Abstract
During virus entry, herpes simplex virus (HSV) glycoprotein D (gD) binds to one of several human cellular receptors. One of these, herpesvirus entry mediator A (HveA), is a member of the tumor necrosis factor receptor (TNFR) superfamily, and its ectodomain contains four characteristic cysteine-rich pseudorepeat (CRP) elements. We previously showed that gD binds the ectodomain of HveA expressed as a truncated, soluble protein [HveA(200t)]. To localize the gD-binding domain of HveA, we expressed three additional soluble forms of HveA consisting of the first CRP [HveA(76t)], the second CRP [HveA(77–120t)], or the first and second CRPs [HveA(120t)]. Biosensor and enzyme-linked immunosorbent assay studies showed that gD bound to HveA(120t) and HveA(200t) with the same affinity. However, gD did not bind to HveA(76t) or HveA(77–120t). Furthermore, HveA(200t) and HveA(120t), but not HveA(76t) or HveA(77–120t), blocked herpes simplex virus (HSV) entry into CHO cells expressing HveA. We also generated six monoclonal antibodies (MAbs) against HveA(200t). MAbs CW1, -2, and -4 bound linear epitopes within the second CRP, while CW7 and -8 bound linear epitopes within the third or fourth CRPs. None of these MAbs blocked the binding of gD to HveA. In contrast, MAb CW3 recognized a discontinuous epitope within the first CRP of HveA, blocked the binding of gD to HveA, and exhibited a limited ability to block virus entry into cells expressing HveA, suggesting that the first domain of HveA contains at least a portion of the gD binding site. The inability of gD to bind HveA(76t) suggests that additional amino acid residues of the gD binding site may reside within the second CRP.
The herpes simplex virus (HSV) genome codes for at least 11 glycoproteins, most of which are present in the virion envelope (34). Infection of susceptible cells is initiated by the attachment of virions, via glycoprotein C (gC) and/or gB, to cell surface heparan sulfate proteoglycans (11, 12, 43). This is followed by the interaction of gD with one of several cellular receptors. Then, pH-independent fusion occurs between the virus envelope and the host cell plasma membrane; gB, gD, and the gH-gL complex have all been implicated in this step (35, 38, 42).
Recently, several mediators of HSV-1 and/or HSV-2 entry into human cells have been identified (4, 7, 22, 30, 39). These molecules, which serve as receptors for HSV gD, are HveA, HveB, HveC, and 3-O-sulfotransferase-3-modified heparan sulfate. HveA (herpesvirus entry mediator [HVEM]) is a member of the tumor necrosis factor receptor (TNFR) superfamily of proteins (22). HveB (also called PRR2 and nectin-2) and HveC (also called PRR1 and nectin-1) are related members of the immunoglobulin (Ig) superfamily (5, 19). A splice variant of HveC, called “HIgR,” also mediates HSV entry through its interaction with gD (4). Truncated, soluble forms of gD, lacking the transmembrane and cytoplasmic domains, bind directly to truncated, soluble forms of each of these receptors (3, 16, 17, 40, 41). In addition, antibodies to HveA, HveB, and HveC block HSV infection in various cell lines (4, 22, 39). Thus, it is clear that HSV can utilize several different and structurally unrelated cell surface proteins as receptors.
Expression of HveA appears to be most abundant in hematopoietic cells and lymphoid tissues such as the spleen and thymus (9, 14, 20). The natural ligands for HveA that have been identified are LIGHT and lymphotoxin alpha (Ltα) (21). Both ligands are structurally related to TNF, exist as trimers (8, 27), and presumably signal by inducing or altering receptor aggregation on the cell surface (2, 18, 31, 33, 36). In response to ligand binding, the cytoplasmic domain of HveA interacts with a subset of adapter proteins in the TRAF family leading to activation of the NF-κB and JNK/AP-1 pathways (9, 14, 20). Monoclonal antibodies (MAbs) against the extracellular domain of HveA block several aspects of T-cell activation, such as proliferation and cytokine production, suggesting that HveA is directly involved in this process (10).
Although the roles of each gD receptor in HSV pathogenesis and survival within the host remain to be elucidated, one possible use of HveA as a receptor in lymphocytes has been proposed. A recent study by Raftery et al. (26) showed that HSV infection of murine T cells leads to viral antigen presentation in the context of major histocompatibility complex (MHC) class I molecules. This is in contrast to fibroblasts, where transport of MHC class I molecules to the surface of HSV-infected cells is blocked (13, 44). HSV-infected T cells then become targets for killing by viral antigen-specific cytotoxic T lymphocytes (fratricide). The authors propose that the elimination of T cells infiltrating a viral lesion by this mechanism constitutes an immune evasion strategy employed by HSV.
HveA was originally identified as a receptor which functions for many strains of HSV-1 and HSV-2. However, HveA failed to mediate the entry of certain laboratory strains (such as rid1 and rid2) with mutations in gD (22). Furthermore, an antiserum against HveA was shown to block virus infection of certain cell types (22). Thus, gD was implicated in HveA-mediated virus entry. Subsequently, it was reported that the ectodomain of HSV gD bound specifically to the ectodomain of HveA, while the ectodomain of gD from the rid1 strain of HSV-1 did not bind HveA (40, 41). These results confirmed that HveA functions in HSV entry through a direct interaction with virion gD.
In this study, we utilized two complementary approaches to localizing the gD-binding domain(s) of HveA. First, we constructed a set of truncated forms of HveA and found that gD binding requires the two N-terminal cysteine-rich pseudorepeat (CRP) domains. Second, we generated a panel of MAbs against the HveA ectodomain, mapped their epitopes, and tested their abilities to block gD binding to HveA and to block HSV infection of HveA-expressing cells. One of these MAbs (CW3) recognized an epitope within the first CRP, blocked gD binding to HveA, and exhibited a limited ability to block virus infection. We propose that the first CRP domain of HveA contains at least a portion of the gD binding site. Furthermore, the second CRP of HveA is necessary for gD binding, either because it contains binding residues or because it affects the presentation of the binding site within the first CRP.
MATERIALS AND METHODS
Cells and virus.
HeLa and Vero cells were grown in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 5% fetal bovine serum (FBS). CHO-HVEM12 cells (22) were grown in Ham's F-12 medium supplemented with 10% FBS and 200 μg of G418 per ml. Sf9 (Spodoptera frugiperda) cells (GIBCO BRL) were grown in Sf900II medium (GIBCO BRL). The HSV-1 β-galactosidase recombinant virus KOS/tk12 (39) was propagated in Vero cells, and its titer was determined.
Construction of baculovirus recombinants expressing truncated forms of HveA.
The strategy employed for construction of baculovirus recombinants has been described previously (32, 40). Briefly, PCR primers were designed and synthesized to amplify and modify the HveA ectodomain coding region contained in plasmid pBEC10 (22) for cloning into the pVT-Bac transfer vector plasmid and expression in a recombinant baculovirus. The upstream primer for amplification of the HveA(120t) and HveA(76t) open reading frames (ORFs) was 5′-GCGAGATCTGCCATCATGCAAGGAGGACGAGTA-3′ and was hybridized to the noncoding strand of the HveA ORF immediately beyond the predicted signal sequence coding region. This primer incorporated a BglII restriction enzyme cleavage site (underlined). The upstream primer for amplification of the HveA(77–120t) ORF was 5′-GGCGGATCCCTGCCCTCCAGGCACCT-3′ and hybridized to the noncoding strand of the HveA ORF at the start of the coding region for the second CRP of the HveA ectodomain. This primer incorporated a BamHI restriction enzyme cleavage site (underlined). The downstream primer used to amplify the PCR fragment for HveA(120t) and for HveA(77–120t) cloning and expression was 5′-CGGGAATTCAGTGGTGGTGGTGGTGGTGACCACACACGGCGTTCTCTGT-3′, and incorporated an EcoRI restriction enzyme cleavage site (underlined). The downstream primer used to amplify the PCR fragment for HveA(76t) cloning and expression was 5′-GCCGAATTCAATGGTGGTGGTGGTGATGTTCACACACTGTGCCCGTCA-3′ and also incorporated an EcoRI restriction enzyme cleavage site (underlined). The PCR-amplified DNA fragments coded for portions of the HveA ORF without its signal sequence so that the mellitin signal sequence, coded for by pVT-Bac, would replace the missing HveA signal sequence. The downstream PCR primers were also designed to append six histidine codons prior to the termination codon to allow for purification of the recombinant proteins by nickel agarose chromatography. The PCR-amplified products were then digested with BglII or BamHI and EcoRI and cloned into pVT-Bac, which had been digested with BamHI and EcoRI. Individual baculovirus recombinants were generated, screened, and plaque purified as described previously (32, 40). The baculovirus recombinant selected for production of HveA truncated after residue 120 was named “bac-HveA(120t).” The soluble protein produced by bac-HveA(120t) is referred to as “HveA(120t).” The nomenclature for HveA(76t) followed the same pattern. The baculovirus construct expressing HveA(77–120t), consisting of HveA residues 77 to 120 (the second CRP element), was named “bac-HveA(77–120t).”
Production and purification of recombinant baculovirus-produced proteins.
The methods for production and DL6 affinity purification of gD-1(306t) (25), as well as production and nickel-agarose purification of HveA(200t) (40), have been described previously. The production and purification of HveA(120t), HveA(76t), and HveA(77–120t) were carried out as described previously for HveA(200t).
Antibodies.
R7 is a rabbit polyclonal antiserum raised against native, full-length gD-2 isolated from virus-infected cells (15). R140 is a rabbit polyclonal antiserum raised against HveA(200t) (37). DL6 is a mouse MAb which binds an epitope within gD residues 272 to 279 (15). A tetra-His-specific MAb (Qiagen) was used to bind the six-histidine tag present on the recombinant baculovirus proteins.
Production of MAbs against HveA(200t).
Mice were immunized with HveA(200t) until suitable serum antibody titers were achieved. Hybridoma production was performed according to standard procedures, and stable hybridomas secreting IgG reactive with HveA(200t) by enzyme-linked immunosorbent assay (ELISA) were subcloned twice. IgG was purified from mouse ascitic fluid by protein G chromatography (HiTrap; Amersham Pharmacia) according to the manufacturer's instructions. Following purification, MAbs were dialyzed against phosphate-buffered saline (PBS). MAb isotype determination was carried out with a mouse hybridoma subtyping kit (Roche) according to the manufacturer's instructions. All of the CW MAbs were determined to be of the IgG1 heavy-chain isotype and kappa light-chain isotype.
Competition ELISA (HveA truncations).
HveA(200t) in PBS was bound to a 96-well ELISA plate for 3 h at 25°C. The plate was washed three times with PBS–0.2% Tween 20 (PBST) and incubated in blocking solution (PBS, 5% nonfat milk, 0.2% Tween 20) for 30 min at 25°C. The plate was then washed three times with PBST and incubated with a fixed concentration of gD-1(306t) (500 nM) combined with various concentrations of the truncated forms of HveA. To control for the variability in purity of HveA protein preparations, the concentrations of HveA(120t), HveA(76t), and HveA(77–120t) were normalized against a standard concentration curve of HveA(200t) via densitometric scanning of a Western blot probed with a MAb which binds the six-histidine tag (present in all forms of HveA). Plates were then washed three times with PBST and incubated for 1 h with a rabbit polyclonal antiserum raised against gD (R7) diluted 1/1,000 in blocking solution. Plates were then washed three times with PBST and incubated for 30 min in horseradish peroxidase-conjugated goat anti-rabbit antibody (Boehringer Mannheim) diluted 1/1,000 in blocking solution. Plates were washed three times with PBS–0.2% Tween 20 and then once with 20 mM sodium citrate (pH 4.5). After removal of the citrate buffer, ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate solution (Moss, Inc.) was added, and the A405 in individual wells was read with a Perkin-Elmer HTS 7000 Bio Assay Reader. Finally, absorbance was plotted against the concentration of HveAt used.
Competition ELISA (CW MAbs and R140).
Competition ELISA experiments with CW MAbs and R140 were performed as described above for the competition ELISA by using HveA truncations, but with the following exceptions. Various concentrations of CW MAb or R140 IgG in blocking solution were added to wells coated with HveA(200t) prior to the addition of gD-1(306t). In experiments in which R140 IgG was used to compete gD binding, gD was detected with MAb DL6 (IgG) at a concentration of 50 μg/ml in blocking solution. DL6 was then detected by incubation with horseradish peroxidase-conjugated goat anti-mouse antibody (Boehringer Mannheim) diluted 1/1,000 in blocking solution.
Blocking of HSV-1 entry into CHO-HVEM12 cells by HveA truncations.
Blocking studies were carried out as previously described (40), except that the HSV-1 β-galactosidase reporter virus KOS/tk12 was used, and plates were read with a Perkin-Elmer HTS 7000 Bio Assay Reader. To control for the variability in purity of HveA protein preparations, the concentrations of HveA(120t), HveA(76t), and HveA(77–120t) were normalized against a standard concentration curve of HveA(200t) via densitometric scanning of a Western blot probed with a MAb which binds the six-histidine tag (present in all forms of HveA).
SDS-PAGE.
Purified glycoproteins were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by incubation at 100°C for 10 min in 2.5% SDS–350 mM β-mercaptoethanol. Samples were loaded onto precast Tris-glycine gels (Novex), electrophoresed, and then either stained with silver nitrate (Pharmacia) or transferred to nitrocellulose (Western blot). Proteins on Western blots were probed with antibodies and visualized by the Amersham ECL (enhanced chemiluminescence) system. For silver-stained gels, protein molecular size standards (Broad Range) were obtained from Bio-Rad. For Western blots, prestained protein molecular size standards (BenchMark) were obtained from Life Technologies. For endoglycosidase H (endo H) and glycopeptidase F (glyco F) studies, purified glycoproteins were digested with glycosidases as previously described (28, 32) prior to SDS-PAGE and Western blot analysis. Reduction and alkylation of HveA(200t) were carried out by a previously described method (28).
Production and use of synthetic peptides.
Eight peptides, encompassing HveA residues 39 to 120 (CRP1 and -2), were synthesized on cellulose membranes by using a spot synthesizer (6). Each peptide overlapped the adjacent peptides by five residues. All but one of the peptides were 15 amino acid (aa) residues in length; the remaining peptide (peptide 8 [see below]) was 12 residues in length. The residues contained in each peptide are as follows: peptide 1, residues 39 to 53; peptide 2, 49 to 63; peptide 3, 59 to 73; peptide 4, 69 to 83; peptide 5, 79 to 93; peptide 6, 89 to 103; peptide 7, 99 to 113; and peptide 8, 109 to 120. Membrane strips containing each peptide were probed for 16 h at 4°C with CW MAb IgG at 1 μg/ml. Reactive peptide spots were visualized by ECL (Amersham).
Biosensor analysis of gD-1(306t) binding to HveA(120t).
Surface plasmon resonance experiments were carried out with a BIACORE X optical biosensor (Biacore AB) at 25°C. Truncated forms of HveA were directly coupled to flow cell 2 (Fc2) of a CM5 research-grade chip (Biacore AB) at pH 5 as previously described (29, 41). The data for gD1(306t) binding to HveA(120t) were collected and analyzed with a 1:1 Langmuir binding model for the global fitting analysis.
Biosensor analysis of the effect of CW3 on gD binding to HveA.
HveA(200t) was directly coupled to Fc2 of a CM5 chip, as previously described (29, 41), except that the chip surface was activated for 4 min with EDC/NHS instead of 7, and the coupling buffer was 10 mM NaOAc buffer at pH 6 instead of pH 4. Approximately 450 response units (RU) of HveA(200t) was coupled to the chip surface this way. Fc1 was activated and blocked without the addition of protein. MAb CW3 (200 μg/ml) flowed over the chip surface until the signal no longer increased. A 0.5 μM solution of gD(306t) was then injected, and its binding was assessed. The experiment was also carried out with MAb CW1 (20 μg/ml).
CW MAb competition experiments (biosensor).
For CW MAb competition experiments with the biosensor, the running buffer was HBS-EP (Biacore AB). A MAb which binds a tetra-histidine epitope (Qiagen) was covalently coupled to a research-grade CM5 chip (Biacore AB) via primary amines by standard coupling techniques (29, 41) to a final surface density on both Fc1 and Fc2 of approximately 4,000 RU. HveAt (1 μM) flowed over Fc2 until approximately 400 RU was captured. Flow was then directed over both flow cells; Fc1 served as the control surface. Each CW MAb flowed across the chip surface to assess binding. The surface was regenerated to baseline after each experiment with short pulses (1 to 2 μl) of 20 mM sodium carbonate (pH 11)–0.5 M NaCl. For the blocking experiments, 200 RU of HveA was captured onto Fc2 as described above. One of the CW MAbs flowed over the chip surface until the signal no longer increased. A second CW MAb was injected, and its level of binding assessed. The concentrations of MAbs used were 20 μg/ml for CW1 and CW2 and 50 μg/ml for CW3.
RESULTS
Construction of baculovirus recombinants expressing truncated forms of HveA.
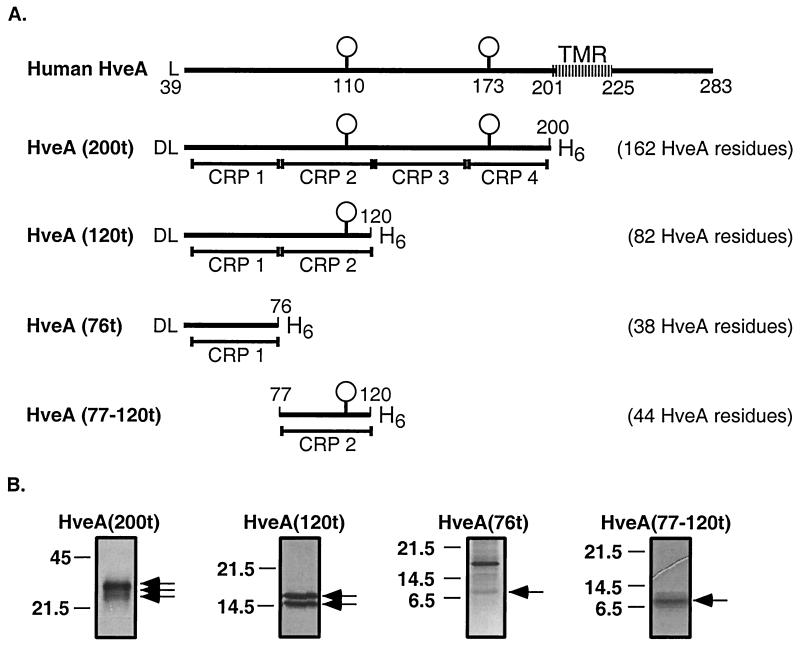
We previously described HveA(200t) (162 aa residues) (Fig. 1A), which lacks the transmembrane and cytoplasmic domains of HveA (40). HveA(200t) bound directly to truncated forms of HSV gD (16, 40, 41). This protein also bound to purified HSV virions (24) and blocked virus infection of cells expressing HveA (40). To determine which portion of HveA is responsible for gD binding and blocking of virus infection, we generated three additional baculovirus constructs expressing smaller forms of HveA (Fig. 1A). In generating these constructs, we kept the CRP domains intact to avoid disrupting the predicted pattern of disulfide bond formation within each domain (23). HveA(120t) consists of the first two CRP elements (82 aa) of the HveA ectodomain. HveA(76t) consists of the first CRP domain (38 aa), and HveA(77–120t) consists of the second CRP domain (44 aa). These recombinant proteins were purified from the culture supernatant of baculovirus-infected Sf9 cells by nickel-agarose chromatography. Figure 1B shows each of the purified HveA proteins following SDS-PAGE and visualization by silver staining. As previously reported (40), HveA(200t) migrates as three distinct bands which differ in the extent of N glycosylation. It is unclear what the additional protein copurifying with HveA(76t) is. However, it does not appear to be an oligomeric form of HveA(76t), since it did not react with an antibody directed against the six-histidine tag (see Fig. 7).
FIG. 1.
Recombinant baculovirus proteins used in this study. (A) Diagrams of full-length human HveA and baculovirus constructs examined in this study. The first amino acid residue of HveA after signal peptide removal is a leucine (L), which is the 39th residue of the predicted HveA ORF. An additional aspartic acid residue (D), encoded by the pVT-Bac transfer vector, is present at the N terminus of the recombinant baculovirus proteins. The two predicted N glycosylation sites of HveA are indicated by balloons and occur at residues 110 and 173. The predicted transmembrane region (TMR) of HveA includes residues 201 to 225. Truncated forms of HveA contained one, two, or four of the CRP elements which constitute the HveA ectodomain. The boundaries of the CRP elements are indicated below each truncated form of HveA. Each recombinant protein was constructed such that six histidine residues (H6) were appended to the C terminus of the protein. The number of HveA amino acid residues present in the recombinant proteins is indicated to the right of each diagram. (B) Silver-stained, purified, recombinant baculovirus proteins following SDS-PAGE. The positions of molecular size markers (in kilodaltons) are shown to the left of each gel. Arrows indicate the bands representing each recombinant protein.
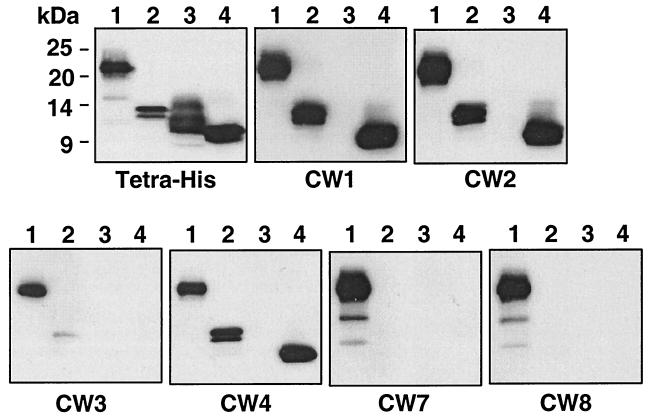
FIG. 7.
Mapping of the CW MAb and epitopes by using HveA truncations. Identical Western blots containing each of the four HveA truncations were probed with the indicated MAbs. The positions of molecular size markers are shown to the left of the first panel. Lanes: 1, HveA(200t); 2, HveA(120t); 3, HveA(76t); 4, HveA(77–120t). The first blot was probed with a MAb which recognizes the histidine tag at the C terminus of each protein. The remaining six blots were probed with the CW MAb indicated below each panel.
Glycosidase treatment of HveA(120t), HveA(76t), and HveA(77–120t).
There are two consensus N glycosylation sites in the HveA ectodomain, one at residue 110 (second CRP) and a second at residue 173 (fourth CRP), and we previously showed that HveA(200t) contained endo H-resistant N-linked carbohydrates (40). Here, treatment of HveA(120t), HveA(76t), and HveA(77–120t), with endo H and glyco F revealed that HveA(120t) and HveA(77–120t) were N glycosylated, but that HveA(76t) was not (Fig. 2). This showed that the consensus N glycosylation site within the second CRP is utilized. Additionally, since the N-linked carbohydrates on both HveA(120t) and HveA(77–120t) were resistant to endo H digestion, the carbohydrate moieties on these proteins were processed from the endo H-sensitive form.
FIG. 2.
Glycosidase digestion of HveA recombinant baculovirus proteins. Each purified recombinant protein was either mock digested (−) or incubated with (+) endo H and glyco F. Samples were then separated via SDS-PAGE, blotted onto nitrocellulose, and probed with a murine MAb which binds the histidine tag present at the C terminus of each protein.
gD-binding properties of the HveA truncations.
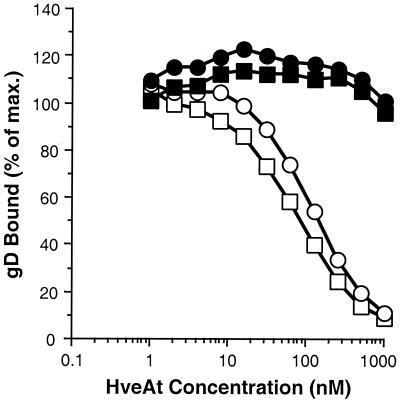
We previously showed by ELISA (40) and by optical biosensor (41) that truncated forms of gD bound to HveA(200t). However, based on previous observations, we were concerned that the structures of some or all of the truncated forms of HveA might be altered, possibly to different extents, following adsorption to an ELISA plate and that such changes might affect gD binding. To avoid this problem, we used a competition ELISA to assess the gD-binding capacity of HveA(120t), HveA(76t), and HveA(77–120t) in comparison with that of HveA(200t) (Fig. 3). HveA(200t) was adsorbed to an ELISA plate and then incubated with a constant amount of gDt in the presence of increasing concentrations of the truncated forms of HveA. Finally, the amount of gDt bound to HveA(200t) on the plate was determined. Thus, this assay measured the ability of the HveAt in solution to compete with the HveA(200t) on the plate for gDt binding. As expected, HveA(200t) competed with itself for gDt binding. HveA(120t) competed for gDt binding as effectively as HveA(200t), indicating that the full gD-binding activity of HveA resides within the first 82 residues of the extracellular domain. In contrast, neither HveA(76t) nor HveA(77–120t) (nor the combination of these two proteins [not shown]) was able to compete with HveA(200t) for gD binding. These data are consistent with the conclusion that residues within both CRP1 and CRP2 are directly involved in gD binding. Alternatively, all of the HveA residues involved in gD binding may reside within one or the other CRP domain, but proper presentation of those residues may require both domains.
FIG. 3.
Competition ELISA. The wells of a microtiter plate were coated with HveA(200t) and then incubated with 500 nM gD-1(306t) in the presence of increasing concentrations of each of the truncated forms of HveA. gD bound to HveA(200t) on the plate was detected with a rabbit polyclonal antiserum (R7) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG. Bound gD was expressed as a percentage of the amount bound in the absence of HveAt in solution and plotted against the concentration of HveAt in solution. □, HveA(200t); ○, HveA(120t); ■, HveA(76t); ●, HveA(77–120t).
Biosensor.
Previously we showed by surface plasmon resonance that gD-1(306t) bound specifically to HveA(200t) with an equilibrium dissociation constant (KD) of 3.2 × 10−6 M (29, 41). Here we used the optical biosensor to examine the binding of gD to HveA(120t), HveA(76t), and HveA(77–120t) (Table 1). Consistent with the results of the competition ELISA, binding of gD-1(306t) to HveA(120t) was evident, whereas binding to HveA(76t) or HveA(77–120t) could not be detected. Furthermore, the KD of gD-1(306t) binding to HveA(120t) was identical to the affinity reported for the binding of gD1(306t) to HveA(200t) (29, 41).
TABLE 1.
Optical biosensor analysis of gD-1(306t) binding to HveA truncations
| Immobilized ligand | kon (M−1 s−1) | koff (s−1) | KD (M [koff/kon]) |
|---|---|---|---|
| HveA(200t)a | (6.1 ± 0.6) × 103 | (2.0 ± 0.2) × 10−2 | (3.2 ± 0.6) × 10−6 |
| HveA(120t)b | (5.3 ± 0.9) × 103 | (1.7 ± 0.4) × 10−2 | (3.2 ± 0.7) × 10−6 |
| HveA(76t)c | Does not bind | ||
| HveA(77–120t)d | Does not bind |
For details, see reference 41.
Values are the average of three experiments.
HveA(76t) was captured on the chip surface via the His tail by an anti-tetra-His MAb.
HveA(77–120t) was captured on the chip surface via the His tail by an anti-tetra-His MAb.
Inhibition of HSV entry into CHO-HVEM12 cells by HveA truncations.
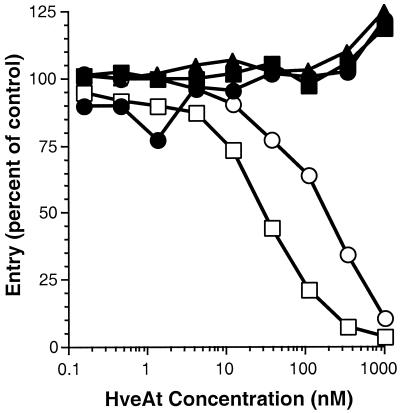
We showed previously that HveA(200t) blocks HSV infection of CHO cells stably expressing HveA (CHO-HVEM12) in a dose-dependent manner (40). This property reflects the ability of the soluble receptor to compete with HveA expressed on cells for binding to virion gD. Here, a constant amount of the HSV-1 β-galactosidase reporter virus KOS/tk12 was incubated with increasing concentrations of HveA(200t), HveA(120t), HveA(76t), HveA(77–120t), or bovine serum albumin (BSA) prior to inoculation of CHO-HVEM12 cells. Infected cells were lysed, and β-galactosidase activity was determined and used as a measure of HSV entry (Fig. 4). Both HveA(200t) and HveA(120t) blocked infection, suggesting that these forms of HveA bind virion gD and compete with HveA expressed on the cell surface for binding to virion gD. This is consistent with the competition ELISA and biosensor results, which demonstrated that gD-1(306t) bound with similar affinity to HveA(200t) and HveA(120t). Neither HveA(76t) nor HveA(77–120t) blocked virus entry. This result was anticipated, since we were unable to detect binding of either of these forms of HveA to gD.
FIG. 4.
Blocking of HSV entry with HveA truncations. HSV β-galactosidase reporter virus KOS/tk12 (105 PFU) was mixed with various concentrations of HveA truncations prior to inoculation of CHO-HVEM12 cells in a 96-well tissue culture plate. After 7 h of infection, the cells were lysed, and β-galactosidase activity was determined. Virus entry is expressed as the level of β-galactosidase activity induced relative to that induced by the reporter virus in the absence of HveAt. Virus entry is plotted against HveAt concentration. □, HveA(200t); ○, HveA(120t); ■, HveA(76t); ●, HveA(77–120t); ▴, BSA.
Examination of the ability of anti-HveA MAbs to block the binding of gD to HveA.
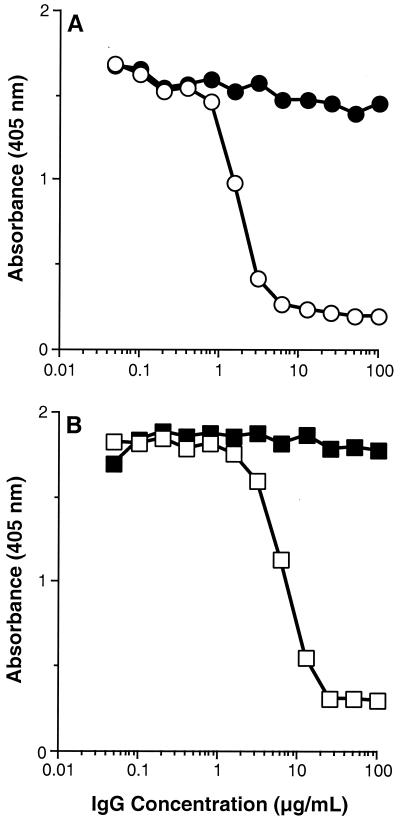
As a second approach to examining the interaction of HveA with HSV gD, we developed a panel of six MAbs against HveA(200t). These MAbs were named CW1, -2, -3, -4, -7, and -8. We used a competition ELISA to determine whether any of these MAbs could block the binding of gD to HveA (Fig. 5A). Here, HveA(200t) was adsorbed to the wells of a 96-well ELISA plate and incubated with increasing concentrations of the anti-HveA MAbs. Then, a constant amount of gD-1(306t) was added to each well. Finally, the amount of gD bound to HveA(200t) on the plate was determined by using a rabbit polyclonal serum against gD. Of the six MAbs tested, only CW3 blocked the binding of gD to HveA in a dose-dependent manner. The blocking activity of CW1 (shown) was similar to that of CW2, -4, -7, and -8 (not shown). This suggests that the CW3 epitope may overlap the gD-binding region of HveA and directly interfere with gD binding. Alternatively, CW3 binding may induce a conformational change in HveA which prevents gD binding.
FIG. 5.
Blocking of gD binding to HveA with CW MAbs and R140. The wells of a microtiter plate were coated with HveA(200t) and then incubated with 500 nM gD-1(306t) in the presence of increasing concentrations of purified IgG. Bound gD-1(306t) was detected with the rabbit polyclonal anti-gD serum R7 (A) or the anti-gD MAb DL6 (B). (A) Purified IgG from MAbs CW1 (●) and CW3 (○). (B) Purified IgG from preimmune (■) and hyperimmune (□) rabbit R140 serum.
Using a competition ELISA, we also examined the ability of a rabbit antiserum raised against HveA(200t) (called “R140”) (37) to block the binding of gD to HveA (Fig. 5B). In this case, the amount of gD bound to HveA(200t) on the plate was determined with a MAb against gD (DL6). The results showed that R140, like CW3, blocked the binding of gD to HveA.
As an additional method of examining the ability of CW3 to block gD binding to HveA, we used the optical biosensor (data not shown). HveA(200t) was covalently attached to Fc2 via primary amines, and CW3 was allowed to flow across the chip surface until saturation was achieved. gD-1(306t) was then injected, and its binding to the chip surface was compared to its binding in the absence of CW3. The binding of gD-1(306t) to HveA(200t) was completely blocked by CW3. In a similar assay, CW1 failed to block the binding of gD-1(306t) to HveA(200t).
Examination of the ability of CW3 and R140 to block HSV entry into CHO-HVEM12 cells.
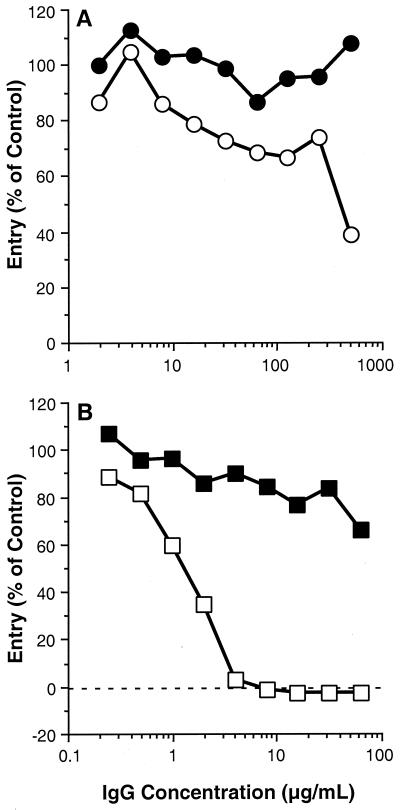
Since CW3 and R140 were able to block the binding of gD to HveA, we next tested the ability of these antibodies to block HSV-1 (KOS/tk12) entry into CHO-HVEM12 cells (Fig. 6). Compared to CW1, which showed no effect on HSV entry, CW3 exhibited a modest reduction of virus entry (Fig. 6A). However, R140 completely blocked virus entry (Fig. 6B), confirming the work of Montgomery et al. (22) showing that HveA is indeed the receptor being used for HSV entry into these cells. The relatively poor entry-blocking activity of CW3 is not due to a failure of CW3 to bind HveA on the cell surface, since this MAb reacted with HveA on the surface of CHO-HVEM12 cells by fluorescence-activated cell sorter analysis as well as by cell ELISA (data not shown).
FIG. 6.
Blocking of HSV infection with CW MAbs and R140. CHO-HVEM12 cells were seeded into a 96-well tissue culture plate and grown overnight. Serial dilutions of the indicated antibodies (purified IgG) were then added to cells in the plates, which had been prechilled to 4°C. Cells were incubated with the antibodies at 4°C for 90 min, after which, 105 PFU of KOS/tk12 was added. Plates were then shifted to 37°C and incubated for 7 h. Finally, cells were lysed and β-galactosidase activity was determined. Virus entry is expressed as the level of β-galactosidase activity induced relative to that induced by the reporter virus in the absence of blocking IgG and is plotted against IgG concentration. (A) Purified IgG from MAbs CW1 (●) and CW3 (○). (B) Purified IgG from preimmune (■) and hyperimmune (□) rabbit R140 serum.
Localization of the CW MAb epitopes.
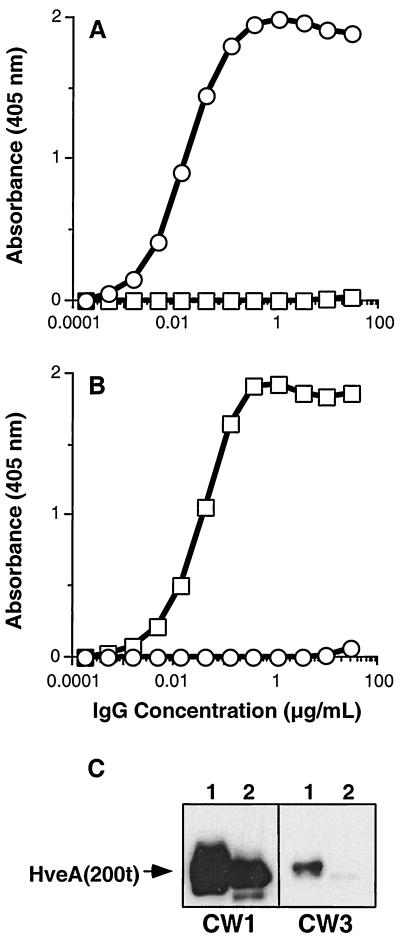
To further characterize the CW MAbs, we examined their binding to the four HveA truncations (described earlier) by Western blot analysis (Fig. 7). CW1, CW2, and CW4 reacted with HveA(200t), HveA(120t), and HveA(77–120t), indicating that these MAbs bind an epitope within the second CRP domain of HveA (residues 77 to 120). CW7 and CW8 bound only to HveA(200t), suggesting that their epitopes are within residues 121 to 200. CW3 reacted weakly with HveA(200t) and to an even lesser extent with HveA(120t). The weak reactivity of CW3 with HveA was not expected, since it bound HveA(200t) as well as CW1 and CW2 by ELISA (data not shown). Because of this, we tested the reactivity of CW3 with HveA(76t) and HveA(77–120t) by ELISA (Fig. 8A and B). CW3 reacted strongly with HveA(76t), but not with HveA(77–120t), indicating that its epitope is within the first CRP domain. Consistent with the epitope-mapping data shown in Fig. 7, CW1 reacted with HveA(77–120t), but not with HveA(76t). Since CW3 failed to react strongly with the HveA truncations on a Western blot, we reasoned that the CW3 epitope may be discontinuous and, as such, may have been completely or partially destroyed by the denaturing conditions. To address this question, we reduced and alkylated an aliquot of HveA(200t) and prepared two Western blots containing equal amounts of the reduced and alkylated HveA(200t) as well as untreated HveA(200t). These blots were reacted with CW1 and CW3 (Fig. 8C). The reactivity of CW1 with HveA was somewhat diminished by reduction and alkylation. However, the reactivity of CW3 was markedly diminished, indicating that the CW3 epitope is stabilized by disulfide bonding.
FIG. 8.
CW3 binds a discontinuous epitope within the first CRP of HveA. CW1 and CW3 were tested for reactivity with HveA(76t) and HveA(77–120t) by ELISA. Twofold dilutions of CW1 and CW3 were added to the wells of ELISA plates coated with HveA(76t) or HveA(77–120t). Bound MAb was detected with horseradish peroxidase-conjugated anti-mouse IgG and horseradish peroxidase substrate. Plates were read at 405 nm, and the absorbance in each well was plotted against the MAb concentration. □, CW1; ○, CW3. (A) ELISA plate coated with HveA(76t). (B) ELISA plate coated with HveA(77–120t). (C) Western blot testing of CW1 and CW3 for reactivity with HveA(200t) or reduced and alkylated HveA(200t). For each blot, lane 1 contained HveA(200t) and lane 2 contained reduced and alkylated HveA(200t). Blots were probed with the indicated MAbs. The position of the band corresponding to HveA(200t) is indicated by an arrow.
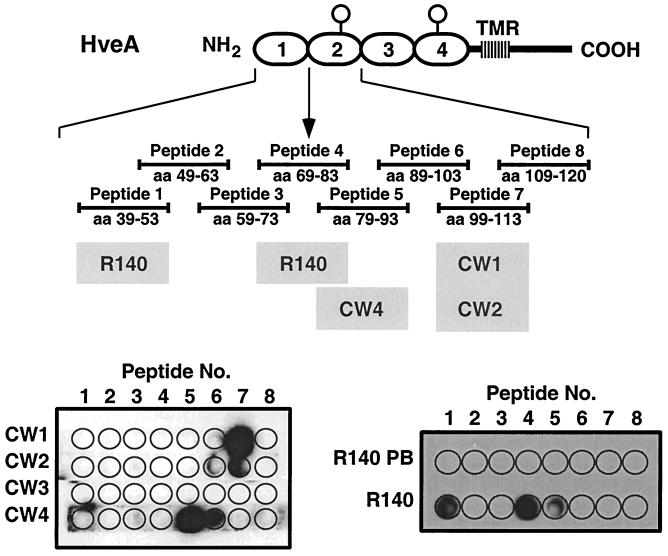
Since the gD-binding region of HveA resides within the first 82 aa residues (CRP1 and -2), we wanted to more precisely define the epitopes of the MAbs which bound within this region. To do this, we generated a set of overlapping 15-aa peptides spanning HveA residues 39 to 120, which were probed with CW1, -2, -3, and -4 (Fig. 9). CW1 and -2 bound to a peptide mimicking HveA residues 99 to 113, thereby localizing the epitopes for both MAbs to this region of HveA (within CRP2). Consistent with this, biosensor studies showed that CW1 and CW2 competed with each other for HveA binding (not shown). CW4 bound strongly to the peptide mimicking HveA residues 79 to 93 and less well to the overlapping peptide mimicking residues 89 to 103. This result suggests that the CW4 epitope is largely or entirely within residues 79 to 93 and that a portion of the CW4 epitope exists within residues 89 to 103 (both peptides are within CRP2). CW3 failed to bind any of these peptides, confirming our previous data showing that CW3 binds a discontinuous epitope.
FIG. 9.
Reactivity of CW MAbs and R140 with HveA(120t) peptides. A diagram of HveA is shown. The four CRP elements comprising the HveA ectodomain are represented by ovals and are numbered 1 to 4. Eight overlapping synthetic peptides (seven 15-mers and one 12-mer [shown as solid bars with the HveA residue numbers indicated]) mimicking residues within the first and second CRP elements of HveA were directly coupled to a cellulose sheet. Peptide 4 spans the junction between CRP1 and CRP2 (indicated by an arrow). Individual strips containing the complete set of peptides were incubated with the indicated CW MAb or the rabbit polyclonal antiserum R140. Reactive peptide spots were then visualized by ECL after incubation with the appropriate horseradish peroxidase-conjugated secondary antibody. TMR, transmembrane region.
We also examined the reactivity of R140 against the same set of HveA peptides (Fig. 9). R140 reacted strongly with two HveA peptides. These mimicked HveA residues 39 to 53 and 69 to 83. R140 also reacted weakly with the peptide mimicking residues 79 to 93, which overlaps the 69-to-83 peptide and may therefore contain a portion of the same epitope present in this peptide. Addition of these peptides to R140 did not diminish its ability to block HSV infection of CHO-HVEM12 cells (not shown). Thus, the antibodies within R140 that react with these peptides do not account for the potent virus-blocking activity of this antiserum.
DISCUSSION
The goal of this study was to localize the gD-binding region of HveA. Our first approach was to produce several new truncated forms of HveA by using the baculovirus expression system. Each truncation consisted of one or more of the four CRP elements which comprise the HveA ectodomain. The HveA constructs studied here consisted of the first CRP alone [HveA(76t)], the second CRP alone [HveA(77–120t)], the first and second CRPs [HveA(120t)], or all four CRPs [HveA(200t)].
Competition ELISA and biosensor analysis showed that HveA(120t) retained full gD binding activity. Indeed, the KD (as determined by biosensor analysis) for the binding of gD-1(306t) to HveA(120t) was identical to that reported for the binding of gD-1(306t) to HveA(200t) (41). In contrast, neither HveA(76t) nor HveA(77–120t) exhibited any capacity to bind gD. This localized the gD binding region to CRP1 and CRP2 of HveA (82 aa residues). Furthermore, HveA(200t) and HveA(120t) blocked HSV entry similarly, whereas neither HveA(76t) nor HveA(77–120t) was able to block virus entry. In addition, the concentrations of HveAt required to block gD binding and virus entry were nearly identical, suggesting that HveAt interacts with virion gD and soluble gD-1(306t) similarly.
Since MAbs had proven to be valuable tools in the localization of receptor-binding regions of gD (16, 24), we reasoned that anti-HveA MAbs might also be useful in identifying gD-binding regions of HveA. We therefore generated six MAbs against HveA and mapped their epitopes by using the HveA truncations as well as synthetic peptides mimicking portions of the deduced HveA amino acid sequence. Two MAbs (CW7 and CW8) recognized linear epitopes within HveA residues 121 to 200 (CRP3 and -4), and three MAbs (CW1, CW2, and CW4) recognized linear epitopes within the second CRP of HveA. The remaining MAb (CW3) recognized a discontinuous epitope within the first CRP of HveA.
Of the six MAbs, only CW3 was able to block the binding of gD to HveA, suggesting that the first CRP is important for gD binding. Interestingly, none of the MAbs which bound within the second CRP (CW1, -2, and -4) blocked gD binding, clearly demonstrating that their epitopes are distinct from HveA residues involved in gD binding.
CW3 was also tested for its ability to block HSV infection of CHO cells expressing HveA. Although it was able to block infection, it did so only at relatively high IgG concentrations. This suggests that HveA expressed on transfected cells is different in some way from HveA(200t). Perhaps HveA behaves differently as an integral membrane protein. Alternatively, the availability of the CW3 epitope on cells may be influenced by the interaction of HveA with itself or with other cell surface molecules. It was recently reported that certain members of the TNFR superfamily self-associate on the surface of cells in the absence of ligand and that this self-association is critical for ligand binding (2, 31).
In conjunction with the MAb studies, we also analyzed the rabbit polyclonal antiserum R140, raised against HveA(200t). We found that this antiserum both blocked gD binding to HveA and completely blocked virus infection of CHO cells expressing HveA. It is not clear what component of R140 is responsible for its blocking activity. Although R140 bound two peptides within the first two CRP elements of HveA, these peptides did not compete its virus-blocking activity, suggesting that the antibodies in R140 which react with these peptides are not responsible for blocking virus entry. Perhaps the virus entry-blocking components of R140 bind a discontinuous epitope(s) on HveA.
Mauri et al. (21) showed that soluble forms of gD and LIGHT competed with each other for HveA binding. If the binding of LIGHT to HveA is similar to the binding of TNF-α to TNFR I, then the ligand contacts within HveA would exist within CRP2 and CRP3 (1). Since gD binding by HveA requires CRP1 and CRP2, it is possible that LIGHT and gD compete for binding to overlapping regions of HveA. Alternatively, each ligand may induce or stabilize a receptor structure that is refractory to binding by the other without directly competing for a common site on HveA. We previously reported that MAb CW8 blocked the binding of LIGHT to HveA (27). The mechanism of blocking in this case is also unclear. However, the fact that CW8 failed to block gD binding to HveA distinguishes the gD-HveA interaction from that of LIGHT and HveA. Indeed, since CW8 binds within HveA residues 121 to 200, this result is consistent with our conclusion that gD binding by HveA involves CRP1 (residues 39 to 76) and possibly CRP2 (residues 77 to 120).
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grant NS-36731 from the National Institute of Neurological Disorders and Stroke (R.J.E. and G.H.C.) and grants AI-18289 (G.H.C. and R.J.E.) and AI-07325 (R.J.E.) from the National Institute of Allergy and Infectious Diseases. C.K. was supported by a fellowship (823A-053464) from the Swiss National Science Foundation.
The production of hybridomas was carried out at the University of Pennsylvania Cell Center Service Facility. We thank Laszlo Otvos for peptide synthesis. We also thank Ann Rux and Richard Milne for helpful discussions.
REFERENCES
- 1.Banner D W, D'Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd receptor-human TNFβ complex: implications for the TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 2.Chan F K, Chun H J, Zheng L, Siegel R M, Bui K L, Lenardo M J. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 3.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberlé F, Dubreuil P, Mattei M-G, Devilard E, Lopez M. The human PRR2 gene, related to the poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 6.Esposito M, Venkatesh V, Otvos L, Weng Z, Vajda S, Banki K, Perl A. Human transaldolase and cross-reactive viral epitopes identified by autoantibodies of multiple sclerosis patients. J Immunol. 1999;163:4027–4032. [PubMed] [Google Scholar]
- 7.Geraghty R J, Krummenacher C, Eisenberg R J, Cohen G H, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 8.Gray P W, Aggarwal B B, Benton C V, Bringman T S, Henzel W J, Jarrett J A, Leung D W, Moffat B, Ng P, Svedersky L P, et al. Cloning and expression of cDNA for human lymphotoxin, a lymphokine with tumour necrosis activity. Nature. 1984;312:721–724. doi: 10.1038/312721a0. [DOI] [PubMed] [Google Scholar]
- 9.Harrop J A, McDonnell P C, Brigham-Burke M, Lyn S D, Minton J, Tan K B, Dede K, Spampanato J, Silverman C, Hensley P, DiPrinzio R, Emery J G, Deen K, Eichman C, Chabot-Fletcher M, Truneh A, Young P R. Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. J Biol Chem. 1998;273:27548–27556. doi: 10.1074/jbc.273.42.27548. [DOI] [PubMed] [Google Scholar]
- 10.Harrop J A, Reddy M, Dede K, Brigham-Burke M, Lyn S, Tan K B, Silverman C, Eichman C, DiPrinzio R, Spampanato J, Porter T, Holmes S, Young P R, Truneh A. Antibodies to TR2 (herpesvirus entry mediator), a new member of the TNF receptor superfamily, block T cell proliferation, expression of activation markers, and production of cytokines. J Immunol. 1998;161:1786–1794. [PubMed] [Google Scholar]
- 11.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 12.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill A B, Barnett B C, McMichael A J, McGeoch D J. HLA class I molecules are not transported to the cell surface in cells infected with herpes simplex virus types 1 and 2. J Immunol. 1994;152:2736–2741. [PubMed] [Google Scholar]
- 14.Hsu S, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 15.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loetscher H, Gentz R, Zulauf M, Lustig A, Tabuchi H, Schlager E-J, Brockhurst M, Gallati H, Manneberg M, Lesslauer W. Recombinant 55-kDa tumor necrosis factor (TNF) receptor. J Biol Chem. 1991;266:18324–18329. [PubMed] [Google Scholar]
- 19.Lopez M, Eberlé F, Mattei M-G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 20.Marsters S A, Ayres T M, Skubatch M, Gray C L, Rothe M, Ashkenazi A. Herpes virus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 21.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 22.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 23.Naismith J H, Sprang S R. Modularity in the TNF-receptor family. Trends Biochem Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- 24.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raftery M J, Behrens C K, Muller A, Krammer P H, Walczak H, Schonrich G. Herpes simplex virus type 1 infection of activated cytotoxic T cells: induction of fratricide as a mechanism of viral immune evasion. J Exp Med. 1999;190:1103–1113. doi: 10.1084/jem.190.8.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney I A, Butrivich K D, Glass A A, Borboroglu S, Benedict C A, Whitbeck J C, Cohen G H, Eisenberg R J, Ware C F. The lymphotoxin-β receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. J Biol Chem. 2000;275:14307–14315. doi: 10.1074/jbc.275.19.14307. [DOI] [PubMed] [Google Scholar]
- 28.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rux A H, Willis S H, Nicola A V, Hou W, Peng C, Lou H, Cohen G H, Eisenberg R J. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J Virol. 1998;72:7091–7098. doi: 10.1128/jvi.72.9.7091-7098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 31.Siegel R M, Frederiksen J K, Zacharias D A, Chan F K, Johnson M, Lynch D, Tsien R Y, Lenardo M J. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 32.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 34.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 35.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 36.Tartaglia L A, Goeddel D V. Tumor necrosis factor receptor signaling. A dominant negative mutation suppresses the activation of the 55-kDa tumor necrosis factor receptor. J Biol Chem. 1992;267:4304–4307. [PubMed] [Google Scholar]
- 37.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R, Cohen G H, Eisenberg R J, Spear P G. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner M S, Martinez W, Geraghty R J, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 40.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittels M, Spear P G. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 1990;18:271–290. doi: 10.1016/0168-1702(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 43.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]