capsid protein.
P3 is required for the association of P2 with CaMV particles.
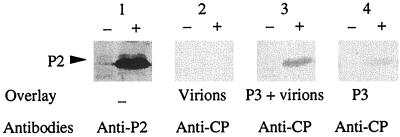
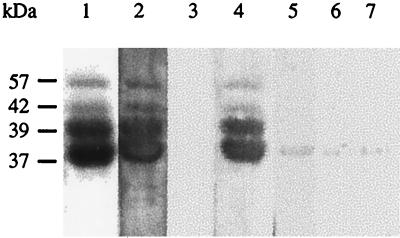
The capacity of P3 to interact with both P2 and CaMV particles suggests that P3 serves as an intermediate in the association of P2 with the virus particles. To investigate this possibility, P2 was produced in insect cells using the baculovirus expression system, fractionated by SDS-PAGE, and blotted onto a nitrocellulose membrane. Strips were incubated with purified CaMV virions that had been either preincubated or not with P3 for 4 h at room temperature, and then the strips were incubated with anti-CP to reveal interactions. At the P2 position, identified by Western analysis (Fig. 3, lane 1+), no band was visible when the virus alone was used as the overlay (Fig. 3, lane 2+), whereas a signal was observed if the overlay was a mixture of purified virus particles and P3 (Fig. 3, lane 3+). When the P2-P3 complex was probed under the same experimental conditions with anti-CP, a faint band appeared, indicating that these antibodies slightly cross-reacted when P3 was used as the overlay (Fig. 3, lane 4+). However, the low intensity of the cross-reaction signal compared to that observed in Fig. 3 (lane 3+) suggests strongly that P2 is unable to interact directly with CaMV in the absence of P3. To confirm this conclusion, we tested the capacity of P2 to interact with CaMV capsid proteins fractionated by SDS-PAGE and blotted onto a membrane in the presence or absence of P3. As shown by staining of the gel, the CaMV capsid preparation consisted of preCP (57 kDa) and of the 42-, 39-, and 37-kDa capsid protein forms, the last two being the predominant components (Fig. 4, lane 1). When P3 was used as the overlay, anti-P3 antibodies revealed bands at the positions of the capsid proteins with intensities proportional to their amounts, thereby confirming that P3 interacts with preCP and its processed forms (Fig. 4, lane 2). As already observed (Fig. 2), anti-P3 did not react with the capsid proteins (Fig. 4, lane 3). An interaction between P2 and the capsid proteins was revealed with anti-P2 when the blot was incubated with P3 for 4 h before the addition of P2 (Fig. 4, lane 4). The slight signal observed when the membrane was incubated with P2 alone (Fig. 4, lane 5) was not due to an interaction of P2 with the capsid proteins, since it was also observed in the control experiments performed with either P3 or mock bacterial extract as the overlay (Fig. 4, lanes 6 and 7).
FIG. 3.
Analysis of the interactions between P2 and virus particles in the presence or in the absence of P3. Insect cells (sf9) extracts containing P2 (+) or not (−) were fractionated by SDS-PAGE (18% polyacrylamide) and transferred onto a nitrocellulose membrane. P2 was detected with anti-P2 antibodies (lanes 1). Strips from the same blot were incubated with purified virions alone (lanes 2), with purified virions mixed with P3 (lanes 3), or with P3 alone (lanes 4). Interactions were revealed with anti-CP antibodies.
FIG. 4.
Analysis of the interactions between P2 and the different capsid protein forms of CaMV. Purified CaMV particles were submitted to SDS-PAGE (18% polyacrylamide), and proteins were either revealed with Coomassie blue (lane 1) or transferred onto a nitrocellulose membrane (lanes 2 to 7). Strips were incubated with P3 (lane 2) or P2 (lane 3) as an overlay and then with anti-P3 or anti-P2 antibodies, respectively. Strips were incubated first with P3, then with P2 (lane 4) or not (lane 5), and finally with anti-P2 antibodies. Strips were also incubated with a mock bacterial extract and subsequently with either anti-P2 (lane 6) or anti-P3 (lane 7) to test the antibody specificity. Molecular masses are indicated to the left.
Taken together, the results indicate that P2 is not able by itself to interact with virus particles and that P3 is likely to ensure a bridging function, allowing formation of a P2-P3-virion complex which is absolutely required in at least one step of the aphid-borne transmission process.
Mapping of the domain of P3 which interacts with virus particles.
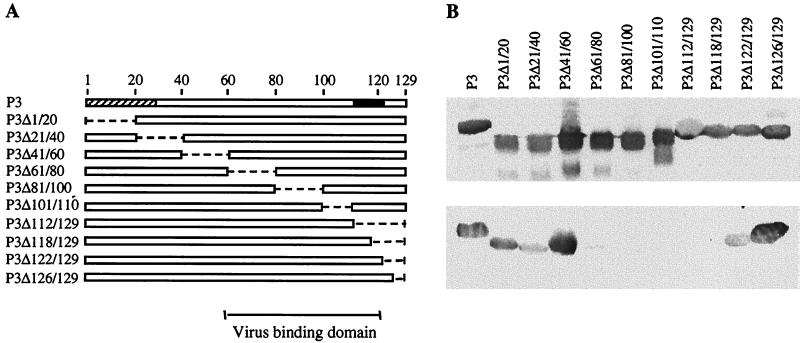
A series of P3 derivative mutants with either internal or C-terminal deletions (Fig. 5A) were tested for their capacity to bind virions in far-Western assays. Equivalent amounts of P3 mutant proteins were used, as shown by Western blotting performed with anti-P3 antibodies (Fig. 5B, top). The faint bands corresponding to polypeptides of lower molecular masses may represent degradation products of P3 recombinant proteins. Twenty-amino-acid deletions in the N-terminal half of P3 (mutants P3Δ1/20, P3Δ21/40, and P3Δ41/60) did not affect its capacity to bind to CaMV particles, nor did the removal of four (P3Δ126/129) or eight (P3Δ122/129) residues at the C terminus (Fig. 5B, bottom). In contrast, internal deletions located between amino acids 61 and 118 either totally abolished the interaction between P3 and purified virus (mutants P3Δ81/100, P3Δ101/110, P3Δ112/129, and P3Δ118/129) or drastically reduced it (P3Δ61/80). The inability of mutants P3Δ81/100 and P3Δ101/110 to interact with virions reinforces the conclusion that neither RNA nor DNA is involved in vitro in the formation of the P3-virion complex, since these mutants still possess the nucleic acid-binding domain. None of the putative degradation products was able to interact with virus particles. We conclude that the virion-binding domain of P3 is located in a 60-amino-acid region spanning most of the C-terminal half of the sequence (amino acids 61 to 122).
FIG. 5.
Mapping of the P3 domain which interacts with CaMV particles. Interaction assays were performed with the mutant P3 proteins presented in panel A, where empty boxes and broken lines correspond to P3 sequence and deleted regions, respectively. At the top, the two P3 domains which interact with P2 and with nucleic acids are indicated by hatched and black boxes, respectively. The location of the virus binding domain is indicated at the bottom. After separation by SDS-PAGE (18% polyacrylamide) and transfer onto a nitrocellulose membrane, P3 mutants were detected with anti-P3 antibodies (top of panel B) or tested for the capacity to interact with purified virions (bottom of panel B). P3-virion interactions were revealed with anti-CP antibodies.
Interaction between P3 and CaMV particles is required for transmission by aphids.
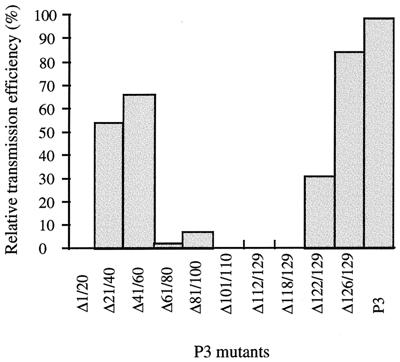
All the P3 deletion mutants were tested for the ability to allow transmission of CaMV by aphids. The insects were first fed baculovirus-expressed P2, then fed each of the different P3 mutant proteins mixed with purified CaMV particles, and finally transferred to turnip plants. Results of the transmission assays are presented in Fig. 6. Comparison of the results in Fig. 5 and 6 shows that the capacity of P3, except for mutant P3Δ1/20, to interact with CaMV particles correlates with its activity in transmission by aphids. Deletions in the C-terminal part of the P3-virion interaction domain (mutants P3Δ101/110, P3Δ112/129, and P3Δ118/129) completely abolished CaMV transmission, whereas removal of sequences in the N-terminal part of the domain (P3Δ61/80 and P3Δ81/100) reduced the transmission rate to 3 and 7%, respectively, of that obtained with wild-type P3. Based on these data, the sequence of P3 between amino acids 101 and 122 appears to be the core interaction domain for transmission. Deletions of sequences flanking this domain (P3Δ41/60, P3Δ61/80, P3Δ81/100, P3Δ122/129, and P3Δ126/129) attenuated transmission but did not inhibit it entirely.
FIG. 6.
Efficiency of CaMV transmission by aphids in the presence of different deleted P3 proteins. Aphids were first fed P2 expressed by recombinant baculovirus in insect cells and then fed a suspension of purified CaMV particles (Cabb-S) mixed with a bacterial extract containing wild-type P3 or a deleted version of P3 (Δ1/20 to Δ126/129). Next, they were placed on test turnip plants, and the percentage of plants developing symptoms was determined 3 weeks postinoculation. The relative transmission efficiency in the presence of each P3 mutant was calculated by taking the value for wild-type P3 as 100%. On average, 51% of the turnip plants were infected when assays for transmission by aphids were carried out with wild-type P3.
As expected, although mutant P3Δ1/20 was perfectly capable of interacting with CaMV particles (Fig. 5), its activity in transmission by aphids was nil due to the absence of the P2-binding motif located mainly in the deleted sequence (15). More surprising was the relatively high transmission rate (53% of that obtained with wild-type P3) observed when P3Δ21/40 was tested. Indeed, in a previous study it was observed that the removal of amino acids 21 through 25 was sufficient to totally abolish transmission because these five residues are part of the P3 N-terminal leucine zipper predicted to interact with P2 (15). This apparent discrepancy may be explained by different structural effects associated with each of the two deletions and is further discussed below.
Mutant P3Δ61/80 was of particular interest because the deleted sequence affects only transmission, whereas all other regions of P3 (except for residues 126 to 129) are required for both transmission by aphids and infectivity. Indeed, CaMV that encodes such a mutated P3 conserves its capacity to systemically infect turnip plants after mechanical inoculation, in contrast to virus that harbors deletions anywhere else in ORF III (13). This allowed us to test the plant-to-plant aphid-borne transmission efficiency of the corresponding CaMV mutant (CaΔ61/80). Aphids were fed directly on turnip plants infected with either CaΔ61/80 or wild-type CaMV. Under our experimental conditions, CaΔ61/80 was transmitted by aphids with a reduced efficiency (42%) compared to that obtained with the wild-type virus (91%). Thus, the effect of the Δ61/80 deletion on P3 activity in transmission by aphids is much more drastic in vitro than in vivo, since little transmission (3% of that with wild-type P3) was observed when P2-loaded aphids were fed a bacterial extract containing P3Δ61/80 and purified CaMV particles (Fig. 6). This might be due to the fact that the conditions for the formation and/or the stability of the transmissible complex are less favorable in vitro than in vivo. Nevertheless, the result obtained in vivo confirms the importance of the P3-P4 interaction domain in forming the viral transmissible complex. This represents the first report of a viable CaMV mutant affected in aphid-borne transmission by alteration of an ORF other than ORF II.
Taken together, these results indicate that the transmission of CaMV by aphids requires interaction between P3 and the CaMV particle and that P3 plays a pivotal role in the formation of a CaMV complex that is transmissible by aphids. Any deletion that weakens this interaction is detrimental to efficient transmission of the virus.
DISCUSSION
Transmission of CaMV from plant to plant by aphids requires the assistance of two viral factors, P2 and P3. In this study, we demonstrated that P3 is able to bind directly to CaMV particles without the mediation of an additional factor and that there is a correlation between the capsid-binding properties of P3 and its activity in aphid-borne transmission, since deletions in the P3-virion interaction domain abolished CaMV transmission.
The domain by which P3 interacts with CaMV or unassembled CPs was mapped to a large region between residues 61 and 122. This domain is distinct from the N-terminal P2-P3 interaction domain (residues 1 to 30) but overlaps the nucleic acid-binding domain at the C terminus of P3 (residues 112 to 122). Recently, a counterpart of CaMV P3, the P2 protein of rice tungro bacilliform badnavirus, was also demonstrated to bind to the CP through its C terminus (9). Moreover, this region of rice tungro bacilliform badnavirus P2 is also able to interact in vitro with nucleic acids in a nonspecific manner (12).
Interaction of P3 with virions probably involves a specific sequence and/or conformation of CP since P3 binds directly to the preCP expressed in bacteria and to its processed forms extracted from purified virions. However, the recognition of a quaternary structure of the capsid (i.e., capsomers) is not completely excluded, as oligomerization of P4 immobilized on the membrane in far-Western experiments could occur at the renaturation step preceding incubation with P3. The domain of P3 which binds to virions is believed to interact with the N terminus of the CP(s) exposed at the outer surface of the capsid (6). Similarly, potyvirus helper component HC-Pro binds to the N terminus of the cognate capsid protein (1, 16) for noncirculative transmission of potyviruses by M. persicae (22).
The results obtained with CaMV harboring P3Δ61/80 transmitted from plant to plant suggest that the C-terminal end of the P3-virion interaction domain (amino acids 101 to 122), which overlaps the nucleic acid-binding domain, might also interact with the CaMV genome. Indeed, when this mutant was transmitted by aphids fed on infected turnip plants, the transmission efficiency was considerably increased (50% of the wild type), in contrast to the transmission observed if feeding was from an artificial mixture through a membrane (3% of the wild type). One explanation for this observation is that the nucleic acid-binding domain allows P3 to be associated through its C terminus with the viral genome during encapsidation, leaving the remaining part of the molecule free for interactions with both P4 and P2. In transmission experiments performed with purified particles, P3Δ61/80 should be unable to interact with the viral DNA already encapsidated. In the absence of such an interaction, formation of the transmissible complex would depend solely on the interaction between the mutated P3 and the capsid, an interaction that was shown to be weak by far-Western experiments. Thus, the inability of externally supplied P3 to access and interact with the viral genome in vitro may explain why P3 mediates CaMV transmission in vitro at a lower rate than in vivo. On the other hand, the 50% decrease in transmission rate with CaMV expressing P3Δ61/80 under natural conditions suggests that the region targeted by this deletion nonetheless has a significant role in the formation of a stable transmissible viral complex.
Deletions outside the P3-virion interaction domain had variable effects on the transmission rate. The reduced efficiency of transmission with P3Δ41/60 (67%) does not result from structural modifications affecting interactions between P3 and either virions (Fig. 5) or P2 (15), since these interactions analyzed separately are strong, but it may reflect the loss of a spacer required for the optimal positioning of these two interaction domains in the transmissible complex. The relatively high transmission rate (53%) obtained with mutant P3Δ21/40 was unexpected, since the deletion introduced into P3 affects a leucine zipper motif between amino acids 4 and 30 which is necessary for the P3-P2 interaction (15). Indeed, this mutant was unable to interact with P2 when it was tested in far-Western experiments (15). However, it appears that an α-helical region is restored at the N terminus of this P3 mutant because of the presence of another leucine zipper motif (23) immediately downstream of the deleted sequence (M. Bergdoll, personal communication). A possible explanation for the 53% transmission rate observed with P3Δ21/40 is that the newly formed secondary structure in this mutant allows the latter to engage in an interaction with P2 which is stable enough for the transmission of CaMV by aphids but which is disrupted under the ionic conditions used in far-Western experiments.
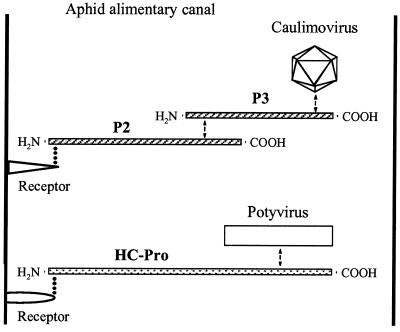
The most straightforward interpretation of our finding is that CaMV particles interact with P2 via P3, i.e., that P3 acts by creating a bridge between P2 and the virion to form a ternary transmissible complex (P2-P3-virion); however, we cannot strictly rule out the possibility that an initial interaction of P2 with P3 alters conformation of P2 so that it becomes competent to interact with the capsid. If, as appears likely, P3 is located between P2 and the virion in the transmissible complex, P2 is hypothesized to be involved in an interaction with a putative receptor located in the alimentary canal of the aphid stylet. Since the C-terminal part of P3 interacts with the viral capsid and its N terminus interacts with the C-terminal region of P2, P2 could bind via its N terminus to the putative receptor as does potyvirus HC-Pro (3). Schematic models of aphid-borne transmissible complexes for potyviruses and caulimoviruses are compared in Fig. 7. The existence of two factors of transmission by aphids in caulimoviruses, while only one is required for potyviruses, further illustrates the fact that different viruses transmitted in a noncirculative manner by the same insect vector may adopt different solutions to comply with the requirement of the “helper strategy” (discussed in reference 18). Caulimovirus proteins P2 and P3 are encoded by two ORFs separated by only one or a few nucleotides. They might have been originally associated in a single protein equivalent to the potyvirus protein HC-Pro and then have split into two separate proteins to allow P3 to ensure independently its functions during the infectious cycle inside the host plant.
FIG. 7.
Models of complexes transmissible by aphids for caulimoviruses and potyviruses. Retention of caulimovirus or potyvirus particles in the alimentary canal of M. persicae occurs at the level of a putative receptor (possibly distinct for each of the two groups of viruses) via a bridge consisting of proteins P2 and P3 for caulimoviruses and of the single protein HC-Pro for potyviruses (reviewed in reference 18). Relevant regions of interaction, known ( ) and hypothetical (
) and hypothetical (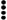 ), are indicated. Relative sizes of the components are not to scale.
), are indicated. Relative sizes of the components are not to scale.
Our tests for transmission by aphids were designed in a way that allowed aphids to acquire P2 first and the P3-virion complex later. The success of these tests is consistent with the fact that P2 can be retained alone in the aphids and can subsequently mediate retention of the P3-virion complex. Testing various combinations in the sequential acquisition of P2, P3, and purified virions will be very valuable for further understanding the mechanisms of CaMV retention in aphid mouthparts and/or its release therefrom and inoculation in a new host plant.
ACKNOWLEDGMENTS
We thank Philippe Hamman for help with automatic sequencing of the plasmid constructions, Takii and Co., Ltd. (Kyoto, Japan), for providing turnip hybrid Just Right, and Richard Wagner for growing plants. We are very grateful to Ken Richards for critical reading of the manuscript.
This work was supported by the Centre National de la Recherche Scientifique, the Université Louis Pasteur of Strasbourg, and the Institut National de la Recherche Agronomique.
REFERENCES
- 1.Atreya P L, Lopez Moya J J, Chu M, Atreya C D, Pirone T P. Mutational analysis of the coat protein N-terminal amino acids involved in potyvirus transmission by aphids. J Gen Virol. 1995;76:265–270. doi: 10.1099/0022-1317-76-2-265. [DOI] [PubMed] [Google Scholar]
- 2.Blanc S, Cerutti M, Usmany M, Vlak J M, Hull R. Biological activity of cauliflower mosaic virus aphid-transmission factor expressed in an heterologous system. Virology. 1993;192:643–650. doi: 10.1006/viro.1993.1080. [DOI] [PubMed] [Google Scholar]
- 3.Blanc S, Ammar E D, Garcia-Lamposona S, Dolja V V, Llave C, Baker J, Pirone T P. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J Gen Virol. 1998;79:3119–3122. doi: 10.1099/0022-1317-79-12-3119. [DOI] [PubMed] [Google Scholar]
- 4.Bonneville J M, Sanfaçon H, Fütterer J, Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- 5.Chapdelaine Y, Hohn T. The cauliflower mosaic virus capsid protein: assembly and nucleic acid binding in vitro. Virus Genes. 1998;17:139–150. doi: 10.1023/a:1008064623335. [DOI] [PubMed] [Google Scholar]
- 6.Cheng R H, Olson N H, Baker T S. Cauliflower mosaic virus: a 420 subunit (T = 7), multilayer structure. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dautel S, Guidasci T, Piqué M, Mougeot J L, Lebeurier G, Yot P, Mesnard J M. The full-length product of cauliflower mosaic virus open reading frame III is associated with the viral particle. Virology. 1994;202:1043–1045. doi: 10.1006/viro.1994.1435. [DOI] [PubMed] [Google Scholar]
- 8.Delseny M, Hull R. Isolation and characterization of faithful altered clones of the genomes of cauliflower mosaic virus isolates Cabb B-JI, CM4–184 and Bari. Plasmid. 1983;9:31–41. doi: 10.1016/0147-619x(83)90029-x. [DOI] [PubMed] [Google Scholar]
- 9.Herzog E, Guerra-Peraza O, Hohn T. The rice tungro bacilliform virus gene II product interacts with the coat protein domain of the viral gene III polyprotein. J Virol. 2000;74:2073–2083. doi: 10.1128/jvi.74.5.2073-2083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hull R, Shepherd R J, Harvey J D. Cauliflower mosaic virus: an improved purification procedure and some properties of the virus particle. J Gen Virol. 1976;31:93–100. [Google Scholar]
- 11.Jacquot E, Hagen L S, Jacquemond M, Yot P. The open reading frame 2 product of cacao swollen shoot badnavirus is a nucleic acid-binding protein. Virology. 1996;225:191–195. doi: 10.1006/viro.1996.0587. [DOI] [PubMed] [Google Scholar]
- 12.Jacquot E, Keller M, Yot P. A short basic domain supports the nucleic acid-binding activity of the open reading II product of rice tungro bacilliform virus. Virology. 1997;239:352–359. doi: 10.1006/viro.1997.8859. [DOI] [PubMed] [Google Scholar]
- 13.Jacquot E, Geldreich A, Keller M, Yot P. Mapping regions of the cauliflower mosaic virus ORF III product required for infectivity. Virology. 1998;242:395–402. doi: 10.1006/viro.1997.8995. [DOI] [PubMed] [Google Scholar]
- 14.Leclerc D, Burri L, Kajava A V, Mougeot J L, Hess D, Lustig A, Kleemann G, Hohn T. The open reading frame III product of cauliflower mosaic virus forms a tetramer through a N-terminal coiled-coil. J Biol Chem. 1998;273:29015–29021. doi: 10.1074/jbc.273.44.29015. [DOI] [PubMed] [Google Scholar]
- 15.Leh V, Jacquot E, Geldreich A, Hermann T, Leclerc D, Cerutti M, Yot P, Keller M, Blanc S. Aphid-transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO J. 1999;18:7077–7085. doi: 10.1093/emboj/18.24.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Moya J J, Wang R Y, Pirone T P. Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J Gen Virol. 1999;80:3281–3288. doi: 10.1099/0022-1317-80-12-3281. [DOI] [PubMed] [Google Scholar]
- 17.Mougeot J L, Guidasci T, Wurch T, Lebeurier G, Mesnard J M. Identification of C-terminal amino acid residues of cauliflower mosaic virus open reading frame III protein responsible for its DNA binding activity. Proc Natl Acad Sci USA. 1993;90:1470–1473. doi: 10.1073/pnas.90.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirone T P, Blanc S. Helper-dependent vector transmission of plant viruses. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 19.Rothnie H, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt I, Blanc S, Esperandieu P, Kuhl G, Devauchelle G, Louis C, Cerutti M. Interaction between the aphid transmission factor and virus particles is part of the molecular mechanism of cauliflower mosaic virus aphid transmission. Proc Natl Acad Sci USA. 1994;91:8885–8889. doi: 10.1073/pnas.91.19.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoelz L, Shepherd R J. Host range control of cauliflower mosaic virus. Virology. 1988;162:30–37. doi: 10.1016/0042-6822(88)90391-1. [DOI] [PubMed] [Google Scholar]
- 22.Thornbury D W, Hellmann G M, Rhoads R E, Pirone T P. Purification and characterization of potyvirus helper component. Virology. 1985;144:260–267. doi: 10.1016/0042-6822(85)90322-8. [DOI] [PubMed] [Google Scholar]
- 23.Tsuge S, Kobayashi K, Nakayashiki H, Mise K, Furusawa I. Cauliflower mosaic virus ORF III product forms tetramer in planta: its implication in viral DNA folding during encapsidation. Microbiol Immunol. 1999;43:773–780. doi: 10.1111/j.1348-0421.1999.tb02469.x. [DOI] [PubMed] [Google Scholar]