Abstract
Duck hepatitis B viruses (DHBV), unlike mammalian hepadnaviruses, are thought to lack X genes, which encode transcription-regulatory proteins believed to contribute to the development of hepatocellular carcinoma. A lack of association of chronic DHBV infection with hepatocellular carcinoma development supports this belief. Here, we demonstrate that DHBV genomes have a hidden open reading frame from which a transcription-regulatory protein, designated DHBx, is expressed both in vitro and in vivo. We show that DHBx enhances neither viral protein expression, intracellular DNA synthesis, nor virion production when assayed in the full-length genome context in LMH cells. However, similar to mammalian hepadnavirus X proteins, DHBx activates cellular and viral promoters via the Raf–mitogen-activated protein kinase signaling pathway and localizes primarily in the cytoplasm. The functional similarities as well as the weak sequence homologies of DHBx and the X proteins of mammalian hepadnaviruses strongly suggest a common ancestry of ortho- and avihepadnavirus X genes. In addition, our data disclose similar intracellular localization and transcription regulatory functions of the corresponding proteins, raise new questions as to their presumed role in hepatocarcinogenesis, and imply unique opportunities for deciphering of their still-enigmatic in vivo functions.
Since the identification of hepatitis B virus (HBV) in humans, several related viruses have been isolated from mammalian and avian species (9, 53, 68). These viruses, known as hepadnaviruses, display high liver tropism, have a narrow host range, and cause acute and chronic hepatitis. Chronic infection with mammalian hepadnaviruses (orthohepadnaviruses) is associated with the development of liver cancer.
The hepadnaviruses are small enveloped DNA viruses with a unique virion ultrastructure. The viral genome is a partially duplexed, relaxed circular DNA molecule (rcDNA) with a size of about 3 kb which upon entry of the cell is converted into a covalently closed circular episome. This covalently closed circular DNA is then transcribed by the host RNA polymerase II, synthesizing subgenomic mRNAs and a greater-than-genome-length RNA known as pregenomic RNA or C-mRNA. Distinctive for all hepadnaviruses is the method of replication by reverse transcription of the pregenomic RNA into rcDNA. Transcripts encoding the envelope proteins, the nucleocapsid protein, the polymerase protein (P protein), and, as exclusively described so far in orthohepadnaviruses, the X protein have been identified. The organization of the viral genome is very compact, with overlapping reading frames and promoters and enhancer elements located within coding regions (20).
The X proteins of orthohepadnaviruses affect signal transduction pathways, transcription, cell transformation, and proliferation (1, 8, 46, 70). The HBV-specific X protein (HBx) is expressed in vivo, as shown by immunohistology and indirectly by identification of an HBx-specific immune response in infected individuals (64). In vivo expression of the X protein (WHx) has been demonstrated most convincingly for woodchuck hepatitis B virus (WHV)-infected animals (14, 32). HBx and WHx are predominantly localized in the cytoplasm, but a small fraction has also been found in the nucleus and associated with the nuclear framework (13, 15, 26, 49, 54, 69). HBx transactivates a wide range of cellular and viral promoters (1, 8, 46, 70). It acts, for instance, as a cytoplasmic activator of known mitogenic signal transduction pathways, in particular the Ras-Raf mitogen-activated protein (MAP) kinase cascade (2, 4, 15, 34, 47, 67). Thus, it can activate a variety of transcription factors and modulate cellular gene expression. HBx also activates transcription directly through protein-protein interactions (11, 26, 27, 44, 50, 51). Further interactions with components of the proteasome (19, 30, 31, 57), a DNA repair protein (33, 40), and a cellular factor with an inhibitory effect on the transactivation properties of HBx (45) have been characterized which offer explanations for the pleiotropic effects of HBx.
In vivo experiments have shown that WHx is required for the establishment of chronic infection (10, 72), but HBx is not essential for replication in hepatoma cells (6). Since HBx has these multiple effects on regulatory cellular pathways, it has been speculated that it might be involved in the development of hepatocellular carcinoma (1, 8, 46, 70). HBx-mediated oncogenic transformation of immortalized rodent cells (29, 59, 61) and development of hepatocellular carcinoma in some strains of transgenic mice (35, 62, 71) support this speculation. The p53-dependent and -independent pro- and antiapoptotic, as well as cell cycle-regulatory, effects of HBx may also contribute to hepatocarcinogenesis (3, 5, 12, 17, 35, 60, 65–67). Despite the many studies of functions of mammalian X proteins, there are so far no data as to the precise structure of X-specific mRNA in infected cells from which this protein is translated (70). Infection of Pekin ducks with duck hepatitis B virus (DHBV) is the most convenient and useful animal model for studies of the life cycle of hepadnaviruses but is less suitable for studies of hepadnavirus-mediated hepatocarcinogenesis because chronic infection of ducks with DHBV does not appear to be associated with the development of liver cancer (16). This is believed to be due to the absence of an X gene in all known DHBV isolates. Although X-protein-like sequences were proposed to be present in the middle of the DHBV nucleocapsid protein (DHBc) (18), functional similarities of DHBc with the mammalian hepadnavirus X proteins have never been identified. Moreover, the existence of an open reading frame (ORF) in the hepadnavirus genomes isolated from grey herons (48), snow geese (9), a Ross goose (48), and white storks (H. J. Netter, S.-F. Chang, and H. Will, unpublished observation) in a position similar to that of the X gene of orthohepadnaviruses argues that an X-like protein may be expressed from avian hepadnavirus genomes.
These findings prompted us to investigate whether avian hepadnaviruses also express an HBx-like regulatory protein. Here, we demonstrate that such a protein is indeed encoded by a hidden DHBV ORF located at a position analogous to those of the X genes of mammalian hepadnaviruses. Furthermore, we show that this protein is expressed in vitro and in vivo and has functions similar to those of the X proteins of mammalian hepadnaviruses.
MATERIALS AND METHODS
Plasmids.
A head-to-tail dimer of the DHBV3 isolate genome was inserted via EcoRI into the vector pUC18, which resulted in plasmid pDHBV3. To prevent the expression of X-like proteins, an analogous plasmid (pDHBV3-X-K.O.) that contains a stop codon in each of the two X-like ORFs of the tandemerized DHBV3 genomes was produced as follows. By oligonucleotide-directed mutagenesis, a G-to-A nucleotide change which converts the codon for tryptophan (TGG) at amino acid position 28 in the X-like ORF into a stop codon (TAG) was introduced at position 2371. To achieve this, DHBV3 DNA was amplified with the primers DHBV2144(+) and DHBV2371GA(−) and, in a separate reaction, with the primers DHBV257(−) and DHBV2371GA(+). The mixture of both amplified products was used as a template in a second PCR performed with the primers DHBV2144(+) and DHBV257(−). The product of this PCR was digested with NcoI and EcoRI, and then the 0.67-kb fragment which contained the mutation was purified. The corresponding wild-type fragment was replaced with the purified PCR fragment, and then the mutated monomeric DHBV genome was head-to-tail dimerized.
Several plasmids for expression of DHBx under the control of the cytomegalovirus (CMV) promoter were constructed by inserting the following XbaI/HindIII PCR fragments into vector pRK5 linearized by the same restriction enzymes. Primers DHBV3-His-M1X and DHBV-X-stop were used to amplify from DHBV3 DNA a fragment coding for a fusion protein which begins with six histidines, followed by a methionine and then the complete DHBx sequence (plasmid pDHBV3-M1X). For the construction of plasmid pDHBV3-M28X, the primers DHBV3-His-M28X and DHBV-X-stop were used to amplify a similar DNA fragment which encodes the same fusion protein except for the first 27 amino acids of DHBx. Plasmid pDHBV3-M28X-stop is almost identical to pDHBV3-M1X but has a stop codon instead of a tryptophan codon at position 28 of DHBx. The corresponding PCR fragment was obtained by using pDHBV3-X-K.O. as a template and primers DHBV3-His-M1X and DHBV3-X-stop.
Constructs used for the analysis of C-gene promoter activity were obtained by amplification of the core promoter regions (nucleotides 1658 to 2520) using pDHBV3 or pDHBV3-X-K.O as a template and the primers DHBV1557(+) and DHBV2520-HindIII. The PCR products were digested with BamHI and HindIII and then cloned into BglII and HindIII sites of the pGL3 basic vector (Promega, Madison, Wis.). This resulted in plasmids pGL3-wt and pGL3-X-K.O. The expression plasmid for the HBV X protein was reported previously (56).
Cell lines and transfection procedures.
The chicken hepatoma-derived LMH cells were maintained in Dulbecco's modified Eagle's medium-F12 medium (Gibco/BRL). Human hepatoma cells (HepG2 and HuH-7), human embryonal kidney cells (293), and African green monkey kidney cells (Cos7) were grown in Dulbecco's modified Eagle's medium. The cell lines were transfected with the help of the FuGene6 reagent; by lipofection, using DOTAP (Roche Molecular Biochemicals, Pentzberg, Germany) according to the manufacturer's instructions; or by calcium phosphate precipitation.
Luciferase, chloramphenicol acetyltransferase (CAT), and β-galactosidase assays.
The transfected cells (HepG2 and LMH) were harvested at day 2 after transfection and lysed in Tris buffer (250 mM; pH 7.8) by five cycles of thawing and freezing. The protein concentration of the lysate was determined by the Bradford assay. Twenty micrograms of protein was used for the luciferase activity assay (Roche Molecular Biochemicals) according to the manufacturer's recommendations. A plasmid expressing β-galactosidase was cotransfected, and the level of expression of this enzyme was used as an internal standard. All assays were done in duplicate, and all experiments were repeated at least once by performing two or more independent transfections.
For the CAT assay, 8 × 105 HepG2 or 293 cells were transfected with the reporter construct pSV2-CAT, p3xAP-1-CAT, or p2xNF-κB-CAT and with the large HBV envelope protein expression plasmid pSVLM-S or the different plasmids expressing HBx or the DHBV X-like proteins. To inhibit the activity of c-Raf-1 kinase, 2.5 μg of the plasmid expressing the transdominant-negative mutant HCR13.1 (36) or pErk2tdn (kindly provided by W. Fantl, San Francisco, Calif.) was used for transfection. Transfection efficiencies were standardized by cotransfection of a luciferase reporter plasmid containing the luciferase gene under the control of the nonstimulatable minimal promoter of pTK-luciferase. The CAT activity was determined by using a commercial enzyme-linked immunosorbent assay system, and the luciferase activity was determined as described above.
Antibodies raised against peptides specific for DHBx.
The sequences of peptides X2 (ILLTAHPGTNRLIGR) and X3 (GYVELKNYTPLLRSC) correspond to DHBx amino acids 46 to 60 and 76 to 90 derived from the DHBV1 genome, respectively. The sequence of peptide p759 (AVVPCDCTFGMYHCL) is identical to the C-terminal end of the predicted X-like protein of HHBV4 (48) and almost identical to the corresponding region of DHBx proteins. All three peptides were conjugated to keyhole limpet hemocyanin and then injected into rabbits (subcutaneously and intramuscularly) to raise the α-X2, α-X3, and α-p759 antibodies. A recombinant protein containing the complete DHBx sequence predicted from the ORF was expressed in Escherichia coli by using the expression vector pQE9 (Qiagen, Düsseldorf, Germany), which places a six-histidine tail at the protein's N terminus. To construct this recombinant plasmid, a DNA fragment encoding DHBx was generated by PCR using the primers DHBV3-2289X and DHBV3-X-stop. This fragment was then digested with BamHI and HindIII and cloned into the vector linearized by the same restriction enzymes. The recombinant protein was purified according to the procedure provided by the plasmid manufacturer and then injected into two rabbits. The resulting antisera (α-DHBV3X) were used as a mixture for immunoblotting.
Indirect immunofluorescence staining.
The Cos7 cells were grown on coverslips and transfected with plasmids expressing the DHBV3-M1X, DHBV3-M28X, and HBx proteins with N-terminal tags of six histidines under the control of the CMV promoter. The cells were fixed in methanol for 5 min at −20°C, and then incubated for 30 s in acetone. The fixed cells were incubated for 1 h at room temperature with an α-six-His monoclonal antibody (Clontech, Palo Alto, Calif.) diluted 1:5,000 in phosphate-buffered saline (PBS). For detection, a fluorescein isothiocyanate (FITC)-conjugated α-mouse immunoglobulin antibody (Dianova, Hamburg, Germany), diluted 1:200 in PBS, was applied.
Immunoblotting.
The cells were rinsed with PBS and then directly lysed in sodium dodecyl sulfate (SDS) loading buffer. The samples were boiled, separated by SDS-polyacrylamide gel electrophoresis, and transfered to a nitrocellulose membrane or a polyvinylidene difluoride membrane. The blots were incubated with rabbit antiserum raised against DHBV-X peptides (α-X2, α-X3, or α-p759) or a mouse antiserum specific for the six-histidine tag (Clontech). This procedure was repeated three to four times to increase the sensitivity of detection. The proteins were then visualized after being incubated with an appropriate peroxidase-coupled secondary antibody followed by incubation with a chemiluminescence substrate.
DNA and protein sequence analyses.
Sequence analysis was performed with the software provided by MacVector (Oxford Molecular Group, Oxford, United Kingdom) and with the package provided by the Wisconsin Genetics Computer Group. The X-protein sequences of all hepadnaviruses used for alignment were taken from GenBank (National Center for Biotechnology Information, Bethesda, Md.). Accession numbers were as follows: DHBV1, X58567; DHBV3 (see reference 63); DHBV26, X58569; snow goose hepatitis B virus 15 (SGHBV15), AF110997; Ross goose hepatitis B virus (RGHV), M95589; heron hepatitis B virus type 4 (HHBV-4), M22056; HBV, J02203; ground squirrel hepatitis virus, K02715; and WHV, M11082.
Synthetic oligonucleotides.
The following oligonucleotides were used: DHBV3-His-M1X; 5′-GCTCTAGATGCATCACCATCACCATCACCATCACTTAAACCTCGATGCCTC-3′; DHBV3-His-M28X, 5′-GCTCTAGATGCATCACCATCACCATCACCATCACTGGCCAAACAGTTGCTC-3′; DHBV3-X-stop, 5′-TCATAAACGATGGTACATACC-3′; DHBV-2371GA(+), 5′-GCTGTGTTAGCCAAACAG-3′; DHBV-2371GA(−), 5′-CTGTTTGGCTAACACAGC-3′; DHBV2520-HindIII, 5′-CCAAGCTTAGCCTGTGTGGAATATATATTGC-3′; DHBV1557(+), 5′-GGCTTGCTGTATCTGACGG-3′; DHBV257(−), 5′-CCACGAGGTTTTCTAGTACC-3′; DHBV2144(+), 5′-CCTTTGCCACGTGTAGC-3′; and DHBV3-2289X, 5′-CGGGATCCATGTTAAACCTCGATGCCTC-3′.
RESULTS
Identification and characterization of the DHBV-specific X-like ORF.
The genomes of all known avihepadnaviruses, except those of DHBV, contain an ORF in a position analogous to that of the X gene of mammalian viruses. It was previously speculated but not experimentally proven that a protein with functions similar to those of mammalian hepadnavirus X protein may be expressed from these ORFs (9, 48). We considered it unlikely that DHBV lacks such an ORF and therefore investigated whether DHBV genomes have X-like ORFs without a conventional translation initiation codon from which an X-like protein could be synthesized. Inspection of the sequences of all DHBV genomes deposited in the GenBank database revealed such an ORF in a position analogous to those of X and X-like ORFs known for all other hepadnaviruses. Provided a conventional translational start codon is used for expression of an X-like protein of HHBV, RGHV, and SGHBV (Fig. 1A) (note that only a selective set of the known DHBV, SGHBV, and HHBV sequences is shown) and a nonconventional codon at the very amino-terminal end is used in the case of DHBV (Fig. 1B), the corresponding DHBV X-like protein could be longer than all other putative avihepadnavirus X-like proteins.
FIG. 1.
Comparison of putative avihepadnavirus X-like proteins and potential translation initiation codons for DHBx. (A) Alignment of predicted X-like protein sequences derived from cloned DHBV1, DHBV3, DHBV26, SGHBV15, HHBV4, and RGHV genomes. For all DHBV strains missing the conventional AUG start codon at the beginning of the corresponding ORFs, the longest possible reading frame is given. Protein sequences conserved among the different avihepadnaviruses are boxed. Identical amino acids are indicated by dark shading, and similar amino acids are indicated by light shading. The conventional translation start codons (M) in SGHBV, HHBV, and RGHV are underlined. (B) Potential nonconventional translation initiation codons in the ORF of DHBx are in boldface and underlined.
There is over 90% sequence identity among the putative X-like protein sequences of all DHBV isolates (data not shown). In addition, the homology of the deduced X-like protein sequences among the various DHBV isolates and the evolutionarily most closely related SGHBV is more than 80% (Fig. 1A). In contrast, the homology between the corresponding sequences of DHBV isolates and the more distantly related HHBV or RGHV ranges only between 40 to 50% (Fig. 1A). In general, sequence identity and similarity among the various X-like proteins are higher in the C-terminal region than at the amino-terminal end (Fig. 1A). This is in part due to the length variation of the X-like ORFs. For instance, all DHBV genomes encode a putative X-like protein with a maximum of 114 amino acids, except the isolates DHBV22 and DHBV26, which lack the first 7 amino acids at the very N terminus (Fig. 1A and data not shown). Moreover, the X-like ORFs of all other known avihepadnaviruses have a conventional AUG translation initiation codon from which translation of the protein could be initiated, but it is located at different positions for each member of the avihepadnaviruses (Fig. 1A). HHBV, which is most distantly related to DHBV, may therefore express the shortest X-like protein of all hepadnaviruses. All of the avian hepadnavirus X-like proteins deduced from the DNA sequences have only very low primary sequence similarity to mammalian X proteins (see below).
Detection of DHBx expression in chronically infected duck liver.
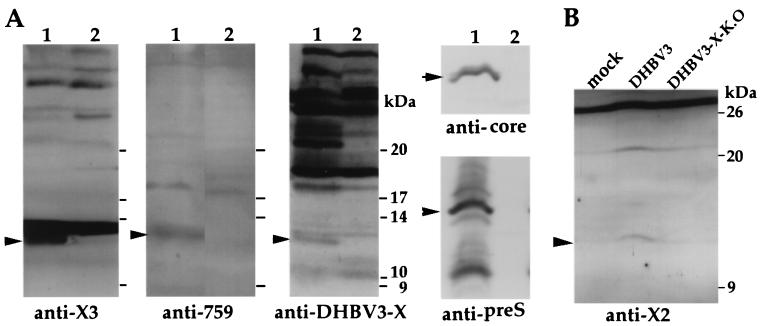
In order to determine whether a protein is expressed from the X-like ORF of DHBV, we produced antibodies against specific synthetic peptides with amino acid sequences from different regions of the predicted protein (α-X2, α-X3, and α-p759) and against a recombinant DHBV X protein expressed in E. coli (α-DHBx) and used them to provide direct evidence for expression of DHBx in vivo. When extracts of livers of chronically infected ducks were analyzed by immunoblotting, three of the four antisera reacted with a protein of 12.5 kDa. No reactivity at the corresponding position was observed in the noninfected liver sample (Fig. 2A). Immunoblots of both liver extracts with polyclonal anti-core and anti-preS antibodies performed as controls showed expression of core and preS proteins in the infected liver only (Fig. 2A). Taken together, these data indicate that a protein is expressed from the X-like ORF of DHBV in the chronically infected liver.
FIG. 2.
Detection of DHBV X-like protein in chronically infected duck liver and in transfected LMH cells by immunoblotting. (A). Protein extracts from a chronically infected duck liver (lanes 1) and a noninfected duck liver (lanes 2). The types of antisera used for blotting are indicated at the bottom, and the DHBV X-like protein-, core-, and major preS protein-specific signals are marked by arrowheads. (B). Protein extracts from LMH cells transfected with pDHBV3 and pDHBV3-X-K.O. DNA or from untransfected LMH cells (mock). The position of the band specifically reacting with antibody from α-X2 peptide antiserum is indicated.
A protein is expressed from the DHBV X-like ORF in LMH cells transfected with full-length DHBV DNA.
Next we tested whether the protein encoded by the X-like ORF of DHBV is also expressed in vitro. Chicken hepatoma (LMH) cells were transfected with plasmid pDHBV3, which contains a dimeric full-length genome of isolate DHBV3, as well as with plasmid pDHBV3-X-K.O, which contains the corresponding knockout mutant with a stop codon in the X-like ORF. The expression of the protein encoded by the DHBV X-like ORF was then analyzed by immunoblotting with the X-specific peptide antisera and cell extracts of the transfected cells. In LMH cells transfected with pDHBV3, a weak but specific band corresponding to a protein with an apparent molecular mass of approximately 12 kDa was identified (data are shown for α-X2 peptide antiserum [Fig. 2B]). These bands were not observed in untransfected cells or in cells transfected with the DHBV-X knockout mutant DNA. These data strongly indicate that a protein, designated DHBx, is expressed from the X-like ORF of the DHBV3 genome in a cell culture system and that is identical in size to that expressed in chronically infected duck liver.
DHBx affects neither viral protein expression nor viral DNA synthesis in LMH cells.
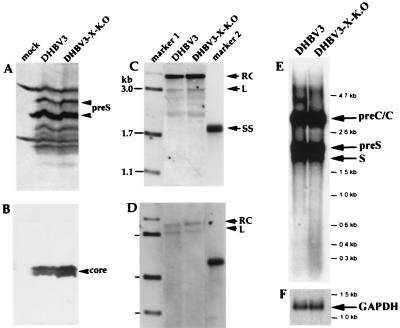
In order to investigate whether DHBx affects the expression of structural proteins of DHBV, we analyzed the expression of the preS and core proteins in LMH cells after transfection with plasmids containing the DHBV3 wild-type genome or the mutated DHBV3 genome, which contains a stop codon within the X ORF. This was done by immunoblotting the cell extracts using DHBV preS- and DHBV core protein-specific antisera. Equal loading of the slots with proteins was controlled by measuring the amount by the Bradford assay (data not shown). Comparison of the lanes of the blot with extracts derived from cells transfected with the wild-type and X-K.O. genomes showed similar levels of preS and core proteins (Fig. 3A and B). The transfection efficiencies obtained with the two genomes were very similar, as was evident from the fact that the same amount of β-galactosidase (variability, less than 10% [data not shown]) was expressed from a cotransfected reporter plasmid. The amounts of intracellular replicative intermediates and of secreted virions were also very similar for all viral genomes, as was evident from the corresponding Southern blots with DNA extracted from the transfected cells and from the secreted virus particles (Fig. 3C and D). Northern blotting of poly(A)-containing RNA from cells transfected with both viral genomes revealed neither a significant difference in the types and amounts of mRNAs expressed nor an mRNA which might be transcribed from only the DHBx coding region (Fig. 3E). These data indicate that DHBx expression affects neither core and preS protein expression nor viral DNA synthesis and virion secretion in cell culture. In addition, these data suggest that DHBx has no strong modulatory effect, if it has any at all, on the steady-state levels of viral transcripts and may itself be translated either from a very minor DHBx-specific mRNA or from any of the other viral mRNAs.
FIG. 3.
Analysis of the role of DHBx for core and preS protein expression and viral DNA synthesis. Immunoblots and Southern blots were obtained from core particles and particles derived from the culture medium of LMH cells, respectively. The LMH cells were transfected with pDHBV3 DNA and pDHBV3-X-K.O. DNA, as well as from nontransfected LMH cells (mock). (A) PreS envelope proteins detected with a polyclonal α-DHBV preS antibody. The positions of the major preS proteins are marked by arrowheads. (B) Core protein detected with a polyclonal α-DHBV core antibody. (C) Intracellular replication intermediates from core particles of transfected cells. (D and E) Viral DNAs from secreted virus particles as well as from poly(A)+ mRNA isolated from transfected cells were detected by Southern (D) and Northern (E) blotting, respectively, with labeled HBV probes. (F) Loading of the Northern blot with equal amounts of cellular RNA was controlled by hybridization with a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe. RC, rcDNA; L, linear DNA; SS, single-stranded DNA.
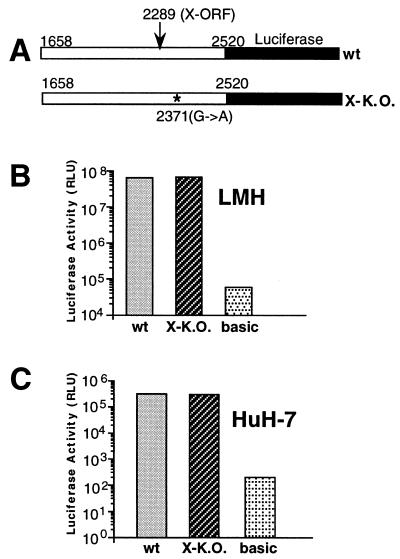
The knockout mutation introduced into the X-like ORF (stop codon TAG at positions 2370 to 2372), which is also located in the core promoter region (43, 55), may have affected core promoter activity, viral mRNA half-life, and viral protein translation efficiency without a notable effect on overall steady-state levels of viral proteins compared to the wild-type genome. In order to examine the possible effect on core promoter activity, we tested the core promoter activities of DNA fragments (nucleotides 1658 to 2520) which cover the X-like ORF of DHBV3 with and without the knockout mutation. These fragments were cloned upstream of the luciferase reporter gene into the eukaryotic promoterless plasmid pGL3 (Fig. 4A). After transfection into LMH and HuH-7 cells, similar promoter activities were measured when comparing the wild-type fragment and the X-K.O. mutant-specific fragment (Fig. 4B and C). However, the activities of two promoter fragments were more than 100-fold higher in LMH cells than in HuH-7 cells. Taken together, these results indicate that the core promoter activity was not altered by the X-K.O. mutation, confirming our conclusion that DHBx has no significant effect on core protein synthesis.
FIG. 4.
Analysis of the core promoter activities of DHBV3 and DHBV3-X-K.O. DNA fragments in transfected cells by luciferase assay. (A) Schematic diagram of the core promoter fragments. The nucleotide positions at the 5′ and 3′ ends of the DHBV-specific sequence are indicated. The positions of the first nucleotide of the DHBV X-like ORF and of the mutation leading to a stop codon in DHBV3-X-K.O. are given, and their locations are marked by an arrow and an asterisk, respectively. (B and C) Luciferase activity of the tested constructs in LMH (B) and HuH-7 (C) cells lysed 2 days after transfection. basic, transfection of the luciferase reporter plasmid without the core promoter, used as a control. The values are the averages of two independent transfection experiments performed in duplicate. The standard deviation was too small to be indicated. wt, wild type; RLU, relative light units.
Subcellular localization and electrophoretic mobility of tagged DHBx proteins.
The low level of DHBx protein expression in full-length DHBV DNA-transfected cells and DHBV-infected livers prevented unequivocal subcellular localization of DHBx by indirect immunofluorescence staining. To circumvent this problem, we expressed DHBx with N-terminal histidine tags under the control of a strong foreign promoter (the CMV IE promoter). In these constructs we forced translation of the tagged DHBx mRNA to initiate at the codon corresponding to amino acid position 1 or 28 or to stop at the codon of the DHBV X-like ORF corresponding to amino acid 28 by introducing a start (AUG) and/or stop codon at the corresponding positions, plasmids pDHBV3-M1X, pDHBV3-M28X, and pDHBV3-M28X-stop, respectively. This was done because the authentic translation initiation codon of DHBx is not known and because analysis of the sizes of these DHBx proteins by immunoblotting may provide hints as to the location of the DHBx translation initiation codon.
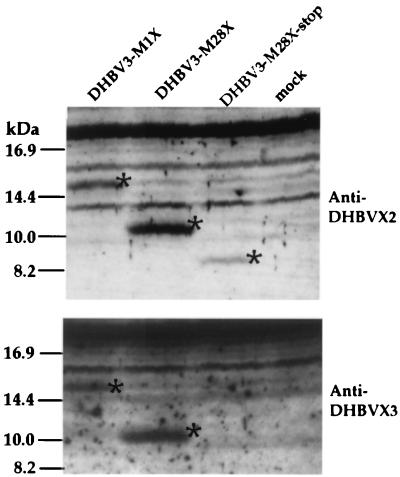
Expression of the tagged DHBx proteins was examined by immunoblotting 2 days after the transfection of different cell lines (LMH, Cos7, and 293) with these constructs. When the amino-terminal X-specific peptide sera α-X2 and α-X3 were used, His-tagged DHBV-M1X and -M28X proteins of the expected sizes (ca. 15 and 10 kDa) were detected (Fig. 5 and data not shown). Since the size of the DHBx expressed under authentic viral promoter control is 12.5 kDa and the polyhistidine tag usually increases the electrophoretic mobility by only approximately 1 kDa, it is possible that the nonconventional translation codon used in vivo is located between amino acid positions 1 and 28. Unexpectedly, immunoblotting of proteins from cells transfected with pDHBV3-M28X-stop performed with α-X2 resulted in a specific, albeit weakly stained, band corresponding to a protein with an apparent molecular mass of 8 kDa (Fig. 5). This band may correspond to an amino-terminally truncated DHBx protein synthesized by translation initiation at a nonconventional start codon downstream of the stop codon at position 28.
FIG. 5.
Full-length and truncated versions of DHBx expressed under the control of the CMV promoter in LMH cells. Protein extracts from mock-transfected LMH cells (mock) and from LMH cells transfected with the plasmid pDHBV3-M1X, pDHBV3-M28X, or pDHBV3-M28X-stop were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted by using anti-DHBx peptide serum X2 (top) or X3 (bottom). The DHBx-specific bands are marked by asterisks.
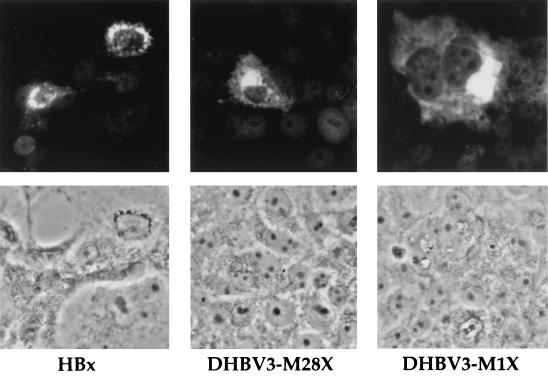
For subcellular localization of the tagged DHBx proteins by indirect immunofluorescence staining, transfected Cos7 cells were examined using a polyhistidine-specific monoclonal antibody and a FITC-labeled anti-mouse antibody. DHBx proteins expressed from pDHBV3-M1X and -M28X proteins were detected predominantly in the cytoplasm (Fig. 6, top), with the most intense staining close to the nuclear membrane, a pattern identical to that seen with HBV-specific histidine-tagged HBx protein (Fig. 6). These results demonstrate that HBx and DHBx have similar subcellular distributions.
FIG. 6.
Localization of DHBx in transfected cells. Indirect immunofluorescence staining of Cos7 cells transfected with constructs expressing His-tagged DHBx (pDHBV3-M1X and pDHBV3-M28X) and HBx. (Top) Recombinant proteins were detected by a monoclonal α-six-His antibody and a FITC-coupled secondary antibody and visualized by fluorescence microscopy. (Bottom) Area corresponding to the top row in phase-contrast microscopy.
DHBx stimulates promoters of cellular and viral origin via the Raf-MAP kinase signaling pathway.
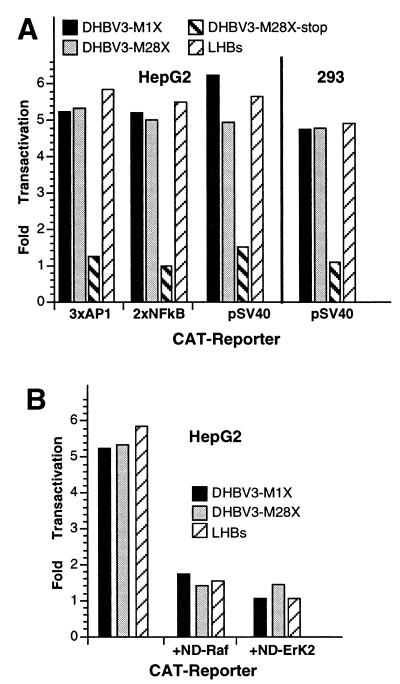
Mammalian hepadnavirus HBx proteins stimulate transcription of a large variety of cellular and viral genes. To investigate whether DHBx has a similar function, some of the CMV-DHBx expression plasmids mentioned above (pDHBV3-M1X, pDHBV3-M28X, and pDHBV3-M28X-stop) were cotransfected into HepG2 and 293 cells with plasmids containing different promoter elements derived from the AP1 gene, the NFκB gene, and the late promoter of simian virus 40 cloned upstream of the CAT reporter gene. As a positive control, a plasmid (pSVLM-S) for expression of HBV large envelope protein (LHBs), known to have a promiscuous transactivation function qualitatively and quantitatively similar to that of HBx (28), was contransfected.
In HepG2 cells, all three promoters tested were stimulated by DHBx proteins expressed from constructs pDHBV-M1X and -M28X, but not by the putative amino-terminally truncated DHBx expressed from construct pDHBV-M28X-stop (Fig. 7A, left). Therefore, the first 28 amino acids encoded by the X-like ORF are dispensable for the transactivation function whereas most of the remaining sequences are needed. All three promoters were transactivated about fivefold by DHBx, which is similar to the value obtained by the expression of the control protein LHBs. Similar transactivation profiles were obtained in 293 cells when the same plasmids were used (Fig. 7A, right). These results demonstrate that DHBx is a promiscuous transactivator of promoters of cellular and viral origin which functions in liver as well as in non-liver-derived cells.
FIG. 7.
Transactivation activity of DHBx analyzed by CAT assay. (A) Activity of AP1, NFκB, and simian virus 40 promoter elements (3xAP1, 2xNFκB, and pSV40) in HepG2 and 293 cells when cotransfected with plasmids expressing DHBx proteins and LHBs, as determined by CAT assay. (B) Activity of the promoter containing three AP1 elements by DHBx proteins and LHBs in HepG2 cells when transdominant-negative mutants of Raf1 (+ND-Raf) or ERK2 (+ND-ErK2) are coexpressed.
The transcription activation function of HBx, and also of LHBs, is mediated by cytoplasmic signaling cascades which lead to activation of the Raf-MAP kinase pathway (2, 28, 47). In order to determine whether the transactivation activity of the DHBx protein is also mediated by the Raf-MAP kinase cascade, the plasmids expressing the DHBV-M1X and DHBV-M28X proteins were cotransfected into HepG2 cells with plasmids expressing dominant-negative mutants of Raf and ERK2 (28). In this assay, the CAT reporter gene was expressed under the control of a promoter containing the AP1 response element known to mediate activation of the Raf-MAP pathway. Both dominant-negative proteins, Raf and ERK2, effectively inhibited both the transactivation activity of the control LHBs and that of the DHBx proteins (Fig. 7B). Expression of luciferase from a cotransfected control plasmid was unaffected, which rules out the possibility that the observed inhibition of activation is due to nonspecific cytotoxicity or squelching (data not shown). These data imply that transactivation of promoters by DHBx depends on the activation of the Raf-MAP kinase cascade pathway.
DISCUSSION
Although the functions of mammalian hepadnavirus X-proteins have been studied in great detail in vitro, there is still much debate about the in vivo relevance of many of these findings. One reason for this is the lack of a convenient animal system, such as DHBV-infected ducks, which would allow detailed in vivo studies to be performed. The presumed lack of an X gene in DHBV has frequently been used as a strong argument for the association of chronic hepadnavirus infection with the development of liver carcinoma in mammals but not in ducks. In our study we challenge this view by demonstrating the existence and expression of a hidden ORF located at a position analogous to that of mammalian hepadnavirus X genes. The corresponding protein, DHBx, was shown to have a transcription-regulatory function and subcellular localization strikingly similar to those documented for the mammalian hepadnavirus X-proteins. The data presented indicate that ducks chronically infected with DHBV provide a convenient animal model to unravel the still-enigmatic in vivo functions of X proteins and suggest that X genes were present in hepadnavirus genomes early in evolution but diverged in sequence through host adaptation. Our data also suggest an in vivo function of X proteins in the life cycles of all hepadnaviruses.
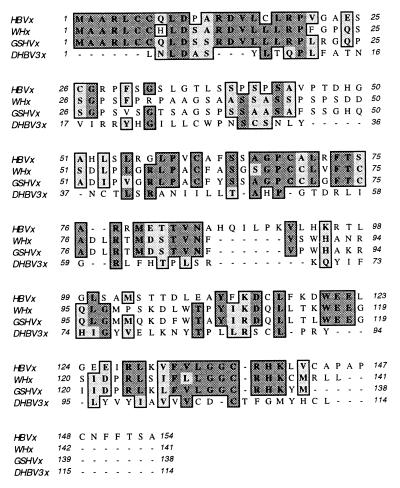
Comparison of the primary protein sequences of DHBx and the X proteins of mammalian hepadnaviruses revealed low similarity when appropriate artificial deletions for optimal alignment are introduced (Fig. 8). This may indicate similar three-dimensional structures of both types of protein even though secondary-structure predictions are not in strong favor of this possibility (data not shown). Alternatively, both types of proteins may have evolved into different structures capable of activating the same signaling pathways. Consistent with this speculation is a recent report of very similar transactivation mechanisms induced by HBx and the regulatory protein Tax of human T-cell leukemia virus, which also have no primary sequence similarity (51).
FIG. 8.
Sequence alignment of HBx, WHx, GHBx, and DHBx proteins. Identical (dark shaded boxes) and similar (light shaded boxes) amino acids are indicated. Gaps introduced for optimal alignment by the ClustalW program of the MacVector software are marked by dashes.
Our data raise the question as to why DHBx and all other putative avian X proteins are smaller than their mammalian hepadnavirus counterparts. One possible answer is that avian X proteins have fewer functions and cellular interaction partners. Alternatively, the avian X proteins may lack only specific putative co-oncogenic functions believed to be associated with the X proteins of mammalian hepadnaviruses. For instance, DHBx may lack a domain functionally homologous to the first 50 amino-terminal amino acids of HBx, which are apparently sufficient for transformation of immortalized cells (23). Recent evidence suggesting that stop codons immediately upstream of the DHBx-encoding ORF (42) may have been lost during evolution supports this speculation. Furthermore, DHBx, unlike HBx, may not upregulate the expression of the proinflammatory cytokines, such as tumor necrosis factor α and interleukin 6 (39, 41), because liver inflammation believed to contribute to HBV-mediated tumor development is virtually absent in chronically infected ducks. We consider this less likely because the transcription-regulatory function of HBx proposed in most previous reports to play a role in hepatocarcinogenesis appears to be quantitatively and qualitatively similar in DHBx. In addition, the minimal sequence length of HBx shown to be required for transcriptional transactivation and interaction with most cellular proteins (22, 30, 38, 50–52) is similar to that of DHBx.
We have shown predominant cytoplasmic localization of DHBx when it is overexpressed under the control of a strong foreign promoter, and most studies of HBx have led to a similar intracellular staining pattern. However, our studies do not exclude the possibility that a proportion of DHBx is also in the nucleus but escaped detection due to its small amount or inaccessibility to our antibodies. The increasing evidence for the existence of nuclear WHx and HBx and the difficulties associated with their detection by immunostaining or cell fractionation (13, 15, 26, 49, 54, 69) are in favor of this possibility. As there is no obvious nuclear localization sequence in DHBx, it is conceivable that a fraction of DHBx is transported to the nucleus by a piggyback mechanism or by passive diffusion and binding to a nuclear protein, similar to what has been demonstrated or speculated previously for mammalian X proteins (13, 15, 26, 49, 54, 69). The low level of DHBx expression in the infected liver is reminiscent of the difficulties in detecting HBx and WHx in infected livers. It was estimated that a WHV-infected woodchuck hepatocyte contains only about 1,000 to 10,000 molecules of WHx (14), and this level probably needs to be kept very low to prevent toxic or proapototic effects (5, 12, 14).
At present, we can only speculate as to how DHBx is produced. While splicing events could create RNA molecules with a conventional AUG start codon for DHBx translation, conserved sequences indicative of intron-exon boundaries are not evident. In contrast to DHBV, all the other avihepadnaviruses contain an X-like ORF with a conserved AUG start codon which may serve as the translation initiation codon for the corresponding proteins. It is conceivable, therefore, that synthesis of DHBx starts at a position corresponding to a nonconventional start codon on an unspliced mRNA. The detection of a strong promoter within a 200-bp-long region upstream of the DHBx ORF in LMH cells transfected with corresponding reporter constructs (preliminary data not shown) supports this assumption. However, corresponding mRNAs extracted from DHBV-infected ducks are neither detectable by Northern blotting (Fig. 3E) nor evident in S1 mapping and primer extension studies (7). It is conceivable that a terminally redundant mRNA very similar in size to and comigrating with the pregenomic RNA is the template for DHBx translation. Similar to what has been shown for HBV, such a DHBx mRNA may be initiated upstream of the DHBx coding region and processed only at the second transit of the single processing or poly(A) site, as reported for an HBx mRNA (25).
There are numerous reports describing initiation of translation at non-AUG codons of both cellular and viral mRNAs (21, 24, 37). Within the DHBx-encoding ORF of DHBV3, there are several in-frame non-AUG start codons (Fig. 1B) which may be used for translation initiation. The sizes of DHBx proteins artificially initiated at codon 1 (15 kDa) or 28 (10 kDa) as expressed under foreign promoter control and those expressed in full-length DHBV DNA-transfected cell lines and in infected livers (12 kDa) would be compatible with DHBx translation initiation between codons 1 and 28 of the hidden ORF. Taking this into account, only 3 of 10 potential alternative translation codons described for eukaryotes and viruses are potential candidates: codons CUG, UUG, and AUU at positions 8 or 26, 12 or 25, and 24, respectively. Mutagenesis of these codons and the corresponding adjacent region should provide a clear answer as to which, if any, of these are used and whether a specific IRES-like structure of the corresponding mRNA is present and required for correct initiation of translation of DHBx. Potential translation of DHBx from a minor undetectable mRNA is an alternative possibility which needs to be investigated by techniques more sensitive than those currently available. Furthermore, our data also do not exclude the possibility that DHBx is synthesized by the processing of a viral fusion protein which may be produced by ribosomal frameshifting or from a spliced mRNA, similar to what is known for other viral proteins.
As pointed out in a recent review, it has been very difficult to define the exact roles of X proteins in vivo because of the necessity of working with woodchucks (70). DHBV infections in ducks represent the most convenient animal system and provide unique opportunities to study the in vivo functions of X proteins in the hepadnavirus life cycle, establishment of chronic infection, and immune surveillance. In addition, studies can also be performed with duck primary hepatocytes derived from embryonated eggs, which are easy to prepare and permissive for infection. Thus, for instance, the hepatocyte differentiation-dependent effect of hepadnavirus X protein reported recently (65) can also be studied without the necessity for an animal facility. Finally, the availability of viral genomes from avian hepadnaviruses from different species allows X-gene-swapping experiments to be performed, which may uncover possible viral genome- and host-specific differences in avian X-protein functions, similar to those initiated for ground squirrel hepatitis virus and WHV (58).
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of Kerstin Reumann. The critical reading of the manuscript by Thomas Macnaughton and Wolfram Gerlich is very much appreciated.
The Heinrich-Pette-Institut is supported by the Bundesministerium für Gesundheit, Berlin, and the Freie und Hansestadt Hamburg. Hans Jürgen Netter was supported in part by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Andrisani O M, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int J Oncol. 1999;15:373–379. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 2.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergametti F, Prigent S, Luber B, Benoit A, Tiollais P, Sarasin A, Transy C. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene. 1999;18:2860–2871. doi: 10.1038/sj.onc.1202643. [DOI] [PubMed] [Google Scholar]
- 6.Blum H E, Zhang Z S, Galun E, von Weizsacker F, Garner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscher M, Reiser W, Will H, Schaller H. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell. 1985;40:717–724. doi: 10.1016/0092-8674(85)90220-x. [DOI] [PubMed] [Google Scholar]
- 8.Caselmann W H, Koshy R. Transactivators of HBV, signal transduction and tumorigenesis. In: Koshy R, Caselmann W H, editors. Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. London, United Kingdom: Imperial College; 1998. pp. 161–181. [Google Scholar]
- 9.Chang S F, Netter H J, Bruns M, Schneider R, Froeich K, Will H. A new avian hepadnavirus infecting snow geese (Anser caerulescens) produces a significant fraction of virions containing single stranded DNA. Virology. 1999;262:39–54. doi: 10.1006/viro.1999.9844. [DOI] [PubMed] [Google Scholar]
- 10.Chen H S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheong J H, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chirillo P, Pagano S, Natoli G, Puri P L, Burgio V L, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dandri M, Petersen J, Stockert R J, Harris T M, Rogler C E. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J Virol. 1998;72:9359–9364. doi: 10.1128/jvi.72.11.9359-9364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duflot A, Mehrotra R, Yu S Z, Barraud L, Trepo C, Cova L. Spectrum of liver disease and duck hepatitis B virus infection in a large series of Chinese ducks with hepatocellular carcinoma. Hepatology. 1995;21:1483–1491. [PubMed] [Google Scholar]
- 17.Elmore L W, Hancock A R, Chang S F, Wang X W, Chang S, Callahan C P, Geller D A, Will H, Harris C C. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707–14712. doi: 10.1073/pnas.94.26.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feitelson M A, Miller R H. X gene-related sequences in the core gene of duck and heron hepatitis B viruses. Proc Natl Acad Sci USA. 1988;85:6162–6166. doi: 10.1073/pnas.85.16.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer M, Runkel L, Schaller H. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 20.Ganem D. Fields et al. (ed.), Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. Hepadnaviridae and their replication; pp. 2703–2737. [Google Scholar]
- 21.Gordon K, Futterer J, Hohn T. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 1992;2:809–813. [PubMed] [Google Scholar]
- 22.Gottlob K, Fulco M, Levrero M, Graessmann A. The hepatitis B virus HBx protein inhibits caspase 3 activity. J Biol Chem. 1998;273:33347–33353. doi: 10.1074/jbc.273.50.33347. [DOI] [PubMed] [Google Scholar]
- 23.Gottlob K, Pagano S, Levrero M, Graessmann A. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 1998;58:3566–3570. [PubMed] [Google Scholar]
- 24.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 25.Guo W T, Wang J, Tam G, Yen T S, Ou J S. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology. 1991;181:630–636. doi: 10.1016/0042-6822(91)90896-j. [DOI] [PubMed] [Google Scholar]
- 26.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 28.Hildt E, Saher G, Bruss V, Hofschneider P H. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology. 1996;225:235–239. doi: 10.1006/viro.1996.0594. [DOI] [PubMed] [Google Scholar]
- 29.Hohne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z, Zhang Z, Doo E, Coux O, Goldberg A L, Liang T J. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol. 1999;73:7231–7240. doi: 10.1128/jvi.73.9.7231-7240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Kwong J, Sun E C, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacob J R, Ascenzi M A, Roneker C A, Toshkov I A, Cote P J, Gerin J L, Tennant B C. Hepatic expression of the woodchuck hepatitis virus X-antigen during acute and chronic infection and detection of a woodchuck hepatitis virus X-antigen antibody response. Hepatology. 1997;26:1607–1615. doi: 10.1002/hep.510260632. [DOI] [PubMed] [Google Scholar]
- 33.Jia L, Wang X W, Harris C C. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875–879. doi: 10.1002/(sici)1097-0215(19990315)80:6<875::aid-ijc13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 35.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 36.Kolch W, Heidecker G, Kochs G, Hummel R, Vahidi H, Mischak H, Finkenzeller G, Marme D, Rapp U R. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature. 1993;364:249–252. doi: 10.1038/364249a0. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 38.Kumar V, Jayasuryan N, Kumar R. A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci USA. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lara P E, Majano P L, Gomez G M, Garcia M C, Moreno O R, Levrero M, Lopez C M. The hepatitis B virus X protein up-regulates tumor necrosis factor alpha gene expression in hepatocytes. Hepatology. 1998;28:1013–1021. doi: 10.1002/hep.510280416. [DOI] [PubMed] [Google Scholar]
- 40.Lee T H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Park U S, Choi I, Yoon S K, Park Y M, Lee Y I. Human interleukin 6 gene is activated by hepatitis B virus-X protein in human hepatoma cells. Clin Cancer Res. 1998;4:1711–1717. [PubMed] [Google Scholar]
- 42.Lin B, Anderson D A. A vestigial X open reading frame in duck hepatitis B virus. Intervirology. 2000;43:185–190. doi: 10.1159/000025037. [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Condreay L D, Burch J B, Mason W. Characterization of the core promoter and enhancer of duck hepatitis B virus. Virology. 1991;184:242–252. doi: 10.1016/0042-6822(91)90841-x. [DOI] [PubMed] [Google Scholar]
- 44.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 45.Melegari M, Scaglioni P P, Wands J R. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol. 1998;72:1737–1743. doi: 10.1128/jvi.72.3.1737-1743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami S. Hepatitis B virus X protein: structure, function and biology. Intervirology. 1999;42:81–99. doi: 10.1159/000024969. [DOI] [PubMed] [Google Scholar]
- 47.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 48.Netter H J, Chassot S, Chang S F, Cova L, Will H. Sequence heterogeneity of heron hepatitis B virus genomes determined by full-length DNA amplification and direct sequencing reveals novel and unique features. J Gen Virol. 1997;78:1707–1718. doi: 10.1099/0022-1317-78-7-1707. [DOI] [PubMed] [Google Scholar]
- 49.Nomura T, Lin Y, Dorjsuren D, Ohno S, Yamashita T, Murakami S. Human hepatitis B virus X protein is detectable in nuclei of transfected cells, and is active for transactivation. Biochim Biophys Acta. 1999;1453:330–340. doi: 10.1016/s0925-4439(99)00004-6. [DOI] [PubMed] [Google Scholar]
- 50.Ohno H, Kaneko S, Lin Y, Kobayashi K, Murakami S. Human hepatitis B virus X protein augments the DNA binding of nuclear factor for IL-6 through its basic-leucine zipper domain. J Med Virol. 1999;58:11–18. doi: 10.1002/(sici)1096-9071(199905)58:1<11::aid-jmv2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 51.Perini G, Oetjen E, Green M R. The hepatitis B pX protein promotes dimerization and DNA binding of cellular basic region/leucine zipper proteins by targeting the conserved basic region. J Biol Chem. 1999;274:13970–13977. doi: 10.1074/jbc.274.20.13970. [DOI] [PubMed] [Google Scholar]
- 52.Runkel L, Fischer M, Schaller H. Two-codon insertion mutations of the HBx define two separate regions necessary for its trans-activation function. Virology. 1993;197:529–536. doi: 10.1006/viro.1993.1626. [DOI] [PubMed] [Google Scholar]
- 53.Schaefer S, Tolle T, Lottmann S, Gerlich W H. Animal models and experimental systems in hepatitis B virus research. In: Koshy R, Caselmann W H, editors. Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. London, United Kingdom: Imperial College Press; 1998. pp. 51–74. [Google Scholar]
- 54.Schek N, Bartenschlager R, Kuhn C, Schaller H. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene. 1991;6:1735–1744. [PubMed] [Google Scholar]
- 55.Schneider R, Will H. Regulatory sequences of duck hepatitis B virus C gene transcription. J Virol. 1991;65:5693–5701. doi: 10.1128/jvi.65.11.5693-5701.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuster R, Gerlich W H, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene. 2000;19:1173–1180. doi: 10.1038/sj.onc.1203417. [DOI] [PubMed] [Google Scholar]
- 57.Seeger C. The hepatitis B virus X protein: the quest for a role in viral replication and pathogenesis. Hepatology. 1997;25:496–498. doi: 10.1053/jhep.1997.v25.pm0009021971. [DOI] [PubMed] [Google Scholar]
- 58.Seeger C, Baldwin B, Hornbuckle W E, Yeager A E, Tennant B C, Cote P, Ferrell L, Ganem D, Varmus H E. Woodchuck hepatitis virus is a more efficient oncogenic agent than ground squirrel hepatitis virus in a common host. J Virol. 1991;65:1673–1679. doi: 10.1128/jvi.65.4.1673-1679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seifer M, Hohne M, Schaefer S, Gerlich W H. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13(Suppl. 4):S61–S65. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 60.Shin E C, Shin J S, Park J H, Kim H, Kim S J. Expression of fas ligand in human hepatoma cell lines: role of hepatitis-B virus X (HBX) in induction of Fas ligand. Int J Cancer. 1999;82:587–591. doi: 10.1002/(sici)1097-0215(19990812)82:4<587::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 61.Shirakata Y, Kawada M, Fujiki Y, Sano H, Oda M, Yaginuma K, Kobayashi M, Koike K. The X gene of hepatitis B virus induced growth stimulation and tumorigenic transformation of mouse NIH3T3 cells. Jpn J Cancer Res. 1989;80:617–621. doi: 10.1111/j.1349-7006.1989.tb01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slagle B L, Lee T H, Medina D, Finegold M J, Butel J S. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 63.Sprengel R, Schneider R, Marion P L, Fernholz D, Wildner G, Will H. Comparative sequence analysis of defective and infectious avian hepadnaviruses. Nucleic Acids Res. 1991;19:4289. doi: 10.1093/nar/19.15.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su Q, Schroder C H, Hofmann W J, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 65.Tarn C, Bilodeau M L, Hullinger R L, Andrisani O M. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 66.Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon M L, Tiollais P, Buendia M A. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 67.Wang X W, Gibson M K, Vermeulen W, Yeh H, Forrester K, Sturzbecher H W, Hoeijmakers J H, Harris C C. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 68.Warren K S, Heeney J L, Swan R A, Heriyanto, Verschoor E J. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weil R, Sirma H, Giannini C, Kremsdorf D, Bessia C, Dargemont C, Brechot C, Israel A. Direct association and nuclear import of the hepatitis B virus X protein with the NFκB inhibitor IκBα. Mol Cell Biol. 1999;19:6345–6354. doi: 10.1128/mcb.19.9.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yen T S B. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 71.Yu D Y, Moon H B, Son J K, Jeong S, Yu S L, Yoon H, Han Y M, Lee C S, Park J S, Lee C H, Hyun B H, Murakami S, Lee K K. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 72.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]