Abstract
In all retroviruses analyzed to date (except for the spumaretroviruses), the Zn2+-coordinating residues of nucleocapsid (NC) perform or assist in crucial reactions necessary to complete the retrovirus life cycle. Six replication-defective mutations have been engineered in the two NC Zn2+ fingers (ZFs) of simian immunodeficiency virus [SIV(Mne)] that change or delete specific Zn2+-interacting Cys residues and were studied by using electron microscopy, reversed-phase high-performance liquid chromatography, immunoblotting, and RNA quantification. We focused on phenotypes of produced particles, specifically morphology, Gag polyprotein processing, and genomic RNA packaging. Phenotypes were similar among viruses containing a point or deletion mutation involving the same ZF. Mutations in the proximal ZF (ZF1) resulted in near-normal Gag processing and full-length genomic RNA incorporation and were most similar to wild-type (WT) virions with electron-dense, conical cores. Mutation of the distal ZF, as well as point mutations in both ZFs, resulted in more unprocessed Gag proteins than a deletion or point mutation in ZF1, with an approximate 30% reduction in levels of full-length genomic RNA in virions. These mutant virions contained condensed cores; however, the cores typically appeared less electron dense and more rod shaped than WT virions. Surprisingly, deletion of both ZFs, including the basic linker region between the ZFs, resulted in the most efficient Gag processing. However, genomic RNA packaging was ∼10% of WT levels, and those particles produced were highly abnormal with respect to size and core morphology. Surprisingly, all NC mutations analyzed demonstrated a significant loss of processed NC in virus particles, suggesting that Zn2+-coordinated NC is protected from excessive proteolytic cleavage. Together, these results indicate that Zn2+ coordination is important for correct Gag precursor processing and NC protein stability. Additionally, SIV particle morphology appears to be the result of proper and complete Gag processing and relies less on full-length genomic RNA incorporation, as dictated by the Zn2+ coordination in the ZFs of the NC protein.
Retroviral nucleocapsid (NC) proteins are typically small basic proteins containing one or two conserved 14-residue peptide segments (-C-[X]2-C-[X]4-H-[X]4-C-) that bind zinc ions, forming structures referred to as zinc fingers (ZFs). NC proteins with native ZFs are required for viral replication and are apparently utilized at many stages of the replication cycle. The biological roles of retroviral NC proteins and their ZFs have been reviewed recently (18, 19) and include functions during viral assembly, maturation, and infection. Regarding particle morphology, the basic residues of NC are required for consistent and proper particle size (21), and the ZFs, when mutated or deleted, result in virions that do not mature properly (7). In fact, a number of the functions of NC are attributed to the ZFs.
All human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) NC proteins contain two ZFs. It is currently believed that each ZF is required for viral replication; however, they are not necessarily equally required for each and every specific function during the replication cycle. In HIV-1, the proximal Zn2+ finger (ZF1) functions in genomic RNA incorporation, viral core morphogenesis, and proviral DNA synthesis (44, 45), while the intact distal Zn2+ finger (ZF2) is required for Gag polyprotein stability during proteolytic processing (35). It is clear in HIV-1 that one finger cannot replace the function of another (25), so there is some degree of independence in ZF activity, but there is also an overlap of many of the NC functions attributed to both ZFs (23, 35, 44, 45). However, these functions may not be consistent among all NC proteins that contain two ZFs.
The functions of HIV-1 and SIV NC ZFs are of particular interest because of their vital roles in viral replication and their potential as targets for viral inactivation (mutational as well as chemical targets). At present, little work has been accomplished to address ZF function in SIV (23). Because these viruses are presently used in many of the in vivo models of HIV-1 disease and vaccination (3, 29, 30, 39, 48) and the retroviral NC protein is presently a recognized target for therapeutic (6) and vaccine (2, 43) development, it is imperative to understand the viral physiology dictated by this protein in the SIV system. To address this, we have mutated the specific Zn2+-binding Cys residues to Ser of each or both fingers of SIV(Mne) NC to eliminate ion binding (8), yet maintain as much of the protein conformation as possible. We have also deleted the first 4 amino acids of each finger or the entire finger-linker sequence to assess finger function differences between the Zn2+-coordinated conformation and primary amino acid sequence conformation. In this report, we have focused only on SIV(Mne) NC function relative to genomic RNA incorporation, Gag polyprotein processing, particle formation, and maturation. Here we provide a detailed analysis of the subtle differences in function attributed to, but not unique to, the individual ZFs of SIV(Mne) NC.
MATERIALS AND METHODS
Mutagenesis and plasmid construction.
SIV(Mne) was isolated and cloned as described previously (4). The generation of the full-length proviral clone and deletion mutants of SIV(Mne) NC has been described previously (23). The subclone pPP1 (23) of SIV(Mne) was used as a template for the generation of point mutants of NC ZFs. The Cys coding sequences in the NC gene were changed to Ser by site-directed mutagenesis (QuickChange site-directed mutagenesis kit; Stratagene, La Jolla, Calif.) with primers (obtained from Life Technologies, Rockville, Md.) designed specifically for each ZF as follows (the mutagenic nucleotides are underlined): ZF1 C→S, plus strand 5′-TAA GAG TTG GAA TAG TGG GAA AGA GGG ACA CTC TGC AAG GCA AAG CAG [5′ end corresponding to nucleotide [nt] position 1705 of the SIV(Mne) sequence; GenBank accession no. M32741], producing pPP1-4.1r; and ZF2 C→S, plus strand 5′-GGC AGC TGG AAA AGT GGA CAA ATG GGC CAT GTT ATG GCC AAA AGC C [5′ end corresponding to nt position 1769 of the SIV(Mne) sequence], producing pPP1-6.15. Cys→Ser changes in both ZFs were introduced by mutagenesis of pPP1-4.1r with the ZF2 C→S mutational oligonucleotide, producing pPP1-4.1.5. All mutations were confirmed by nucleotide sequencing and are noted in the 52-amino-acid SIV(Mne) NC protein sequence (31) depicted in Fig. 1.
FIG. 1.
Protein sequence of WT and mutant SIV(Mne) NC proteins. Zinc-coordinating residues are indicated with residue numbers. Dotted lines indicate Zn2+ coordination. A dashed line indicates deletions.
SIV coding sequences were transferred to mammalian expression constructs that are all based in the pCEP4 expression vector from Invitrogen (Carlsbad, Calif.). A portion of the SIV(Mne) proviral sequence was excised from plasmid pRB130 (23) at the unique BsiEI sites in the U3 region of the long terminal repeats (LTRs), producing a 9.4-kbp fragment. Plasmid pRB130 is an expression plasmid that has a full-length SIV(Mne) proviral clone containing a deletion in the coding sequence for the first four amino acids of NC ZF2 (ΔCys33 to Cys36; ΔZF2) (23). The BsiEI fragment was blunt ended with T4 polymerase (Promega Corp., Madison, Wis.) and cloned into the unique PvuII site of pCEP4, producing pCEP4-ΔZF2. In order to manipulate the viral LTR and promoter sequences, pBks5′LTRmne was constructed containing the SalI-BamHI proviral fragment of SIV(Mne) from pRB130 [the SalI site is contained in the uncharacterized mammalian flanking sequence upstream of the 5′ LTR, and the BamHI site is located in the SIV(Mne) coding sequence at nt position 1329; GenBank accession no. M37241] in the homologous sites of pBluescriptKS+ pBks+; Stratagene). Next, pBks5′LTRmne was digested with StyI and blunt ended with T4 polymerase, and the largest StyI fragment was ligated with the blunt-ended 1,722-bp SalI-BanI cytomegalovirus (CMV) enhancer sequence from pCEP4 (nt 594 to 1316) in the U3 region of the 5′ LTR in pBks5′LTRmne. The resulting plasmid was designated pBks5′LTRmne-eCMVs8, in which the CMV enhancer was ligated in the negative orientation (with respect to the LTR), because gene expression was greatest in this conformation. The final expression vector used in these studies, designated pS8, was generated by replacing the NruI-BamHI fragment of pCEP4-ΔZF2 (which removed the entire original CMV promoter-enhancer) with an EcoRV-BamHI fragment from plasmid pBks5′LTRmne-eCMVs8 containing the CMV immediate-early enhancer sequence (nt 594 to 1316 in pCEP4) at the blunt-ended StyI site in the 5′ LTR of SIV(Mne), thus retaining the known NF-κB and SpI binding sites in the 5′ LTR, but removing the sequence of the 5′ LTR upstream from the StyI site. Each deletion and point mutation from the pPP1 series was introduced into pS8 by exchanging BamHI-Sse8387I fragments containing NC sequence. In the 3′ LTR, the known NF-κB and SpI binding sequences of U3, as well as the R and U5 regions, have been replaced with the simian virus 40 (SV40) poly(A) signal, leaving the nef sequence intact.
Transfections.
293T cells were transfected as previously described (26) with equimolar amounts of proviral plasmids by calcium phosphate precipitation (5′→3′, Inc., Boulder, Colo.). Supernatants were harvested and clarified as previously described (24).
Electron microscopy and particle analysis.
Supernatants from transfected 293T cells were clarified by centrifugation, and virus was pelleted at 100,000 × g through a 20% (wt/vol) sucrose cushion in phosphate-buffered saline (PBS). Virus pellets were fixed in 1.25% (vol/vol) glutaraldehyde in PBS. Electron microscopy was performed with a Hitachi H-7000 microscope at 75 kV as previously described (23). Six photographs were taken of each virus pellet, with an average number of 119 virions per photo. Virus particles (magnification of ×90,000) were digitized with a ScanAce III scanner (Pacific Image Electronics Co., Ltd., Torrance, Calif.) at a 600-dpi resolution and processed with Adobe Photoshop (Adobe Systems, Inc., Mountain View, Calif.).
SIV p28CA antigen capture assay.
Virus was produced and pelleted as described above and then resuspended in PBS. Serial dilutions of sample aliquots and standards were prepared and assayed as previously described for aldrithiol-2-treated HIV p24CA (43), except that the capture antibody (AIDS Vaccine Program, Biological Products Laboratory [AVP-BPL] inventory no. 0089A-9B5-8BE5-6AC10) and detector antibody (AVP-BPL inventory no. DJ-40115, bleed 10263) were specific for SIV(Mne). Assay standards were based on a wild-type (WT) SIV(Mne), clone E11S stock, produced from infected HuT78 cells that had been accurately quantitated by reversed-phase high-performance liquid chromatography (RP-HPLC) and amino acid composition.
Viral protein western analysis.
Virus was produced and pelleted as described above. Samples containing equivalent reverse transcriptase (RT) activity (3.8 × 105 cpm by exogenous template RT activity assay [27]) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 20% Tris-glycine Novex gels (Invitrogen Corp.), immunoblotted as described previously (25), and then incubated with mouse monoclonal anti-p28CA antibodies (AVP-BPL inventory no. 96-0020 89A P9B5 H8B1) and visualized with the Amersham ECL (enhanced chemiluminescence) system (Arlington Heights, Ill.). Membranes were stripped according to manufacturer's recommendations (Amersham ECL system) and then incubated with rabbit polyclonal anti-gp120 and anti-p8NC antibodies (AVP-BPL no. DJ-36603 003545 and DJ-37545 005970, respectively) concomitantly and visualized with ECL as well. Lyophilized fractions from RP-HPLC were fractionated by SDS-PAGE and immunoblotted as described above. Membranes were sequentially incubated no more than three times with rabbit polyclonal anti-p28CA (no. B2119-5), anti-p8NC (no. 62837), anti-p6Gag (no. 63157), and anti-p2Gag (no. 63158) antibodies supplied by Raoul Benveniste, Basic Research Laboratory, National Cancer Institute at Frederick, Frederick, Md. Each hybridization was followed by ECL visualization. Blots were completely stripped of all antisera before being reprobed with the next antibody.
RP-HPLC separation of SIV(Mne) WT and ZF mutant proteins.
Pelleted virus from equivalent supernatant volumes of transfected 293T cultures was first resuspended in 50 μl of PBS and then reduced in 10 mM dithiothreitol (Calbiochem, La Jolla, Calif.) in the presence of 50 μl of 8 M sequenal-grade aqueous concentrate of high-purity guanidine-HCl (Pierce, Rockford, Ill.) and 10% (vol/vol) acetonitrile (EM Science, Gibbstown, N.J.) for 5 min at 37°C and loaded onto a Poros R2/H narrow-bore column (2.1 by 100 mm) (PerSeptive Biosystems, Inc., Framingham, Mass.). RP-HPLC was performed with a Shimadzu system as described previously (36). Fractions were eluted at a flow rate of 0.3 ml/min with a variable gradient of acetonitrile in 0.1% (vol/vol) Spectro-grade trifluoroacetic acid (Applied Biosystems, Warrington, United Kingdom) as follows: from 10 to 36.5% (vol/vol) acetonitrile for 12 min, 36.5 to 37% for 4 min, 37 to 41% for 7 min, 41 to 70% for 12 min, and 70% for 5 min. Peaks were detected by UV absorption at 206, 280, and 260 nm and analyzed by sequencing with an automated Applied Biosystems, Inc., Procise Protein Sequencer, SDS-PAGE, and immunoblot detection with the ECL system (Amersham Life Science). Fractions were collected every 30 s. Selected fractions were also analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry as described previously (36).
Viral RNA analysis.
Viral RNA was prepared as previously described (23). Samples were normalized for equivalent RT activity (4.2 × 106 cpm) determined from pelleted virus samples. RNA was electrophoresed through a formaldehyde-agarose gel, transferred to a nitrocellulose membrane, and treated as described previously (27). WT RNA was additionally analyzed at dilutions of 1:4, 1:16, 1:64, and 1:128. The full-length WT SIV(Mne) proviral plasmid pRB85 (23) was cut with XhoI restriction enzyme, random-prime 32P labeled (DECAprimeII kit; Ambion, Inc., Austin, Tex.), and used as a probe for full-length genomic RNA. RNA blots were hybridized overnight at 42°C in a mixture of 50% (vol/vol) formamide, 120 mM Na2HPO4 (pH 7.2), 250 mM NaCl, 7% (wt/vol) SDS, and the entire Sephadex G-50 (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) spin column-purified labeled probe. Blots were washed as previously described (23). Phosphorimager analysis was performed with bands of full-length genomic RNA with a Bio-Rad Molecular Imager FX phosphorimager with a K-Imaging Screen (Hercules, Calif.).
Quantitative RT-PCR was performed as previously described (23) with aliquots from RNA samples described above.
RESULTS
Virion morphology.
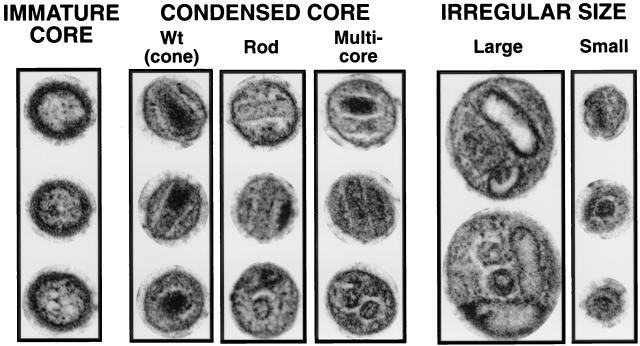
To characterize the effect of the removal of zinc-binding residues of NC on particle formation and maturation, mutations were introduced into the SIV(Mne) viral coding sequence. Specifically, the 52-amino-acid NC domain of Gag was altered to produce the mutants listed in Fig. 1. Virus particles were produced from 293T cells by transfection of DNA plasmids containing the complete coding sequence of WT or the various NC mutants of SIV(Mne). Virions were collected by centrifugation and examined by electron microscopy with positive staining. Electron micrographs were digitized so that individual virions could be sorted according to specific morphological criteria. Virus particles were isolated from two representative photographs of each mutant and categorized as (i) mature morphology if a condensed core and thin capsid membrane were observed; (ii) immature morphology if a noncondensed core and thick capsid membrane were observed, or (iii) abnormally large or small morphology if particles deviated from the average 100-nm diameter of WT particles by greater than 10% (Fig. 2). Particles were excluded if they contained both characteristics and/or cores were not visible because of the plane of thin sectioning (Table 1). Those particles of condensed core morphology were subcategorized as (i) cone, or WT core of high electron density; (ii) rod core of low electron density; or (iii) multiple cores, all of which were rod morphologies.
FIG. 2.
Electron microscopic classification of virion morphology. Three typical examples of each type of morphology (except irregular large) are shown. The upper two virions in each category are typical longitudinal cross-sections, and the lower virion in each category is a typical latitudinal cross-section. Virus was pelleted from clarified supernatants of 293T cells 72 h after transfection with mutant or WT plasmid DNA. Images are digital and were scanned from electron micrographs at a magnification of ×90,000.
TABLE 1.
Morphological analysis of WT and mutant SIV(Mne) virions
| Type of virions | Total no. of virionsa | % of virions
|
|||||
|---|---|---|---|---|---|---|---|
| Immature core | Condensed core
|
Abnormally largeb | Abnormally smallc | ||||
| WT | Rod shaped | Multiple rods | |||||
| WT | 122 | 2 | 88 | 6 | 4 | 0 | 0 |
| ZFI mutation | |||||||
| ZF1 C→S | 215 | 26 | 17 | 21 | 24 | 12 | 0 |
| ΔZF1 | 220 | 24 | 14 | 32 | 13 | 17 | 0 |
| ZF2 mutation | |||||||
| ZF2 C→S | 185 | 30 | 2 | 35 | 19 | 13 | 1 |
| ΔZF2 | 218 | 28 | 3 | 43 | 11 | 14 | 1 |
| Dual mutation | |||||||
| ZF1+2 C→S | 153 | 20 | 1 | 44 | 10 | 20 | 5 |
| ΔZF1+2 | 89 | 7 | 1 | 10 | 2 | 26 | 55 |
Total number of virions minus those of undefinable morphology. Numbers of virions with undefinable morphologies were not significantly different among all mutant and WT virions observed.
Virions >∼110 nm in diameter.
Virions <∼90 nm in diameter.
An examination of the purified particles by electron microscopy showed that the WT virus produced particles with normal dimensions (∼100 nm) (Table 1), whereas all of the mutants had a significant number of abnormally large particles (12 to 26%) (Table 1). Larger-than-normal particles probably result from abnormal function of the Gag precursor during initial phases of assembly and budding. The fact that each mutant shows a distribution of particle dimensions suggests that both normal and abnormal assembly paths are possible. It is interesting to note that point mutants or deletions which affect one ZF but leave the other intact (ZF1 mutants and ZF2 mutants) (Table 1) show about the same proportion of large particles (14% ± 3%). Point mutations destroying both ZFs (ZF1+2 C→S) (Table 1) produce a slightly greater proportion of large particles (20%), whereas deletion of both ZFs (ΔZF1+2) resulted in a significant number of abnormally small particles (55%) in addition to an increased proportion of large particles (26%). These data show that from the limited standpoint of particle dimension, there is little or no difference between the mutants, except for mutant ΔZF1+2 with both ZFs deleted.
All of the mutants exhibited a higher proportion of normal-sized particles with immature morphology (7 to 30%) than was observed with the WT virus (2%). The double deletion mutant, ΔZF1+2, produced very few particles with normal dimensions (Table 1), and it was therefore difficult to assess the proportion of particles with immature morphologies.
The major differences between the normal-sized particles produced by the various mutants examined in Table 1 were in the morphology of condensed cores. At least three distinctly different condensed core morphologies could be identified; condensed cores with a WT cone shape; condensed cores with a rod shape, and condensed core with multiple rods. Mutations in the first ZF (deletion or point) gave the highest proportion of particles with WT cores (14 to 17%), but most of the condensed cores were rod shaped. Mutations in the second ZF (deletion or point) gave an even higher proportion of particles with rod-shaped cores and very few particles with WT-shaped cores.
Taken together, these results show that deletion of both ZFs and the linker region between ZFs (ΔZF1+2) dramatically reduces the number of normal-sized particles and is consistent with previous observations stressing the importance of the linker in particle formation (10, 14, 21, 37). The ZFs seem to play an important role in determining the extent of maturation and morphology of the condensed core. Mutations affecting either ZF increase the number of immature forms and the number of particles with rod-shaped cores.
Virus particle production.
To determine the ratio of particle-associated Gag-Pol polyprotein to Gag produced by the NC mutant and WT transfections, virus was quantitated by RT activity in an exogenous template assay (to estimate Gag-Pol levels) and Gag expression through a p28CA antigen capture assay. Table 2 shows that the Gag-Pol/Gag ratios are relatively constant among the mutant and WT viruses, except for the ΔZF1+2 mutant. RT activity was measured in viral pellets as well as polyethylene glycol (PEG)-precipitated culture supernatants to observe any differences in particle-associated and supernatant RT. These results are also summarized in Table 2. All mutants demonstrate similar ratios of particle-associated to supernatant RT levels.
TABLE 2.
Comparison of RT activity in culture supernatant and viral pellets to that of virus-associated p28CA
| Sample | p28CA concn (ng/ml)a | RT activity (cpm/ml)b
|
RTPP/p28CA ratioc | RTPP/RTVP ratiod | |
|---|---|---|---|---|---|
| Virus pellet | PEG precipitated | ||||
| Mock | 0 | 0 | 0 | ||
| ZF1 C→S | 2,232 | 209,900 | 373,500 | 167 | 1.78 |
| ZF2 C→S | 652 | 49,410 | 141,140 | 216 | 2.86 |
| ZF1+2 C→S | 778 | 109,300 | 171,500 | 220 | 1.57 |
| ΔZF2 | 1,584 | 175,500 | 315,500 | 199 | 1.80 |
| ΔZF1 | 1,543 | 184,300 | 319,000 | 207 | 1.73 |
| ΔZF1+2 | 1,279 | 204,200 | 455,400 | 356 | 2.23 |
| S8-WT | 4,173 | 381,800 | 1,079,400 | 259 | 2.16 |
| WT | 5,418 | 691,700 | 1,203,010 | 222 | 1.74 |
Nanograms of virus-associated p28CA per milliliter of culture fluid measured by antigen capture.
cpm of [3H]TTP incorporated per milliliter of culture fluid.
cpm per nanogram of RT/p28CA in equivalent volumes.
Relative ratio of PEG-precipitated (RTPP) to virus pellet (RTVP) RT activity.
Genomic RNA incorporation.
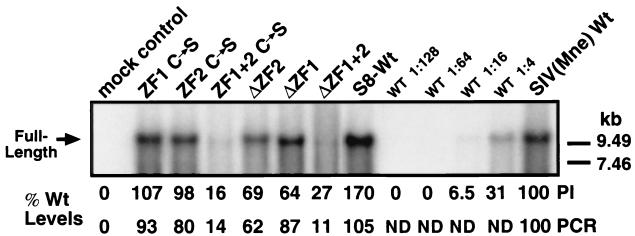
To determine if the morphological differences observed could be attributed to variations of full-length genomic RNA incorporation, particle-associated RNA was isolated and analyzed by Northern blotting and quantitative RT-PCR (Fig. 3) (23). It appears that the alteration of NC ZF1 does not significantly affect genomic RNA incorporation, yet the alteration of NC ZF2 results in reduced incorporation of full-length genomic RNA. Similar to the ΔZF1+2 mutant (23), ZF1+2 C→S incorporates <20% WT levels of full-length genomic RNA. These results suggest that there is no correlation between genomic RNA incorporation and viral core condensation. While it is apparent that loss of zinc coordination results in some reduction of genomic RNA encapsidation—and ZF2 appears to function in this capacity more so than ZF1—the observations with the ΔZF1+2 mutant suggest that it is the linker region and basic charge of NC that influence genomic RNA encapsidation the greatest.
FIG. 3.
Analysis of genomic RNA incorporation into WT and NC mutant SIV(Mne) virions. Virus was pelleted from clarified supernatants of transfected 293T cells, and viral RNA was isolated by phenol-chloroform extraction–ethanol precipitation. Samples, equalized for pelletable RT activity, were probed with the pRB85 vector containing full-length SIV(Mne) that had been linearized with XhoI and then labeled with 32P (23). The mock control lane contains an RNA preparation from carrier DNA-transfected 293T supernatants. Full-length genomic RNA (9.6 kbp) indicated on the left was quantitated by phosphorimaging (PI) and quantitative real-time RT-PCR (PCR). Positions of RNA markers are shown on the right. ND, not determined.
Polyprotein processing.
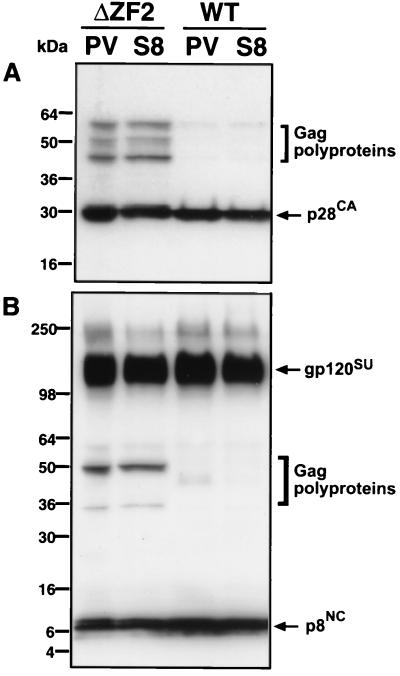
To determine if improper or incomplete proteolytic cleavage of the Gag, Gag-Pol, and Env polyproteins could be associated with the observed morphological differences, WT and mutant viruses were examined by Western analysis and fractionation by RP-HPLC. Because this laboratory focuses on the development of novel vaccines against HIV and SIV, a plasmid vector (pS8) was designed for use as a naked DNA vaccine. pS8 consists of the full-length SIV(Mne) coding sequence, but the LTRs have been partially deleted (see Materials and Methods) for increased in vivo safety purposes. Briefly, the R and U5 regions in the 3′ LTR have been replaced by an exogenous SV40 polyadenylation sequence to prevent first-strand transfer during viral DNA synthesis, yet provide the polyadenylation necessary for proper viral gene expression. The LTR deletions, in combination with the specific NC mutations, render these viruses replication incompetent (data not shown). Because of the changes in LTR structure in the pS8 expression vector, it was necessary to demonstrate that particle assembly and polyprotein processing were not affected. Viral proteins of the WT and an example of one of the NC mutants, ΔZF2, were compared in the context of the pS8 vector or a complete proviral vector (Fig. 4). By gp120SU, p28CA, and p8NC Western analysis, no significant differences in polyprotein processing were observed if the LTR sequences were altered as described for the pS8 vector. While these LTR modifications decrease infectivity at least 3 logs in the clone containing WT NC (data not shown), Fig. 4 demonstrates that the processing differences observed were clearly the result of the mutation present in the NC domain and were not due to the LTR alterations.
FIG. 4.
Immunoblot comparison of viral protein processing with complete or deleted LTR ends. WT or ΔZF2 mutant SIV(Mne) virus was produced by transfection of 293T cells with plasmids containing complete LTRs (provirus [PV]) or deleted LTRs (S8) as described in Materials and Methods. Virus pelleted from clarified supernatants 72 h after transfection was resuspended in SDS-PAGE lysis buffer and then subjected to Western blot analysis. Protein samples were adjusted to equivalent RT activity before SDS-PAGE. Blots were initially incubated with mouse monoclonal p28CA antibody (A) and then stripped and reprobed concomitantly with rabbit polyclonal gp120 and p8NC antibodies (B). Molecular markers are indicated on the left, and immunodetected proteins are indicated on the right.
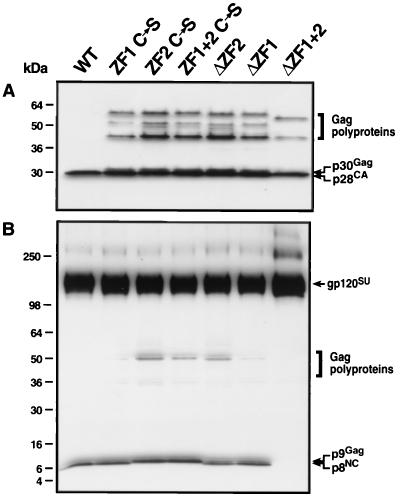
To compare WT and mutant polyprotein processing, immunoblot analysis was performed with pelleted virus (Fig. 5). All mutants were able to process Env polyprotein similar to the WT (Fig. 5B). However, the degree of Gag processing was subtly different, depending on the ZF mutation (Fig. 5). Deletion and point mutations in ZF2 resulted in the highest accumulation of unprocessed Gag, while mutations in ZF1 resulted in less accumulation of unprocessed Gag. Point mutations in both ZFs resulted in accumulation of unprocessed Gag similar to ZF2 mutations. All mutants except for ΔZF1+2 produced a significant amount of slower-migrating p28CA (designated p30Gag; Fig. 5A), which is presumably unprocessed p28CA-p2Gag. Also, an additional NC-reactive band was observed (p9Gag; Fig. 5B) that is presumably unprocessed p8NC-p1Gag. Surprisingly, the smallest amount of unprocessed Gag was observed with ΔZF1+2; however, this mutant also contained significantly higher levels of virus-associated gp120SU (Fig. 5B) and no detectable NC protein (Fig. 5B and 6), possibly due to the loss of reactive epitopes in the deleted sequence. Interestingly, only point mutants of ZF2 or ZF1+2 produced NC-containing proteins of slower mobility. From these data, it can be summarized that alteration of ZF2 results in more unprocessed Gag than alteration of ZF1, but the removal of both ZFs and the basic linker results in very efficient Gag cleavage.
FIG. 5.
Immunoblot analysis of viral protein processing in WT and NC mutant SIV(Mne). Virus was pelleted as described in Materials and Methods and resuspended in SDS-PAGE lysis buffer. Samples of equivalent RT activity (3.8 × 105 cpm by exogenous template RT assay) were heat denatured, subjected to 4 to 20% SDS-PAGE, and transferred to Immobilon-P (Millipore Corp.). Membranes were probed with mouse monoclonal p28CA antibodies and visualized with the Amersham ECL system (A). Membranes were stripped and reprobed with rabbit anti-gp120 and p8NC concomitantly and visualized with ECL as well (B). Molecular mass markers are shown on the left, and immunolabeled proteins are shown on the right.
FIG. 6.
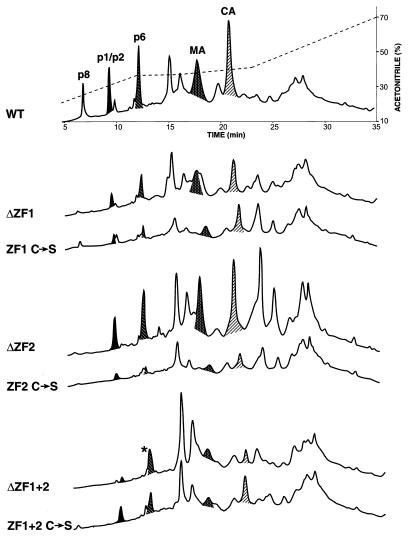
RP-HPLC analysis of WT and NC mutant SIV(Mne). Virus was pelleted from 30 ml of clarified culture supernatant of 293T cells transfected with WT or NC mutant expression plasmids and then resuspended in PBS in the presence of 8 M guanidine-HCl, 10% acetonitrile, and 10 mM dithiothreitol. Equivalent volumes were loaded on a microbore RP-HPLC column and eluted as described in Materials and Methods. Chromatographs are grouped according to the ZF mutation for ease of comparison. The acetonitrile concentration is superimposed over the WT chromatograph. Viral protein peaks were determined by Coomassie staining, immunoblotting, and protein sequencing analyses of eluted fractions and are labeled accordingly: NC, nucleocapsid protein; p1/p2, spacer protein 1 and spacer protein 2 of Gag polyprotein; MA, matrix; and CA, capsid. An asterisk indicates contamination of the p6Gag peak with an unidentified cellular protein in the ΔZF1+2 chromatograph.
To quantitate relative amounts of viral proteins and determine the nature of unprocessed Gag polyproteins, WT and mutant virus was pelleted and subjected to RP-HPLC fractionation and various peptide analyses. RP-HPLC fractions were collected and subsequently analyzed by Western blotting with antisera to specific Gag proteins. The ability to separate and identify viral proteins in this manner allows much more accurate quantitation than conventional Western analysis of whole viral preparations. Chromatographs comparing WT and mutant virus are shown in Fig. 6. Two significant observations were noted. First, the peak height of the eluted NC protein relative to other processed Gag proteins (p1Gag/p2Gag, p6Gag, p16MA, and p28CA) is reduced and shifted to the left in chromatographs of the mutants where NC is detectable, compared to that in the WT. Mutations in ZF2 or both ZFs resulted in less NC protein than mutations in ZF1. Confirming the results presented in Fig. 5B, NC protein is undetectable in ΔZF1+2 by RP-HPLC. These results show that there is a significant loss of the processed p8NC protein in all mutants.
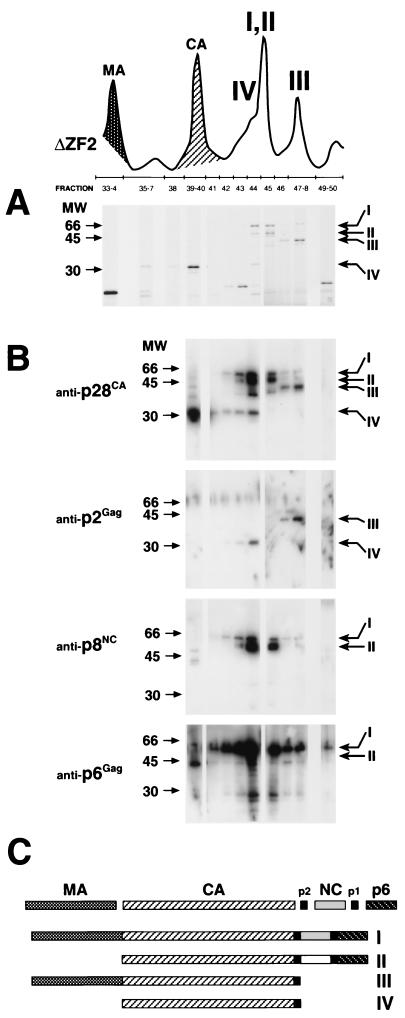
The second significant observation in the chromatographs in Fig. 6 is the relative increase of two peaks following elution of p28CA (e.g., ΔZF2). Because these peaks might be the accumulated unprocessed Gag polyproteins observed in the Western analyses (Fig. 5), fractions were collected, fractionated by SDS-PAGE, and analyzed by Coomassie staining and Western analysis. Figure 7 shows an example of an analysis of a representative mutant, ΔZF2, showing the portion of the chromatograph following elution of p6Gag. Fractionated proteins in the peaks were separated by SDS-PAGE, visualized by Coomassie staining, and determined to consist of four major proteins (I to IV in Fig. 7A). Next, the same fractions were blotted and tested for reactivity to antisera against p28CA, p2Gag, p8NC, and p6Gag (Fig. 7B). Because available anti-p16MA antibodies did not recognize p16MA epitopes in the Gag precursor, the presence of the p16MA domain was confirmed in some fractions by proteolytic digestion and peptide sequencing. Also, the anti-p2Gag antibodies only recognized p2Gag epitopes in completely processed p2Gag or when the Gag precursor was cleaved at the p2Gag-p8NC junction (i.e., a p2Gag with a free C terminus). For example, protein III in Fig. 7A reacted with antibodies to p2Gag and p28CA, but not to p8NC or p6Gag (Fig. 7B); it contained p16MA peptide sequence; and it was approximately 44 kDa in size. Therefore, it was concluded that protein III was a partially processed Gag precursor consisting of MA, CA, and p2Gag (Fig. 7C). Fractions across the entire RP-HPLC were analyzed for Gag precursors by Western blotting, but no peaks other than I to IV contained partially processed or uncleaved Gag polyproteins. From these results, the elution and consistency of partially processed and uncleaved viral Gag polyproteins were identified and diagrammed as proteins I to IV in Fig. 7C. All mutant particles contained the same molecules of Gag proteins I to IV, but in different proportions, as observed by peak heights of proteins I to IV relative to the other, fully processed Gag proteins within the same chromatograph (Fig. 6).
FIG. 7.
SDS-PAGE and immunoblot analysis of incomplete Gag polyprotein processing. (A) A section of the chromatograph from Fig. 6 (ΔZF2) showing an example of the NC mutant, ΔZF2, RP-HPLC, and numbered protein fractions that were fractionated by SDS-PAGE and visualized with Coomassie stain. Four distinct proteins (I to IV) were noted in peaks more prominent in the NC mutant than in the WT SIV(Mne) chromatographs. (B) Immunoblot analyses of fractions for anti-p28CA, anti-p2Gag, anti-p8NC, and anti-p6Gag antibodies. Molecular markers are shown on the left; the positions of labeled proteins I to IV are indicated on the right. Note that the p2Gag antibody only recognizes a cleaved or C-terminal epitope. (C) Identity of incompletely processed Gag proteins I to IV based on Coomassie, immunoblot, and partial amino acid sequence analyses. The same Gag polyproteins I to IV were observed in all NC mutants, only in different amounts.
To quantify the relative loss of NC protein in the mutant virions, the peak areas of fully processed (Fig. 6) and partially processed NC (Fig. 7, peaks I and II) were compared to those of processed (Fig. 6) and unprocessed CA (Fig. 7, peaks I to IV) within each of the mutants and the WT. The results were expressed as a ratio of processed NC to processed CA and p6Gag to CA in Table 3. The majority of detectable mutant NC that is present in the virion is in the form of uncleaved and partially processed Gag precursor (72 to 99%), whereas the majority of detectable WT NC is fully processed (86%). In the mutant virions, CA is present approximately equally in fully and partially processed forms (41 to 70%). The mutant virions contained a greater variation in the levels of processed and unprocessed p6Gag. As expected, the relative ratio of processed CA to processed NC in WT is nearly 1:1 (0.94), as is processed CA to processed p6Gag; however, NC/CA ratios in the mutants demonstrate a significant loss of processed NC. At present, it cannot be stated whether this apparent loss of NC protein is due to proteolytic degradation or alternative NC processing into smaller fragments, but no detectable fragments of mutant NC protein were eluted from the RP-HPLC column under the conditions described.
TABLE 3.
Quantitative analysis of cleaved p6Gag, NC, and CA proteins
| Type of virus | Amt of protein
|
NC/CA ratio | p6Gag/CA ratio | ||
|---|---|---|---|---|---|
| CAcleaveda/CAtotalb | NCcleavedc/NCtotalb | p6Gagcleavedd/p6Gagtotalb | |||
| WT | 0.92 | 0.86 | 0.86 | 0.94 | 0.94 |
| ZF1 C→S | 0.70 | 0.16 | 0.15 | 0.23 | 0.22 |
| ΔZF1 | 0.62 | 0.15 | 0.51 | 0.24 | 0.82 |
| ZF2 C→S | 0.41 | 0.06 | 0.17 | 0.15 | 0.41 |
| ΔZF2 | 0.49 | 0.07 | 0.39 | 0.14 | 0.80 |
| ZF1+2 C→S | 0.65 | 0.28 | 0.57 | 0.43 | 0.88 |
| ΔZF1+2 | 0.52 | 0.01 | NDe | 0.03 | ND |
DISCUSSION
While previous studies of SIV have focused on genomic RNA encapsidation and general viral protein production and processing in relation to NC ZFs, we show for the first time that subtle differences in virion morphology can be attributed to the Zn2+ coordination of each finger. Specifically, proper core condensation is severely disrupted by mutation of ZF2, whereas mutation of ZF1 disrupts this process to a lesser extent (Table 1 and Fig. 2). We utilized SIV(Mne) NC mutants (Fig. 1) that inhibit zinc binding by either changing specific zinc-coordinating Cys residues to Ser or deleting the first four amino acids in each or both fingers, including the basic linker sequence. Our results support previous reports that the two finger arrays of retroviral NC proteins exert different activities, yet do not function independently (9, 13, 25, 35). However, our analyses of SIV NC ZF function differ from those reported for other retroviruses, such as HIV, murine leukemia virus, and Rous sarcoma virus (16, 41, 44, 45). In agreement with previous reports (12, 40), we found that genomic RNA incorporation (Fig. 3) is less dependent on Zn2+ coordination, but is more dependent on the basic charge of NC. However, contrary to observations with HIV-1, we found that it is ZF2 that affects RNA encapsidation more than ZF1.
The results from the RP-HPLC analyses (Fig. 6) support those of the immunoblot assays (Fig. 5 and 7): complete proteolytic cleavage of Gag is associated with proper core condensation. The RP-HPLC results go two steps further to indicate that (i) the removal of the Zn2+ from the NC protein results in a reduction in processed NC protein when compared to all possible Gag cleavage products in each mutant (Fig. 6, compare relative peak heights of Gag cleavage products containing NC with peaks of other cleavage products that do not contain NC) and (ii) the unprocessed Gag polyproteins occur in four major forms. Gag cleavage is an ordered process that is necessary to produce morphologically mature virions (17, 38, 50). Incomplete processing of Gag due to conformation, protein-protein, and/or protein-nucleic acid interactions resulting from the loss of Zn2+ in our NC ZF mutants may be responsible for the inability of virion cores to properly condense as has been suggested previously in HIV studies (28, 32, 33, 47). To our knowledge, Gag-Gag interaction domains (I domains) (10) have not been studied in SIV. While it is possible that the mutations discussed here may alter protein conformation, the number of basic residues of the NC in the ZF1+2 C→S mutant is the same as that in the WT NC, yet this mutant produces a significant amount of partially processed and uncleaved Gag with some full-length genomic RNA encapsidation. Therefore, effects on the I domains would have to be due to altered conformation of the protein. An altered conformation could result from (i) a “loose” folding of Gag in the NC domain due to the lack of Zn2+ coordination of the ZFs or (ii) a “reconfigured” folding of the NC domain due to (a) possible Zn2+ coordination between the remaining His and Cys residues in the deleted ZF and those in the complete ZF and/or (b) possible disulfide bond formation in the single-finger deletion mutants. Disulfide bonds do indeed form when the Zn2+ ions are ejected from the ZF of WT NC in HIV and SIV (2, 43).
NC in an altered conformation resulting from the loss of the Zn2+ in either or both fingers may be open to extensive proteolytic cleavage, most likely from the viral protease. The significant loss of mutant NC protein observed in the RP-HPLC (Fig. 6 and Table 2) suggests this scenario. Previous studies have demonstrated protease-specific NC cleavage in HIV-1 and equine infectious anemia virus (42, 46) and that the removal of the Zn2+ is necessary for this cleavage (51). The destruction of NC early in virus formation would surely have a profound effect on downstream events in the life cycle that are known to require NC, including RNA stability, reverse transcription, and, possibly, integration of the viral genome.
NC protein is present in isolated mature conical HIV-1 cores (49) and has been shown to be required for the condensation of viral protein with nucleic acid to a conical core phenotype in in vitro assembly systems (11, 20). Our results show that genomic RNA incorporation in ZF2 C→S and ΔZF2 is not severely reduced, but relative levels of unprocessed Gag are much higher than those in the ZF1 C→S and ΔZF1 mutants (Fig. 3). Yet there is a striking difference in the number of particles with properly condensed cores between virions with ZF1 and ZF2 mutations, suggesting the differences in core morphology are dictated more by Gag cleavage than full-length genomic RNA encapsidation. However, these associations cannot be stated independently, because at least one other factor, the basic charge of NC, is known to affect virion morphology. Although ΔZF1+2 virions contained more fully processed Gag, it contained very little detectable full-length genomic RNA by Northern blotting (23), whereas the ZF1+2 C→S mutant produced a profile of Gag processing similar to those of ZF2 C→S and ΔZF2 and also had severely reduced levels of virion-associated full-length genomic RNA. Cimarelli et al. (12) demonstrated the relationship between the basic charge of NC, RNA incorporation, and virion morphology in HIV-1. Their HIV-1 mutants BR and M1-2/BR contained little and no viral or cellular RNA, respectively, and produced irregular large and small virions with multiple rod-shaped cores similar to those produced by the ΔZF1+2 mutant. The requirement for nucleic acid in virion core morphology suggests that mutants lacking viral RNA may substitute nonspecific cellular RNA to achieve a condensed core (5).
Previous studies have only demonstrated that mutations in NC protein inhibit virion maturation (6, 7, 23, 35, 45). The negative effect of the ZF mutations on complete Gag processing may suggest an upstream role for NC in the complete process of virion maturation, since Gag mutations outside of the NC domain that result in improper or incomplete proteolytic processing also affect virion core condensation (1, 15, 22, 34, 47, 50). Indeed, there is a correlation between the presence of p30Gag (Fig. 5A) and an increase in immature virions (Table 1). Whether the NC protein in this process is active (Zn2+ transfer or release) or passive (structural conformation or dimerization) remains to be elucidated.
ACKNOWLEDGMENTS
Kunio Nagashima at SAIC Electron Microscopy Laboratory, Frederick, Md., performed electron microscopy. We thank Raoul Benveniste (NCI, Basic Research Laboratory) for the SIV(Mne) antisera used for immunoblot analyses, Terra Schaden for technical expertise with p28CA antigen capture analysis, and Michael Piatak, Jr., who performed the real-time RT-PCR measurements with virion RNA. We also thank David Ott, Dexter Poon, and Larry Arthur for constructive criticism in writing the manuscript.
This work was supported by NIH contract NO1-CO-56000 with SAIC-Frederick.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur L O, Bess J W, Jr, Chertova E N, Rossio J L, Esser M T, Benveniste R E, Henderson L E, Lifson J D. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res Hum Retrovir. 1998;14(Suppl. 3):S311–S319. [PubMed] [Google Scholar]
- 3.Arthur L O, Bess J W, Jr, Urban R G, Strominger J L, Morton W R, Mann D L, Henderson L E, Benveniste R L. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J Virol. 1995;69:3117–3124. doi: 10.1128/jvi.69.5.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benveniste R E, Arthur L O, Tsai C-C, Sowder R, Copeland T D, Henderson L E, Oroszlan S. Isolation of a lentivirus from a macaque with lymphoma: comparison with HTLV-III/LAV and other lentiviruses. J Virol. 1986;60:483–490. doi: 10.1128/jvi.60.2.483-490.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 6.Berthoux L, Péchoux C, Darlix J-L. Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication. J Virol. 1999;73:10000–10009. doi: 10.1128/jvi.73.12.10000-10009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthoux L, Péchoux C, Ottmann M, Morel G, Darlix J-L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bess J W, Jr, Powell P J, Issaq H J, Schumack L J, Grimes M K, Henderson L E, Arthur L O. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J Virol. 1992;66:840–847. doi: 10.1128/jvi.66.2.840-847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowles N E, Damay P, Spahr P-F. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67:623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 15.Dorfman T, Bukovsky A, Öhagen Å, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupraz P, Oertle S, Meric C, Damay P, Spahr P-F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison C A D, Loeb D D, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retrovir. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 18.Frankel A D, Young J A. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 20.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 21.Garnier L, Ratner L, Rovinski B, Cao S-X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gay B, Tournier J, Chazal N, Carriere C, Boulanger P. Morphopoietic determinants of HIV-1 Gag particles assembled in baculovirus-infected cells. Virology. 1998;247:160–169. doi: 10.1006/viro.1998.9237. [DOI] [PubMed] [Google Scholar]
- 23.Gorelick R J, Benveniste R E, Gagliardi T D, Wiltrout T A, Busch L K, Bosche W J, Coren L V, Lifson J D, Bradley P J, Henderson L E, Arthur L O. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology. 1999;253:259–270. doi: 10.1006/viro.1998.9513. [DOI] [PubMed] [Google Scholar]
- 24.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick R J, Gagliardi T D, Bosche W J, Wiltrout T A, Coren L V, Chabot D J, Lifson J D, Henderson L E, Arthur L O. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 27.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haigwood N L, Pierce C C, Robertson M N, Watson A J, Montefiori D C, Rabin M, Lynch J B, Kuller L, Thompson J, Morton W R, Benveniste R E, Hu S L, Greenberg P, Mossman S P. Protection from pathogenic SIV challenge using multigenic DNA vaccines. Immunol Lett. 1999;66:183–188. doi: 10.1016/s0165-2478(98)00156-4. [DOI] [PubMed] [Google Scholar]
- 30.Hanke T, Samuel R V, Blanchard T J, Neumann V C, Allen T M, Boyson J E, Sharpe S A, Cook N, Smith G L, Watkins D I, Cranage M P, McMichael A J. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J Virol. 1999;73:7524–7532. doi: 10.1128/jvi.73.9.7524-7532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson L E, Benveniste R E, Sowder R, Copeland T D, Schultz A M, Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kräusslich H G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang C, Rong L, Cherry E, Kleiman L, Laughrea M, Wainberg M A. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J Virol. 1999;73:6147–6151. doi: 10.1128/jvi.73.7.6147-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mammano F, Öhagen Å, Höglund S, Göttlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno A, Ido E, Goto T, Kuwata T, Nakai M, Hayami M. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effects on proteolytic processing of Gag precursors and particle formation. AIDS Res Hum Retrovir. 1996;12:793–800. doi: 10.1089/aid.1996.12.793. [DOI] [PubMed] [Google Scholar]
- 36.Ott D E, Chertova E N, Busch L K, Coren L V, Gagliardi T D, Johnson D G. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J Virol. 1999;73:19–28. doi: 10.1128/jvi.73.1.19-28.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottmann M, Gabus C, Darlix J-L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polacino P S, Stallard V, Klaniecki J E, Pennathur S, Montefiori D C, Langlois A J, Richardson B A, Morton W R, Benveniste R E, Hu S-L. Role of immune responses against the envelope and the core antigens of simian immunodeficiency virus SIVmne in protection against homologous cloned and uncloned virus challenge in macaques. J Virol. 1999;73:8201–8215. doi: 10.1128/jvi.73.10.8201-8215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts M M, Copeland T D, Oroszlan S. In situ processing of a retroviral nucleocapsid protein by the viral proteinase. Protein Eng. 1991;4:695–700. doi: 10.1093/protein/4.6.695. [DOI] [PubMed] [Google Scholar]
- 43.Rossio J L, Esser M T, Suryanarayana K, Schneider D K, Bess J W, Jr, Vasquez G M, Wiltrout T A, Chertova E, Grimes M K, Sattentau Q, Arthur L O, Henderson L E, Lifson J D. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz M D, Fiore D, Panganiban A T. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J Virol. 1997;71:9295–9305. doi: 10.1128/jvi.71.12.9295-9305.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanchou V, Decimo D, Péchoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J-L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tozer J, Friedman D, Weber I, Blaha I, Oroszlan S. Studies on the substrate specificity of the proteinase of equine infectious anemia virus using oligopeptide substrates. Biochemistry. 1993;32:3347–3353. doi: 10.1021/bi00064a018. [DOI] [PubMed] [Google Scholar]
- 47.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S-W, Schmelz G, Kozlowski P A, Manson K, Wyand M S, Glickman R, Montefiori D, Lifson J D, Johnson R P, Neutra M R, Aldovini A. Effective induction of simian immunodeficiency virus-specific systemic and mucosal immune responses in primates by vaccination with proviral DNA producing intact but noninfectious virions. J Virol. 2000;74:10514–10522. doi: 10.1128/jvi.74.22.10514-10522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welker R, Heinrich H, Tessmer U, Huckhagel C, Kräusslich H-G. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations in individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wondrak E M, Sakaguchi K, Rice W G, Kun E, Kimmel A R, Louis J M. Removal of zinc is required for processing of the mature nucleocapsid protein of human immunodeficiency virus, type 1, by the viral protease. J Biol Chem. 1994;269:21948–21950. [PubMed] [Google Scholar]