Abstract
Background
Liquid biopsy-based biomarkers, including circulating tumor DNA (ctDNA) and blood tumor mutational burden (bTMB), are recognized as promising predictors of prognoses and responses to immune checkpoint inhibitors (ICIs), despite insufficient sensitivity of single biomarker detection. This research aims to determine whether the combinatorial utility of longitudinal ctDNA with bTMB analysis could improve the prognostic and predictive effects.
Methods
This prospective two-center cohort trial, consisting of discovery and validation datasets, enrolled unresectable locally advanced non-small-cell lung cancer (LA-NSCLC) patients and assigned them to chemoradiotherapy (CRT) or CRT + consolidation ICI cohorts from 2018 to 2022. Blood specimens were collected pretreatment, 4 weeks post-CRT, and at progression to assess bTMB and ctDNA using 486-gene next-generation sequencing. Dynamic ∆bTMB was calculated as post-CRT bTMB minus baseline bTMB levels. Decision curve analyses were performed to calculate Concordance index (C-index).
Results
One hundred twenty-eight patients were enrolled. In the discovery dataset (n = 73), patients treated with CRT and consolidation ICI had significantly longer overall survival (OS; median not reached [NR] vs 20.2 months; P < 0.001) and progression-free survival (PFS; median 25.2 vs 11.4 months; P = 0.011) than those without ICI. Longitudinal analysis demonstrated a significant decrease in ctDNA abundance post-CRT (P < 0.001) but a relative increase with disease progression. Post-CRT detectable residual ctDNA correlated with significantly shorter OS (median 18.3 months vs NR; P = 0.001) and PFS (median 7.3 vs 25.2 months; P < 0.001). For patients with residual ctDNA, consolidation ICI brought significantly greater OS (median NR vs 14.8 months; P = 0.005) and PFS (median 13.8 vs 6.2 months; P = 0.028) benefit, but no significant difference for patients with ctDNA clearance. Dynamic ∆bTMB was predictive of prognosis. Patients with residual ctDNA and increased ∆bTMB (∆bTMB > 0) had significantly worse OS (median 9.0 vs 23.0 months vs NR; P < 0.001) and PFS (median 3.4 vs 7.3 vs 25.2 months; P < 0.001). The combinatorial model integrating post-CRT ctDNA with ∆bTMB had optimal predictive effects on OS (C-index = 0.723) and PFS (C-index = 0.693), outperforming individual features. In the independent validation set, we confirmed residual ctDNA predicted poorer PFS (median 50.8 vs 14.3 months; P = 0.026) but identified more consolidation ICI benefit (median NR vs 8.3 months; P = 0.039). The combined model exhibited a stable predictive advantage (C-index = 0.742 for PFS).
Conclusions
The multiparameter assay integrating qualitative residual ctDNA testing with quantitative ∆bTMB dynamics improves patient prognostic risk stratification and efficacy predictions, allowing for personalized consolidation therapy for LA-NSCLC.
Keywords: Non-small-cell lung cancer, Circulating tumor DNA, Tumor mutational burden, Immune checkpoint inhibitor, Liquid biopsy
1. Introduction
Lung cancer remains the most common reason for globally cancer-related mortality, and the number of locally advanced non-small cell lung cancer (LA-NSCLC) cases is rising annually.1,2 Previously, definitive chemoradiotherapy (CRT) was the standard-of-care regimen for patients diagnosed as unresectable LA-NSCLC. Since 2017, the PACIFIC trial has showed impressive survival benefit from consolidation durvalumab therapy after concurrent CRT.3,4 However, the RTOG 0617 trial demonstrated that a certain proportion of patients can be cured by definitive CRT,5 and this subset of patients may suffer more toxicities if given additional immune checkpoint inhibitor (ICI) therapy,6,7 while many others may develop disease progression even after one year of consolidation ICI, on the basis of the long-term results from the PACIFIC trial.4 Patients have heterogeneous responses to ICI treatments, and thus identifying effective biomarkers to tailor personalized ICI therapy is necessary.
Liquid biopsy-based biomarkers, including circulating tumor DNA (ctDNA) and blood tumor mutational burden (bTMB), have emerged as predictors of prognosis and therapeutic efficacy in multiple advanced cancers.8, 9, 10 Previous studies suggested that ctDNA detection could reflect molecular residual disease after radical radiotherapy (RT) or surgery in early-stage or localized NSCLC.11, 12, 13 bTMB, derived from all tumor niches and released into the peripheral circulation, is also a promising and readily-available biomarker.14,15 Though pretreatment bTMB at 10 and 16 mutations per megabase (mut/Mb) cutoffs failed to predict therapeutic response to ICIs in the phase 3 BFAST and phase 2 B-F1RST trials, bTMB exhibited an incremental predictive effect as the cutoff threshold increased.16,17 The exploratory analyses of the randomized controlled phase 3 MYSTIC study indicated that a baseline bTMB point of ≥20 mut/Mb could predict improved efficacy of ICI treatment.18,19 Therefore, the optimal cutoff and prediction efficiency of bTMB warrant further investigation.
Moreover, though ctDNA and bTMB have been found effective in predicting the prognoses of patients with advanced cancers receiving ICI therapy, given that RT could remodel the tumor microenvironment and alter tumor immunophenotypes, the prediction effects of liquid biopsy in LA-NSCLC patients undergoing RT and ICIs remain uncertain and need more explorations.20,21 Additionally, prior studies indicated combining ctDNA with bTMB could further improve prognostic predictions in lung cancer.22,23 Since ctDNA represents the systemic disease burden, and bTMB correlates with tumor neoantigens and responses to ICIs, the combination of ctDNA and bTMB may reflect both tumor load and treatment efficacy, with improved prognostic and predictive performance.24, 25, 26 Hence, this cohort study sought to explore and validate the role of dynamic bTMB and ctDNA detection in predicting the survival outcome of LA-NSCLC patients with curative-intent CRT and determining the therapeutic response to subsequent consolidation ICI. We further investigated the combinatorial clinical utility of bTMB integrated with ctDNA, which is hypothesized to be a more efficient co-predictor for LA-NSCLC patients.
2. Materials and methods
2.1. Study design and eligibility criteria
This two-center, prospective, non-randomized, cohort trial, consisting of the discovery dataset from the Cancer Hospital of Chinese Academy of Medical Sciences (CAMS) in Beijing, China, and the external independent validation dataset from Shenzhen Hospital of CAMS in Shenzhen, China, was conducted between July 2018 and November 2022. The study is registered at ClinicalTrials.gov (NCT04014465).
Consecutive treatment-naïve patients with pathologically diagnosed unresectable stage II or III NSCLC according to the 8th edition of the American Joint Committee on Cancer staging system were included. Key exclusion criteria were patients having driver gene mutations, namely epidermal growth factor receptor or anaplastic lymphoma kinase mutations. In the discovery and validation sets, eligible patients were assigned to the CRT cohort (concurrent or sequential curative-intent CRT) or CRT + ICI cohort (CRT followed by consolidation ICI) in a non-randomized way. Therapeutic decisions on whether to use consolidation ICI were made by the shared-decision making mechanism,27 specifically, based on patients’ disease conditions, performance status, socioeconomic backgrounds, personal willingness, and other essential factors, after careful discussions and consultations among physicians, patients, and caregivers. Peripheral blood specimens of all patients were collected at diagnosis (baseline), 4 weeks after completing radical CRT (post-CRT), and at progressive disease (progression).
2.2. Treatments, follow-up, and blood collection
All patients adopted four-dimensional computed tomography (CT) simulation and used intensity modulated RT techniques. Prescription radiation doses ranged 56–66 Gy, combined with 2 or more cycles of concurrent or sequential platinum-based doublet chemotherapy. Patients in the CRT + ICI cohort further received consolidation programmed cell death ligand-1 (PD-L1) or programmed cell death-1 inhibitor therapy after CRT. Consolidation ICIs were administrated for one year or early discontinued for unacceptable toxicities or progression.
Participants were subjected to follow-up evaluations every 3 months during the initial 3 years, thereafter every 6 months from the 3rd to 5th year, and annually from the 5th year onwards. Follow-up assessments were physical examinations, hematological tests, and imaging examinations. Tumor treatment responses were evaluated based on images from positron emission tomography-CT or contrast-enhanced (preferred) CT, using Response Evaluation Criteria in Solid Tumors (RECIST) v1.1.
For all included patients in both the discovery and validation datasets, peripheral blood specimens were collected pretreatment (baseline) and 4 weeks after the finish of radical CRT (post-CRT). If patients had progressive disease, plasma samples were further collected at the first local-regional-failure or distant-metastasis time point (progression). Patients whose blood plasma was not collected at prespecified time points were excluded from the final analysis. Plasma specimens were assessed for ctDNA and bTMB using next-generation sequencing technology covering 486 cancer-associated genes (Supplementary Table 1) by Nanjing Geneseeq Technology Inc.
2.3. DNA extraction and library construction
The plasma fraction of peripheral blood samples (8–10 mL), which were centrifuged within two hours after specimen collection to separate plasma, was subjected to circulating free DNA (cfDNA) extraction. Purified cfDNA samples were qualified using Nanodrop2000 and quantified with the Qubit 2.0 dsDNA HS Assay Kit. Approximately 50 ng of cfDNA was processed, with end repair, A-tailing, and ligation with indexed adapters performed according to the optimized manufacturer's instructions of the KAPA Hyper Prep Kit for library preparation. The samples were subsequently performed size selection, followed by polymerase chain reaction (PCR) amplification. Target enrichment was conducted by utilizing previously tailored xGen lockdown probes targeting 486 cancer-associated and radiosensitivity-related genes (Supplementary Table 1). Next, capture hybridizations were completed using Dynabeads M-279. Library purification and quantification were carried out by Agencourt AMPure XP beads and quantitative PCR, respectively. The sizes of library fragment were confirmed with the Agilent Bioanalyzer 2100 system.
2.4. Next-generation sequencing and quality control
Libraries were sequenced using Illumina HiSeq4000 platform. For data quality control, Trimmomatic was used to trim out low-quality reads from the FASTQ files. Sequencing reading was aligned to human reference genome (hg19) using default parameters of Burrows-Wheeler Aligner (BWA). Duplicate reads were subsequently removed using Picard tools to improve the data accuracy. We employed Genome Analysis Toolkit to conduct local realignment around insertion/deletions and recalibrate base scores. To ensure the reliable identification of tumor-specific mutations in cfDNA, several quality-control measures were implemented. We established a threshold for variant allele frequency of at least 0.2% and a requirement of at least 3 unique mutant reads to report a mutation, and excluded common variants with a population frequency threshold of >1% reported in any public database, such as the 1000 Genomes Project (1000 G) or Exome Aggregation Consortium (ExAC). We also applied CH-filtering and sequenced white blood cells to manage the confounding factors of clonal hematopoiesis, and removed recurrent sequencing artifacts by filtering the mutation list against an in-house database generated from normal pooled blood samples. Gene fusions were detected and enumerated by FACTERA. After removing PCR duplicates, we achieved a medium depth of coverage of >2,000X for plasma-derived cfDNA samples. ctDNA and bTMB levels were calculated based on previous studies: ctDNA concentration (hGE/mL) = mean ctDNA allele frequency × cfDNA concentration (ng/mL) × 1,000 ∕ 3.3, and ctDNA abundance (ng/mL) = max ctDNA allele frequency × cfDNA concentration (ng/mL).12,28 bTMB (mut/Mb) = absolute mutation count × 1,000,000 ∕ panel exonic base num.29 Dynamic bTMB after CRT (∆bTMB) = post-CRT bTMB level−baseline bTMB level.
2.5. Endpoints and sample size justification
The primary endpoint was progression-free survival (PFS), defined as the time from the initiation of therapy to the first documented event of progressive disease or death in the absence of progression, according to RECIST v1.1. The secondary endpoint was overall survival (OS), defined as the time between the start of therapy and death. Based on the findings of prior studies,30 we hypothesized that residual ctDNA after CRT could predict more therapeutic benefit from consolidation ICI, that is, patients with post-CRT detectable ctDNA receiving consolidation ICI therapy could have improved PFS than those patients without consolidation ICI. Chaudhuri et al.12 demonstrated median PFS for LA-NSCLC patients who had residual ctDNA after CRT but did not receive consolidation ICI was 4.0 months. We estimated that 66% of patients could have detectable ctDNA after CRT, and 34% with undetectable ctDNA.30,31 Thus, a two-sided log-rank test with a sample size of 45 subjects with detectable ctDNA (22 with CRT alone and 23 with CRT plus ICI) could achieve 80.4% power at a 0.05 significance level to detect a hazard ratio (HR) of 0.40 when median PFS for CRT alone is 4.0 months. Proportionally, 23 patients were expected to have undetectable ctDNA, including 11 with CRT and 12 with CRT plus ICI. The study lasts for 30 months, of which subject accrual occurs in the first 12 months. In summary, we aimed for an overall sample size of 68 patients, involving 33 subjects in the CRT cohort and 35 patients in the CRT + ICI cohort, to account for an 80.4% probability of detecting PFS benefit from consolidation ICI therapy in the residual ctDNA population.
2.6. Statistical analyses
In terms of survival data, the Kaplan-Meier method was employed to calculate the survival time. Log-rank tests were used to compared the survival outcomes. For multiple comparisons between subgroups, the Holm correction method was performed to adjust P values to reduce type I errors or potential false positive rates. Cox regression was performed by time-dependent method to minimize guarantee-time bias and to estimate HRs and 95% confidence intervals (CIs). Univariate and multivariate Cox proportional hazard modeling was used to identify potential predictors or covariables associated with the probability of survival. Furthermore, proportions of categorical variables were compared using Fisher's exact tests; and distributions of continuous variables were compared using Mann-Whitney tests. Wilcoxon signed-rank tests were performed to analyze paired data, such as ctDNA concentrations of individual patients before and post CRT. We conducted Spearman's correlation tests to identify potential correlations, which were expressed as R coefficients. Time-dependent receiver operating characteristic curves (ROCs) were utilized to calculate the areas under the curve (AUCs), in order to assess and compare the predictive power of models. Concordance index (C-index), calculated by decision curve analysis (DCA), was employed to determine the usefulness of various prediction models for clinical decision-making benefit. P value <0.05 was considered statistically significant. Statistics were analyzed by R software.
3. Results
3.1. Patient characteristics and survival outcomes
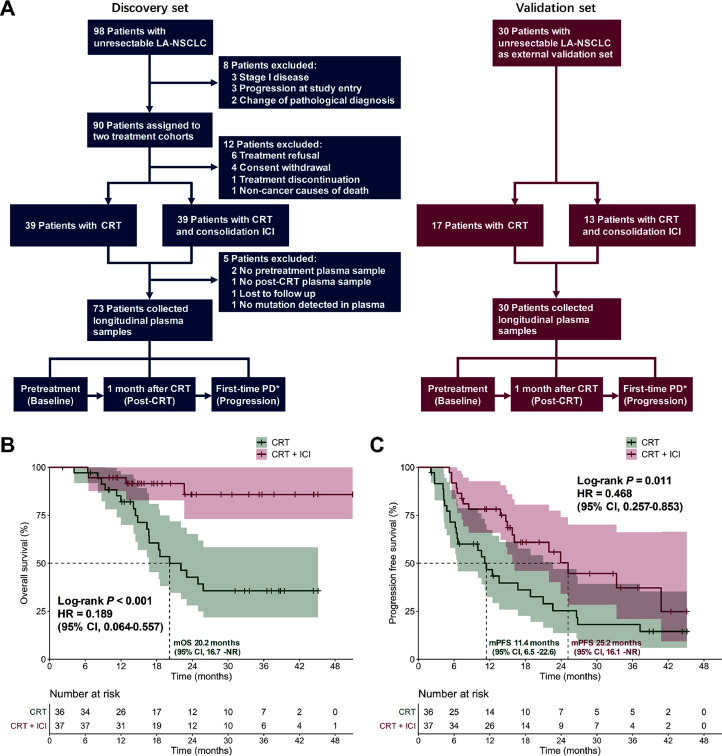
A total of 128 patients with pathologically confirmed unresectable LA-NSCLC without driver gene mutations were enrolled, and 103 patients were finally analyzed, including the discovery set and independent validation set (Fig. 1A). Eligible patients were assigned to the CRT or the CRT + consolidation ICI cohort. Among the 73 patients in the discovery set, 36 (49.3%) patients were administered CRT alone, and 37 (50.7%) were administered CRT plus consolidation ICI. Peripheral plasma samples of all these patients were collected at baseline, post-CRT, and progressive time points to profile longitudinal ctDNA and bTMB dynamics. Patient characteristics were balanced across two cohorts (Table 1). In the CRT + ICI cohort, the majority of patients (n = 21, 56.8%) were treated with consolidation durvalumab. Other ICI agents were pembrolizumab (n = 9, 24.3%), sintilimab (n = 3, 8.1%), toripalimab (n = 3, 8.1%), and camrelizumab (n = 1, 2.7%). However, we observed no significant differences in either OS (P = 0.203) or PFS (P = 0.326) between patients using durvalumab versus other ICI agents (Supplementary Fig. 1A and 1B). After the median follow-up of 28.8 (95% CI, 20.3–35.7) months, median OS and PFS was not reached (NR) and 16.7 (95% CI, 13.6–26.6) months, respectively (Supplementary Fig. 1C and 1D). Patients with consolidation ICI therapy exhibited significantly improved OS (median NR vs 20.2 [95% CI, 16.7–NR] months; HR, 0.189 [95% CI, 0.064–0.557]; P < 0.001; Fig. 1B) and PFS (median 25.2 [95% CI, 16.1–NR] vs 11.4 [95% CI, 6.5–22.6] months; HR, 0.468 [95% CI, 0.257–0.853]; P = 0.011; Fig. 1C) than patients without consolidation ICI. Cox regression modeling further determined that clinical-pathological features failed to predict survival (Supplementary Table 2), while post-CRT ctDNA detection (P = 0.049) and dynamic bTMB (P = 0.001) significantly correlated with outcomes (Supplementary Table 3).
Fig. 1.
Study design. (A) The CONSORT diagram showing the patient enrollment, treatment assignment, and the time points for liquid biopsy biomarker analysis in the discovery and validation sets. OS (B) and PFS (C) of all patients in the discovery set stratified by CRT ± consolidation ICI. ⁎ Evaluation of radiological PD is according to the Common Terminology Criteria for Adverse Events version 5.0. CI, confidence interval; CRT, chemoradiotherapy; HR, hazard ratio; ICI, immune checkpoint inhibitor; LA-NSCLC, locally advanced non-small-cell lung cancer; mOS, median overall survival; mPFS, median progression-free survival; NR, not reached; PD, progressive disease.
Table 1.
Baseline characteristics of patients in the discovery set.
| Discovery cohort | Overall | CRT | CRT + ICI | P⁎ |
|---|---|---|---|---|
| (No. = 73) | (No. = 36) | (No. = 37) | ||
| Age, median (IQR), years | 64 (58–67) | 64 (55–66) | 64 (44–68) | 0.595 |
| Sex, No. (%) Male Female |
61 (84) 12 (16) |
31 (86) 5 (14) |
30 (81) 7 (19) |
0.562 |
| Smoking, No. (%) Current/Former Never |
54 (74) 19 (26) |
26 (72) 10 (28) |
28 (76) 9 (24) |
0.737 |
| Histology, No. (%) Squamous Adenocarcinoma Others |
38 (52) 30 (41) 5 (7) |
19 (53) 14 (39) 3 (8) |
19 (51) 16 (43) 2 (5) |
0.852 |
| ECOG performance status, No. (%) 0 1 |
12 (16) 61 (84) |
3 (8) 33 (92) |
9 (24) 28 (76) |
0.065 |
| Stage, No. (%) II IIIA IIIB IIIC |
3 (4) 15 (21) 43 (59) 12 (16) |
2 (6) 7 (19) 20 (56) 7 (19) |
1 (3) 8 (22) 23 (62) 5 (14) |
0.818 |
| PD-L1 status¶, No. (%) ≥1% <1% |
20 (54) 17 (46) |
2 (33) 4 (67) |
18 (58) 13 (42) |
0.383 |
| Baseline ctDNA, median (IQR) ctDNA abundance, ng/mL ctDNA concentration, hGE/mL |
0.2 (0–0.8) 43.8 (0–116.4) |
0.3 (0–1.2) 67.8 (0–143.8) |
0.2 (0–0.5) 37.4 (0–69.8) |
0.341 0.164 |
| Baseline bTMB, median (IQR), mut/Mb | 2.9 (0–9.2) | 2.3 (0–7.2) | 3.8 (0–11.8) | 0.610 |
Comparisons of baseline characteristics between patients treated with CRT and patients with CRT plus consolidation ICI.
A total of 36 patients had unknown PD-L1 expression data, including 30 in the CRT cohort and 6 in the CRT plus consolidation ICI cohort.
Abbreviations: bTMB, blood tumor mutational burden; CRT, chemoradiotherapy; ctDNA, circulating tumor DNA; ECOG, Eastern Cooperative Oncology Group; ICI, immune checkpoint inhibitor; PD-L1, programmed death ligand 1; No., number.
3.2. Post-CRT residual ctDNA predicts poorer survival
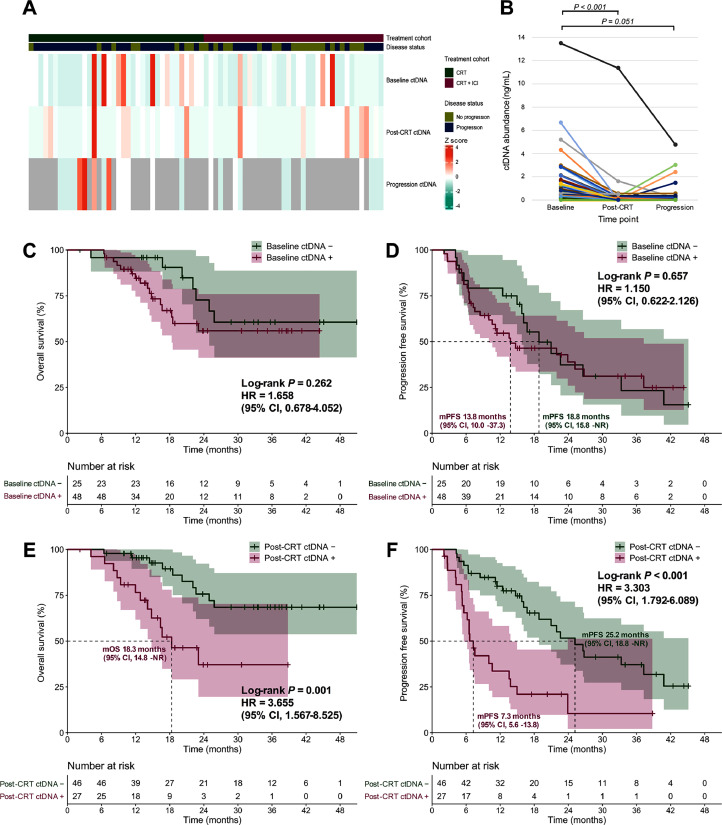
Of all patients in the discovery set, baseline ctDNA was detected in 48 (65.8%) patients with a median concentration of 43.8 hGE/mL, but post-CRT ctDNA significantly decreased (detected in 27 [37.0%] patients with a median concentration of 0 hGE/mL; P < 0.001 for both percentage detected and paired concentration; Fig. 2A). Longitudinal ctDNA analysis (Fig. 2B) also indicated a significant decrease (P < 0.001) in ctDNA abundance post-CRT but a relative increase (P = 0.051) at the progressive time point.
Fig. 2.
Predictive value of ctDNA in the discovery set. (A) Heatmap depicting ctDNA concentration of patients with different disease status before treatment (baseline), 4 weeks after CRT completion (post-CRT), and at the first progressive disease (progression). (B) Paired scatter plot illustrating longitudinal ctDNA abundance dynamics. OS (C) and PFS (D) according to baseline ctDNA status. OS (E) and PFS (F) according to post-CRT ctDNA testing. CI, confidence interval; CRT, chemoradiotherapy; ctDNA, circulating tumor DNA; HR, hazard ratio; ICI, immune checkpoint inhibitor; mOS, median overall survival; mPFS, median progression-free survival; NR, not reached.
In addition, no significant differences in either OS (median NR vs NR; P = 0.262) or PFS (median 13.8 [95% CI, 10.0–37.3] vs 18.8 [95% CI, 15.8–NR] months; P = 0.657) between patients with detectable versus undetectable ctDNA at baseline was observed (Fig. 2C and D). However, residual detectable ctDNA post-CRT was related to significantly poorer OS (median 18.3 [95% CI, 14.8–NR] months vs NR; HR, 3.655 [95% CI, 1.567–8.525]; P = 0.001) and PFS (median 7.3 [95% CI, 5.6–13.8] vs 25.2 [95% CI, 18.8–NR] months; HR, 3.303 [95% CI, 1.792–6.089]; P < 0.001), compared with ctDNA clearance (Fig. 2E and F). Meanwhile, post-CRT residual ctDNA predicted significantly worse PFS in both the CRT (median 6.2 [95% CI, 4.2–13.6] vs 21.0 [95% CI, 12.4–NR] months; P = 0.002) and the CRT + ICI cohorts (median 13.8 [95% CI, 7.3–NR] vs 33.3 [95% CI, 21.9–NR] months; P = 0.008; Supplementary Fig. 2).
3.3. Residual ctDNA predicts benefit from consolidation ICI
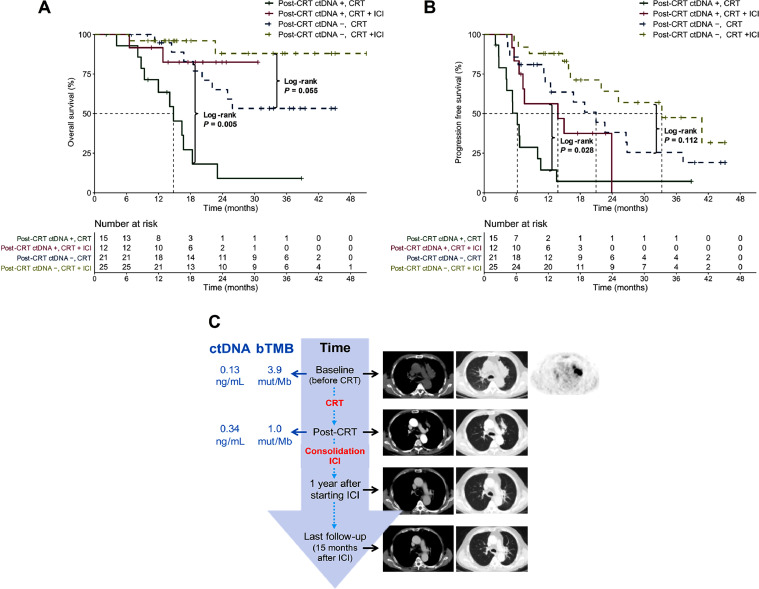
We further analyzed the interactive effect of consolidation ICI therapy and post-CRT ctDNA status on survival outcomes. Strikingly, in patients with residual ctDNA detected post-CRT, consolidation ICI brought significantly greater OS benefit than CRT alone (median NR vs 14.8 [95% CI, 12.0–NR] months; P = 0.005; Fig. 3A). However, among subjects with undetectable ctDNA post-CRT, despite the trend of better OS in patients with CRT + ICI, no statistically significance was observed (median NR vs NR; P = 0.055; Fig. 3A). Likewise, among patients with residual ctDNA post-CRT, those undergoing CRT and consolidation ICI exhibited significantly longer PFS than those with CRT alone (median 13.8 [95% CI, 7.3–NR] vs 6.2 [95% CI, 4.2–13.6] months; P = 0.028; Fig. 3B), whereas no significant PFS difference between the CRT+ ICI versus CRT cohort among patients with ctDNA clearance (median 33.3 [95% CI, 21.9–NR] vs 21.0 [95% CI, 12.4–NR] months; P = 0.112; Fig. 3B). In one representative case, a patient with post-CRT residual ctDNA, after receiving 1-year consolidation ICI therapy, exhibited favorable survival and long-term disease control (Fig. 3C). Hence, although residual ctDNA detected post-CRT was predicative of poorer survival, it effectively identified the patient subgroup most likely to benefit from consolidation ICI.
Fig. 3.
Post-CRT residual ctDNA identifies benefit from consolidation ICI therapy. Kaplan-Meier plots of OS (A) and PFS (B) according to post-CRT ctDNA detection and treatment regimens. (C) A representative case of a patient with detectable ctDNA and bTMB after CRT still exhibited favorable survival and long-term disease control after one year of consolidation ICI therapy. bTMB, blood tumor mutational burden; CRT, chemoradiotherapy; ctDNA, circulating tumor DNA; ICI, immune checkpoint inhibitor.
3.4. Increased ∆bTMB is predictive of worse survival
We then investigated the predictive effect of baseline bTMB at the cutoff thresholds of 10, 16, and 20 mut/Mb, which had been underlined by previous studies,16, 17, 18 but neither OS nor PFS could be significantly predicted by bTMB at these cutoffs (Supplementary Fig. 3). Moreover, baseline bTMB failed to predict OS or PFS at the cutoff points ranging from 4 to 20 mut/Mb (Supplementary Fig. 4).
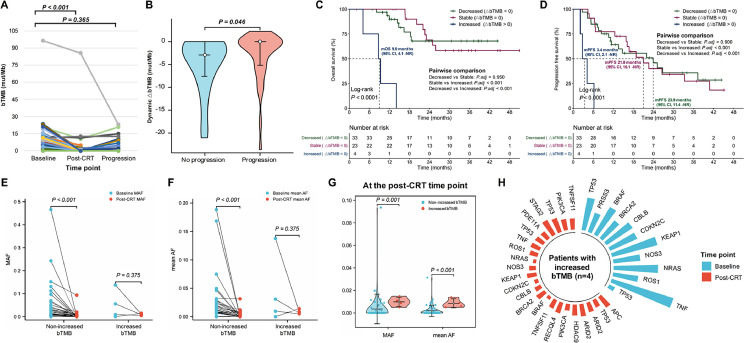
Nevertheless, longitudinal bTMB profiling demonstrated that bTMB significantly decreased at the post-CRT time point (P < 0.001) but increased as the disease progressed (P = 0.365; Fig. 4A). Dynamic ∆bTMB was calculated by subtracting baseline bTMB level from post-CRT bTMB level. Decreased ∆bTMB (∆bTMB < 0) represents effective treatment response after CRT, stable ∆bTMB (∆bTMB = 0) represents the same bTMB level before and after CRT, and increased ∆bTMB (∆bTMB > 0) indicates worse post-treatment scenarios. Median ∆bTMB after CRT for all patients was −1.6 mut/Mb. Patients without progression had significantly more reductions in ∆bTMB after CRT than patients who developed disease progression (P = 0.046; Fig. 4B). Hence, we analyzed the predictive value of ∆bTMB on survival. Though no significant difference in OS between the decreased (∆bTMB < 0) and stable ∆bTMB (∆bTMB = 0) groups (median NR vs NR; P = 0.950), patients with increased ∆bTMB (∆bTMB > 0) had the significantly shortest OS (median 9.0 [95% CI, 4.1–NR] months vs NR vs NR; P < 0.001; Fig. 4C). Similarly, we did not observe significant PFS difference between patients with decreased versus stable ∆bTMB (median 23.9 [95% CI, 11.4–NR] vs 21.0 [95% CI, 16.1–NR] months; P = 0.900), whereas increased ∆bTMB was related to the significantly worst PFS (median 3.4 [95% CI, 2.1–NR] vs 21.0 [95% CI, 16.1–NR] vs 23.9 [95% CI, 11.4–NR] months; P < 0.001; Fig. 4D). Taken together, instead of baseline bTMB at various cutoffs, dynamic ∆bTMB after CRT is predictive of survival outcomes.
Fig. 4.
Dynamic ∆bTMB predicts survival outcomes. (A) Paired scatter plot showed that bTMB significantly decreased after CRT but increased at progression. (B) Violin plot comparing the distribution of reductions in ∆bTMB between patients with and without disease progression. Kaplan-Meier analyses of OS (C) and PFS (D) according to dynamic ∆bTMB suggested that, though no significant difference in survival between the decreased (∆bTMB < 0) and stable ∆bTMB (∆bTMB = 0) groups, patients with increased ∆bTMB (∆bTMB > 0) had the significantly worst survival outcomes. Patients with non-increased bTMB had significantly decreased in MAF (E) and mean AF (F) after CRT, while patients with increased bTMB did not. (G) Patients with increased bTMB had significantly higher MAF and mean AF at the post-CRT time point those with non-increased bTMB. (H) Radial bar chart representing variants detected at baseline and post-CRT time points in patients with increased bTMB (n = 4). AF, allele frequency; bTMB, blood tumor mutational burden; CI, confidence interval; CRT, chemoradiotherapy; MAF, max allele frequency; mOS, median overall survival; mPFS, median progression-free survival; NR, not reached.
We further observed patients with non-increased ∆bTMB had significantly reduced max allele frequency (MAF) at post-CRT than baseline time points (P < 0.001), while no significant decrease in MAF after CRT among patients with increased ∆bTMB (P = 0.375; Fig. 4E). Likewise, patients with non-increased ∆bTMB showed significantly lower mean allele frequency (AF) after CRT (P < 0.001), whereas no significant reduction in mean AF among patients with increased ∆bTMB (P = 0.375; Fig. 4F). Patients with increased ∆bTMB also had significantly higher levels of MAF (P = 0.001) and mean AF (P < 0.001) at the post-CRT time point, compared to those with non-increased ∆bTMB (Fig. 4G). More importantly, in patients with post-CRT increased ∆bTMB, we detected new variants that were absent pretreatment and had lower percentages of reads than original variants (Fig. 4H). Overall, instead of an increase in original variant abundance, patients with increased ∆bTMB after CRT exhibited increasing heterogeneity, which may be the crucial reason for worse prognosis and poorer efficacy of this patient population.
3.5. ∆bTMB is an independent stratification factor of post-CRT ctDNA
Next, we compared the predictive effects of baseline, post-CRT, and dynamic ∆bTMB levels on survival. Time-dependent ROCs were used to quantify the predictive power. For patients in the discovery set, neither baseline bTMB, nor post-CRT bTMB, nor ∆bTMB levels, could effectively predict 1-year PFS rate (all AUCs < 0.75; Supplementary Fig. 5A). Strikingly, in patients with residual ctDNA, ∆bTMB levels had the best predictive power at the 1-year PFS time point (AUC = 0.815; Supplementary Fig. 5B). For patients with ctDNA clearance, no effective predictor with an AUC greater than 0.75 was found (Supplementary Fig. 5C). Subgroup survival analyses (Supplementary Fig. 6) showed that, among patients with residual ctDNA, increased ∆bTMB correlated with significantly worse OS (median 9.0 [95% CI, 4.1–NR] vs 23.0 [95% CI, 16.4–NR] months; P < 0.001) and PFS (median 3.4 [95% CI, 2.1–NR] vs 7.3 [95% CI, 6.5–NR] months; P = 0.002). As a result, dynamic ∆bTMB might be a further survival stratification factor for patients with post-CRT residual ctDNA.
Furthermore, we investigated the correlation between bTMB and ctDNA. Baseline bTMB was highly associated with baseline ctDNA (P < 0.001, R = 0.890; Supplementary Fig. 7A). A strong correlation was also identified between post-CRT bTMB and post-CRT ctDNA (P < 0.001, R = 0.920; Supplementary Fig. 7B). However, the correlation between dynamic ∆bTMB and post-CRT ctDNA levels was negligible (P = 0.743, R = −0.041; Supplementary Fig. 7C). ∆bTMB was also independent of post-CRT ctDNA status (P = 0.977), in the both CRT (P = 0.694) and CRT + ICI cohorts (P = 0.455; Supplementary Fig. 7D and 7E). Overall, dynamic ∆bTMB was independent of post-CRT ctDNA but could further differentiate the prognosis of patients with residual ctDNA post-CRT.
3.6. Predictive model combing post-CRT ctDNA with ∆bTMB
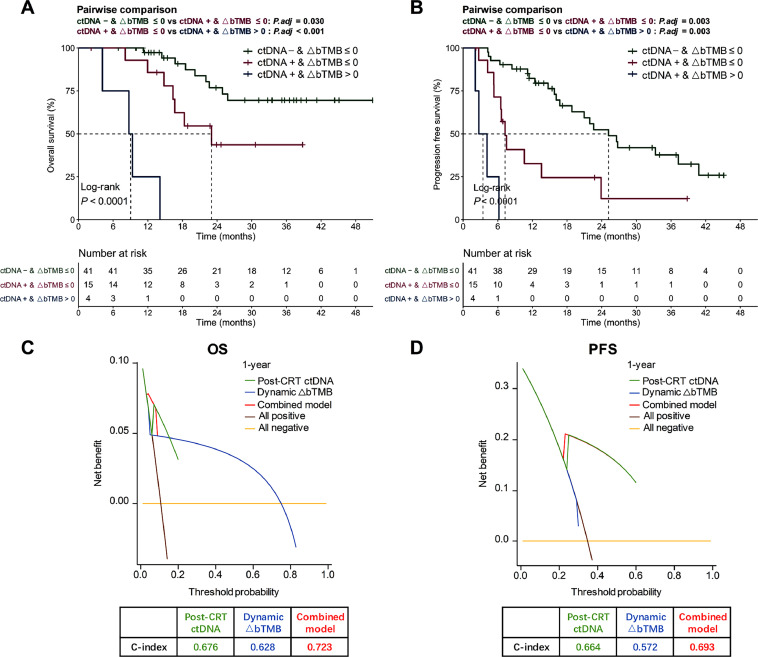
Multivariate Cox regression indicated that, among all qualitative or quantitative liquid biopsy biomarkers, qualitative post-CRT ctDNA detection and quantitative longitudinal bTMB dynamics were significantly associated with survival outcomes (Supplementary Table 3). All patients with undetectable ctDNA post-CRT had non-increased ∆bTMB (∆bTMB ≤ 0), whereas those with detectable ctDNA post-CRT might have non-increased (∆bTMB ≤ 0) or increased ∆bTMB (∆bTMB > 0). Therefore, subjects were divided into 3 groups on the basis of their post-CRT ctDNA detection status and ∆bTMB (Fig. 5A and B). Patients with undetectable ctDNA and non-increased ∆bTMB exhibited the longest OS and PFS, while patients with residual ctDNA and increased ∆bTMB had the significantly shortest OS (median 9.0 [95% CI, 4.1–NR] vs 23.0 [95% CI, 16.4–NR] months vs NR; P < 0.001; Fig. 5A) and PFS (median 3.4 [95% CI, 2.1–NR] vs 7.3 [95% CI, 6.5–NR] vs 25.2 [95% CI, 18.8–NR] months; P < 0.001; Fig. 5B). During the pairwise comparisons of these 3 groups, significant differences in OS and PFS were further observed (Fig. 5A and B).
Fig. 5.
The combinatorial model of residual ctDNA and ∆bTMB. Kaplan-Meier analyses of OS (A) and PFS (B) stratified by post-CRT ctDNA detection together with dynamic ∆bTMB. Decision curve analyses and concordance index (C-index) are described to compare clinical usefulness of post-CRT ctDNA detection, ∆bTMB, and the combined model, for predicting OS (C) and PFS (D). bTMB, blood tumor mutational burden; CI, confidence interval; C-index, concordance index; CRT, chemoradiotherapy; ctDNA, circulating tumor DNA; PFS, progression-free survival; OS, overall survival.
In addition, DCA and C-index were employed to compare the clinical usefulness of different predictive models. DCA curves reported the combined model integrating residual ctDNA detection with ∆bTMB had optimal predictive effects on OS (C-index = 0.723) and PFS (C-index = 0.693), outperforming individual features (Fig. 5C and D). Cox regression analysis also suggested that combining post-CRT ctDNA with ∆bTMB could refine survival predictions into 3 groups with significant differences (Supplementary Fig. 8) and thus might be a more effective co-predictor. Overall, the combinatorial utility of residual ctDNA detection and ∆bTMB improved the prediction of survival and outperformed individual factors.
3.7. External independent validation
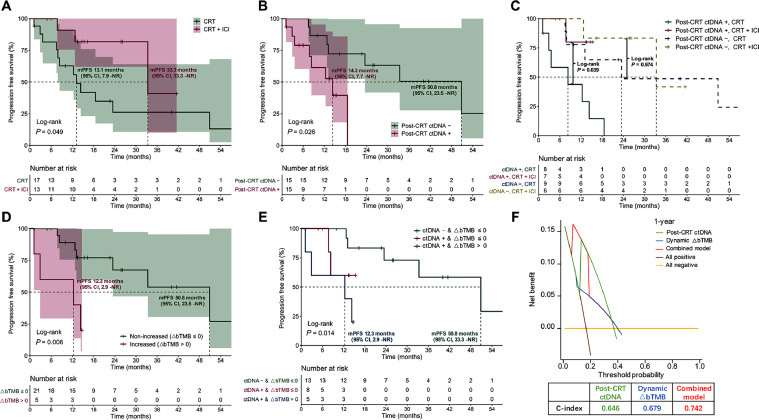
To verify the above findings, the external independent validation set was employed. Thirty patients were analyzed, including 17 (56.7%) with CRT and 13 (43.3%) with CRT + consolidation ICI. Baseline characteristics were well balanced across treatment cohorts (Supplementary Table 4). Most patients (n = 5, 38.5%) received consolidation durvalumab. Other ICI agents included tislelizumab (n = 4, 30.8%), toripalimab (n = 2, 15.4%), pembrolizumab (n = 1, 7.7%), and sintilimab (n = 1, 7.7%). Despite a non-randomized assignment design, patients used the identical shared decision-making mechanism to be assigned to two treatment regimens in both discovery and validation sets. Comparisons of patient characteristics also showed no significant differences between the discovery and validation cohorts (Supplementary Table 5). Patients treated with CRT and consolidation ICI showed significantly improved PFS than patients without ICI (median 33.3 [95% CI, 33.3–NR] vs 13.1 [95% CI, 7.9–NR] months; HR, 0.298 [95% CI, 0.083–1.073]; P = 0.049; Fig. 6A). Post-CRT residual ctDNA was correlated with significantly shorter PFS (median 14.3 [95% CI, 7.7–NR] vs 50.8 [95% CI, 23.5–NR] months; HR, 3.770 [95% CI, 1.088–13.060]; P = 0.026; Fig. 6B). Among subjects with residual ctDNA, significantly greater PFS benefit from consolidation ICI therapy was observed (median NR vs 8.3 [95% CI, 2.9–NR] months; P = 0.039), but no such significant difference in patients with ctDNA clearance post-CRT (median 33.3 [95% CI, 33.3–NR] vs 23.5 [95% CI, 13.1–NR] months; P = 0.574; Fig. 6C). Meanwhile, for all patients in the validation set, increased ∆bTMB (∆bTMB > 0) was related to significantly shorter PFS (median 12.3 [95% CI, 2.9–NR] vs 50.8 [23.5–NR] months; P = 0.006; Fig. 6D). Significant differences in PFS were further observed when stratified by both post-CRT ctDNA detection and ∆bTMB (median 12.3 [95% CI, 2.9–NR] vs NR vs 50.8 [95% CI, 33.3–NR] months; P = 0.014; Fig. 6E). DCA curves also illustrated that this combinatorial model of residual ctDNA plus ∆bTMB had the optimal prediction effect on PFS (C-index = 0.742), superior to individual features (Fig. 6F).
Fig. 6.
External validation of the predictive effect of combining residual ctDNA with ∆bTMB. (A) In the validation cohort, Kaplan-Meier comparison of PFS according to patients receiving CRT versus CRT + consolidation ICI. (B) PFS stratified by post-CRT ctDNA status. (C) PFS stratified by both post-CRT ctDNA status and treatment regimen. (D) PFS stratified by ∆bTMB dynamics. (E) PFS stratified by the combinatorial model integrating residual ctDNA detection with ∆bTMB. (F) Decision curve analyses and C-index to evaluate the clinical benefit of each model for predicting PFS. bTMB, blood tumor mutational burden; CI, confidence interval; C-index, concordance index; CRT, chemoradiotherapy; ctDNA, circulating tumor DNA; ICI, immune checkpoint inhibitor; mPFS, median progression-free survival; NR, not reached; PFS, progression-free survival; OS, overall survival.
4. Discussion
Identification of effective predictive biomarkers is of crucial importance in developing personalized cancer therapies. In this prospective cohort trial, we not only determined the predictive value of residual ctDNA detection in LA-NSCLC, but discovered that in patients with residual ctDNA, dynamic ∆bTMB was a significant factor for further patient risk stratification. Thus, we proposed the multiparameter model integrating ctDNA detection with ∆bTMB, and validated its improved prediction effects. To our knowledge, this trial provides the first evidence for combinatorial utility of residual ctDNA together with ∆bTMB in LA-NSCLC, highlighting the complementary effect of various liquid biopsy-based biomarkers in promoting treatment personalization.
Considering the difficulty of obtaining sufficient tumor specimens in patients with unresectable LA-NSCLC, non-invasive, easily-accessible, and repeatable liquid biopsies, such as ctDNA and bTMB, provide an alternative.32, 33, 34, 35 Previous research explored some landmark time points to deploy the predictive ability of ctDNA for NSCLC. Gale et al. reported residual ctDNA detection within 2 weeks to 4 months after surgery or CRT could predict relapse in early-stage NSCLC.13 Yang et al. concluded that ctDNA at one month after definitive RT was predictive of survival.36 Our findings agreed that residual ctDNA detected at one month after CRT could predict poorer outcomes in LA-NSCLC patients receiving CRT ± consolidation ICI. Furthermore, we found patients with residual ctDNA could benefit more from consolidation ICI therapy, consistent with prior work.30 For patients with post-CRT ctDNA clearance, a trend of improved survival in the CRT + ICI cohort was observed, albeit with no statistically significant differences. Our results emphasized that for patients with residual ctDNA after CRT, it may be necessary to increase the intensity of following treatments, such as consolidation ICI for more than one year, to ameliorate potentially inferior prognosis.
Of note, in addition to qualitative ctDNA testing after CRT, we found quantitative changes in bTMB during treatment played a pivotal role in further stratification of patient outcomes. In line with our conclusions, an exploratory analysis of the IMpower150 trial suggested that the joint model of multiple ctDNA metrics, including both quantitative and qualitative assessments, could improve risk stratification and prediction in the metastatic NSCLC setting.37 Herein, in LA-NSCLC patients without driver gene mutation, we developed this integrative modal of dynamic ∆bTMB together with residual ctDNA, and validated the favorable clinical utility of this combinatorial model (C-index = 0.742), allowing for optimal survival predication. Unlike previous studies mostly focusing on single biomarker,12,30,38 our findings established the feasibility of multiparameter liquid biopsy model for LA-NSCLC. In advanced NSCLC, Nabet et al. also supported that incorporating bTMB and ctDNA into the integrative assay could improve the predictive power.21
We identified that dynamic ∆bTMB, rather than baseline bTMB at various cutoffs, could effectively predict survival. bTMB has been considered as an indicator for tumor neoantigen load and ICI efficacy.39,40 Lebow et al. retrospectively reported that tissue-based TMB ≥ 10 mut/Mb prior to therapy identified improved local-regional control and PFS in LA-NSCLC patients receiving CRT and ICI therapy,38 yet sufficient tumor samples for high-quality tissue-based biomarker assessments are rarely available for unresectable LA-NSCLC and are difficult to repeat, leading to greater value of bTMB in clinical practice. Nonetheless, bTMB at 10 mut/Mb failed to predict survival in the phase III BFAST trial, presumably due to the continuous nature of bTMB.16 Accordingly, bTMB dynamics may better accommodate the characteristics of bTMB as a continuous biomarker and its nonlinear prediction effects.16,41,42 Our findings suggested that increased ∆bTMB could predict worse outcomes, and patients with increased ∆bTMB after CRT had new variants that were absent at baseline and had lower percentages of reads than original variants, implying increasing tumor heterogeneity. The rationale mainly lies in that, after the selective pressure of radical CRT, increased ∆bTMB may reflect an increase in radiation-resistant subclonal mutations resulting from intratumor heterogeneity and clonal evolution, implying inferior therapeutic responses and poorer survival.26,43, 44, 45, 46 We also found that in patients with residual ctDNA, dynamic ∆bTMB could identify further poorer prognosis and imply the potential need of consolidation therapy intensification. However, in order to address the ever-increasing clinical need for efficient biomarkers, biological mechanisms underlying the interaction between dynamic bTMB and ctDNA warrant more explorations. We envision that this co-predictive model may be optimized through further in-depth elucidation of functions and relationships between multiple biomarkers, such as comprehensive immunophenotyping.
This research has several limitations, including relatively small sample size and non-randomized study design, which may introduce potential confounding factors and limit the generalizability of our findings. As a non-randomized cohort study, there is likely an imbalance between two treatment arms, while baseline features are well-balanced across cohorts in the both discovery and validation sets, presumable due to the small sample size. Therefore, as the first prospective cohort trial exploring longitudinal ctDNA and ∆bTMB in LA-NSCLC, our results need further validation in future larger-scale studies, because larger sample sizes are essential to increase the generalizability of the conclusions and improve the representativeness of the study population. Second, PD-L1 data are not fully accessible, as most patients were diagnosed before 2020, when the study centers did not routinely examine the PD-L1 expression levels for LA-NSCLC patients. Third, biological mechanisms of the interaction between ctDNA and ∆bTMB are unclear, and a direct mechanism linking liquid biopsy biomarkers to survival outcomes remains lacking, which highlights the necessity of more basic research. Besides, patients were treated with somewhat heterogeneous consolidation ICI regimens, which might be a potential confounding factor and impact survival prognoses for LA-NSCLC patients, because durvalumab was not officially approved in China until December 2019. Further investigation in future clinical trials is warranted.
5. Conclusions
This prospective two-center cohort trial demonstrates that residual ctDNA detected after CRT portends poorer prognosis of LA-NSCLC but identifies more survival benefit from following consolidation ICI treatment. Moreover, in patients with residual ctDNA and predictable unfavorable prognosis, dynamic ∆bTMB can further distinguish their outcomes, allowing for more tailored patient risk stratification. The multiparameter non-invasive model integrating residual ctDNA detection with ∆bTMB dynamics effectively improves the prognostic and predictive effect, outperforming each individual feature. Given the difficulty to date of predicting the survival for LA-NSCLC patients receiving curative-intent RT and ICIs, our study sheds light on the liquid biopsy-based survival prediction and risk-adaptive cancer precision treatment.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Ethics statement
This trial was reviewed and approved by the ethics committee of CAMS (approval number: 20/453-2649), and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants. Only de-identified patient images were used to protect patient privacy.
Availability of data
The raw data are not publicly available because of participant privacy or consent. To minimize the risk of patient re-identification, anonymized individual patient-level datasets, sequencing data, and other information required to reanalyze the results of this study will be available from the corresponding authors upon reasonable request.
Acknowledgments
This work was funded by National Natural Sciences Foundation Key Program (grant number: 82173348), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant number: 2021-1-I2M-1-012), and the Special Research Fund for Central Universities, Peking Union Medical College (grant number: 3332023133).
Author contributions
Y.W., T.Z., Y.Y., J.Y.W. and C.J.L. performed the data curation, methodology, software, and formal analysis; W.W., X.X., Y.Q.W., Y.J. and J.H.D. provided resource and conducted the data curation; Y.W. conducted visualization and writing - original draft; N.B and L.H.W. acquired funding and conducted the supervision, validation, and writing - review & editing. All authors participated in the conceptualization, project administration, and investigation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jncc.2024.01.008.
Contributor Information
Luhua Wang, Email: wlhwq@yahoo.coms.
Nan Bi, Email: binan_email@163.com.
Appendix. Supplementary materials
References
- 1.Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–436. doi: 10.3322/caac.21731. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Zhang S, Wang S, et al. Lung cancer incidence and mortality in China: updated statistics and an overview of temporal trends from 2000 to 2016. J Natl Cancer Cent. 2022;2:139–147. doi: 10.1016/j.jncc.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auperin A, Le Pechoux C, Rolland E, et al. Meta-analysis of concomitant versus sequential radiochemotherapy in locally advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:2181–2190. doi: 10.1200/JCO.2009.26.2543. [DOI] [PubMed] [Google Scholar]
- 4.Spigel DR, Faivre-Finn C, Gray JE, et al. Five-year survival outcomes from the PACIFIC trial: durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. J Clin Oncol. 2022;40:1301–1311. doi: 10.1200/jco.21.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley JD, Hu C, Komaki RR, et al. Long-term results of NRG Oncology RTOG 0617: standard- versus high-dose chemoradiotherapy with or without cetuximab for unresectable stage III non-small-cell lung cancer. J Clin Oncol. 2020;38:706–714. doi: 10.1200/jco.19.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Zhang T, Huang Y, et al. Real-world safety and efficacy of consolidation durvalumab after chemoradiation therapy for stage III non-small cell lung cancer: a systematic review and meta-analysis. Int J Radiat Oncol Biol Phys. 2022;112:1154–1164. doi: 10.1016/j.ijrobp.2021.12.150. [DOI] [PubMed] [Google Scholar]
- 7.Shintani T, Kishi N, Matsuo Y, et al. Incidence and risk factors of symptomatic radiation pneumonitis in non–small-cell lung cancer patients treated with concurrent chemoradiotherapy and consolidation durvalumab. Clin Lung Cancer. 2021;22(5):401-410 doi: 10.1016/j.cllc.2021.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–881. doi: 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura Y, Taniguchi H, Ikeda M, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: sCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–1864. doi: 10.1038/s41591-020-1063-5. [DOI] [PubMed] [Google Scholar]
- 10.Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J Clin. 2021;71:176–190. doi: 10.3322/caac.21650. [DOI] [PubMed] [Google Scholar]
- 11.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15:577–586. doi: 10.1038/s41571-018-0058-3. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.Cd-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale D, Heider K, Ruiz-Valdepenas A, et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann Oncol. 2022;33:500–510. doi: 10.1016/j.annonc.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulz P, Thallinger GG, Auer M, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet. 2016;48:1273–1278. doi: 10.1038/ng.3648. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters S, Dziadziuszko R, Morabito A, et al. Atezolizumab versus chemotherapy in advanced or metastatic NSCLC with high blood-based tumor mutational burden: primary analysis of BFAST cohort C randomized phase 3 trial. Nat Med. 2022;28:1831–1839. doi: 10.1038/s41591-022-01933-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim ES, Velcheti V, Mekhail T, et al. Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat Med. 2022;28:939–945. doi: 10.1038/s41591-022-01754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol. 2020;6:661–674. doi: 10.1001/jamaoncol.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si H, Kuziora M, Quinn KJ, et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: results from the MYSTIC study. Clin Cancer Res. 2021;27:1631–1640. doi: 10.1158/1078-0432.Ccr-20-3771. [DOI] [PubMed] [Google Scholar]
- 20.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 21.Nabet BY, Esfahani MS, Moding EJ, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell. 2020;183:363–376.e13. doi: 10.1016/j.cell.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie W, Wang ZJ, Zhang K, et al. ctDNA-adjusted bTMB as a predictive biomarker for patients with NSCLC treated with PD-(L)1 inhibitors. BMC Med. 2022;20:170. doi: 10.1186/s12916-022-02360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Wang Z, Zhang J, et al. Genomic landscape and tumor mutational burden determination of circulating tumor DNA in over 5,000 Chinese patients with lung cancer. Clin Cancer Res. 2021;27:6184–6196. doi: 10.1158/1078-0432.Ccr-21-1537. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Chang L, Yang Y, et al. The correlations of tumor mutational burden among single-region tissue, multi-region tissues and blood in non-small cell lung cancer. J Immunother Cancer. 2019;7:98. doi: 10.1186/s40425-019-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae YK, Davis AA, Agte S, et al. Clinical implications of circulating tumor DNA tumor mutational burden (ctDNA TMB) in non-small cell lung cancer. Oncologist. 2019;24:820–828. doi: 10.1634/theoncologist.2018-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann TC, Montori VM, Del Mar C. The connection between evidence-based medicine and shared decision making. JAMA. 2014;312:1295–1296. doi: 10.1001/jama.2014.10186. [DOI] [PubMed] [Google Scholar]
- 28.Mao X, Zhang Z, Zheng X, et al. Capture-based targeted ultradeep sequencing in paired tissue and plasma samples demonstrates differential subclonal ctDNA-releasing capability in advanced lung cancer. J Thorac Oncol. 2017;12:663–672. doi: 10.1016/j.jtho.2016.11.2235. [DOI] [PubMed] [Google Scholar]
- 29.Jiang T, Chen J, Xu X, et al. On-treatment blood TMB as predictors for camrelizumab plus chemotherapy in advanced lung squamous cell carcinoma: biomarker analysis of a phase III trial. Mol Cancer. 2022;21:4. doi: 10.1186/s12943-021-01479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moding EJ, Liu Y, Nabet BY, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small cell lung cancer. Nat Cancer. 2020;1:176–183. doi: 10.1038/s43018-019-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley JD, Hu C, Komaki RU, et al. Long-term results of RTOG 0617: a randomized phase 3 comparison of standard dose versus high dose conformal chemoradiation therapy +/- cetuximab for stage III NSCLC. Int J Radiat Oncol Biol Phys. 2017;99:S105. doi: 10.1016/j.ijrobp.2017.06.250. [DOI] [Google Scholar]
- 32.Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26:1415–1421. doi: 10.1093/annonc/mdv208. [DOI] [PubMed] [Google Scholar]
- 33.Varlotto JM, Sun Z, Ky B, et al. A review of concurrent chemo/radiation, immunotherapy, radiation planning, and biomarkers for locally advanced non-small cell lung cancer and their role in the development of ECOG-ACRIN EA5181. Clin Lung Cancer. 2022;23:547–560. doi: 10.1016/j.cllc.2022.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Chen K, Kang G, Zhao H, et al. Liquid biopsy in newly diagnosed patients with locoregional (I-IIIA) non-small cell lung cancer. Expert Rev Mol Diagn. 2019;19:419–427. doi: 10.1080/14737159.2019.1599717. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Zhang T, Wang J, et al. Induction immune checkpoint inhibitors and chemotherapy before definitive chemoradiation therapy for patients with bulky unresectable stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2023;116:590–600. doi: 10.1016/j.ijrobp.2022.12.042. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Zhang T, Wang J, et al. The clinical utility of dynamic ctDNA monitoring in inoperable localized NSCLC patients. Mol Cancer. 2022;21:117. doi: 10.1186/s12943-022-01590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assaf ZJF, Zou W, Fine AD, et al. A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat Med. 2023;29:859–868. doi: 10.1038/s41591-023-02226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebow ES, Shepherd A, Eichholz JE, et al. Analysis of tumor mutational burden, progression-free survival, and local-regional control in patents with locally advanced non-small cell lung cancer treated with chemoradiation and durvalumab. JAMA Netw Open. 2023;6 doi: 10.1001/jamanetworkopen.2022.49591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie W, Qian J, Xu MD, et al. A non-linear association between blood tumor mutation burden and prognosis in NSCLC patients receiving atezolizumab. Oncoimmunology. 2020;9 doi: 10.1080/2162402x.2020.1731072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montesion M, Murugesan K, Jin DX, et al. Somatic HLA Class I loss is a widespread mechanism of immune evasion which refines the use of tumor mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov. 2021;11:282–292. doi: 10.1158/2159-8290.Cd-20-0672. [DOI] [PubMed] [Google Scholar]
- 43.Niknafs N, Balan A, Cherry C, et al. Persistent mutation burden drives sustained anti-tumor immune responses. Nat Med. 2023;29:440–449. doi: 10.1038/s41591-022-02163-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21:1258–1266. doi: 10.1158/1078-0432.Ccr-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf Y, Bartok O, Patkar S, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell. 2019;179:219–235.e21. doi: 10.1016/j.cell.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Dong Y, Bai H, et al. Therapeutic guidance of tumor mutation burden on immune checkpoint inhibitors in advanced non-small cell lung cancer: a systematic review and comprehensive meta-analysis. J Natl Cancer Cent. 2022;2:41–49. doi: 10.1016/j.jncc.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data are not publicly available because of participant privacy or consent. To minimize the risk of patient re-identification, anonymized individual patient-level datasets, sequencing data, and other information required to reanalyze the results of this study will be available from the corresponding authors upon reasonable request.