Abstract
The relationship between psoriasis and site-specific cancers remains unclear. Here, we aim to investigate whether psoriasis is causally associated with site-specific cancers. We use observational and genetic data from the UK Biobank, obtaining GWAS summary data, eQTL analysis data, TCGA data, and GTEx data from public datasets. We perform PheWAS, polygenic risk score analysis, and one-sample and two-sample Mendelian randomization analyses to investigate the potential causal associations between psoriasis and cancers. In the unselected PheWAS analysis, psoriasis is associated with higher risks of 16 types of cancer. Using one-sample Mendelian randomization analyses, it is found that genetically predicted psoriasis is associated with higher risks of anal canal cancer, breast cancer, follicular non-Hodgkin’s lymphoma and nonmelanoma skin cancer in women; and lung cancer and kidney cancer in men. Our two-sample Mendelian randomization analysis indicates that psoriasis is causally associated with breast cancer and lung cancer. Gene annotation shows that psoriasis-related genes, such as ERAP1, are significantly different in lung and breast cancer tissues. Taken together, clinical attention to lung cancer and breast cancer may be warranted among patients with psoriasis.
Subject terms: Genetic association study, Cancer genetics, Psoriasis
The relationship between psoriasis and cancer remains unclear. Here, the authors use PheWAS, polygenic risk scores, and Mendelian randomization analyses to demonstrate that psoriasis is causally associated with lung and breast cancer.
Introduction
Psoriasis is an immune-system-mediated inflammatory skin disease that affects approximately 3% of the population1. Cancer has been reported to be one of the leading causes in patients with psoriasis2, but the association between psoriasis and the risk of site-specific cancers remains unclear. A meta-analysis that included cohort or case-control studies indicated that psoriasis was associated with an increased risk of developing 11 types of site-specific cancers, including colon, colorectal, kidney, laryngeal, liver, lymphoma, keratinocyte, esophageal, oral cavity, pancreatic cancers, and non-Hodgkin lymphoma3. However, another meta-analysis including cohort studies found that patients with psoriasis or psoriatic arthritis had higher risks of keratinocyte cancer, lymphoma, lung cancer, and bladder cancer4. These inconsistent results might be ascribed, at least partially, to limited sample sizes, varied diagnostic criteria of psoriasis, and insufficient adjustments for potential confounders.
Psoriasis has a significant hereditary predisposition5. Genome-wide association studies (GWASs) have identified several susceptibility loci of psoriasis involved in immune-mediated inflammatory disorders and innate/acquired host defense6, leading to a higher possibility of developing cancer for patients with psoriasis1,7,8. Although integrating genetic data into analyses of the polygenic risk score (PRS) or Mendelian randomization (MR) is an effective approach to exploring causal effects on outcomes9,10, different MR studies lead to controversial results regarding the causal association between psoriasis and cancer11–13.
To date, systematic explorations based on a large-scale cohort with genetic data examining the relationship between psoriasis and the incidence of malignancy are lacking. In this study, we systematically explored associations between psoriasis and 89 types of cancers in a large population-based cohort. Then, we used PRS and MR analyses to investigate whether psoriasis is causally associated with site-specific cancers. The biological functions and expression levels of psoriasis-related genetic variants in cancers were further explored to uncover potential molecular correlations.
Results
Baseline characteristics
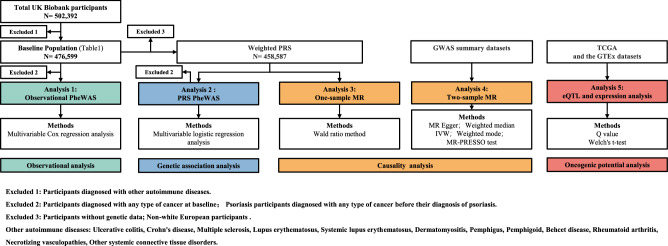
For analyses 1–3, a total of 13,463 patients with psoriasis and 463,136 participants without psoriasis were included, and their characteristics at baseline are summarized in Table 1. In analysis 4, we obtained GWAS summary data from public datasets, and the genotype and gene expression data in analysis 5 were collected from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) datasets. We illustrate the flow chart for the study design in Fig. 1. The research question, data sources utilized, and strengths and limitations of each analysis are presented in Table 2.
Table 1.
Baseline characteristics of participants after exclusion of other autoimmune diseases
| Non-PSO | PSO | SMD | |
|---|---|---|---|
| N = 463136 | N = 13463 | ||
| Age (years) | 56.9 (8.11) | 57.4 (7.98) | 0.057 |
| Male (N, %) | 212843 (46.0%) | 6840 (50.8%) | 0.097 |
| BMI (kg/m2) | 27.4 (4.76) | 28.3 (5.09) | 0.178 |
| SBP (mmHg) | 138 (18.50) | 139 (18.20) | 0.052 |
| FBG (mmol/L) | 5.12 (1.23) | 5.20 (1.39) | 0.064 |
| Smoking status (%) | 0.228 | ||
| Never | 256841 (55.5%) | 6001 (44.6%) | |
| Previous | 158186 (34.2%) | 5428 (40.3%) | |
| Current | 48109 (10.4%) | 2034 (15.1%) | |
| Alcohol intake frequency (%) | 0.048 | ||
| Daily or almost daily | 94455 (20.4%) | 2992 (22.2%) | |
| Three or four times a week | 107973 (23.3%) | 3086 (22.9%) | |
| Once or twice a week | 120155 (25.9%) | 3314 (24.6%) | |
| One to three times a month | 51509 (11.1%) | 1491 (11.1%) | |
| Special occasions only | 52594 (11.4%) | 1520 (11.3%) | |
| Never | 36450 (7.9%) | 1060 (7.9%) | |
| Physical activity (%) | 0.047 | ||
| Low intensity | 86288 (18.6%) | 2759 (20.5%) | |
| Moderate intensity | 189459 (40.9%) | 5420 (40.3%) | |
| High intensity | 187389 (40.5%) | 5284 (39.2%) | |
| Medication use | |||
| Glucocorticoids (N, %) | 4780 (1.03%) | 282 (2.09%) | <0.001 |
| Methotrexate (N, %) | 305 (0.07%) | 403 (2.99%) | <0.001 |
| Cyclosporine (N, %) | 68 (0.01%) | 15 (0.11%) | <0.001 |
PSO psoriasis, BMI body mass index, SBP systolic blood pressure, FBG fasting blood glucose, SMD, standardized mean difference.
Fig. 1. Flow chart for study design.
Excluded 1: participants diagnosed with other autoimmune diseases. Excluded 2: participants diagnosed with any type of cancer at baseline and psoriasis participants diagnosed with any type of cancer before their diagnosis of psoriasis. Excluded 3: participants without genetic data; non-white European participants. Other autoimmune diseases: ulcerative colitis, Crohn’s disease, multiple sclerosis, lupus erythematosus, systemic lupus erythematosus, dermatomyositis, pemphigus, pemphigoid, Behcet disease, rheumatoid arthritis, necrotizing vasculopathy, other systemic connective tissue disorders. Different colors correspond to different types of analysis. PheWAS phenome-wide association study, PRS polygenic risk score, MR Mendelian randomization. MR-Egger mendelian randomization-Egger, IVW inverse variance-weighted method, MR-PRESSO MR pleiotropy residual sum, and outlier test, GWAS Genome-wide Association Study, eQTL expression quantitative trait loci, TCGA the Cancer Genome Atlas, GTEx genotype-tissue expression.
Table 2.
Summary of the study design, research question, data sources utilized, and strengths and limitations of each methodological approach applied in the present study
| Study design | Research question | Data sources | Key strengths | Key limitations |
|---|---|---|---|---|
| Analysis 1: observational PheWAS | Are patients with psoriasis associated with a higher risk of site-specific cancers? | Individual-level data from the UK Biobank. | A large population-based dataset; low heterogeneity; prospective data collection; large availability of confounding data. | Observational design with unknown confounding factors; cannot assess causality. |
| Analysis 2: PRS PheWAS | Is genetic susceptibility to psoriasis associated with an elevated risk of site-specific cancers? | GWAS summary data from public datasets, and individual-level genotype and phenotype data from the UK Biobank. | Quantifies the latent genetic predisposition for psoriasis, regardless of clinical diagnosis; potentially mitigating exposure misclassification within the observational study; information on the outcome phenotype which is collected prospectively. | Horizontal pleiotropy; causal inference is inferior to MR because of the pleiotropy. |
| Analysis 3: one-sample MR | Does genetic predisposition to psoriasis have a causal impact on the risk of developing site-specific cancers? | Individual-level genotype and phenotype data from the UK Biobank. | Causal effects can be determined; akin to a natural randomized controlled trial; minimally affected by measurement error, confounding factors, and reverse causality. | False-positive results due to weak instruments. |
| Analysis 4: two-sample MR | The same as one-sample MR. | GWAS summary data from public datasets. | The same as one-sample MR but more powerful and is less prone to false-positive bias. | Investigation of a subset of participants requires new GWAS to be performed. |
| Analysis 5: eQTL and expression analysis | Do psoriasis-related SNPs potentially influence cancer development? | Genotype and gene expression data were collected from TCGA and the GTEx datasets. | Offering mechanistic insights. | Lack of causative links and clinical significance. |
MR Mendelian randomization, GWAS genome-wide association study, eQTL expression quantitative trait loci, TCGA the cancer genome atlas, GTEx genotype-tissue expression.
Observational PheWAS
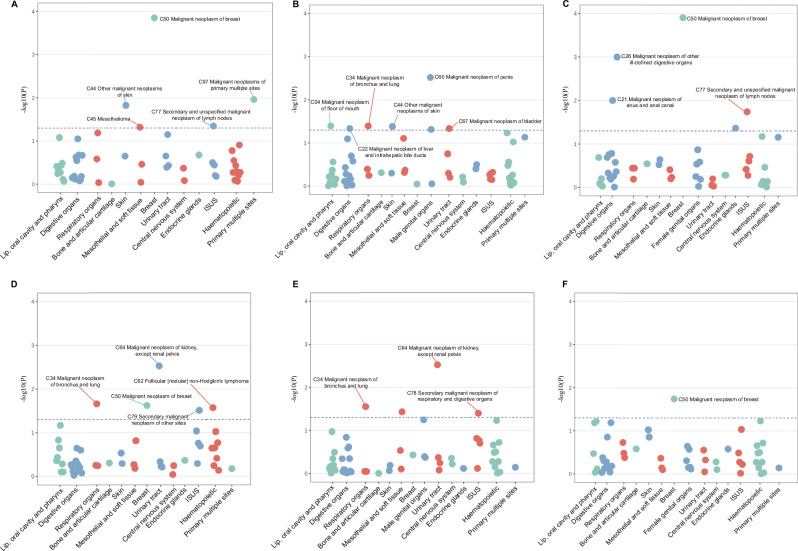
In observational phenome-wide association study (PheWAS), psoriasis was significantly associated with a higher risk of breast cancer (the tenth version of the International Classification of Diseases [ICD-10] code C50, hazard ratio [HR] 1.23, 95% CI: 1.11–1.38), NMSC (nonmelanoma skin cancer, C44, HR 1.11, 95% C:I 1.02–1.21), PMS (malignant neoplasms of independent primary multiple sites, C97, HR 1.43, 95% CI: 1.08–1.88), SLN (secondary and unspecified malignant neoplasm of lymph nodes, C77, HR 1.12, 95% CI: 1.00–1.25), and mesothelioma (C45, HR 1.54, 95% CI: 1.00–2.37) (Fig. 2A). In males, the association of psoriasis with NMSC remained consistent with the overall population. We further observed significantly higher risks of lung cancer (C34, HR 1.22, 95% CI: 1.01–1.48), penile cancer (C60, HR 3.02, 95% CI: 1.45–6.29), liver cancer (C22, HR 1.47, 95% CI: 1.01–2.14), bladder cancer (C67, HR 1.23, 95% CI: 1.00–1.51) and mouth cancer (C04, HR 2.94, 95% CI: 1.05–8.27) among patients with psoriasis (Fig. 2B). In females, risk of breast cancer was found to be significantly elevated (C50, HR 1.24, 95% CI: 1.11–1.38). We further observed a higher risk of developing OIDO (malignant neoplasm of other and ill-defined digestive organs, C26, HR 3.17, 95% CI: 1.63–6.18), SLN (C77, HR 1.20, 95% CI: 1.03–1.39) and anal canal cancer (C21, HR 2.27, 95% CI: 1.21–4.23) among patients with psoriasis (Fig. 2C). No association between psoriasis and risk of other site-specific cancers (not mentioned above) was observed in the total population or in each sex (Supplementary Tables 1–3). After excluding patients who had multiple cancer diagnoses, psoriasis is still significantly associated with a higher risk of lung cancer (C34, HR 1.56, 95% CI: 1.14–2.12) and breast cancer (C50, HR 1.28, 95% CI: 1.09–1.49) (Supplementary Tables 4–6).
Fig. 2. Manhattan plot of phenome-wide association studies.
A–C The adjusted p value (−log10(p value)) from multi-variable Cox regression models for site-specific cancers, according to psoriasis in all participants (A), male participants (B), and female participants (C). D–F The adjusted p value (−log10(p value)) from multi-variable logistic regression models for site-specific cancers, according to the PRSs for psoriasis in all participants (D), male participants (E) and female participants (F). Dots represent site-specific cancers, grouped into systemic categories denoted by different colors. The horizontal hatched line indicates the statistical significance (p < 0.05). All statistical tests are two-sided and unadjusted for multiple comparisons. ISUS ill-defined, secondary, unspecified sites. Source data are provided as a Source Data file.
PheWAS of psoriasis PRS
A higher PRS of psoriasis was associated with an increased risk of lung cancer (C34, odds ratio [OR] 1.01, 95% CI: 1.00–1.01), breast cancer (C50, OR 1.01, 95% CI: 1.00–1.01), kidney cancer (C64, OR 1.02, 95% CI: 1.01–1.03), SOS (secondary malignant neoplasm of other sites, C79, OR 1.01, 95% CI: 1.00–1.01) and follicular non-Hodgkin’s lymphoma (C82, OR 1.02, 95% CI: 1.00–1.04) (Fig. 2D and Supplementary Table 7). In males, a higher PRS of psoriasis was associated with an increased risk of lung cancer (C34, OR 1.01, 95% CI: 1.00–1.02), kidney cancer (C64, OR 1.02, 95% CI: 1.01–1.03) and SRD (secondary malignant neoplasm of respiratory and digestive organs, C78, OR 1.01, 95% CI: 1.00–1.01). In females, a higher PRS of psoriasis was associated with an increased risk of breast cancer in females (C50, OR 1.01, 95% CI: 1.00–1.01) (Fig. 2E, F and Supplementary Tables 8 and 9).
One-sample MR
In one-sample MR analysis, genetically predicted psoriasis was significantly associated with anal canal cancer (C21, OR 1.38, 95% CI: 1.01–1.87), lung cancer (C34, OR 1.12, 95% CI: 1.03–1.21), breast cancer (C50, OR 1.06, 95% CI: 1.01–1.12), kidney cancer (C64, OR 1.25, 95% CI: 1.09–1.42), SOS (C79, OR 1.10, 95% CI: 1.03–1.17) and follicular non-Hodgkin’s lymphoma (C82, OR 1.32, 95% CI: 1.07–1.64) in the total population (Table 3). In males, genetically predicted psoriasis was associated with a higher risk of lung cancer (C34, OR 1.17, 95% CI: 1.04–1.32), kidney cancer (C64, OR 1.34, 95% CI: 1.13–1.58), and SRD (C78, OR 1.10, 95% CI: 1.01–1.20). In females, genetically predicted psoriasis was significantly associated with a higher risk of anal canal cancer (C21, OR 1.61, 95% CI: 1.12–2.32), NMSC (C44, OR 1.07, 95% CI: 1.01–1.14), breast cancer (C50, OR 1.06, 95% CI: 1.02–1.11), SOS (C79, OR 1.12, 95% CI: 1.02–1.22) and follicular non-Hodgkin’s lymphoma (C82, OR 1.50, 95% CI: 1.13–1.98). No causal relationship was observed between psoriasis and mouth cancer, liver cancer, OIDO, mesothelioma, penile cancer, bladder cancer, SLN, or PMS.
Table 3.
One-sample MR estimates of psoriasis on the risk for site-specific cancers
| Cancer | Sex | Case/total | OR (95% CI) | p value |
|---|---|---|---|---|
| C04 malignant neoplasm of floor of mouth | Male and female | 83/458,587 | 1.17 (0.61, 2.23) | 0.648 |
| Male | 61/209,587 | 1.86 (0.86, 4.04) | 0.117 | |
| Female | 22/249,000 | 0.36 (0.11, 1.19) | 0.093 | |
| C21 malignant neoplasm of anus and anal canal | Male and female | 369/458,587 | 1.38 (1.01, 1.87) | 0.041 |
| Male | 124/209,587 | 0.97 (0.56, 1.68) | 0.909 | |
| Female | 245/249,000 | 1.61 (1.12, 2.32) | 0.010 | |
| C22 malignant neoplasm of liver and intrahepatic bile ducts | Male and female | 973/458,587 | 1.00 (0.83, 1.21) | 0.970 |
| Male | 597/209,587 | 1.10 (0.85, 1.41) | 0.474 | |
| Female | 376/249,000 | 0.88 (0.66, 1.18) | 0.406 | |
| C26 malignant neoplasm of other and ill-defined digestive organs | Male and female | 340/458,587 | 1.19 (0.86, 1.63) | 0.303 |
| Male | 195/209,587 | 1.10 (0.71, 1.71) | 0.688 | |
| Female | 145/249,000 | 1.30 (0.81, 2.09) | 0.276 | |
| C34 malignant neoplasm of bronchus and lung | Male and female | 5228/458,587 | 1.12 (1.03, 1.21) | 0.008 |
| Male | 2690/209,587 | 1.17 (1.04, 1.32) | 0.009 | |
| Female | 2538/249,000 | 1.07 (0.95, 1.19) | 0.278 | |
| C44 other malignant neoplasms of skin | Male and female | 23072/458,587 | 1.04 (0.99, 1.08) | 0.092 |
| Male | 12476/209,587 | 1.00 (0.95, 1.06) | 0.941 | |
| Female | 10596/249,000 | 1.07 (1.01, 1.14) | 0.017 | |
| C45 mesothelioma | Male and female | 496/458,587 | 0.92 (0.71, 1.21) | 0.578 |
| Male | 399/209,587 | 0.84 (0.62, 1.14) | 0.267 | |
| Female | 97/249,000 | 1.33 (0.75, 2.37) | 0.339 | |
| C50 malignant neoplasm of breast | Male and female | 16276/458,587 | 1.06 (1.01, 1.12) | 0.012 |
| Male | 132/209,587 | 0.79 (0.46, 1.36) | 0.402 | |
| female | 16144/249,000 | 1.06 (1.02, 1.11) | 0.010 | |
| C60 malignant neoplasm of penis | Male and female | NA | NA | NA |
| Male | 114/209,587 | 0.61 (0.34,1.08) | 0.090 | |
| Female | NA | NA | NA | |
| C64 malignant neoplasm of kidney, except renal pelvis | Male and female | 2102/458,587 | 1.25 (1.09, 1.42) | 0.001 |
| Male | 1348/209,587 | 1.34 (1.13, 1.58) | 0.001 | |
| Female | 754/249,000 | 1.11 (0.91, 1.37) | 0.312 | |
| C67 malignant neoplasm of bladder | Male and female | 4092/458,587 | 1.00 (0.91, 1.09) | 0.954 |
| Male | 3068/209,587 | 0.98 (0.88, 1.10) | 0.791 | |
| Female | 1024/249,000 | 1.03 (0.86, 1.24) | 0.735 | |
| C77 secondary and unspecified malignant neoplasm of lymph nodes | Male and female | 11433/458,587 | 1.04 (0.98, 1.10) | 0.182 |
| Male | 4587/209,587 | 1.06 (0.96, 1.16) | 0.257 | |
| Female | 6846/249,000 | 1.03 (0.96, 1.10) | 0.432 | |
| C78 secondary malignant neoplasm of respiratory and digestive organs | Male and female | 11621/458,587 | 1.05 (0.99, 1.11) | 0.086 |
| Male | 5442/209,587 | 1.10 (1.01, 1.20) | 0.028 | |
| Female | 6179/249,000 | 1.01 (0.94, 1.09) | 0.777 | |
| C79 secondary malignant neoplasm of other sites | Male and female | 9300/458,587 | 1.10 (1.03, 1.17) | 0.004 |
| Male | 4807/209,587 | 1.08 (0.98, 1.18) | 0.114 | |
| Female | 4493/249,000 | 1.12 (1.02, 1.22) | 0.012 | |
| C82 follicular [nodular] non-Hodgkin’s lymphoma | Male and female | 793/458,587 | 1.32 (1.07, 1.64) | 0.009 |
| Male | 379/209,587 | 1.14 (0.83, 1.57) | 0.412 | |
| Female | 414/249,000 | 1.50 (1.13, 1.98) | 0.005 | |
| C97 malignant neoplasms of primary multiple sites | Male and female | 1335/458,587 | 1.02 (0.87, 1.20) | 0.805 |
| Male | 786/209,587 | 1.02 (0.82, 1.27) | 0.870 | |
| Female | 549/249,000 | 1.03 (0.80, 1.31) | 0.845 |
All statistical tests are two-sided and unadjusted for multiple comparisons.
MR Mendelian randomization, OR odds ratio, CI confidence interval.
Two-sample MR analysis
We conducted a two-sample MR analysis as it is less prone to false-positive bias than a one-sample MR analysis. Psoriasis was causally associated with breast cancer (inverse variance weighted [IVW] OR 1.02, 95% CI: 1.01–1.03) and lung cancer (IVW OR 1.12, 95% CI: 1.02–1.22) (Table 4). Pleiotropy robust methods (weighted median and weighted mode) showed results similar to those of the IVW method (Table 4). No outliers were identified by the MR-pleiotropy residual sum and outlier (MR-PRESSO) method. We observed no evidence of heterogeneity in the association between psoriasis and site-specific cancers (Supplementary Table 10). We found no evidence of directional pleiotropy by using Mendelian randomization-Egger (MR-Egger) intercepts (p = 0.512 for breast cancer and p = 0.815 for lung cancer) or MR-PRESSO (p = 0.111 for breast cancer and p = 0.160 for lung cancer) (Supplementary Table 11). Most of the psoriasis susceptibility loci included as instrument variables in the two-sample MR analysis were not significantly associated with breast or lung cancer outcomes (Supplementary Table 12). These instrumental variables were also not associated with common risk factors for breast cancer or lung cancer, including body mass index (BMI), oestradiol, sex hormone-binding globulin (SHBG), testosterone, and smoking (Supplementary Table 13). In leave-one-out analysis, the main results remained robust when we removed individual single-nucleotide polymorphism (SNP) from the main two-sample MR analysis (Supplementary Fig. 1). We present additional visualizations of the causal effect of psoriasis on the risk of lung cancer and breast cancer in Supplementary Figs. 2–4.
Table 4.
Two-sample MR analysis for the causal relationship between psoriasis and site-specific cancers
| Outcome | Methods | OR (95% CI) | p value |
|---|---|---|---|
| Anal canal cancer | MR-Egger | 1.05 (0.61, 1.78) | 0.874 |
| Weighted median | 1.01 (0.65, 1.57) | 0.958 | |
| Inverse variance weighted | 1.00 (0.70, 1.44) | 0.989 | |
| Weighted mode | 1.03 (0.67, 1.58) | 0.902 | |
| Lung cancer | MR-Egger | 1.13 (0.99, 1.29) | 0.113 |
| Weighted median | 1.12 (1.03, 1.23) | 0.012 | |
| Inverse variance weighted | 1.12 (1.02, 1.22) | 0.013 | |
| Weighted mode | 1.13 (1.03, 1.24) | 0.034 | |
| Non-melanoma skin cancer | MR-Egger | 1.00 (1.00, 1.00) | 0.499 |
| Weighted median | 1.00 (1.00,1.00) | 0.820 | |
| Inverse variance weighted | 1.00 (1.00, 1.00) | 0.500 | |
| Weighted mode | 1.00 (1.00, 1.00) | 0.914 | |
| Breast cancer | MR-Egger | 1.02 (1.01, 1.04) | 0.024 |
| Weighted median | 1.02 (1.01, 1.03) | 0.008 | |
| Inverse variance weighted | 1.02 (1.01, 1.03) | 0.002 | |
| Weighted mode | 1.02 (1.01, 1.03) | 0.006 | |
| Kidney cancer | MR-Egger | 1.15 (0.93, 1.42) | 0.236 |
| Weighted median | 1.12 (0.99, 1.26) | 0.075 | |
| Inverse variance weighted | 1.06 (0.91, 1.23) | 0.426 | |
| Weighted mode | 1.12 (0.99, 1.27) | 0.110 | |
| Secondary malignant neoplasm of respiratory and digestive organs | MR-Egger | 1.00 (1.00, 1.00) | 0.572 |
| Weighted median | 1.00 (1.00, 1.00) | 0.669 | |
| Inverse variance weighted | 1.00 (1.00, 1.00) | 0.391 | |
| Weighted mode | 1.00 (1.00, 1.00) | 0.666 | |
| Secondary malignant neoplasm of other sites | MR-Egger | 1.00 (1.00, 1.00) | 0.044 |
| Weighted median | 1.00 (1.00, 1.00) | 0.033 | |
| Inverse variance weighted | 1.00 (1.00, 1.00) | 0.071 | |
| Weighted mode | 1.00 (1.00, 1.00) | 0.047 | |
| Follicular [nodular] non-Hodgkin’s lymphoma | MR-Egger | 1.36 (1.03,1.79) | 0.060 |
| Weighted median | 1.21 (0.97, 1.51) | 0.088 | |
| Inverse variance weighted | 1.09 (0.89, 1.33) | 0.406 | |
| Weighted mode | 1.22 (0.96, 1.54) | 0.129 |
All statistical tests are two-sided and unadjusted for multiple comparisons.
MR Mendelian randomization, OR odds ratio, CI confidence interval, MR-Egger Mendelian randomization-Egger.
Gene annotation for the molecular association between psoriasis and cancer
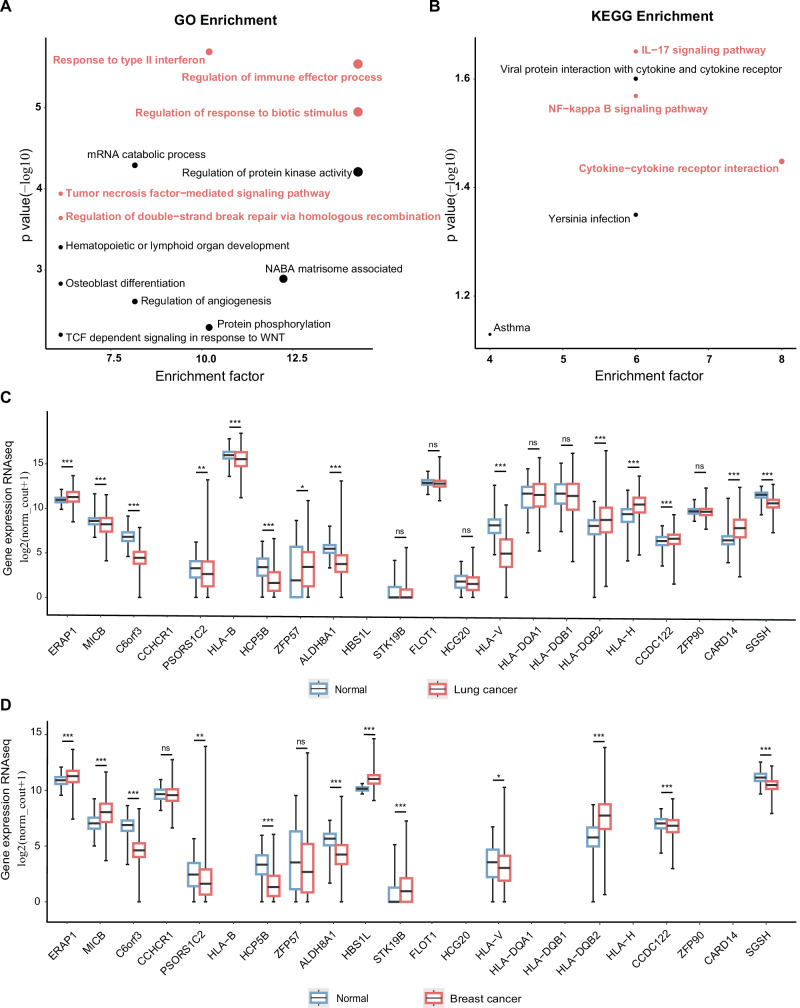
We performed gene annotation on the aforementioned 74 SNPs used in the study and subsequently conducted enrichment analysis on the related 50 genes. Gene Ontology (GO) enrichment analysis revealed that the psoriasis-related genes (41 genes in the output) were enriched in ‘Response to type II interferon’, ‘Regulation of immune effector process’, ‘Regulation of response to biotic stimulus’, ‘Tumor necrosis factor-mediated signaling pathway’, and ‘Regulation of double-strand break repair via homologous recombination’ (Fig. 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (eight genes in the output) indicated that ‘IL-17 signaling pathway’, ‘NF-kappa B signaling pathway’ and ‘cytokine‒cytokine receptor interaction’ are involved in psoriasis (Fig. 3B). cis-Expression quantitative trait locus (cis-eQTL) analysis revealed associations between psoriasis susceptibility loci and multiple genes in the normal lung (Supplementary Table 14) and breast (Supplementary Table 15) tissues. We then evaluated the gene expression of those eQTL-related genes in TCGA and GTEx. Among 287 normal tissues and 1013 lung cancer tissues, we observed a marked increase in expression levels of ERAP1, ZFP57, HLA-DQB2, HLA-H, CCDC122, and CARD14 in lung cancer compared to normal tissues from GTEx (Fig. 3C). Expression levels of MICB, C6orf3, PSORS1C2, HLA-B, HCP5B, ALDH8A1, HLA-V, and SGSH were significantly lower in lung cancer tissues than in normal tissues from GTEx (Fig. 3C). When comparing breast cancer tissues (n = 1099) to normal tissues (n = 179), we observed significantly higher expression levels of ERAP1, MICB, HBS1L, STK19B, and HLA-DQB2 and significantly lower expression levels of C6orf3, PSORS1C2, HCP5B, ALDH8A1, HLA-V, CCDC122, and SGSH (Fig. 3D).
Fig. 3. Psoriasis-related genetic variants on gene biological functions and expression patterns in tumors.
A, B Enriched GO terms and KEGG pathways in genes mapped from psoriasis-related SNPs. Scatter diagram illustrating the distribution of the −log10(p values) and the enrichment factor (the ratio of enriched genes to the total input genes). The red dots on the diagram indicate associations with the inflammatory response and DNA repair pathways. The size of the dots corresponds to the enrichment factor, with larger dots indicating higher enrichment levels. C The expression distribution of genes in lung cancer tissues (n = 1013) and normal tissues (n = 287). D The expression distribution of genes in breast cancer tissues (n = 1099) and normal tissues (n = 179). The abscissa represents different genes, and the ordinate represents the expression distribution of miRNA (log2(normalized count + 1)). Box plots display the median (center line), 25th and 75th percentiles (box bounds), and minimum and maximum values (whiskers). Different colors represent distinct groups. The statistical difference between the two groups was compared through Welch’s t-test. Exact p values for each gene are provided in Supplementary Tables 28 and 29. *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant. All statistical tests are two-sided and unadjusted for multiple comparisons. GO gene ontology, KEGG Kyoto Encyclopedia of Genes and Genomes. Source data are provided as a Source Data file.
Discussion
Based on unselected observational PheWAS analysis, we found psoriasis to be associated with higher risks of 12 types of cancer. In PRS PheWAS analysis, genetically predicted psoriasis was associated with breast cancer, kidney cancer, lung cancer, SOS, and follicular non-Hodgkin’s lymphoma. However, MR analysis only verified the causal relationship between psoriasis, lung cancer, and breast cancer, and gene annotation indicated that psoriasis-related genes (such as ERAP1) might be mediators linking psoriasis to lung or breast cancer. This is a systematic analysis of the association between psoriasis and site-specific cancers based on a large population sample. Our data confirmed that psoriasis is causally associated with lung cancer and breast cancer.
Using unselected PheWAS analysis, our observational data not only confirmed previous reports that psoriasis is associated with higher risks of site-specific cancers in the lung, kidney, liver, bladder, nonmelanoma skin, oral cavity, lymph nodes, and non-Hodgkin’s lymphoma3,4,7 but also revealed associations between psoriasis and cancers of the breast, penis, anal canal and mesothelioma. Furthermore, our genetic analysis (including PRS and MR analyses) confirmed the causal relationship between psoriasis and lung cancer/breast cancer. Previous meta-analyses have several shortcomings that may lead to biased results, including significant heterogeneity across the included studies, inconsistent diagnostic criteria for psoriasis, and insufficient adjustment for potential confounders linked to cancer. Hence, previous reports on the multiple sites of malignancy related to psoriasis3,4 should be interpreted cautiously. It should be noted that not all results from one-sample MR and two-sample MR were consistent. In one-sample MR analyses, we found causal associations between psoriasis and several cancers, including anal canal cancer, lung cancer, breast cancer, kidney cancer, SOS, and follicular non-Hodgkin’s lymphoma. In two-sample MR analyses, we only observed causal associations between psoriasis, lung cancer, and breast cancer. One-sample MR may generate a false positive result due to its weak instrument variables14. For two-sample MR without sample overlap, bias caused by weak instrument variables is towards the null, which rarely leads to false positive result14. Moreover, two-sample MR often refers to summary data from a larger sample size, which enhances the power to detect potential causal relationships. However, only having access to summary data limits the flexibility in conducting subgroup analysis. To cautiously and flexibly interpret the causal association between psoriasis and cancers, we used both one-sample MR and two-sample MR analyses. The consistent results from both methods strengthen the robustness of the findings on the association between psoriasis, lung cancer, and breast cancer.
Two studies have performed two-sample MR analysis to investigate the association between psoriasis and lung cancer, but the results were inconsistent. Luo et al12., based on data from UK Biobank (3871 cases and 337,159 total), reported that psoriasis was causally associated with a 6% increased risk of lung cancer. In contrast, Wang et al11. showed no significant causal relationship between psoriasis and lung cancer, either with regard to subtypes of squamous cell lung cancer or pulmonary adenocarcinoma. Such conflicting evidence may be attributed to the insufficiencies in methodology in MR analysis. Wang et al11. used genetic variants with obvious heterogeneity, and Luo et al12. selected genetic datasets for psoriasis and lung cancer both from the same population (UK Biobank), which may influence the reliability and accuracy of results15. Compared to these two meta-analyses, we were able to identify an association between psoriasis and site-specific cancers with a relatively low degree of cohort heterogeneity. In our study, measurement errors, confounding factors, and false-positive bias were gradually controlled from observational PheWAS, PRS PheWAS, and one-sample analysis to two-sample MR analysis14,16,17. Although the genetically increased risks are not as high as observational results, our data warrant clinical attention to lung cancer and breast cancer among patients with psoriasis.
Psoriasis is a chronic inflammatory disease, and the link between chronic inflammation and cancers has been reported in many studies18,19, which is similar to other immune-mediated inflammatory diseases, such as inflammatory bowel diseases, rheumatoid arthritis, and sarcoidosis20–22. On the other hand, psoriasis is also closely associated with immune dysfunction, and the proteins encoded by these susceptibility genes play important roles in immune and signaling pathways, especially interferon, tumor necrosis factor, the NF-kB pathway, and the IL-23/Th17 axis6,23. These upregulated cytokines and activated pathways in psoriasis are also involved in the development of breast cancer. TNF-ɑ is reported to promote the growth and metastasis of breast cancer by activating the NF-kB pathway24. Excessive infiltration of Th-17 cells in the breast tumor microenvironment releases large amounts of IL17A and promotes the development of breast cancer25. By integrating public datasets of eQTL, TCGA, and GTEx, our gene annotation revealed a potential molecular association between psoriasis and lung or breast cancer. Among them, we fortuitously found that ERAP1, an endoplasmic reticulum aminopeptidase, may be one of the promising candidates. On one hand, several GWASs and exome sequencing identified ERAP1 as an important susceptibility locus harbored gene for psoriasis26–28. On the other hand, patients with breast cancer or lung cancer often have abnormal expression levels of ERAP129, which may lead to abnormal antigen processing, and subsequently facilitate tumor immune escape and malignant progression30. However, the exact role of ERAP1 in the link between psoriasis and lung/breast cancer remains to be studied. The possible mechanisms of cancer development in psoriasis patients are summarized in Supplementary Fig. 5.
The strength of our study is its comprehensive study design from PheWAS to MR analyses. Large-scale sample sizes and genomic data from UK Biobank and public datasets ensured that the causal relationship between psoriasis and breast/lung cancer was robust. There are also several limitations of the current study. First, the populations included were exclusively of European ancestry, and our findings may not be generalizable to other populations, including those of Asian or African ancestry. Second, the level of evidence provided by an MR analysis is second to randomized controlled trials. Future interventional studies that confirm a causal relationship between psoriasis and site-specific cancers will provide more compelling evidence.
In conclusion, psoriasis is causally associated with lung cancer and breast cancer. However, other previously reported psoriasis-related cancers may not be major concerns of psoriasis patients.
Methods
Ethics
This study was approved under UK Biobank Project 103654. The National Information Governance Board for Health and Social Care and the National Health Service North West Multicenter Research Ethics Committee (reference 13/NW/0382) approved the UK Biobank ethical application. All participants provided informed consent through electronic signatures at the first assessment. This study was conducted in accordance with the principles of the Declaration of Helsinki.
UK Biobank
UK Biobank is a prospective population-based cohort involving more than 500,000 participants aged between 40 years and 70 years from the United Kingdom31. Information on psoriasis, cancer status (prevalent and incident cases), relevant confounding factors, and genetic data are available in the UK Biobank. We set the cohort baseline (2006–2010) as our analysis baseline, and the end of the follow-up period was set as 2019.
Study setting
In the current study, we conducted five analyses to investigate the causal relationship between psoriasis and site-specific cancers. Analysis 1 was designed as a prospective cohort study, and we used observational data from the UK Biobank. Analysis 2 was a PRS study, and we used GWAS summary data from public datasets and genetic data from the UK Biobank. Analysis 3 was a one-sample MR analysis, and we used genotype and phenotypic data from the UK Biobank. Analysis 4 was a two-sample MR analysis, and we used GWAS summary data from public datasets. For analysis 5, eQTL and differential expression analyses between tumor samples and normal samples were evaluated based on GTEx and TCGA data.
We used the PheWAS method to explore the association between all site-specific cancers and psoriasis (analysis 1) or the PRS of psoriasis (analysis 2). In analysis 3, we performed a one-sample MR analysis including the site-specific cancer types with p values < 0.05 in PheWAS of analysis 1 or 2. Two-sample MR analysis (analysis 4) was performed if the p value of the site-specific cancer type was <0.05 in one-sample MR analysis. We performed analysis 5 for site-specific cancer types if their p value < 0.05 in two-sample MR analysis.
Study population
In the UK Biobank, we included unrelated individuals of European ancestry who had a medical history and genetic data that passed the quality control steps described previously32. We excluded participants if they were diagnosed with other autoimmune diseases that might potentially influence the association between psoriasis and cancers, including ulcerative colitis, Crohn’s disease, multiple sclerosis, lupus erythematosus, systemic lupus erythematosus, dermatomyositis, pemphigus, pemphigoid, Behcet’s disease, rheumatoid arthritis, necrotizing vasculopathy and other systemic connective tissue disorders. We excluded participants who had been diagnosed with any type of cancer at baseline or before the diagnosis of psoriasis in Analysis 1 and Analysis 2. For PRS and MR analyses, we further excluded participants who were not white Europeans and those who lacked genetic data. We excluded cancer types with <50 cases in the total population or <25 cases in subgroups of women and men. We also excluded the types of cancers with cases less than 6 in the psoriasis patients.
Diagnoses of psoriasis and cancers
Participants from the UK Biobank were registered with the National Health Service (NHS) and agreed to link their medical records. The UK Biobank tracks participants’ electronic medical or health-related records, including hospital inpatient admissions and deaths. All disease types (including psoriasis and site-specific cancers) are recorded and analyzed according to ICD-10. Individual codes of psoriasis and site-specific cancers are summarized in Supplementary Table 16.
Covariates adjusted for in multivariable regression analysis
Covariates were selected based on their potential associations with both psoriasis and cancer risk (Supplementary Table 17). Sex from UK Biobank data was primarily based on self-reporting. Smoking status was categorized into three levels never, previous, and current smoking. Alcohol intake frequency was categorized into six levels, including daily or almost daily, three or four times a week, once or twice a week, one to three times a month, special occasions only, and never. Physical activity was categorized into three levels, specifically low intensity, moderate intensity, and high intensity. Medication use (including glucocorticoids, methotrexate, and cyclosporine) was defined as whether the corresponding medication was used or not.
We further included additional covariates for selected cancer outcomes. For skin cancer (C43–C44), we additionally adjusted for sun exposure33. Sun exposure was defined as the time spent outdoors in summer and in winter. For female reproductive system malignancies and breast cancer (C50–C58), we further adjusted for age at menarche, number of live births, and menopausal status34–37. For colorectal cancer (C18–C20), we further adjusted for red meat and fiber intake levels38,39. Red meat intake levels were defined as the median data of beef, lamb/mutton, and pork intake. Fiber intake levels were defined as the median data of cooked vegetable and salad/raw vegetable intake.
PRS of psoriasis
We extracted the genetic instruments from published GWAS summary data of European ancestry6 (Supplementary Data 1, GWAS Catalog inquiry code: GCST005527). We selected SNPs based on the following criteria: (1) SNPs significantly associated with psoriasis (p ≤ 5 × 10−8); (2) SNPs with minor allele frequencies >0.01; and (3) no insertion or deletion or ambiguous SNPs. Clump-based linkage disequilibrium pruning was performed with a physical distance threshold of 250 kb and an LD threshold (R2) < 0.001. We identified a total of 74 SNPs and present their information in Supplementary Table 18. PRS was calculated using the PRSice software40, which derives the PRS as a weighted sum of the risk alleles carried by an individual at each SNP and multiplied by the corresponding effect size obtained from the primary GWAS.
Data information of MR analysis
We used the PRS of psoriasis to conduct a one-sample MR analysis using UK Biobank data. For a two-sample MR analysis, all public GWAS data information was shown in (Supplementary Data 1).
We obtained summary-level data from public GWAS sources from FinnGen (https://www.finngen.fi/fi) via the IEU-OpenGWAS project (inquiry code: fnn-b-L12_PSORIASIS). Individuals from the FinnGen project were genotyped using Illumina and Affymetrix chip arrays. A total of 4510 psoriasis cases were identified by ICD-10 code (L40) and 212,242 healthy controls were included. GWAS data for carcinoma in situ of the anus are derived from 107 European ancestry cases and 456,241 European ancestry controls (GWAS Catalog inquiry code: GCST90043915)41. GWAS data for breast cancer are derived from 76,192 female cases and 63,082 controls of European ancestry (IEU-OpenGWAS project inquiry code: ebi-a-GCST004988), and most SNPs were reported in a previous GWAS42. We obtained GWAS summary data for lung and kidney cancer from the GWAS Catalog (inquiry code: GCST90011812 and GCST90011818), which included 2485 cases of lung cancer, 1338 cases of kidney cancer (identified by ICD-9 or ICD-10 codes in the UK Biobank, and ICD-O-3 codes in Kaiser Permanente Genetic Epidemiology Research on Adult Health and Aging cohorts) and 410,350 healthy controls43. We obtained GWAS summary data of NMSC from the IEU-OpenGWAS project (inquiry code: ieu-b-4959), with sequencing data sourced from UK Biobank, which comprised 23,694 cases and 372,016 control individuals. GWAS summary data of secondary malignant neoplasm of respiratory and digestive organs, secondary malignant neoplasm of other sites, and follicular non-Hodgkin’s lymphoma was obtained from the IEU-OpenGWAS project (inquiry code: ukb-d-C78 and ukb-d-C79) and GWAS Catalog (inquiry code: GCST90042739).
Annotation and enrichment analysis
We annotated the 74 SNPs included in the PRS analysis by using gProfiler (http://biit.cs.ut.ee/gprofiler/)44. The 74 SNPs were mapped to 50 genes (Supplementary Table 19). To assess the potential biological functions of the mapped genes, GO and KEGG pathway enrichment analyses were performed in Metascape45 and DAVID46, respectively.
eQTL and TCGA
We used the GTEx project to conduct a single-tissue eQTL analysis. The GTEx project is supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. We acquired detailed data from the GTEx portal on 08/01/2023. We obtained gene expression RNA-seq data and clinical data from TCGA and GTEx RNA datasets, as accessed via the UCSC XENA database (http://xena.ucsc.edu/)47. TCGA data has been co-analyzed with GTEx data using the UCSC bioinformatic pipeline48. In eQTL analysis, cis-eQTLs were defined if the SNP was in a +/− 1 Mb cis window around the transcription start site. Significance was determined using a Q value threshold, and protein-coding genes with a p value < 5 × 10−10 were selected for subsequent analysis. For TCGA analysis, all data were transformed by log2(x + 1), where x represents the value normalized by the upper quartile method. We compared gene expression in normal tissues from GTEx with various cancer types from TCGA, with Welch’s t-test to test any difference.
Statistical analysis
Categorical variables are presented as percentages, and normally distributed continuous variables are presented as the mean ± SD. We imputed missing data of covariates by multivariate imputation with a chained equations algorithm. A p value < 0.05 was considered statistically significant. Data were analyzed with the use of R software, version 4.0.3, with the following packages: data.table, Rcpp, RcppParrallel, future, survival, ggplot2, TwoSampleMR and MR-PRESSO. We conducted all statistical analyses on the supercomputer platform (inspur M5).
PheWAS for associations between psoriasis and incident site-specific cancers (the observational PheWAS) was estimated by Cox regression. In the main analysis, we adjusted for potential confounders including age, sex, race, BMI, smoking status, alcohol intake frequency, physical activity, and medication use. PheWAS for associations between the PRS of psoriasis and site-specific cancer (the PRS PheWAS) was estimated by logistic regression while adjusting for age, sex, BMI, smoking status, alcohol intake frequency, physical activity and top ten principal components and medication use.
In one-sample MR analysis, we implemented a Wald ratio method to estimate the causal effect of psoriasis on site-specific cancers. We determined the association between the PRS and psoriasis (βX|G), as well as the association between the PRS and site-specific cancers (βY|G), by using logistic regression adjusted for age, sex, and the top ten principal components of the genetic information. The ratio estimate of the causal effect is calculated as βIV = βY|G/βX|G. The detailed calculation methods for the standard error of ratio estimates were previously described49, which can be expressed as: se(βIV) . We further conducted subgroup analyses by sex with the same methodology.
We conducted a two-sample MR analysis by using the ‘TwoSampleMR’ package in R to extract instrumental variables with p ≤ 5 × 10−8, and we then clumped these SNPs on the basis of the European ancestry reference panel with an R2 < 0.001 within a 10,000-kb window. The SNPs used in the two-sample MR analysis are presented in Supplementary Table 20 to Supplementary Table 27. We used four distinct methods to evaluate causal effects, namely, MR-Egger, weighted median, IVW, and weighted mode. Each method makes different assumptions regarding the effectiveness of instrumental variables, and the IVW method is more reliable when there is less potential violation in MR assumptions. For sensitivity analysis, we used the MR-PRESSO method to remove the potential influence of outliers50. We used Cochrane’s Q-test for IVW analyses and Rücker’s Q-test for MR-Egger analyses to assess heterogeneity. We used the MR-Egger intercept method and MR-PRESSO method to test the horizontal pleiotropy of instrumental variables. We performed leave-one-out analyses using MR-Egger or fixed effects inverse variance methods to investigate whether the overall results were driven by any individual variant51. We have further examined the association between SNPs used in two-sample MR analysis and cancers or known risk factors for lung cancer and breast cancer, including smoking, hormone levels (oestradiol, SHBG, and testosterone), and obesity (BMI) by using logistic regression adjusted for age, sex, and the top ten principal components of the genetic information.
In our study, R.L. and W.L. independently repeated all analyses once, and the results were further validated by X.C. They all successfully obtained the same conclusions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
The computing work in this paper was partly supported by the Supercomputing Center of Chongqing Medical University. The authors thank the participants and the staff of the UK Biobank. This research has been conducted using the UK Biobank Resource under Application Number 103654. Thanks to Home for Researchers (www.home-for-researchers.com). This work was supported by the National Natural Science Foundation of China (A.C.: 81874238 and J.H.: 82270878) and the Natural Science Foundation of Chongqing (A.C.: CSTB2024NSCQ-LZX0068).
Author contributions
J.H. and A.C. designed the study. Data analysis was done by R.L., W.L., and P.W. Data interpretation was done by R.L., W.L., X.C., Q.Z., P.W., J.H., S.Y., and A.C. Data was validated by QLZ and XJC. RLL, XJC, and JBH wrote the first draft of the manuscript. S.Y. and A.C. reviewed and edited the first draft of the manuscript. This report was approved for publication by all co-authors. The authors affirm the integrity and precision of the data and analyses. They had complete access to the study’s data and held ultimate responsibility for the decision to submit it for publication.
Peer review
Peer review information
Nature Communications thanks Han Wang, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
The current analysis was approved by the UK Biobank in August 2020 with the ID 103654. The raw UK Biobank data are protected and are not available due to data privacy laws. The UK Biobank data are available through a standard application protocol (https://www.ukbiobank.ac.uk/register-apply/). Summary-level data from publicly available GWAS can be obtained through the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/) and the GWAS Catalog (https://www.ebi.ac.uk/gwas/). The specific GWAS summary datasets used in PRS calculation and Two-sample MR analyses are listed in Supplementary Data 1. Gene expression RNA-seq data and clinical data from TCGA and GTEx RNA datasets can be accessed via the UCSC XENA database (dataset ID: TcgaTargetGtex_RSEM_Hugo_norm_count [https://xenabrowser.net/datapages/?dataset=TcgaTargetGtex_RSEM_Hugo_norm_count&host=https%3A%2F%2Ftoil.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443]). The cis-eQTL data can be obtained through the following link: https://www.gtexportal.org/home/snp/[rs_number], where [rs_number] should be replaced with the rs number of each SNP listed in Supplementary Table 18. Source data are provided with this paper.
Code availability
Customed codes for the analyses are available at https://github.com/Luowenjin826/psoriasis_study/ (10.5281/zenodo.12783539).
Competing interests
The authors have disclosed no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ruolin Li, Wenjin Luo.
Contributor Information
Jinbo Hu, Email: hujinbo@cqmu.edu.cn.
Aijun Chen, Email: chenaijun@hospital.cqmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51824-6.
References
- 1.Lowes, M. A., Suárez-Fariñas, M. & Krueger, J. G. Immunology of psoriasis. Annu. Rev. Immunol.32, 227–255 (2014). 10.1146/annurev-immunol-032713-120225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colaco, K. et al. Trends in mortality and cause-specific mortality among patients with psoriasis and psoriatic arthritis in Ontario, Canada. J. Am. Acad. Dermatol.84, 1302–1309 (2021). 10.1016/j.jaad.2020.10.031 [DOI] [PubMed] [Google Scholar]
- 3.Trafford, A. M., Parisi, R., Kontopantelis, E., Griffiths, C. E. M. & Ashcroft, D. M. Association of psoriasis with the risk of developing or dying of cancer: a systematic review and meta-analysis. JAMA Dermatol.155, 1390–1403 (2019). 10.1001/jamadermatol.2019.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaengebjerg, S., Skov, L., Egeberg, A. & Loft, N. D. Prevalence, incidence, and risk of cancer in patients with psoriasis and psoriatic arthritis: a systematic review and meta-analysis. JAMA Dermatol.156, 421–429 (2020). 10.1001/jamadermatol.2020.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lønnberg, A. S. et al. Heritability of psoriasis in a large twin sample. Br. J. Dermatol.169, 412–416 (2013). 10.1111/bjd.12375 [DOI] [PubMed] [Google Scholar]
- 6.Tsoi, L. C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet.44, 1341–1348 (2012). 10.1038/ng.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balda, A. et al. Psoriasis and skin cancer—Is there a link? Int. Immunopharmacol.121, 110464 (2023). 10.1016/j.intimp.2023.110464 [DOI] [PubMed] [Google Scholar]
- 8.Srivastava, A. K. et al. Insights into interplay of immunopathophysiological events and molecular mechanistic cascades in psoriasis and its associated comorbidities. J. Autoimmun.118, 102614 (2021). 10.1016/j.jaut.2021.102614 [DOI] [PubMed] [Google Scholar]
- 9.Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ375, n2233 (2021). 10.1136/bmj.n2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konuma, T. & Okada, Y. Statistical genetics and polygenic risk score for precision medicine. Inflamm. Regen.41, 18 (2021). 10.1186/s41232-021-00172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, X. et al. Association between psoriasis and lung cancer: two-sample Mendelian randomization analyses. BMC Pulm. Med.23, 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, Q. et al. Psoriasis may increase the risk of lung cancer: a two-sample Mendelian randomization study. J. Eur. Acad. Dermatol. Venereol.36, 2113–2119 (2022). 10.1111/jdv.18437 [DOI] [PubMed] [Google Scholar]
- 13.Yin, Q. et al. Mendelian randomization analyses of chronic immune-mediated diseases, circulating inflammatory biomarkers, and cytokines in relation to liver cancer. Cancers15, 2930 (2023). 10.3390/cancers15112930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemani, G., Bowden, J. & Davey Smith, G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet.27, R195–R208 (2018). 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess, S. et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res.4, 186 (2019). 10.12688/wellcomeopenres.15555.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawlor, D. A., Harbord, R. M., Sterne, J. A. C., Timpson, N. & Davey Smith, G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med.27, 1133–1163 (2008). 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 17.Richmond, R. C. et al. Investigating causal relations between sleep traits and risk of breast cancer in women: mendelian randomisation study. BMJ365, l2327 (2019). 10.1136/bmj.l2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elinav, E. et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer13, 759–771 (2013). 10.1038/nrc3611 [DOI] [PubMed] [Google Scholar]
- 19.Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity51, 27–41 (2019). 10.1016/j.immuni.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen, N. et al. Risk of extra-intestinal cancer in inflammatory bowel disease: meta-analysis of population-based cohort studies. Am. J. Gastroenterol.105, 1480–1487 (2010). 10.1038/ajg.2009.760 [DOI] [PubMed] [Google Scholar]
- 21.Smitten, A. L., Simon, T. A., Hochberg, M. C. & Suissa, S. A meta-analysis of the incidence of malignancy in adult patients with rheumatoid arthritis. Arthritis Res. Ther.10, 1–8 (2008). 10.1186/ar2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boffetta, P., Rabkin, C. S. & Gridley, G. A cohort study of cancer among sarcoidosis patients. Int. J. Cancer124, 2697–2700 (2009). 10.1002/ijc.24261 [DOI] [PubMed] [Google Scholar]
- 23.Yin, X. et al. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat. Commun.6, 6916 (2015). 10.1038/ncomms7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, Y. & Zhou, B. P. TNF-α/NFκ-B/snail pathway in cancer cell migration and invasion. Br. J. Cancer102, 639–644 (2010). 10.1038/sj.bjc.6605530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffelt, S. B. et al. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature522, 345–348 (2015). 10.1038/nature14282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet.42, 985–990 (2010). 10.1038/ng.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng, Y. et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat. Commun.5, 4331 (2014). 10.1038/ncomms5331 [DOI] [PubMed] [Google Scholar]
- 28.Tang, H. et al. A large-scale screen for coding variants predisposing to psoriasis. Nat. Genet.46, 45–50 (2014). 10.1038/ng.2827 [DOI] [PubMed] [Google Scholar]
- 29.Compagnone, M., Cifaldi, L. & Fruci, D. Regulation of ERAP1 and ERAP2 genes and their disfunction in human cancer. Hum. Immunol.80, 318–324 (2019). 10.1016/j.humimm.2019.02.014 [DOI] [PubMed] [Google Scholar]
- 30.Stratikos, E., Stamogiannos, A., Zervoudi, E. & Fruci, D. A role for naturally occurring alleles of endoplasmic reticulum aminopeptidases in tumor immunity and cancer pre-disposition. Front. Oncol.4, 363 (2014). 10.3389/fonc.2014.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conroy, M. C. et al. UK Biobank: a globally important resource for cancer research. Br. J. Cancer128, 519–527 (2023). 10.1038/s41416-022-02053-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes, A., Albers, P. K., Dendrou, C. A., Fugger, L. & McVean, G. Identifying cross-disease components of genetic risk across hospital data in the UK Biobank. Nat. Genet.52, 126–134 (2020). 10.1038/s41588-019-0550-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasan, N. et al. Skin cancer: understanding the journey of transformation from conventional to advanced treatment approaches. Mol. Cancer22, 168 (2023). 10.1186/s12943-023-01854-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong, T.-T., Wu, Q.-J., Vogtmann, E., Lin, B. & Wang, Y.-L. Age at menarche and risk of ovarian cancer: a meta-analysis of epidemiological studies. Int. J. cancer132, 2894–2900 (2013). 10.1002/ijc.27952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong, T.-T., Wang, Y.-L. & Ma, X.-X. Age at menarche and endometrial cancer risk: a dose-response meta-analysis of prospective studies. Sci. Rep.5, 14051 (2015). 10.1038/srep14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Urso, S. et al. Mendelian randomization analysis of factors related to ovulation and reproductive function and endometrial cancer risk. BMC Med.20, 419 (2022). 10.1186/s12916-022-02585-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menarche menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol.13, 1141–1151 (2012). 10.1016/S1470-2045(12)70425-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farvid, M. S. et al. Consumption of red meat and processed meat and cancer incidence: a systematic review and meta-analysis of prospective studies. Eur. J. Epidemiol.36, 937–951 (2021). 10.1007/s10654-021-00741-9 [DOI] [PubMed] [Google Scholar]
- 39.Celiberto, F. et al. Fibres and colorectal cancer: clinical and molecular evidence. Int. J. Mol. Sci.24, 13501 (2023). 10.3390/ijms241713501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience8, giz082 (2019). 10.1093/gigascience/giz082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang, L., Zheng, Z., Fang, H. & Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat. Genet.53, 1616–1621 (2021). 10.1038/s41588-021-00954-4 [DOI] [PubMed] [Google Scholar]
- 42.Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature551, 92–94 (2017). 10.1038/nature24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rashkin, S. R. et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun.11, 4423 (2020). 10.1038/s41467-020-18246-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolberg, L. et al. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res.51, 207–212 (2023). 10.1093/nar/gkad347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.10, 1523 (2019). 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.4, 44–57 (2009). 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 47.Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol.38, 675–678 (2020). 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vivian, J. et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol.35, 314–316 (2017). 10.1038/nbt.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birney, E. Mendelian randomization. Cold Spring Harb. Perspect. Med.12, a041302 (2022). [DOI] [PMC free article] [PubMed]
- 50.Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet.50, 693–698 (2018). 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess, S., Bowden, J., Fall, T., Ingelsson, E. & Thompson, S. G. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology28, 30–42 (2017). 10.1097/EDE.0000000000000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
The current analysis was approved by the UK Biobank in August 2020 with the ID 103654. The raw UK Biobank data are protected and are not available due to data privacy laws. The UK Biobank data are available through a standard application protocol (https://www.ukbiobank.ac.uk/register-apply/). Summary-level data from publicly available GWAS can be obtained through the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/) and the GWAS Catalog (https://www.ebi.ac.uk/gwas/). The specific GWAS summary datasets used in PRS calculation and Two-sample MR analyses are listed in Supplementary Data 1. Gene expression RNA-seq data and clinical data from TCGA and GTEx RNA datasets can be accessed via the UCSC XENA database (dataset ID: TcgaTargetGtex_RSEM_Hugo_norm_count [https://xenabrowser.net/datapages/?dataset=TcgaTargetGtex_RSEM_Hugo_norm_count&host=https%3A%2F%2Ftoil.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443]). The cis-eQTL data can be obtained through the following link: https://www.gtexportal.org/home/snp/[rs_number], where [rs_number] should be replaced with the rs number of each SNP listed in Supplementary Table 18. Source data are provided with this paper.
Customed codes for the analyses are available at https://github.com/Luowenjin826/psoriasis_study/ (10.5281/zenodo.12783539).