Abstract
The evolutionary dynamics of cancer, characterized by its profound heterogeneity, demand sophisticated tools for a holistic understanding. This review delves into tumor phylogenetics, an essential approach bridging evolutionary biology with oncology, offering unparalleled insights into cancer's evolutionary trajectory. We provide an overview of the workflow, encompassing study design, data acquisition, and phylogeny reconstruction. Notably, the integration of diverse data sets emerges as a transformative step, enhancing the depth and breadth of evolutionary insights. With this integrated perspective, tumor phylogenetics stands poised to redefine our understanding of cancer evolution and influence therapeutic strategies.
Keywords: Tumor phylogenetics, Cancer evolution, Phylogenetic trees, Data integration
1. Introduction
Cancer continues to be a pressing global health crisis, causing millions of deaths annually.1 According to the International Agency for Research on Cancer, it was reported that there were 19.3 million new cancer cases and 10.04 million cancer-related deaths in 2020. The projection for 2040 estimates a 47% increase in global cancer cases, bringing the total to 28.4 million.2 This high incidence, coupled with the absence of universally effective treatments, emphasizes the urgent need for a deeper understanding of the underlying mechanisms of cancer.
As researchers strive to discover more effective therapeutic strategies, several challenges consistently surface, including metastasis, drug resistance, and relapse. Metastasis is the spread of cancer cells from their primary site to distant body parts.3 Drug resistance, especially secondary resistance, arises when cancer cells adapt to neutralize anti-cancer drugs.4,5 Relapse, the post-treatment resurgence of cancer, often exacerbates therapeutic challenges.6 These phenomena are formidable obstacles to successful cancer treatment.7, 8, 9 Crucially, all can be traced back to one common root – the evolutionary nature of cancer.10, 11, 12, 13, 14 Understanding the dynamics of tumor evolution is fundamental to addressing metastasis, drug resistance, and relapse, highlighting the critical need to comprehend cancer evolution.
To gain a better understanding of the complex nature of cancer evolution, researchers have been employing a variety of analytical tools and methodologies. One method that has proven particularly useful is phylogenetics, originally developed to study the evolutionary relationships between species.15 It facilitates the reconstruction of tumor evolutionary trajectories, thereby equipping researchers with actionable insights that could pave the way for tailored. Since its inception, the field of therapeutic interventions has rapidly advanced and gained significant popularity, becoming a crucial branch of oncology studies.16,17
This review aims to provide a comprehensive introduction to the workflow in tumor phylogenetics, covering aspects from study design and data collection to the methods used for reconstructing phylogenetic trees and their various applications. Finally, this review will highlight the significance of integrating external data to enhance the interpretability and applicability of phylogenetic analyses in cancer research.
2. Tumor phylogenetics: an overview
The field of tumor phylogenetics has evolved considerably since Peter Nowell first introduced the Clonal Evolution Model in 1976.18 This model suggests that tumors originate from a single abnormal cell. Subsequent theories and models, including branching and linear evolution, neutral and adaptive evolutionary models, and the concept of punctuated evolution, have added depth to our understanding of intratumoral heterogeneity and evolutionary dynamics.19,20
In general, tumor evolution is a process driven by the acquisition of genetic and epigenetic mutations, resulting in a complex and continually evolving mosaic of cell populations within each tumor—known as subclones. Each subclone, with its unique genetic and phenotypic profile, contributes to the tumor's overall heterogeneity. This heterogeneity is driven by the increasing instability of the cancer genome, which leads to the continuous generation of genetic diversity.18,21, 22, 23, 24, 25, 26 This form of heterogeneity is an universal feature present across all cancer types,19,27,28 leading to the emergence of drug-resistant subclones or induced metastasis, complicating the task of effective therapy.29,30
Although this diversity of tumor cells presents formidable challenges in cancer treatment, it also provides a unique opportunity. The diverse genetic profile of subclones, resulting from mutation accumulation during tumor evolution, can serve as a historical record of the tumor's evolutionary journey, essentially acting as a 'tumor history recorder'. Studying these genetic variations enables us to backtrack the course of tumor evolution.28 Specifically, mutations common to all cancer cells in a tumor can be used to delineate the ‘trunk’ of the tumor's evolutionary tree, while mutations unique to specific subclones represent the ‘branches’, thereby providing a comprehensive picture of the tumor's evolutionary landscape and diversity.
In this context, tools originally designed for evolutionary biology prove to be suitable for reconstructing these evolutionary trees from the heterogeneous cancer genetic data. Notably, phylogenetics stands out as a highly developed and successful evolutionary approach,15 offering a profound perspective on the intricate dynamics of tumor evolution. Within this framework, tumor phylogenetics, or phylooncology, emerged, providing an innovative approach to understanding tumor progression.
Essentially, both tumor phylogenetics and species phylogenetics share the same goal of utilizing genetic variation to construct phylogenetic trees that reveal evolutionary history. However, there are still some significant differences between these two fields that we need to be aware of. Unlike species evolution, in which chromosome structures and numbers evolve over millions of years, chromosomes change in cancer evolution can occur within hours due to chromosomal instability (CIN).31 This, combined with the extremely large populations of tumor cells, may lead to the emergence of grossly altered clones that could possess adaptive advantages, a phenomenon rarely seen in species evolution.19 Moreover, the dynamic change of the microenvironment provides a catalyst for tumors to generate heterogeneity and adapt to external pressures. Importantly, in tumor evolution, tumor epigenetic plasticity can cause phenotypic changes without genetic alterations, serving as a fundamental force guiding tumor adaptation. On the other hand, species phylogenetics seeks to uncover the evolutionary relationships and common ancestries among different species and to understand the history of speciation. Tumor phylogenetics focuses on understanding the dynamics of cancer evolution and tracking down tumor clonal evolution history to identify key genetic events to improve clinical outcomes, including the formulation of accurate prognoses and personalized treatment strategies. Despite these differences, the conceptual framework of species phylogenetics still provides valuable insights into tumor phylogenetics. For instance, methods derived from phylogeography can be adapted to analyze tumor metastasis by tracing the geographical spread of tumor cells within the body.32, 33, 34, 35, 36
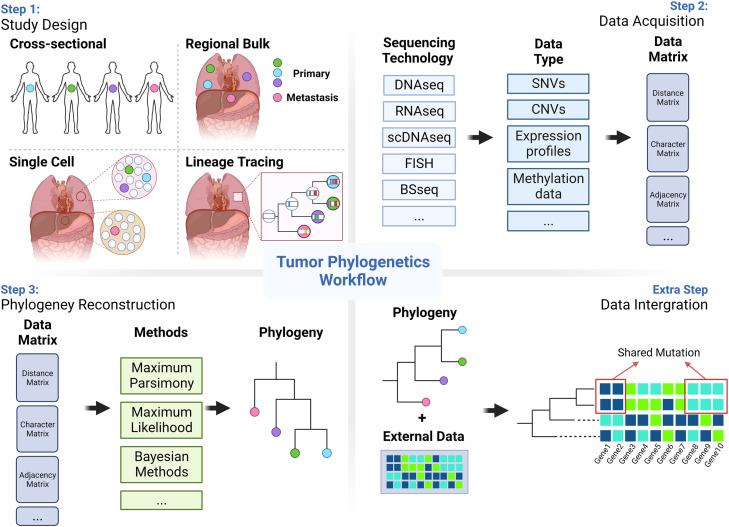
Since its inception, the field of tumor phylogenetics has garnered increasing attention and has rapidly developed. Today, tumor phylogenetics has evolved into a highly diversified field. This diversity manifests itself in every facet of the workflow—from the initial study design to the types of data collected, to the methods and tools used for reconstructing phylogenies, as well as the applications of tumor phylogenetics in the real world. In the following sections, we will offer an overview that encapsulates this multifaceted nature, guided by the commonly used workflow in the field (Fig. 1).
Fig. 1.
Workflow for tumor phylogenetics. Step 1 involves selecting an appropriate study design, such as cross-sectional, regional bulk sequencing, single-cell sequencing, or lineage tracing. Step 2 focuses on collecting and preprocessing the data. Step 3 utilizes the preprocessed data to choose an appropriate method for reconstructing the tumor phylogeny. An optional fourth step integrates the phylogeny with external data to uncover specific patterns, such as subclone-specific mutations.
3. Study designs and data in tumor phylogenetics
The workflow for reconstructing a phylogenetic tree begins with two critical steps: selecting an appropriate study design and acquiring the requisite data. In the field of tumor phylogenetics, researchers commonly utilize one of four study designs: cross-sectional studies, regional bulk sequencing, single-cell sequencing, and lineage tracing. Each design comes with its own set of advantages and limitations. The chosen study design then determines the types of data used for reconstructing the phylogeny, which can vary from genomic data like single nucleotide variants (SNVs) and copy number variants (CNVs) to transcriptomic and epigenomic data. These different types of data have been proved useful in phylogenetic and tumor analysis.16,37, 38, 39, 40, 41 For example, it has been shown that epigenetic passenger variations are effective in reconstructing the unperturbed biology of human cancer.40 Therefore, each data type is able to provide unique insights, making the resulting phylogenies trees more robust and informative.
3.1. Cross-sectional tumor phylogenetics
As the field of tumor phylogenetics began to take shape, researchers initially reconstructed evolutionary trees based on tumor data from different patients. The central idea behind this approach was rooted in the perspectives of Fearon and Vogelstein, two prominent researchers in the field. They proposed that by sampling and analyzing tumor specimens from multiple patients in bulk, researchers could infer certain characteristic mutation sequences or tumor evolutionary paths.42 These pathways could be considered evolutionary trajectories that most tumors commonly follow during their development.
Building upon this concept, researchers started to develop what is known as the "oncogenetic tree".43 By sampling different patients and treating their tumor samples as distinct "species," these trees were created. Within such phylogenies, tips represent tumor samples gathered from various patients, and they are categorized into different subtypes based on the clades they belong to, indicating their shared mutation patterns. The edges signify genetic divergence between different tumors. This oncogenetic tree reveals the evolutionary relationships between different tumor samples and helps identify standard tumor evolution trajectories.
In order to glean evolutionary evidence accumulated during tumor development from these samples, a variety of sequencing methods have been employed in cross-sectional tumor phylogeny. Before the widespread adoption of next-generation sequencing (NGS), researchers initially relied on comparative genomic hybridization (CGH)44 to identify CNVs by comparing fluorescence intensity between tumor and normal DNA samples on a DNA array.43 Thereafter, some researchers began to incorporate both expression data, acquired through cDNA microarrays,45 and methylation data46,47 into their efforts to reconstruct tumor phylogenies. With the advent of NGS, whole exome sequencing (WES) and whole genome sequencing (WGS) have become the techniques of choice.48, 49, 50, 51 These methods either sequence the entire set of protein-coding genes (exome) or the full genome in individual tumor samples from different patients. Specialized bioinformatics tools like GATK,52 VarScan,53 and CNVnator54 are used to identify SNVs and CNVs in the sequence data. This information is then utilized as the foundation for subsequent tumor phylogenetic reconstruction. A minority of studies also use RNAseq to analyze different tumor subtypes, generating gene expression data for subsequent phylogenetic analyses.55
However, the cross-sectional method has its drawbacks. Since it primarily relies on tumor samples from different patients, the inherent tumor heterogeneity among patients can considerably impact the results. It might fail to capture the subtle variations within a single patient's tumor, leading to inaccuracies in the reconstructed tree. Moreover, while this approach may reveal general evolutionary paths in tumors, the specific evolutionary trajectory of a tumor in an individual patient could differ due to intertumoral heterogeneity. Nonetheless, the cross-sectional research method has laid a solid foundation for the development of tumor phylogenetics and has provided significant insights and directions for subsequent studies.
3.2. Regional bulk tumor phylogenetics
As the understanding of intratumoral genetic heterogeneity grows, researchers have turned to a multi-sampling strategy for individual tumors. This led to the emergence of regional bulk sequencing methods. In this approach, multiple regions within the same tumor, or various tumor sites from a single patient, are sampled and analyzed separately. The technique affords a comprehensive view of the tumor landscape, enabling the reconstruction of phylogenetic trees that elucidate relationships among the sampled regions. Due to the nature of bulk sequencing, data is gathered from the heterogeneous population of cancer cells in each tumor sample and sequenced collectively. The result is a composite profile that captures the averaged characteristics of the cancer cell population within the sampled tumor site.
Nonetheless, this generalized view can cause some problems in identifying different subclones within the tumor. To address this issue, an important process of deconvolution to parse the complex data obtained from bulk sequencing is required. This computational technique elucidates the tumor's subclonal architecture through identifying SNVs, indels, and CNAs to assess tumor heterogeneity. This process calculates the variant allele frequency (VAF) to infer the cellular prevalence (CP) and cancer cell fraction (CCF) of mutations, distinguishing clonal mutations from subclonal ones. Copy number states are then reconstructed by analyzing local read depth and B-allele frequency (BAF), categorizing CNAs as either clonal or subclonal. Finally, SNVs are clustered into distinct clones based on CP and CCF. Utilizing the resulting detailed subclonal architecture, researchers can then infer the lineage relationships among subclones through integrating the genetic profiles of subclones with phylogenetic models.56 And this deconvolution can provide us with some critical insights. For instance, the analysis of the subclone composition of biopsies before and after aromatase inhibitor treatment in breast cancer reveals subclonal dynamics, indicating varied responses to treatment and highlighting the subclone possibly responsible for therapy resistance.57
Several tools have been specifically developed for deconvolution. Clomial leverages a binomial mixture model on VAF data to dissect mixed populations into distinguishable subclonal elements, Applies phylogenetic principles to chart subclonal diversification through reconstructing mutation lineage trees. Unmixing utilizes statistical strategies to unravel mixed genomic signals, highlighting distinct subclonal entities. PyClone adopts Bayesian clustering to navigate through extensive copy-number variations, identifying subclones with precision.58 SciClone, optimized for copy-number neutral scenarios, combines VAF and copy-number data to accurately trace clonal evolution across samples.57 Additionally, some tools even integrate deconvolution with phylogenetic tree reconstruction. Further discussion on this aspect will be provided in the upcoming phylogeny reconstruction session.
In regional bulk tumor phylogenetics, the data collection strategy generally aligns with the methodologies used in cross-sectional tumor phylogenetics. Initially, microsatellite markers59 and aCGH60 were employed for capturing CNVs, and then transitioned to utilizing WES and WGS-derived SNVs and CNVs for a more comprehensive and accurate analysis of tumor genetics.
Yet, the bulk sequencing-based approach is not without limitations. It fails to capture the full extent of intratumor heterogeneity due to the averaging effect of bulk sequencing. Additionally, some key mutations with low frequency may be overlooked. Moreover, the number of sequenced tumor regions significantly impacts the ability to distinguish genuine clonal mutations.61,62 Despite these challenges, the bulk sequencing-based approach laid a solid foundation for the development of more refined techniques in tumor phylogenetics. With its affordability and comprehensive view of tumors derived from multi-region sampling, bulk sequencing continues to be a widely used method in the field of cancer research.
3.3. Single-cell tumor phylogenetics
The advent of single-cell sequencing technologies has revolutionized tumor phylogenetics by providing researchers with significantly higher resolution for analyzing tumor evolution.63,64 This technology enables the dissection of tumors at the cellular level by individually sequencing each cell's genome, thereby offering a detailed view of the tumor's genetic diversity. By comparing mutation profiles across many individual cells, a phylogenetic tree can be reconstructed with different tumor cells as tip nodes and distinct clades representing subclones. This allows for a clear visualization of the clonal evolution of cancer cells within the tumor.
Before the emergence of single-cell sequencing technologies, researchers utilized various methods to dissect tumor evolution at the cellular level. For instance, fluorescence in situ hybridization (FISH)65 was employed to acquire CNVs for specific genes at the single-cell level. Methods like single-cell qPCR were utilized to obtain single-cell-level genetic markers, including SNVs, CNVs, and microsatellites.66,67 While these methods contributed valuable insights, they had limitations in terms of throughput, accuracy, and the range of detectable genomic features. The advent of single-cell sequencing has substantially mitigated these challenges, heralding a new era in tumor phylogenetics. One of the most common techniques is single-cell DNA sequencing, which allows for high-throughput and high-resolution genomic analyses, identifying SNVs and CNVs at single-cell levels.68,69 Single-cell RNA sequencing is used to collect expression matrices and to estimate copy number variations,70,71 both of which can be utilized in subsequent phylogenetic reconstructions.
Despite its tremendous potential, single-cell sequencing-based tumor phylogenetics faces several challenges. Technical issues like allelic dropout and amplification noise are difficult to eliminate, even with specialized correction tools, thereby obscuring the tumor's true genetic landscape.72, 73, 74 Additionally, the current high cost and labor-intensive nature of single-cell sequencing techniques limit the number of cells that can be feasibly analyzed in a single experiment.72,74 This may lead to an underrepresentation of the tumor's genetic diversity, potentially missing rare but clinically significant subclones.
3.4. Lineage tracing tumor phylogenetics
Lineage tracing, a technique widely used in biological studies, provides insight into cell origin, differentiation, and movement within an organism. By mapping the ancestral lineage of cells, it provides a lens to visualize developmental processes and has been instrumental in fields ranging from developmental biology and neuroscience to regenerative medicine and oncology. In cancer research, lineage tracing offers an invaluable tool for understanding tumor development, growth, and evolution.66,75
Traditional lineage tracing techniques rely on marking specific cells with dyes or fluorescent proteins, radioactive labels, or genetic markers such as microsatellites. With these markers, the behavior and destiny of the cells and their progeny can be observed. Traditional lineage tracing methods provide us with some significant insights into tumor phylogenetics.66,75, 76, 77, 78 However, these traditional methods falter in tracking large cell populations over time and struggle with the heterogeneity often found in tumors.79,80
Single cell dynamic lineage tracing, a newer advancement in the field, employs cutting-edge technologies such as the CRISPR-Cas9 system81, 82, 83 and recombinase systems84,85 to create exogenous barcodes. Specifically, the CRISPR-Cas9 method leverages the gene-editing capability of the system to create unique, inheritable marks in the genomic DNA of individual cells, while recombinase systems, like the Polylox system, use site-specific DNA recombination events to generate permanent and unique genetic marks (reviewed in79,80). Using the single-cell sequencing technique, we can identify unique genetic marks for high-resolution lineage tracking, enabling the study of numerous cells over extended periods within their genomic context. Its robust performance in interpreting zebrafish lineage trees82,86,87 has led to its application in tumor phylogenetics, providing a more detailed and comprehensive understanding of tumor heterogeneity and the dynamics of tumor evolution.75,88, 89, 90, 91
CRISPR-Cas9-based dynamic lineage tracing methods employ short-read sequencing and enable simultaneous capture of transcriptomes and lineage recordings via single-cell RNA sequencing.88,91 In contrast, recombinase approaches necessitate long-read sequencing technologies, such as PacBio SMRT sequencing, to obtain barcode DNA sequences, thereby limiting their compatibility with single-cell methods.
Dynamic lineage tracing faces considerable challenges, including technical limitations like incomplete barcode capture, potentially affecting the accuracy of downstream analysis.80 The computational burden of interpreting barcode data to reconstruct phylogenetic trees and significant costs hinder its widespread adoption.79,80 Despite these drawbacks, its unique ability to track cells at single-cell resolution and in real-time offers unparalleled insights into tumor heterogeneity and evolution.
4. Phylogeny reconstruction
In the previous session, we delved into various study designs pertinent to tumor phylogenetics and elucidated the types of sequencing data associated with each strategy. After acquiring and processing this data, the next challenge is utilizing it effectively to reconstruct phylogenetic trees.
Phylogeny reconstruction methods mainly fall into two categories: distance-based and character-based. Distance-based methods, such as UPGMA,92 neighbor-joining (NJ),93 and minimum evolution,94 primarily operate by estimating morphological or genetic distances between species or sequences, based on observed differences or similarities. Specifically, UPGMA and NJ cluster species hierarchically and minimum evolution aims for a tree that minimizes the sum of branch lengths. Although they may be less accurate than character-based methods due to the potential information loss during the creation of the distance matrix, their computational efficiency and straightforwardness make them valuable for handling large datasets. Generally, when applying distance-based methods to reconstruct a tumor phylogeny, the calculation of a distance matrix demonstrating the pairwise evolutionary distance between samples is required. Data obtained from previous steps are often binarized or categorized, such as gene expression being classified into upregulated, downregulated, or normal states, and CNVs being denoted as gain, loss, or neutral states. Techniques like Euclidean distance are then employed to generate this matrix. The generated distance matrix effectively quantifies the genetic dissimilarity between individual samples, serving as the foundational substrate upon which the aforementioned distance-based algorithms act to reconstruct the phylogenetic tree.
Conversely, character-based methods are a more sophisticated approach in phylogenetics that considers unique attributes or characteristics of organisms to elucidate their evolutionary relationships. These methods include Maximum Parsimony (MP),95 which aims to identify the tree that requires the least number of evolutionary changes. Maximum Likelihood (ML),96 on the other hand, seeks to find the tree that maximizes the probability of observing the given data under specific evolutionary models. Another important character-based method is the Bayesian Markov Chain Monte Carlo (BMCMC).97 It differs from Maximum Likelihood as it considers the model's parameters as random variables, merging prior knowledge with observed data. This allows for direct and clearer interpretations of the probabilities for different phylogenetic trees. Character-based methods utilize the sequencing data as unique attributes for each sample. Unlike distance-based methods, which require a pre-calculated distance matrix, these attributes are categorized and then used directly in the algorithms we mentioned above to reconstruct phylogenetic trees.
Many studies in tumor phylogenetics continue to leverage traditional evolutionary biology tools like PHYLIP,98, 99 Beast,100, 101 and phangorn102, 103, 104for tree reconstruction. In cross-sectional tumor phylogenetics, traditional phylogenetics methods have played a significant role, largely because this study design itself is among the earlier avenues of exploration within tumor phylogenetics. Initially, tools like oncotrees were employed, which primarily used aCGH-derived large-scale CNVs and relied on MP-based tree reconstruction.43 However, due to the limitations of MP's assumptions in capturing the intricacies of tumor evolution, more recent approaches have pivoted toward optimization techniques based on ML and BMCMC algorithms,105 such as Mtreemix106 and Rtreemix.107 In order to handle large datasets, distance-based methods are also gaining traction.108 The range of data types applied has expanded as well, encompassing DNAseq-derived CNVs,48 SNVs,49 and expression data.45
Regarding regional bulk tumor phylogenetics, the trajectory of algorithmic and data type developments is somewhat parallel to that of cross-sectional studies. Yet, because bulk sequencing has its limitations in pinpointing subclones accurately, the focus of many software solutions has shifted toward integrating deconvolution techniques with phylogenetic frameworks, such as TCS,109 TrAp,110 TITAN,111 SubcloneSeeker112 and CITUP.113 The goal is to accurately infer the subclonal frequency of tumors and use this refined framework for subsequent phylogenetic tree reconstruction. For example, TrAp was used to identify specific mutation co-localizations and temporal orders in melanoma metastases through the reconstruction of subclone architecture and lineage relationship, offering critical insights into the mechanisms of tumor progression and chemoresistance.110 Some specialized tools like phyloWGS114 and Canopy115 are even exploiting the differing rates and mechanisms of SNVs and CNVs, synthesizing them into a unified analysis based on BMCMC methods.
For single-cell phylogenetics, traditional methods falter due to issues like technical artifacts, false positives, and false negatives inherent to scRNAseq data. Newly developed methods like OncoNEM116 and SCITE117 tackle this by incorporating these uncertainties directly into their likelihood calculations with SNV data. However, they still make the infinite-sites assumption (ISA), which is often violated in tumor evolution. To address this, newer models like SiFit118 and SphyR119 have emerged, adopting finite-site evolution models that better fit the nuances of cancer data.
In lineage-tracing studies, the unique challenges posed by barcode site data are not well-handled by traditional algorithms. Unlike typical phylogenetic studies that deal with a wide and short matrix of nucleotide changes across a limited number of species, lineage-tracing barcodes produce a tall and narrow matrix, capturing thousands of cells with limited mutable sites. This dramatic increase in computational complexity has led to the development of specialized methods. The Cassiopeia suite offers three different algorithms tailored for varying dataset sizes and consistently outperforms traditional methods, proving the need for customized approaches in lineage tracing data.120
In addition to the tools discussed earlier, there are various other methods specifically designed for reconstructing tumor phylogenies. For readers' convenience, we have compiled a brief summary of key tools in Supplementary Table 1. Additionally, several articles also offer more comprehensive reviews of these tools.16,17,121
5. Insights obtained from tumor phylogenetics
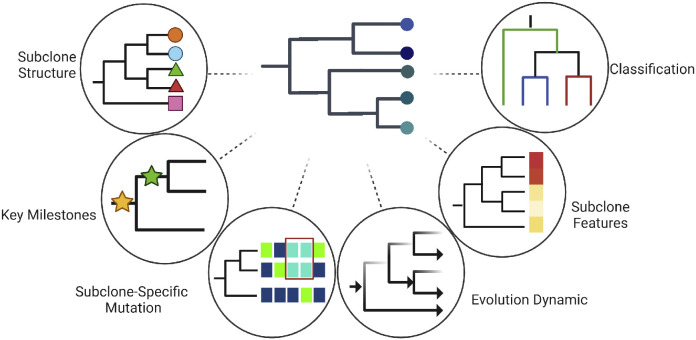
After examining the methods for reconstructing tumor phylogenetic trees, this section pivots to the critical insights these trees offer into tumor evolution, heterogeneity, and classification (Fig. 2).
Fig. 2.
Insights from tumor phylogeny. Utilizing tumor phylogenetics enables the extraction of critical insights in areas including, but not limited to, the following: subclonal architecture, key milestones in tumor evolution, subclone-specific mutations, tumor evolutionary dynamics, subclone features, and tumor classification.
5.1. Tracing tumor evolutionary history
Originally conceptualized to articulate the evolutionary history of biological species, tumor phylogenetics naturally excels in tracing the evolutionary paths of cancers. In the context of oncology, reconstructing the evolutionary history of a tumor can yield invaluable insights. Early research in this arena primarily focused on identifying distinct modes of tumor evolution68,122,123 and determining the timing of key evolutionary milestones.124, 125, 126 For example, a study that employed whole-exome sequencing and various phylogenetic methods on 40 matched samples—normal, primary, and metastatic—discovered that the genetic divergence of metastatic lineages often predated the initial diagnosis.127 Recent advances in research have allowed for more comprehensive explorations of tumor evolutionary dynamics combining multi-omics data.88 For instance, one study using a CRISPR-Cas9-based KP-tracer mouse model with the Cassiopeia-Hybrid algorithm found that specific rare subclones with unique transcriptional programs drive tumor evolution and metastasis following a period of increased cellular plasticity.91 Another study delved into the genomic and immune landscapes of multi-regional metastases in 10 therapy-resistant breast cancer autopsies, revealing intricate evolutionary dynamics within metastatic genomes.128 These retrospective analyses furnish critical insights into the tumor's origins, progression patterns, and potential future behaviors, thereby informing treatment strategies and potentially unveiling new therapeutic targets.
A key application of tracing tumor evolutionary history is that it can help researchers resolve the formation of therapy resistance. It often arises from genetic mutations and cellular adaptations that enable cancer cells to evade the effects of therapeutic agents.4,5 Particularly under the pressure of anti-tumor therapy, tumor cells may be selected for subclones with high metastatic potential. This metastasis can be monoclonal or polyclonal, indicating the presence of potentially diverse selection patterns. Cells from metastatic tumors may recirculate and seed new metastatic lesions, which, under the pressure of treatment, can develop varying degrees of drug resistance.129, 130, 131 Diverging from the traditional non-phylogenetic bioinformatics that offers a static analysis of drug target mutation sequences,132 tumor phylogenetics provides us with a dynamic view of tumor evolution. It enables an in-depth analysis of the tumor's internal clonal diversity and evolutionary history, enabling researchers to pinpoint the timing and causes of the emergence of therapy resistance. This can identify the origins of different treatment-resistant cells, guide clinical decisions, and determine the choice of treatment and their order. For example, phylogenetic analysis of lung adenocarcinoma reveals that cisplatin therapy induced a localized surge of mutations, leading to the evolution of EGFR T790M resistance to erlotinib.133
5.2. Resolving tumor heterogeneity
Tumor phylogenetics stands as a robust tool for unraveling intratumoral heterogeneity. By doing so, it illuminates the relationships between distinct subclones, reconstructing a more complete picture of the tumor's internal landscape. Utilizing tumor phylogenetics, researchers can investigate the structure of subclones within a tumor,134, 135, 136, 137 and study their changes in frequency during tumor evolution.138 Furthermore, the mutation profiles of individual subclones can be integrated to showcase subclone-specific mutations.139 Lastly, features of these subclones can be highlighted to expose phenotypic differences among them.48,91 The knowledge gained from resolving intratumoral heterogeneity holds the potential for the improvement of patient prognosis and the development of more effective therapies.
5.3. Tumor classification
Beyond its applications in studying tumor evolutionary history, tumor phylogenetics also functions as a clustering tool. It offers another avenue for predicting and classifying tumor types. It can be employed to group tumors based on their mutation or expression profiles, effectively segmenting them into distinct subtypes. Compared to traditional clustering, phylogenetic classification trees provide a more precise analysis by avoiding geometric assumptions and the arbitrary selection of cluster numbers, as well as capturing information on the degree of separation between various clusters that hierarchical clustering lacks.45,55,126 A study utilizes phylogenetic methods to classify tumors based on gene expression levels, demonstrating success in separating small round blue-cell tumors into four major groups and distinguishing breast tumors with BRCA1 and BRCA2 mutations in two specific datasets.45 A precise classification of tumor types can offer valuable insights for targeted therapies and prognostic evaluations, as each subtype may respond differently to various treatments.
In summary, tumor phylogenetics has deeply influenced oncology, offering crucial insights into tumor classification, intratumoral heterogeneity, and evolutionary pathways. These findings pave the way for more personalized and effective treatments. As the field matures, we can expect tumor phylogenetics to increasingly inform both cancer research and clinical practice.
6. The power of data integration in tumor phylogenetics
From the application examples mentioned in the previous chapters, it's evident that as tumor phylogenetics has evolved, newer studies rarely rely solely on tumor phylogenetic trees for basic evolutionary analysis. Reconstructing the tumor phylogeny is a key step, but it's not the endpoint. An extra step of integrating external data into the tumor phylogeny is often required to elucidate complex biological phenomena. For tumor phylogeny, this integration enriches tree interpretation, revealing deeper insights into tumor evolution. For the external data, this integration enriches the analysis by adding an evolutionary context, thereby providing a new dimension for interpreting the data within an evolutionary framework.
However, the current practice of integrating external data with phylogenetic trees remains rudimentary and infrequent. While some studies attempt to unify different types of data with phylogenetic analysis, these initial steps are typically simplistic and lack depth, often limited to straightforward combination methods, like using symbolic points and text on trees to indicate cancer clone and mutation profiles respectively,137 or simply display them in different figures.126 These preliminary combinations, although a step in the right direction, yield only a marginal increase in interpretative value and leave much room for improvement.
The simplicity of current methods for integrating external data with phylogenetic trees is largely due to the substantial challenges present in the endeavor. Firstly, there's a wide variety of formats for phylogenetic trees, making the task of reading these pre-reconstructed tumor evolution tree files far from straightforward. Additionally, the heterogeneity of external data introduces significant obstacles during the integration process, with variability spanning from genomic to transcriptomic scales, diverse data structures like continuous or categorical forms, and different sources such as clinical records or molecular assays. Lastly, visualizing the integrated data brings its own set of challenges. While presenting and annotating tumor phylogeny is already demanding, adding multidimensional external data only heightens the complexity.
Looking ahead, the maturation of tumor phylogenetics anticipates a new era where the integration of multi-omics data unveils the intricate genetic, epigenetic, and transcriptomic interplays that propel cancer evolution. Paving the way for these advances, pioneering studies are setting the stage for comprehensive analysis. Some studies have already begun to delve into deeper layers of data integration, harnessing a broader array of datasets for a multidimensional display and analysis that enriches the understanding of tumor phylogeny, seeking to uncover the full spectrum of evolutionary forces at work within tumors. For instance, in the examples mentioned in our previous session, subclonal mutations were identified by integrating mutation profiles,139 and tumor evolutionary dynamics were investigated through the integration of multi-omics data.91 Based on these examples, it becomes evident that incorporating multidimensional data into tumor phylogenetic trees is a powerful means that leads to new discoveries. Compared to a basic combination of data, such integration provides a multi-faceted perspective to phylogenetic constructs, proving that synergistic data convergence has a promising potential for revealing the underlying mechanisms of tumor genesis and progression.
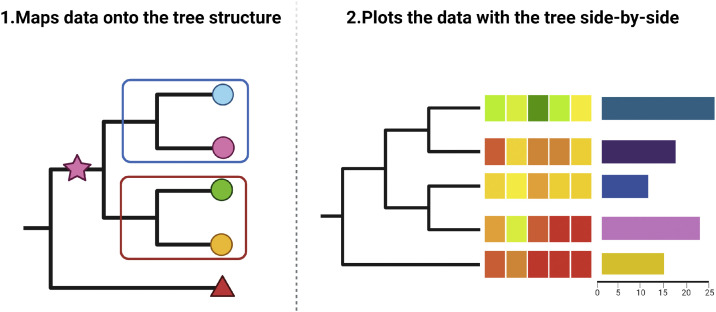
To better elucidate the complex interplay between genetic alterations and tumor evolution, two general approaches have been highlighted for the integration of external data with phylogenetic trees (Fig. 3).140 These methods were brought up with the emergence of the ggtree package in order to provide a versatile way of combining external data with phylogenetic trees. The first method maps external data directly onto the tree structure, annotating the tree with visual characteristics like symbolic points or labels. In the context of tumor phylogenetics, this can be invaluable in emphasizing specific evolutionary events141 or highlighting important subclones.142 Secondly, we can generate side-by-side visual comparisons by aligning graphs, like mutation profiles or subclone features, with the structure of the phylogenetic tree. In tumor evolution research, it can potentially spotlight concurrent events or patterns, like mutations shared by specific subclones139,143 or the consistency of genetic variations.70 This approach excels at displaying multidimensional data alongside the tree. It's also useful to combine both methods, enhancing the richness and depth of analytical insight. In a study on human triple-negative breast cancer (TNBC), an analysis combining the annotated tree and copy number profiles revealed a complex hierarchy of many subclones and fewer superclones, all from a common evolutionary root. Key early events include TP53 mutations and genome doubling that occurred before a common ancestor, followed by a burst of subclonal diversification that stabilized during tumor growth.125 Through these examples, it's clear that these two methods are universally applicable and effective in representing complex data layers. Utilizing such methods enhances our ability to display a comprehensive view of the tumor's evolutionary trajectory, paving the way for richer insights and potential therapeutic interventions.
Fig. 3.
Methods of plotting tree with data. Method 1 involves directly mapping data onto the tree structure by adding visual annotations such as symbolic points or labels. Method 2 consists of plotting the data adjacent to the tree after proper alignment.
An expanding range of tools and methodologies sharing this concept are being developed to assist researchers in elucidating the complex interplay between genetic alterations and tumor evolution. For instance, the ggtree packages suite that we mentioned above stands out by bringing integration of phylogeny data into R, offering a set of data importation and manipulation functions via treeio and tidytree, and enhancing annotated phylogenies with multi-dimensional data through ggtree and ggtreeExtra, which support the flexible combination of both visualization methods.144, 145, 146 Besides ggtree, phytools is an R package well-known for its comprehensive range of phylogenetic analysis functions, also offering advanced tree visualization functions.147,148 EvolView presents an interactive, user-friendly interface for annotating and customizing phylogenetic trees, making it accessible for users with limited coding experience.149 On the other hand, iTOL stands out for its robust online platform for visualizing, editing, and sharing detailed phylogenetic trees.150 Additionally, E-scape offers an interactive browser-based visualization suite specifically designed to present tumor phylogeny.151 Utilizing these tools enhances our capacity to display a comprehensive view of the tumor's evolutionary narrative, paving the way for richer insights and potential therapeutic interventions.
In conclusion, the essence of fully understanding tumor evolution lies not just in the tree itself but in its intricate interplay with external data. By integrating this data, researchers are empowered with a more holistic view, allowing them to uncover hidden evolutionary patterns that might otherwise remain obscured.
7. Discussion and conclusions
Throughout this review, we have undertaken an extensive exploration of the various study designs employed in tumor phylogenetics. The utilization of cross-sectional tumor phylogenetics (TP) has diminished in recent research, largely due to the widespread adoption of NGS. Even when comparing tumors from different patients, bulk sequencing TP and single-cell TP are more commonly used to reconstruct individual phylogenies that can later be compared.91,139,152 Both bulk sequencing TP and single-cell TP come with their respective drawbacks—lower resolution for the former and computational complexity for the latter. The merging of these two approaches has been proposed as a complementary strategy.153 Lineage tracing TP offers granular details of tumor evolutionary dynamics, but its high cost and computational intensity often discourage widespread use.
We have also discussed various data types used for reconstructing tumor phylogenies. While genomic-derived SNVs and CNVs have long been the mainstays, the development, and incorporation of other 'omics' data, such as transcriptomics and epigenomics, are equally essential, providing another aspect to inspect tumor evolution.45, 46, 47,71 The attempt to combine these diverse data types could also enrich the quality and depth of the resulting phylogenies.115,134
Tumor phylogenetics has grown substantially over the past several decades. Its advantage in capturing the evolutionary history and complex structure of tumor heterogeneity has brought our understanding of tumor evolution to another level. In current tumor phylogenetics, an increasing number of researchers are now leveraging the multi-omics characteristics of tumor heterogeneity by integrating data across genomic, transcriptomic, and epigenetic for comprehensive phylogenetic analysis, including methods for deconvolution and tumor phylogenetic reconstruction that synergize multi-omic data.91,154, 155, 156, 157, 158 As technology advances, cutting-edge techniques like spatial transcriptomics are also being used to explore the spatial dimensions of tumor evolution, marking a significant step forward in the field.143,159,160 Meanwhile, the utilization of machine learning methods for tumor phylogeny topology inference has provided new angles for tumor phylogenetic reconstruction.161, 162, 163, 164 The advancement of tumor phylogenetics promises access to increasingly multidimensional and multimodal data, enhancing our understanding of tumors. By integrating these complex data sets with tumor phylogenetics using tools such as the ggtree package suite, we are able to analyze tumor evolution from a more comprehensive perspective, as discussed earlier. Therefore, it's compelling to say that the future potential of this field may well lie in the integration of tumor phylogenetics with external data.
In conclusion, tumor phylogenetics serves as a critical lens through which we can unravel the complexities of cancer evolution and heterogeneity. This multidisciplinary field holds the potential not only to elucidate fundamental mechanisms of tumor growth and spread but also to inform patient-specific treatment strategies. By providing insights into the origins, progression, and diverse evolutionary paths of tumors, phylogenetics is poised to make a transformative impact on cancer research and patient prognosis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (approval number: 32270677).
Author contributions
All authors contributed to writing the manuscript and designed and prepared the figures and legends.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jncc.2024.03.001.
Appendix. Supplementary materials
References
- 1.Frick C, Rumgay H, Vignat J, et al. Quantitative estimates of preventable and treatable deaths from 36 cancers worldwide: a population-based study. Lancet Glob Health. 2023;11(11):e1700–e1712. doi: 10.1016/S2214-109X(23)00406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 4.Mansoori B, Mohammadi A, Davudian S, et al. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Housman G, Byler S, Heerboth S, et al. Drug resistance in cancer: an overview. Cancers. 2014;6(3):1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aktipis CA, Kwan VS, Johnson KA, et al. Overlooking evolution: a systematic analysis of cancer relapse and therapeutic resistance research. PLoS One. 2011;6(11):e26100. doi: 10.1371/journal.pone.0026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duprez F, Berwouts D, De Neve W, et al. Distant metastases in head and neck cancer. Head Neck. 2017;39(9):1733–1743. doi: 10.1002/hed.24687. [DOI] [PubMed] [Google Scholar]
- 8.Monteiro J, Fodde R. Cancer stemness and metastasis: therapeutic consequences and perspectives. Eur J Cancer. 2010;46(7):1198–1203. doi: 10.1016/j.ejca.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 9.Rueff J, Rodrigues AS. Cancer drug resistance: a brief overview from a genetic viewpoint. Methods Mol Biol. 2016;1395:1–18. doi: 10.1007/978-1-4939-3347-1_1. [DOI] [PubMed] [Google Scholar]
- 10.SN Aleksakhina, Kashyap A, Imyanitov EN. Mechanisms of acquired tumor drug resistance. Biochim Biophys Acta Rev Cancer. 2019;1872(2) doi: 10.1016/j.bbcan.2019.188310. [DOI] [PubMed] [Google Scholar]
- 11.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 12.Rogiers A, Lobon I, Spain L, Turajlic S. The genetic evolution of metastasis. Cancer Res. 2022;82(10):1849–1857. doi: 10.1158/0008-5472.CAN-21-3863. [DOI] [PubMed] [Google Scholar]
- 13.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352(6282):169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 14.Yates LR, Knappskog S, Wedge D, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2):169–184. doi: 10.1016/j.ccell.2017.07.005. e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Rannala B. Molecular phylogenetics: principles and practice. Nat Rev Genet. 2012;13(5):303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- 16.Somarelli JA, Ware KE, Kostadinov R, et al. PhyloOncology: understanding cancer through phylogenetic analysis. Biochim Biophys Acta Rev Cancer. 2017;1867(2):101–108. doi: 10.1016/j.bbcan.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz R, Schäffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet. 2017;18(4):213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowell PC. The clonal evolution of tumor cell populations: acquired genetic lability permits stepwise selection of variant sublines and underlies tumor progression. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 19.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20(7):404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 20.Beerenwinkel N, Schwarz RF, Gerstung M, Markowetz F. Cancer evolution: mathematical models and computational inference. Syst Biol. 2015;64(1):e1–e25. doi: 10.1093/sysbio/syu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakhoum SF, Landau DA. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb Perspect Med. 2017;7(6) doi: 10.1101/cshperspect.a029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrell RA, Swanton C. The evolution of the unstable cancer genome. Curr Opin Genet Dev. 2014;24:61–67. doi: 10.1016/j.gde.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Huang S. Genetic and non-genetic instability in tumor progression: link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev. 2013;32(3–4):423–448. doi: 10.1007/s10555-013-9435-7. [DOI] [PubMed] [Google Scholar]
- 24.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32(3–4):341–352. doi: 10.1007/s10555-013-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun XX, Yu Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol Sin. 2015;36(10):1219–1227. doi: 10.1038/aps.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Zhou XA, Zhang N, Wang J. Evolving insights: how DNA repair pathways impact cancer evolution. Cancer Biol Med. 2020;17(4):805–827. doi: 10.20892/j.issn.2095-3941.2020.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dentro SC, Leshchiner I, Haase K, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184(8):2239–2254. doi: 10.1016/j.cell.2021.03.009. e2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 29.Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015;21(5):440–448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 30.Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5(8):806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald C, McClelland SE. Chromosome instability through the ages: parallels between speciation and somatic (cancer) evolution. Trends Genet. 2021;37(8):691–694. doi: 10.1016/j.tig.2021.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Alves JM, Prado-Lopez S, Cameselle-Teijeiro JM, Posada D. Rapid evolution and biogeographic spread in a colorectal cancer. Nat Commun. 2019;10(1):5139. doi: 10.1038/s41467-019-12926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chroni A, Miura S, Hamilton L, et al. Clone phylogenetics reveals metastatic tumor migrations, maps, and models. Cancers. 2022;14(17):4326. doi: 10.3390/cancers14174326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chroni A, Miura S, Oladeinde O, et al. Migrations of cancer cells through the lens of phylogenetic biogeography. Sci Rep. 2021;11(1):17184. doi: 10.1038/s41598-021-96215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chroni A, Vu T, Miura S, Kumar S. Delineation of tumor migration paths by using a Bayesian biogeographic approach. Cancers. 2019;11(12):1880. doi: 10.3390/cancers11121880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Chroni A, Tamura K, et al. PathFinder: Bayesian inference of clone migration histories in cancer. Bioinformatics. 2020;36(Suppl_2):i675–i683. doi: 10.1093/bioinformatics/btaa795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso C, Pérez R, Bazaga P, Herrera CM. Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms. Front Genet. 2015;6:4. doi: 10.3389/fgene.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazor T, Pankov A, Johnson BE, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas P, Kim KM, Shibata D, Tavaré S. The stem cell population of the human colon crypt: analysis via methylation patterns. PLoS Comput Biol. 2007;3(3):e28. doi: 10.1371/journal.pcbi.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegmund KD, Marjoram P, Woo YJ, et al. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A. 2009;106(12):4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skinner MK, Gurerrero-Bosagna C, Haque MM, et al. Epigenetics and the evolution of Darwin’s finches. Genome Biol Evol. 2014;6(8):1972–1989. doi: 10.1093/gbe/evu158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 43.Desper R, Jiang F, Kallioniemi OP, et al. Inferring tree models for oncogenesis from comparative genome hybridization data. J Comput Biol. 1999;6(1):37–51. doi: 10.1089/cmb.1999.6.37. [DOI] [PubMed] [Google Scholar]
- 44.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(Suppl):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 45.Desper R, Khan J, Schäffer AA. Tumor classification using phylogenetic methods on expression data. Theor Biol. 2004;228(4):477–496. doi: 10.1016/j.jtbi.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 46.Brocks D, Assenov Y, Minner S, et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8(3):798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 47.Sottoriva A, Spiteri I, Shibata D, et al. Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res. 2013;73(1):41–49. doi: 10.1158/0008-5472.CAN-12-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz RF, Ng CK, Cooke SL, et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12(2) doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bashashati A, Ha G, Tone A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231(1):21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng PC, Kirkness EF. Whole genome sequencing. Methods Mol Biol. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 51.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59(1):5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 52.Van der Auwera GA, BD O’Connor. O’Reilly Media; 2020. Genomics in the cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 53.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riester M, Stephan-Otto Attolini C, Downey RJ, et al. A differentiation-based phylogeny of cancer subtypes. PLoS Comput Biol. 2010;6(5) doi: 10.1371/journal.pcbi.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarabichi M, Salcedo A, Deshwar AG, et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat Methods. 2021;18(2):144–155. doi: 10.1038/s41592-020-01013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller CA, White BS, Dees ND, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10(8) doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khalique L, Ayhan A, Whittaker JC, et al. The clonal evolution of metastases from primary serous epithelial ovarian cancers. Int J Cancer. 2009;124(7):1579–1586. doi: 10.1002/ijc.24148. [DOI] [PubMed] [Google Scholar]
- 60.Navin N, Krasnitz A, Rodgers L, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20(1):68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17(3):175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 64.Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116(Pt 14):2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 66.Frumkin D, Wasserstrom A, Itzkovitz S, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68(14):5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 67.Shlush LI, Chapal-Ilani N, Adar R, et al. Cell lineage analysis of acute leukemia relapse uncovers the role of replication-rate heterogeneity and microsatellite instability. Blood. 2012;120(3):603–612. doi: 10.1182/blood-2011-10-388629. [DOI] [PubMed] [Google Scholar]
- 68.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qi M, Pang J, Mitsiades I, et al. Loss of chromosome Y in primary tumors. Cell. 2023 doi: 10.1016/j.cell.2023.06.006. S0092-8674(23)00646-3. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Griffiths JI, Bishara I, et al. Phylogenetic inference from single-cell RNA-seq data. Sci Rep. 2023;13(1):12854. doi: 10.1038/s41598-023-39995-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt F, Efferth T. Tumor heterogeneity, single-cell sequencing, and drug resistance. Pharmaceuticals. 2016;9(2):33. doi: 10.3390/ph9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hou Y, Song L, Zhu P, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 74.Tang X, Huang Y, Lei J, et al. The single-cell sequencing: new developments and medical applications. Cell Biosci. 2019;9:53. doi: 10.1186/s13578-019-0314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Humphries A, Cereser B, Gay LJ, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proc Natl Acad Sci U S A. 2013;110(27):E2490–E2499. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guest RV, Boulter L, Kendall TJ, et al. Cell lineage tracing reveals a biliary origin of intrahepatic cholangiocarcinomacholangiocarcinoma can arise from cholangiocytes. Cancer Res. 2014;74(4):1005–1010. doi: 10.1158/0008-5472.CAN-13-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamprecht S, Schmidt EM, Blaj C, et al. Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer. Nat Commun. 2017;8(1):1406. doi: 10.1038/s41467-017-00976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin S, Wangensteen KJ, Teta-Bissett M, et al. Genetic lineage tracing analysis of the cell of origin of hepatotoxin-induced liver tumors in mice. Hepatology. 2016;64(4):1163–1177. doi: 10.1002/hep.28602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C, Liao Y, Peng G. Connecting past and present: single-cell lineage tracing. Protein Cell. 2022;13(11):790–807. doi: 10.1007/s13238-022-00913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKenna A, Gagnon JA. Recording development with single cell dynamic lineage tracing. Development. 2019;146(12) doi: 10.1242/dev.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baron CS, van Oudenaarden A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat Rev Mol Cell Biol. 2019;20(12):753–765. doi: 10.1038/s41580-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 82.McKenna A, Findlay GM, Gagnon JA, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353(6298):aaf7907. doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spanjaard B, Hu B, Mitic N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat Biotechnol. 2018;36(5):469–473. doi: 10.1038/nbt.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pei W, Feyerabend TB, Rössler J, et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature. 2017;548(7668):456–460. doi: 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peikon ID, Gizatullina DI. Zador AM. In vivo generation of DNA sequence diversity for cellular barcoding. Nucleic Acids Res. 2014;42(16):e127. doi: 10.1093/nar/gku604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raj B, Wagner DE, McKenna A, et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol. 2018;36(5):442–450. doi: 10.1038/nbt.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wagner DE, Weinreb C, Collins ZM, et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018;360(6392):981–987. doi: 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quinn JJ, Jones MG, Okimoto RA, et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science. 2021;371(6532):eabc1944. doi: 10.1126/science.abc1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simeonov KP, Byrns CN, Clark ML, et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell. 2021;39(8):1150–1162. doi: 10.1016/j.ccell.2021.05.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vinuesa-Pitarch E, Ortega-Álvarez D, Rodilla V. How lineage tracing studies can unveil tumor heterogeneity in breast cancer. Biomedicines. 2021;10(1):3. doi: 10.3390/biomedicines10010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang D, Jones MG, Naranjo S, et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell. 2022;185(11):1905–1923. doi: 10.1016/j.cell.2022.04.015. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sokal RR. A statiscal method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 93.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 94.Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol. 2002;9(5):687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 95.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol. 1971;20(4):406–416. [Google Scholar]
- 96.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 97.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 1981;17(6):304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 98.Felsenstein J. Joseph Felsenstein; 1993. PHYLIP (Phylogeny Inference Package), Version 3.5 c. [Google Scholar]
- 99.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kostadinov RL, Kuhner MK, Li X, et al. NSAIDs modulate clonal evolution in Barrett’s esophagus. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Formenti SC, Rudqvist NP, Golden E, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat Med. 2018;24(12):1845–1851. doi: 10.1038/s41591-018-0232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao R, Davis A, McDonald TO, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48(10):1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.vonHeydebreck A, Gunawan B, Füzesi L. Maximum likelihood estimation of oncogenetic tree models. Biostatistics. 2004;5(4):545–556. doi: 10.1093/biostatistics/kxh007. [DOI] [PubMed] [Google Scholar]
- 106.Beerenwinkel N, Rahnenführer J, Kaiser R, et al. Mtreemix: a software package for learning and using mixture models of mutagenetic trees. Bioinformatics. 2005;21(9):2106–2107. doi: 10.1093/bioinformatics/bti274. [DOI] [PubMed] [Google Scholar]
- 107.Bogojeska J, Alexa A, Altmann A, et al. Rtreemix: an R package for estimating evolutionary pathways and genetic progression scores. Bioinformatics. 2008;24(20):2391–2392. doi: 10.1093/bioinformatics/btn410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Desper R, Jiang F, Kallioniemi OP, et al. Distance-based reconstruction of tree models for oncogenesis. J Comput Biol. 2000;7(6):789–803. doi: 10.1089/10665270050514936. [DOI] [PubMed] [Google Scholar]
- 109.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 110.Strino F, Parisi F, Micsinai M, Kluger Y. TrAp: a tree approach for fingerprinting subclonal tumor composition. Nucleic Acids Res. 2013;41(17):e165. doi: 10.1093/nar/gkt641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ha G, Roth A, Khattra J, et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014;24(11):1881–1893. doi: 10.1101/gr.180281.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qiao Y, Quinlan AR, Jazaeri AA, et al. SubcloneSeeker: a computational framework for reconstructing tumor clone structure for cancer variant interpretation and prioritization. Genome Biol. 2014;15(8):443. doi: 10.1186/s13059-014-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malikic S, McPherson AW, Donmez N, Sahinalp CS. Clonality inference in multiple tumor samples using phylogeny. Bioinformatics. 2015;31(9):1349–1356. doi: 10.1093/bioinformatics/btv003. [DOI] [PubMed] [Google Scholar]
- 114.Deshwar AG, Vembu S, Yung CK, et al. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16(1):35. doi: 10.1186/s13059-015-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang Y, Qiu Y, Minn AJ, Zhang NR. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci U S A. 2016;113(37):E5528–E5537. doi: 10.1073/pnas.1522203113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jahn K, Kuipers J, Beerenwinkel N. Tree inference for single-cell data. Genome Biol. 2016;17:86. doi: 10.1186/s13059-016-0936-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zafar H, Tzen A, Navin N, et al. SiFit: inferring tumor trees from single-cell sequencing data under finite-sites models. Genome Biol. 2017;18(1):178. doi: 10.1186/s13059-017-1311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.El-Kebir M. SPhyR: tumor phylogeny estimation from single-cell sequencing data under loss and error. Bioinformatics. 2018;34(17):i671–i679. doi: 10.1093/bioinformatics/bty589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones MG, Khodaverdian A, Quinn JJ, et al. Inference of single-cell phylogenies from lineage tracing data using Cassiopeia. Genome Biol. 2020;21(1):92. doi: 10.1186/s13059-020-02000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schwartz R. Bioinformatics and Phylogenetics. Computational Biology, Vol 29. Springer; Cham: 2019. Computational models for cancer phylogenetics. [DOI] [Google Scholar]
- 122.Cresswell GD, Apps JR, Chagtai T, et al. Intra-tumor genetic heterogeneity in Wilms tumor: clonal evolution and clinical implications. EBioMedicine. 2016;9:120–129. doi: 10.1016/j.ebiom.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brown D, Smeets D, Székely B, et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun. 2017;8:14944. doi: 10.1038/ncomms14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Attolini CSO, Cheng YK, Beroukhim R, et al. A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci U S A. 2010;107(41):17604–17609. doi: 10.1073/pnas.1009117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Minussi DC, Nicholson MD, Ye H, et al. Breast tumours maintain a reservoir of subclonal diversity during expansion. Nature. 2021;592(7853):302–308. doi: 10.1038/s41586-021-03357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abu-Asab MS, Abu-Asab N, Loffredo CA, et al. Identifying early events of gene expression in breast cancer with systems biology phylogenetics. Cytogenet Genome Res. 2013;139(3):206–214. doi: 10.1159/000348433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhao ZM, Zhao B, Bai Y, et al. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci U S A. 2016;113(8):2140–2145. doi: 10.1073/pnas.1525677113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.De Mattos-Arruda L, Sammut SJ, Ross EM, et al. The genomic and immune landscapes of lethal metastatic breast cancer. Cell Rep. 2019;27(9):2690–2708. doi: 10.1016/j.celrep.2019.04.098. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Comen E, Norton L, Massague J. Clinical implications of cancer self-seeding. Nat Rev Clin Oncol. 2011;8(6):369–377. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 130.Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turajlic S, Xu H, Litchfield K, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173(3):581–594. doi: 10.1016/j.cell.2018.03.057. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gautam A, Chaudhary K, Kumar R, et al. Managing drug resistance in cancer: role of cancer informatics. Methods Mol Biol. 2016;1395:299–312. doi: 10.1007/978-1-4939-3347-1_17. [DOI] [PubMed] [Google Scholar]
- 133.Fisk JN, Mahal AR, Dornburg A, et al. Premetastatic shifts of endogenous and exogenous mutational processes support consolidative therapy in EGFR-driven lung adenocarcinoma. Cancer Lett. 2022;526:346–351. doi: 10.1016/j.canlet.2021.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raynaud F, Mina M, Tavernari D, Ciriello G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet. 2018;14(9) doi: 10.1371/journal.pgen.1007669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chai S, Ruiz-Velasco C, Naghdloo A, et al. Identification of epithelial and mesenchymal circulating tumor cells in clonal lineage of an aggressive prostate cancer case. NPJ Precis Oncol. 2022;6(1):41. doi: 10.1038/s41698-022-00289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Campbell PJ, Pleasance ED, Stephens PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105(35):13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Potter NE, Ermini L, Papaemmanuil E, et al. Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res. 2013;23(12):2115–2125. doi: 10.1101/gr.159913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Anderson K, Lutz C, Van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 139.Tang J, Tu K, Lu K, et al. Single-cell exome sequencing reveals multiple subclones in metastatic colorectal carcinoma. Genome Med. 2021;13(1):148. doi: 10.1186/s13073-021-00962-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu G, Lam TT, Zhu H., Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol. 2018;35(12):3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao ZM, Zhao B, Bai Y, et al. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci U S A. 2016;113(8):2140–2145. doi: 10.1073/pnas.1525677113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang D, Jones MG, Naranjo S, et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell. 2022;185(11):1905–1923. doi: 10.1016/j.cell.2022.04.015. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu HJ, Temko D, Maliga Z, et al. Spatial intra-tumor heterogeneity is associated with survival of lung adenocarcinoma patients. Cell Genom. 2022;2(8) doi: 10.1016/j.xgen.2022.100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu S, Dai Z, Guo P, et al. ggtreeExtra: compact visualization of richly annotated phylogenetic data. Mol Biol Evol. 2021;38(9):4039–4042. doi: 10.1093/molbev/msab166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu G, Smith DK, Zhu H, et al. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 146.Wang LG, Lam TT, Xu S, et al. Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol Biol Evol. 2020;37(2):599–603. doi: 10.1093/molbev/msz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 148.Revell LJ. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things) PeerJ. 2024;12:e16505. doi: 10.7717/peerj.16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.He Z, Zhang H, Gao S, et al. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44(W1):W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Smith MA, Nielsen CB, Chan FC, et al. E-scape: interactive visualizattr`ion of single-cell phylogenetics and cancer evolution. Nat Methods. 2017;14(6):549–550. doi: 10.1038/nmeth.4303. [DOI] [PubMed] [Google Scholar]
- 152.Zhang Y, Chang L, Yang Y, et al. Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol Cancer. 2019;18(1):7. doi: 10.1186/s12943-019-0939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Malikic S, Mehrabadi FR, Ciccolella S, et al. PhISCS: a combinatorial approach for subperfect tumor phylogeny reconstruction via integrative use of single-cell and bulk sequencing data. Genome Res. 2019;29(11):1860–1877. doi: 10.1101/gr.234435.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu X, Wang W, Liu X, et al. Multi-omics analysis of intra-tumoural and inter-tumoural heterogeneity in pancreatic ductal adenocarcinoma. Clin Transl Med. 2022;12(1):e670. doi: 10.1002/ctm2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet. 2021;22(1):3–18. doi: 10.1038/s41576-020-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Song Y, Zhou J, Zhao X, et al. Lineage tracing for multiple lung cancer by spatiotemporal heterogeneity using a multi-omics analysis method integrating genomic, transcriptomic, and immune-related features. Front Oncol. 2023;13 doi: 10.3389/fonc.2023.1237308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ortega MA, Poirion O, Zhu X, et al. Using single-cell multiple omics approaches to resolve tumor heterogeneity. Clin Transl Med. 2017;6(1):46. doi: 10.1186/s40169-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun Y, Wu P, Zhang Z, et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell. 2024;42(1):135–156. doi: 10.1016/j.ccell.2023.11.010. e17. [DOI] [PubMed] [Google Scholar]
- 159.Erickson A, Figiel S, Rajakumar T, et al. Clonal phylogenies inferred from bulk, single cell, and spatial transcriptomic analysis of cancer. bioRxiv. 2023 2023.2002. 2026.530145. [Google Scholar]
- 160.Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee D, Park Y, Kim S. Towards multi-omics characterization of tumor heterogeneity: a comprehensive review of statistical and machine learning approaches. Brief Bioinform. 2021;22(3):bbaa188. doi: 10.1093/bib/bbaa188. [DOI] [PubMed] [Google Scholar]
- 162.Azer ES, Ebrahimabadi MH, Malikić S, et al. Tumor phylogeny topology inference via deep learning. iScience. 2020;23(11) doi: 10.1016/j.isci.2020.101655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Caravagna G, Heide T, Williams MJ, et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nat Genet. 2020;52(9):898–907. doi: 10.1038/s41588-020-0675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Edrisi M, Ogilvie HA, Li M, Nakhleh L. In: Comparative Genomics. RECOMB-CG 2023. Lecture Notes in Computer Science. Jahn K., Vinař T., editors. Springer; Cham: 2023. MoTERNN: classifying the mode of cancer evolution using recursive neural networks. vol 13883. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.