Abstract
Several retroviruses have recently been shown to promote translation of their gag gene products by internal ribosome entry. In this report, we show that mRNAs containing the human immunodeficiency virus type 1 (HIV-1) gag open reading frame (ORF) exhibit internal ribosome entry site (IRES) activity that can promote translational initiation of Pr55gag. Remarkably, this IRES activity is driven by sequences within the gag ORF itself and is not dependent on the native gag 5′-untranslated region (UTR). This cap-independent mechanism for Pr55gag translation may help explain the high levels of translation of this protein in the face of major RNA structural barriers to scanning ribosomes found in the gag 5′ UTR. The gag IRES activity described here also drives translation of a novel 40-kDa Gag isoform through translational initiation at an internal AUG codon found near the amino terminus of the Pr55gag capsid domain. Our findings suggest that this low-abundance Gag isoform may be important for wild-type replication of HIV-1 in cultured cells. The activities of the HIV-1 gag IRES may be an important feature of the HIV-1 life cycle and could serve as a novel target for antiretroviral therapeutic strategies.
Like other complex retroviruses, human immunodeficiency virus type 1 (HIV-1) employs a variety of mechanisms to express numerous proteins from a single-genomic-length RNA transcript. One such mechanism is the regulated splicing of the primary transcript into more than 30 distinct mRNA species (65, 74–76). Other mechanisms for the production of different polypeptides from mature viral mRNA species rely on translational events that are atypical of normal host protein translation.
The initiation of translation of most eukaryotic mRNAs is thought to occur by a process involving ribosome scanning. In this model, the 40S ribosomal subunit binds to the 5′ end of a capped mRNA (80, 81), commences scanning toward the 3′ end, and initiates translation upon encountering an AUG codon in suitable context (often referred to as a “Kozak” context) (47, 49, 50). The 60S ribosomal subunit is then recruited, and translation of a single polypeptide begins.
Translation of some HIV-1 mRNAs can depart from this basic model in several ways. Translation of the pol open reading frame (ORF) requires a −1 frameshift at a “slippery” sequence within the gag ORF. This frameshift allows roughly 5 to 10% of translating ribosomes to bypass the normal gag stop codon, enabling translation of a Gag-Pol precursor protein (reviewed in reference 32). Other HIV-1 gene products, including Env and Nef, have been shown to be translated by “leaky” scanning of ribosomes past the vpu and rev AUGs, respectively (74, 75, 77).
Another alternative mechanism for translational initiation is driven by RNA structural elements referred to as internal ribosome entry sites (or segments) (IRESs). IRESs are thought to promote initiation of translation by directly binding ribosomes, with participation of other host cell factors, in a manner independent of the mRNA cap or of scanning through upstream sequences. Since the initial discovery of an IRES in the 5′ untranslated region (UTR) of picornaviruses (43, 66), IRES activity has been described in a wide variety of viral and cellular mRNAs (reviewed in references 41 and 71). Details of the molecular mechanisms through which these RNA structures promote the internal entry of ribosomes are the subject of much current interest.
IRESs have been described in several retroviruses (4, 5, 19, 63, 91), leading to speculation that HIV-1 mRNAs might exhibit IRES activity. The data presented in this report indicate that mRNAs containing the HIV-1 gag ORF exhibit IRES activity that can drive production of both the familiar Gag precursor protein, Pr55gag, and a novel 40-kDa N-terminally truncated isoform of Gag (p40). We demonstrate that this novel Gag isoform is present in cultured cells infected with HIV-1. Our findings suggest that expression of p40 may be required for wild-type viral growth kinetics.
Unlike the majority of other IRESs, which have been found partially or wholly within the 5′ UTRs of mRNAs, the IRES activity reported here is driven by sequences entirely within the gag ORF. This surprising arrangement is similar to the IRES of Moloney murine leukemia virus (MMLV) (91). The observation of IRES activity in a variety of retroviral species suggests that such activity may be an important feature of retroviral biology. The HIV-1 gag IRES described here might thus serve as a novel target for viral attenuation or antiretroviral therapeutics.
MATERIALS AND METHODS
Vaccinia virus expression vectors.
Vaccinia virus vvB2 was generated by ligation of a PCR product of the HXB2 gag gene from plasmid pHXB2neo (kindly provided by Xiao Fang Yu, Johns Hopkins University) into the KpnI-NheI sites of vector pSC11MCS1 (32). This placed gag ATG1 8 nucleotides (nt) downstream of an ATG codon in strong Kozak context (AGT ACC ATG G) found in the multicloning site of this vector, resulting in somewhat diminished Pr55gag expression compared to unobstructed Pr55gag. Virus vvENVGAG was generated by first transferring the XmaI-XbaI fragment of plasmid vector pENVGAG (see below) into pSC11MCS1 XmaI-NheI vector species. The KpnI-EcoNI env fragment of the resulting plasmid was replaced with the KpnI-EcoNI env fragment of plasmid pPE15 (kindly provided by Patricia Earl, National Institutes of Health) in order to remove the two vaccinia virus early transcription termination signals found in this portion of env (20). The resulting construct, pvEG, encodes a hybrid env ORF derived from two closely related env sequences, HIV-1 LAI (pENVGAG) and HIV-1 BH8 (pPE15). Recombinant vaccinia viruses were generated by recombination into strain vVWR-LVar and subsequent plaque purification according to a method described previously (21).
Plasmid expression vectors.
Plasmid construction was accomplished by standard methods. All PCR inserts and splice junctions were verified by sequencing. The vectors were based on the Pr55gag gene of HIV-1 isolate HXB2 (HIV Sequence Database, 1997 [http://hiv-web.lanl.gov/], Los Alamos National Laboratory Theoretical Biology and Biophysics, Los Alamos, N.Mex.). The Pr55gag gene was silently modified to introduce an XhoI site near its 5′ end (G9→T, A10→C) and to attenuate the slippery sequence which promotes ribosomal frameshifting to the pol reading frame as described earlier (70). The Kozak context surrounding the start codon Pr55gag in these vectors is CACACCATG G. The HIV-1 env gene was derived from provirus pLAI.
Vectors pEMCV-CAT, pCAT, pCAT-GAG, pENVGAG, pENV, pGAG, pENV-ΔUTR-GAG, pENV-Ψ-GAG, pΔENV-GAG, pENV-ΔΔGAG, and pENVCAT were all expressed from the backbone of pCI (Promega). Vectors pLACZ, pIGAG, pΔ40, pΔ55, pFS, and pFSΔ40 were expressed from the backbone of plasmid pCMV6C.p55gag-pre, which was kindly provided by Chiron Corp. (Emeryville, Calif.). All vectors contain cytomegalovirus (CMV) promoter and an intron in the 5′ UTR to boost expression. All of the vectors also bear the hepatitis B virus (HBV) posttranscriptional regulatory element (PRE) (corresponding to the roughly 570-bp SphI-FspI fragment of HBV strain adw 7) in the 3′ UTR to help overcome the Rev-dependency of gag and env mRNAs (35, 36).
Plasmids pIGAG and pGAG contain the silently modified HXB2 Pr55gag gene. pΔ40 bears an ATG142→ATC mutation generated by PCR-directed mutagenesis. pΔ55 was made by intramolecular ligation of blunted NheI and XhoI sites surrounding ATG1, resulting in a 7-nt truncation at the 5′ end of Pr55gag. pFS bears a single G deletion from a string of five G residues at gag bases 28 to 32.
The control vector pCAT was generated by ligating the KpnI-XbaI chloramphenicol acetyltransferase (CAT) gene fragment from vector pCDNA3.1CAT (Invitrogen) into vector pGAG such that CAT replaced Pr55gag between the intron and the HBV PRE. Vector pENVGAG was generated by three-way ligation of a pLAI env gene fragment and a short PCR product into vector pCIGAG such that the env stop codon was replaced by a GGC CAC ACC (Gly-His-Thr) linker between the two genes. pENV-ΔUTR-GAG was generated by replacing the AvrII-BstBI Pr55gag fragment of pENVGAG with an NheI-BstBI fragment of pCIGAG, resulting in a +1 frameshift between the env and gag ORFs. In this construct, the env ORF terminates at a stop codon found in the gag +1 frame at bp 23 to 25. To create vector pΔENV-GAG, a frameshift was introduced into vector pENV-GAG by blunting an Acc65I site found at nt 123 to 128 of the env ORF. Vector pENV-UTR-GAG was generated by ligating a PCR product bearing the entire 335-bp gag 5′ UTR upstream of the gag gene in the context of pENVGAG such that the env ORF terminates at a TGA codon at positions −308 to −310 bp upstream of Pr55gag ATG1. Vector pENV-Ψ-GAG was generated by transferring the BssHII-SpI fragment of the HXB2 provirus into pENVGAG, thereby replacing a fragment of the gag 5′ UTR starting from position −78 such that the env ORF terminates at positions −23 to −25. The BssHII site used for making pENV-Ψ-GAG interrupts the kissing loop found in stem-loop 1 of the packaging (Ψ) signal but leaves intact the 5′ major splice donor and Ψ signal stem-loops 2, 3, and 4. pENVCAT was generated by removing gag from vector pENVGAG by replacing its XhoI-XbaI gag fragment with the BsrBI-XbaI CAT fragment of pCDNA3.1CAT. Initiation of translation at the remaining Pr55gag AUG1 would thus generate a slightly amino-extended version of CAT predicted to weigh 27.5 kDa (as opposed to the predicted 25.6 kDa of wild-type CAT). Vector pCAT-GAG was generated by first transferring the NheI-XbaI CAT fragment of pCDNA3.1CAT (which includes the native CAT stop codon) into an NheI site upstream of gag in vector pCIGAG and the replacing the XhoI-XbaI fragment of the resulting vector with the XhoI-XbaI Ψ-gag fragment of pENVΨGAG. pEMCV-CAT was generated by first transferring the SpeI-HincII fragment of pEP2AmetWT (a gift from Richard Lloyd, Baylor College of Medicine) into pCI XbaI-SmaI and then ligating the HindIII (blunted)-Bsp120 fragment pCDNA3.1CAT into the resulting vector trimmed TfiI (blunted)-NotI.
p40sac stable expression vector pSAC was generated by a series of ligations involving fragments from pLAI and pHygro. pHygro was generated by removing the HindIII fragments from vector pIRES1-Hygro (Clontech), thus removing hybrid intron and encephalomyocarditis virus (EMCV) IRES sequences. Intermediate vector pLΔRH was generated by ligating the SalI-PvuII fragment of pHygro into the SalI-AatII (blunted) vector fragment of pVATG (see below). A second intermediate vector, pLRH, was generated by transferring the Rev-responsive element (RRE)-bearing BsaBI-XhoI fragment of pLAI into the StuI-SalI vector fragment of pLΔRH. Finally, vector pSAC was generated by ligating an enhanced green fluorescent protein (EGFP) (Clontech)-bearing PCR fragment into the SfoI-ClaI vector fragment of pLRH. This PCR product contained a Pr55gag ATG1→ATC mutation and inserted an AG dinucleotide between the EGFP stop codon and the mutated Pr55gag ATC1. The finished vector thus contains, in 5′ to 3′ order, 5′ long terminal repeat (LTR), deletion of 5′ splice donor and Ψ signal, EGFP, Pr55gag ATG1→ATC mutation, sac-pol, RRE, CMV promoter, hygromycin B resistance gene, and bovine growth hormone poly(A) signal. Control vector pΔSAC was generated by cutting vector pSAC BsrGI-AccI, blunting, and religating, thus removing all gag and pol coding regions.
Northern blotting probe vector pBHg was generated by transferring the HindIII-MfeI fragment of gag from vector pGAG into pBluescript trimmed with EcoRI-HindIII. This vector was linearized with XhoI prior to transcription with T7 RNA polymerase (see Northern blotting section, below). The resulting probe is homologous to bases 923 to 1183 of the Pr55gag ORF and is also homologous to a 28-bp section of the 3′ UTR of all pCI-based vectors (including pCAT).
Large-scale plasmid preps for transfection purposes were made by EndoFree Plasmid Maxi kit (Qiagen).
Proviral plasmids.
The SapI-SphI fragment of ATG142→ATC mutant plasmid pΔ40 was transferred into plasmid-cut pLAI SapI-SphI to create provirus pVATC. Because this SapI-SphI fragment bears a nonsilent SerLAI→AsnHXB2 mutation at codon 126, an isogenic control provirus, pVATG, was generated using the SapI-SphI fragment of unmutated pGAG.
Cell culture and transfection.
Peripheral blood mononuclear cells (PBMC) were obtained by Ficoll purification of blood from laboratory volunteers, and maintained in STCM (RPMI, 10% fetal calf serum [FCS], interleukin-2 [100 U/ml], T-cell growth factor). PBMC were activated by treatment for 3 days with 0.25 mg of phytohemagglutinin (PHA) per ml. Activated PBMC cultures were CD8+ depleted by treatment with anti-CD8 magnetic beads (Dynal). Epstein-Barr virus (EBV)-transformed B cells were derived from patient A42 (32). COS-7 and 293T cell lines were obtained from the American Type Culture Collection (ATCC) and maintained according to conditions described in the ATCC catalog.
COS-7 cells were transfected in 12-well plates with Lipofectamine Plus (Life Technologies, Inc.) and were subjected to radioimmunoprecipitation assay (RIPA) or p24 enzyme-linked immunosorbent assay (ELISA) at 24 h posttransfection. For poliovirus infection experiments, 3.5 × 106 293T cells were preplated overnight in a 25-cm2 flask and then transfected with Lipofectamine 2000 (Life Technologies). At 24 h posttransfection, cells were divided into eight wells of a 12-well plate (4 cm2/well) and then infected with poliovirus beginning 40 h posttransfection. Infectious HIV-1 supernatants were collected from provirus-transfected COS-7 cells 3 days after transfection.
The stable Jurkat lines Jurkat+sac and JurkatΔsac were generated by electroporation of Jurkat cells with 10 μg of BspHI-linearized pSAC or pΔSAC in RPMI at 270 mV and 960 μF. The transfected cells were incubated for 2 days and then subjected to selection in 350 μg of hygromycin B (Boehringer) per ml for 3 weeks. Cells were then sorted using a fluorescence-activated cell sorter for EGFP expression, and the brightest 10% of FL1+ cells were collected (data not shown). The resulting polyclonal populations were maintained in the presence of 350 μg of hygromycin B per ml.
Immunoprecipitation.
Pulse-chase experiments on vaccinia virus-infected cells were performed as described elsewhere (86). Briefly, 1 million A42 EBV cells were vaccinia virus infected for 4 h, starved for 30 min in starvation medium (Cys− Met− Dulbecco modified Eagle medium [DMEM], 5% dialyzed FCS [Life Technologies]), and then radiolabeled for 30 min in a small volume of starvation medium supplemented with 1.5 mCi of [35S]Cys-Met ProMix (Amersham) per ml. Cells were chased with chase medium (DMEM, 10% FCS, and 450 mg of cysteine and 300 mg of methionine per ml) and then disrupted in Triton lysis buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 0.1% sodium dodecyl sulfate [SDS]; 1 mM EDTA; 1% Triton X-100; protease inhibitor cocktail). Lysates were precleared with normal human immunoglobulin G (IgG; Sigma). Immunoprecipitation was accomplished using ∼2 μg of anti-HIV IgIV, a human polyclonal antibody stock derived from HIV-1-seropositive individuals, and protein G-Sepharose beads (Pharmacia), followed by five washes with Triton lysis buffer.
RIPA of transfected COS-7 cells was performed in a similar manner. Approximately 5 × 105 transfected cells were suspended by treatment with phosphate-buffered saline (PBS)–EDTA (PBS, 0.5 mM EDTA, 5% dialyzed FCS) at 37°C for 15 min. Cells were then radiolabeled for 30 min in 50 μl of 1.5 mCi of [35S]Cys-Met ProMix per ml, washed once with Tris-saline (50 mM Tris [pH 8.0], 150 mM NaCl), and then lysed in RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail). The lysates were precleared with 50 μg normal human immunoglobulin G and immunoprecipitated with anti-HIV IgIV. The resulting beads were washed with two washes of IP wash (50 mM Tris [pH 8.0], 150 mM NaCl, 0.1% NP-40), one wash of Tris-saline (50 mM Tris [pH 8.0], 150 mM NaCl), and one wash of 50 mM Tris (pH 6.8). We chose to assess the presence or absence of Pr55gag and p40 in transfected cells by RIPA rather than by Western blot because the HIV-IgIV antibody stock performs better in RIPA. However, similar results were also obtained by Western blotting (data not shown).
Detection of p40 in HIV-1-infected PBMC cultures was performed by pretreating 2 × 106 PHA-activated, CD8+-depleted PBMC infected with vLAI, vATG, or vATC with or without indinavir for 2 h. Indinavir was provided by The Merck Company. Cells were starved for 30 min in starvation medium with or without indinavir and then radiolabeled with 200 μCi of ProMix (with or without indinavir) for 1 h. Cells were washed and lysed using the same procedure as for COS-7 cells above, except that 50 μM indinavir was added to the lysis buffer and the lysate was immunoprecipitated with a polyclonal rabbit anti-p24 serum (NIAID ARRRP catalog no. 384) instead of anti-HIV IgIV to avoid possible detection of gp41.
In order to minimize background poliovirus bands, more stringent denaturing immunoprecipitation conditions were used to detect Pr55gag in transfected 293T cells infected with poliovirus. Transfected cells (see above) were infected with poliovirus by the addition of amplified poliovirus stock for the times shown in Fig. 1. Cells were then detached and radiolabeled in suspension for 45 min at 37°C. After radiolabeling, the cells were washed once in Tris-saline and then lysed by the addition of 35 μl of 2× SDS loading buffer (125 mM Tris, pH 6.8; 20% glycerol; 2% β-mercaptoethanol; 4% SDS; 50 μg of bromophenol blue per ml). The lysate was incubated for 5 min at 100°C and then brought up in 1.5 ml of RIPA buffer. Samples were precleared with 50 μg of normal human immunoglobulin and then cleared with a mixture of 2 μg of anti-HIV IgIV and 1 μg of polyclonal rabbit anti-CAT (Eppendorf 5′, Inc.). The beads were then washed once with RIPA buffer, twice with EE wash (0.5 M NaCl; 50 mM Tris, pH 8.0; 5 mM EDTA; 5% sucrose; 1% NP-40), once with IP wash, once with Tris-saline, and once with 50 mM Tris (pH 6.8).
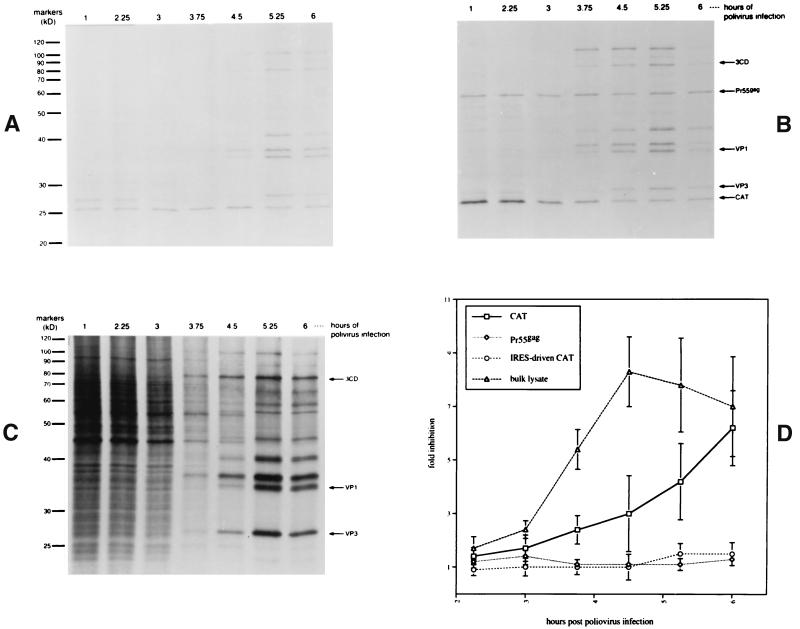
FIG. 1.
RIPA analysis of Pr55gag from plasmid-transfected and poliovirus-infected 293T cells. Cells were transfected with pEMCV-CAT (A) or pCAT-GAG (B) for 40 h and then infected with freshly amplified poliovirus stock for the times shown. Cells were then detached, radiolabeled with [35S]Cys-Met for 45 min, lysed, and subjected to immunoprecipitation with anti-HIV–anti-CAT antibody mixture. Unprecipitated bulk lysate of cells transfected with pCAT-GAG is shown in panel C. Panel D presents a summary of PhosphorImager analysis of five independent transfection-infection experiments. The y axis represents the fold inhibition of band intensity with respect to the band seen at 1 h post-poliovirus infection for the protein indicated. In two of the five experiments summarized in panel D, the CAT and Pr55gag bands were quantitated in cells cotransfected with pCAT and pENV-ΔUTR-GAG instead of pCAT-GAG. The intensity of signal of lysate bands was determined by examining a 28- to 33-kDa window found between prominent poliovirus VP3 and VP1 bands.
Immunoprecipitated samples were analyzed by comparison to The Benchmark Unstained Protein Ladder (Life Technologies) by SDS-polyacrylamide gel electrophoresis (PAGE) on 8.5 or 12% gels.
Northern blotting.
Cytoplasmic RNA was isolated (RNeasy kit; Qiagen) from 293T cells transfected for 40 h with the plasmid vector indicated in Fig. 2. Each RNA (5 μg) was subjected to Northern blotting (NorthernMax-Gly Kit; Ambion) according to the kit instructions. Blots were probed with a biotinylated RNA probe generated by T7 transcription of vector pBHg (Maxiscript Kit; Ambion) with a 60:40 ratio of UTP to biotin-16-UTP (Boehringer). Detection was carried out using Ambion's Brightstar Biodetect Kit according to manufacturer's instructions.
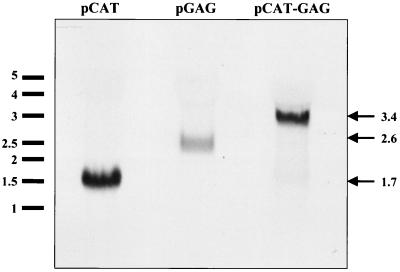
FIG. 2.
Northern blotting of bicistronic vector pCAT-GAG. 293T cells were transfected with the plasmid expression vector indicated above each lane and subjected to Northern blotting. Kilobase RNA size markers are shown on the left. The predicted sizes the full-length transcript of each vector are given on the right.
p24 ELISA.
A commercially available anti-p24 ELISA kit (Coulter/Immunotech) was used to determine the levels of Gag in cell lysates and supernatants. Cells were washed twice with PBS-EDTA and then lysed in 300 μl of the lysis buffer provided with the kit. Cell lysates were diluted appropriately (typically, 1:100) to fall within the 7- to 125-pg/ml range of the assay. Lysates were standardized by quantitation of total protein using a detergent-compatible protein assay kit (Bio-Rad).
Edman microsequencing.
Microsequencing of p40sac was performed by the Johns Hopkins University School of Medicine Biosynthesis and Sequencing Facility using a Perkin-Elmer/Applied Biosystems Procise 492 Protein Microsequencer. p40sac was isolated for microsequencing by immunoprecipitation of 5 × 107 vvB2-infected A42 EBV cells with HIV-IgIV (see above). The immunoprecipitated sample was electrophoresed by SDS-PAGE on a 12% gel and blotted overnight at 15 V onto a polyvinylidene difluoride membrane (Bio-Rad). The membrane was stained with Ponceau S (Bio-Rad), and the p40sac band was excised and subjected to sequencing. Attempts to microsequence the 55-kDa species produced by vvB2-infected cells were performed in a similar manner except that the HIV-IgIV antibody was covalently conjugated to protein G-Sepharose beads prior to immunoprecipitation to avoid elution of interfering immunoglobulin heavy chain.
Virus culture.
For visualization of p40sac in HIV-1-infected cells, 2 × 106 activated/depleted PBMC were treated overnight with 1 ml of infectious supernatant collected from provirus-transfected COS-7 cells, incubated for 5 days, and then subjected to RIPA. For viral growth curves, provirus-transfected COS-7 supernatants were normalized by p24 ELISA so that roughly 50-ng equivalents (roughly 50 μl of supernatant) of p24 were added to 105 Jurkat cells or the stable cell lines Jurkat+sac or JurkatΔsac in a 1-ml volume in a 24-well plate. Cells were washed twice with 10 ml of culture medium 24 h postinfection. Supernatants were sampled periodically and subjected to p24 ELISA. Infected cultures were split as necessary.
Cesium-banded high-titer Mahoney type 1 poliovirus stock was the generous gift of Ellie Ehrenfeld (National Institutes of Health). In addition to being more easily transfectable than the HeLa cells typically used in poliovirus shutoff experiments, 293T cells were found to exhibit more-protracted shutoff kinetics than HeLa cells (data not shown). Amplified infectious poliovirus stocks were generated by adding ∼0.1 μl of high-titer poliovirus stock (1011 PFU/ml) to a confluent T-75 flask of 293T cells in 15 ml of culture medium for 24 h. The resulting cell lysate was clarified by centrifugation at 300 × g for 10 min, and 0.5 ml of the clarified lysate was used to infect confluent 4-cm2 wells of transfected 293T cells.
RESULTS
Expression of Pr55gag in poliovirus-infected cells.
To investigate the hypothesis that the HIV-1 gag ORF exhibits IRES activity, we undertook a series of experiments involving gag expression in poliovirus-infected cells. Because poliovirus mediates a rapid inhibition of cap-dependent translation in permissive host cells (23, 56), it has been widely used in the investigation of the cap dependency of translation of various mRNAs.
Figure 1 depicts a time course experiment in which 293T cells were first transfected with the indicated plasmid expression vector and then infected with poliovirus for various times. Bicistronic vector pCAT-GAG encodes the CAT reporter gene upstream of the Pr55gag gene, including a 77-bp segment of the native Pr55gag 5′ UTR. Control vector pEMCV-CAT encodes CAT under control of the EMCV IRES. After transfection and poliovirus infection, cells were radiolabeled with [35S]Cys-Met for 45 min and subjected to immunoprecipitation with an anti-HIV–anti-CAT antibody mixture (Fig. 1A and B). Using this method, it was found that translation of EMCV IRES-driven CAT (Fig. 1A) and Pr55gag (Fig. 1B) were not significantly impacted by poliovirus-mediated shutoff of cap-dependent translation. In contrast, the cap-dependent translation of the upstream CAT ORF from vector pCAT-GAG declined substantially over the course of the infection. Bulk lysates of the transfected cells (Fig. 1C) displayed a pattern in which the translation of host proteins diminished with duration of poliovirus infection, while a characteristic pattern of poliovirus-specific bands began to appear.
A summary of PhosphorImager (Molecular Dynamics) quantitation of five independent poliovirus time course experiments is shown in Fig. 1D. In two of these experiments, cells were cotransfected with a CAT expression vector, pCAT, and a different bicistronic vector, pENV-ΔUTR-GAG (see below), instead of pCAT-GAG. The results suggest that the expression of Pr55gag from the constructs pCAT-GAG and pENV-ΔUTR-GAG was cap independent, a finding which supports the hypothesis that the HIV-1 gag gene encodes an IRES.
Northern blotting of pCAT-GAG.
In addition to IRES activity, several possible explanations can be postulated to explain the expression of the second cistron of bicistronic constructs such as pCAT-GAG. These alternative scenarios include the existence of alternate splicing variants, RNA species derived from a cryptic promoter just upstream of the second cistron, or species of RNAs broken apart between the two cistrons. To rule out these possible explanations for the production of Pr55gag from construct pCAT-GAG, we performed Northern blots on cells transfected with pCAT-GAG (Fig. 2). This Northern blotting shows that pCAT-GAG produces only one major RNA which is the predicted size (3.4 kb) of the full-length bicistronic CAT-GAG mRNA. This result argues against the possibility that the production of Pr55gag is the result of a cryptic RNA species.
Env-Gag bicistronic vectors.
To further investigate the IRES activity of the gag gene, we constructed a panel of bicistronic vectors. In these constructs, the HIV-1 env gene was used as an upstream reporter cistron, and the gag gene served both as the sequence to be tested for IRES activity and as a downstream reporter cistron. The env gene was chosen as an upstream reporter gene because it contains a large number of ATG codons and regions of highly structured RNA which would be expected to significantly disrupt scanning by any ribosomes bypassing the ATG codons within env (24). Expression of a novel 40-kDa gag isoform (p40), which results from translational initiation at an internal gag AUG codon, was also monitored in these experiments and will be discussed in greater detail below.
We first sought to determine what portion of the Pr55gag 5′ UTR might be necessary for potentiating the translation of gag as a second cistron. Vector pENV-ΔUTR-GAG encodes only the env and gag reporter ORFs with the gag 5′ UTR entirely omitted. This construct was found to express Pr55gag efficiently in poliovirus-infected cells (see above), suggesting that the native gag 5′ UTR is dispensable for the IRES activity of the gag ORF. In order to determine what impact the native gag 5′ UTR might have on gag's translation as a second cistron, we constructed vector pENV-UTR-GAG, which contains the entire Pr55gag 5′ UTR between the two ORFs. We also constructed vector pENV-Ψ-GAG, which contains a 77-bp fragment of the gag 5′ UTR encompassing most of the HIV-1 packaging (Ψ) signal. All three vectors were found to drive significant amounts of Pr55gag expression when compared to vector pGAG, which encodes only Pr55gag with an unobstructed vector-derived 5′ UTR (Fig. 3A). This result confirms the observation that the native gag 5′ UTR is not necessary to potentiate the expression of Pr55gag as a second cistron. Indeed, placement of the entire gag 5′ UTR upstream of Pr55gag (in vector pENV-UTR-GAG) resulted in somewhat decreased levels of Pr55gag expression relative to the expression seen from vector pENV-ΔUTR-GAG. This decrease could have been due to activity of HIV-1 polyadenylation signal (found in the R domain of the HIV-1 LTRs), the presence of the primer binding site, or the presence of the viral dimerization initiation signal, since removal of these sequences (in pENV-Ψ-GAG) resulted in higher Pr55gag expression levels.
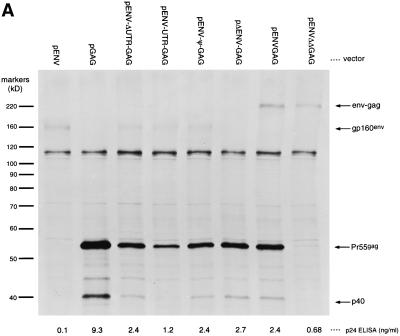
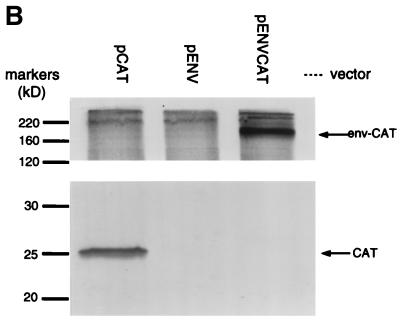
FIG. 3.
Analysis of Gag expression using bicistronic Env and Gag vectors. RIPA analysis of COS-7 cells transfected with the indicated plasmid expression vector was done using either anti-HIV-1 antibody stock (A) or anti-CAT antibody stock (B). The numbers below each lane in panel A represent the p24 ELISA signal from lysates of a separatedly transfected population of COS-7 cells.
We generated several other vectors to investigate the conditions under which gag might function as a downstream cistron. In vector pΔENV-GAG, a frameshift was introduced near the 5′ end of the env ORF of vector pENV-ΔUTR-GAG in order to demonstrate that translation of the env ORF is not required for translation of the downstream gag ORF. Vector pENVGAG encodes an in-frame fusion of the env ORF (lacking the native env stop codon) to the gag ORF. Expression of Pr55gag from this vector (Fig. 3A) argues against the possibility that the expression of Pr55gag from vector pENVGAG is due to translational reinitiation following env translation, since the env-gag and gag ORFs are coterminal. A control vector, pENV-ΔΔGAG, encodes the same Env-Gag fusion protein as vector pENVGAG except that the Pr55gag and p40 initiator codons have been mutated to ATC, a noninitiator Ile codon (79). As seen in Fig. 3A, mutation of these ATG codons abrogates Pr55gag and p40 production, suggesting that their expression is the result of translation initiation at these internal sites.
In a control vector, pENVCAT, all but the first 12 bp of the gag sequence of pENVGAG were replaced with the CAT reporter gene. As seen in Fig. 3B, pENVCAT does not drive expression of any CAT-sized species. This result suggests that the internal translational initiation at Pr55gag AUG1 observed in vector pENVGAG is dependent on sequences within the gag ORF, since the CAT ORF was not sufficient to direct this activity.
In order to further exclude the possibility that Pr55gag and p40 expression from vector pENVGAG was the result of cryptic splicing or cryptic promoter usage, we generated a recombinant vaccinia virus vector, vvENVGAG, which encodes the same Env-Gag fusion protein as plasmid pENVGAG. Because vaccinia viruses replicate in the cytoplasm of infected cells and utilize virus-encoded transcription factors (59), it is unlikely that cryptic host promoters would have any activity in the context of a recombinant vaccinia virus. Splicing of vaccinia virus RNAs has not been reported. COS-7 cells infected with vvENVGAG exhibited Env-Gag, Pr55gag, and p40 bands (data not shown), indicating that expression of Pr55gag and p40 from these vectors was not due to cryptic splicing or cryptic promoter usage.
These results confirm that sequences within the gag ORF can promote internal initiation of translation of Pr55gag even when the gag ORF is placed downstream of large numbers of ATG codons and highly structured RNA sequences. This activity is a feature of sequences within the gag ORF and functions even in the absence of the native gag 5′ UTR.
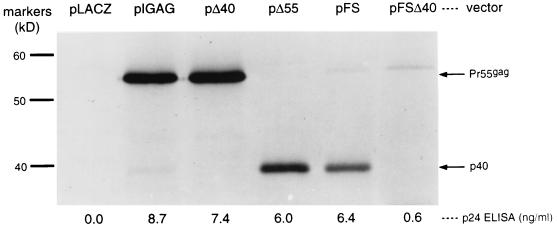
Characterization of a 40-kDa gag gene product.
A wide variety of plasmid and vaccinia virus expression vectors encoding Pr55gag consistently display a minor gag gene-specific 40-kDa band (Fig. 3 and 4 and data not shown). Because the gag gene contains an isolated AUG at codon 142 that could initiate translation of a 40-kDa protein, we considered the possibility that p40 was the result of an internal initiation event at this AUG. To help determine its origins, we immunoprecipitated p40 from cells infected with vvB2, a vaccinia virus vector encoding Pr55gag, and subjected it to Edman microsequencing. This sequencing yielded the amino acid sequence NH2-VHQAISPRT, a sequence identical to the predicted amino acid sequence of a polypeptide initiated at AUG142, assuming its initiator methionine was cleaved away (82; reviewed in reference 1). Pulse-chase experiments were also performed on cells infected with vvB2. Pr55gag and p40 exhibited no precursor-product relationship, arguing against the concept that p40 was derived through proteolytic cleavage of Pr55gag (data not shown).
FIG. 4.
Manipulation of p40 expression. Each lane represents comparable numbers of Cos-7 cells transfected with the indicated plasmid vector and subjected to RIPA with anti-HIV antibody stock. The numbers below each lane represent p24 ELISA signal from lysates of a separatedly transfected population of Cos-7 cells.
We also examined a panel of plasmid expression vectors (Fig. 4) to verify that p40 was the result of an independent translation initiation event and not simply a by-product of Pr55gag. In two vectors, we independently mutated the Pr55gag and p40 initiator ATG codons to ATC. We found that mutation of ATG142 to ATC abrogated p40 expression, suggesting that this codon is used as an initiator codon for p40. Although the mutation of Pr55gag ATG1 to ATC prevented Pr55gag expression, it did not abolish p40 expression. Similarly, introduction of a frameshift downstream of ATG1 did not abolish p40 expression. Thus, p40 can be expressed independently of Pr55gag through translational initiation at gag codon 142.
p40 expression in HIV-1-infected cells.
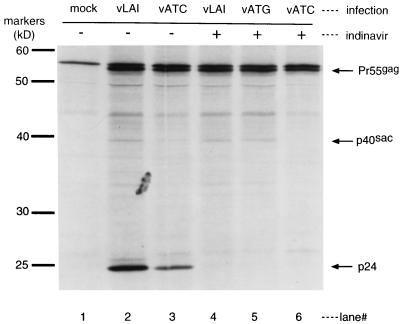
To determine whether internal initiation at AUG142 occurs during viral replication, we examined HIV-1-infected cells for the presence of p40. Answering this question was complicated by the activity of the HIV-1 protease. Met142 lies only six amino acids downstream of the eight-amino-acid protease recognition site (codons 129 to 136) at the matrix/capsid boundary. Thus, a single HIV-1 protease cleavage of Pr55gag yields a protein nearly identical in size to p40. Similarly, a single cleavage of Pr55gag at the capsid-p2 boundary results in a protein of approximately 40 kDa. For clarity, we will hereafter refer to the 40-kDa protein generated by initiation at AUG142 as p40sac for “start at capsid.” To determine whether p40sac is produced in infected cells, we used indinavir (an HIV-1 protease inhibitor) to prevent the generation of Pr55gag protease cleavage products (89). PHA-activated, CD8+-depleted PBMC were infected with supernatants from COS-7 cells transfected with wild-type provirus pLAI, p40sac knockout provirus pVATC (carrying an ATG142→ATC [Met→Ile] mutation), or provirus pVATG, an isogenic wild-type control for pVATC (see Materials and Methods). Five days after infection, PBMC cultures were pretreated with 50 mM indinavir and then radiolabeled with [35S]Cys-Met. Thus, all radiolabeled gag gene products in the indinavir-treated cells were generated in the functional absence of HIV-1 protease. The labeled cells were then lysed and subjected to immunoprecipitation with anti-p24 serum.
Cells infected with wild-type viruses vLAI and vATG expressed 40-kDa products both in the presence and in the absence of indinavir (Fig. 5). The absence of a p24 band in cultures treated with indinavir suggests a strong inhibition of HIV-1 protease in these cultures. Although some 40-kDa Pr55gag protease cleavage products accumulated in vATC-infected cells in the absence of indinavir, the ATG142→ATC mutation in this virus completely ablated expression of p40sac, as shown by the absence of a 40-kD band in the presence of indinavir. The results support the conclusion that HIV-1-infected cells produce small amounts of a novel 40-kDa protein, p40sac, through internal initiation at AUG142.
FIG. 5.
p40sac expression in HIV-1-infected PBMCs. PHA-activated, CD8+-depleted PBMC were mock infected (lane 1) or were infected with wild-type viruses vLAI or vATG (lanes 2, 4, and 5) or with p40sac knockout virus vATC for 5 days. Cells were treated with (+) or without (−) indinavir, an HIV-1 protease inhibitor, and then radiolabeled and subjected to RIPA with polyclonal anti-p24 serum.
Rescue of impaired replication of a p40sac-deficient HIV-1 mutant.
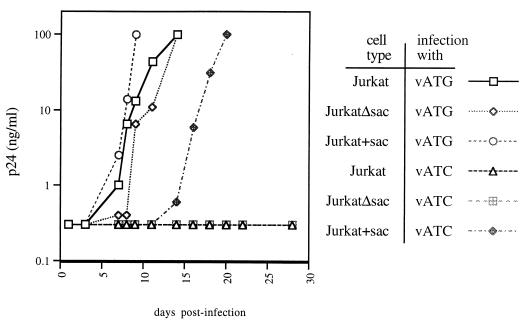
In order to investigate whether p40sac might play a role in viral replication in culture, we examined the replication kinetics of the p40sac knockout virus vATC. This mutant virus displayed dramatically impaired replication compared to wild-type vLAI or isogenic wild-type control vATG in Jurkat and CD8-depleted PBMC cultures (Fig. 6; also data not shown). In light of the fact that ATG142 and the region surrounding it are highly conserved (60; HIV Sequence Database), even relatively conservative nonsilent mutations (such as the Met142→Ile mutation found in vATC) might be expected to be deleterious to viral growth through disruption of the normal functions of Pr55gag.
FIG. 6.
Rescue of codon 142 mutant virus vATC by expression of p40sac and p145sac-pol in trans. Infectious supernatants from 293T cells transfected with codon 142 mutant provirus pVATC or isogenic wild-type control provirus pVATG were normalized for p24 content and added to Jurkat cells, the p40sac and p145sac-pol stable Jurkat cell line Jurkat+sac, or the control Jurkat cell line JurkatΔsac. Cells were washed at 24 h postinfection, and the supernatants were sampled at the times shown and subjected to p24 ELISA.
To determine whether the delayed replication kinetics of vATC were due to its inability to produce p40sac or due to the amino acid substitution it carries in Pr55gag, we generated a polyclonal stably transfected Jurkat line which expresses p40sac in trans. In the context of full-length HIV-1 mRNAs, translational initiation at Pr55gag codon 142 would be expected to result in production of p40sac and a 145-kDa protein, p145sac-pol, by virtue of frameshifting at the HIV-1 “slippery” site. In an effort to provide appropriate levels of p40sac and p145sac-pol, we made a Rev- and Tat- inducible sac-pol vector, pSAC, based on the pLAI provirus. The vector expresses p40sac and p145sac-pol from the HIV-1 LTR. To block ribosome scanning and to monitor expression, we inserted the EGFP gene upstream of Pr55gag codon 1, which was mutated from ATG to ATC to abrogate Pr55gag expression. A fragment containing the HIV-1 RRE was placed downstream of the pol stop codon, as was a fragment carrying the CMV promoter, hygromycin B phosphotransferase, and a poly(A) signal. A Jurkat line stably carrying a control vector, pΔSAC, in which the sac-pol gene was removed from pSAC, was also made. In the presence of Tat and Rev provided by an infecting virus, transcription and nuclear export of EGFP-p40sac-p145sac-pol mRNA expressed from these vectors should be induced. Consistent with this prediction, uninfected Jurkat cultures stably carrying these constructs displayed constitutive low levels of fluorescence (and low p24 ELISA signal for vector pSAC), while infected cultures of the lines showed a substantial population of highly fluorescent cells (data not shown). We named these cell lines Jurkat+sac and JurkatΔsac.
Although p40sac knockout virus vATC failed to grow to high titers in Jurkat and JurkatΔsac cultures, it was able to grow in the Jurkat+sac line (Fig. 6). When cultures were infected with much higher inoculums, vATC could eventually grow to high titers in Jurkat and JurkatΔsac cultures but with dramatically slower kinetics than seen in Jurkat+sac cultures (data not shown). Although vATC grew well in the Jurkat+sac line, growth was somewhat attenuated compared to wild-type virus vATG. This could reflect some impact of the Pr55gag ATG142→ATC mutation in vATC, inappropriate timing of p40sac and p145sac-pol expression, or inappropriate levels of p40sac and p145sac-pol in the Jurkat+sac line.
These results are consistent with the concept that the defective growth phenotype of mutant virus vATC can be overcome, at least in part, by expression of p40sac and p145sac-pol in trans. Thus, expression of p40sac (or p145sac-pol) may be required for normal growth kinetics of HIV-1 in culture.
DISCUSSION
Our results demonstrate that the HIV-1 gag gene encodes an IRES. One line of evidence that Pr55gag can be translated in a cap-independent fashion via an IRES-like mechanism comes from experiments involving poliovirus-infected cells. Poliovirus mediates a rapid reduction in cap-dependent translation shortly after infection of permissive cells. This host cell translational shutoff coincides with the cleavage by poliovirus 2A protease of the eIF4G subunit of eIF4F (also known as the cap-binding complex), and the cleavage of poly(A) binding protein by poliovirus proteases 2A and 3C (44, 46). The IRES-mediated translation of poliovirus RNAs under these conditions is either unimpaired or enhanced. Poliovirus-infected cells (45) and extracts of cells infected with poliovirus have been widely used to demonstrate the presence of IRES activity within various mRNA sequences (23). As seen in Fig. 1, the translation of Pr55gag in cells transfected with bicistronic vector pCAT-GAG was insensitive to poliovirus-mediated shutdown. This supports the concept that Pr55gag can be translated through a cap-independent translation mechanism, such as an IRES, located within the gag ORF. Northern blots of cells transfected with pCAT-GAG were found to exhibit only one full-length mRNA species, arguing against the possibility that this cap-independent expression of Pr55gag was due to alternative splicing, cryptic promoter usage, or broken RNA species.
A separate line of evidence which suggests that gag encodes an IRES comes from a series of bicistronic vectors where cap-dependent ribosome scanning was obstructed by placement of the HIV-1 env ORF upstream of gag. As seen in Fig. 3A, placement of the env ORF upstream of gag does not abrogate Pr55gag expression. This result held true whether the native gag 5′ UTR was present or not, whether the env ORF was translated or not, and also when the env ORF was translated as an in-frame fusion with the gag ORF. Utilization of Pr55gag AUG1 as an internal initiator codon required the presence of the gag ORF itself, as evidenced by the absence of expression of any CAT-length products by vector pENVCAT, in which CAT replaces most of the gag ORF downstream of env (Fig. 3B). These results argue against modified ribosome scanning mechanisms for Pr55gag or p40sac expression such as leaky scanning (49, 77), translational reinitiation (28, 54), ribosome shunting (72, 96), broken RNA, or ribosome “stutter” (18, 95). The expression of Pr55gag both from vaccinia virus vector vvENVGAG and plasmid vector pENVGAG argues against the possibility that expression of these proteins is due to cryptic promoter usage or cryptic splicing. Thus, two separate lines of evidence indicate that Pr55gag can be translated as an internal or downstream cistron by virtue of an IRES within the gag ORF.
In addition to potentiating the translation of Pr55gag, the HIV-1 gag IRES also facilitates translation of a novel 40-kDa protein, p40sac, whose translation initiates at an AUG codon near the 5′ end of the p24capsid domain of Pr55gag. Several features of HIV-1 biology may have prevented the previous identification of this novel Gag isoform. As demonstrated in Fig. 5, intermediate products of the cleavage of Pr55gag by the HIV-1 protease are roughly 40 kDa in mass and can obscure the presence of p40sac (26). In past instances when infected cells treated with HIV-1 protease inhibitors have exhibited a 40-kDa band, the band may have been discounted as reflecting incomplete inhibition of protease activity by the drug (89, 92). The fact that p40sac is expressed at much lower levels than Pr55gag has also perhaps hidden it from identification even when Pr55gag has been expressed in the absence of HIV-1 protease (25, 57, 73).
Despite its low abundance, our results suggest that p40sac may play a vital role in viral replication in vitro. HIV-1 mutant virus vATC, which fails to generate p40sac, and, presumptively, p145sac-pol replicates with dramatically impaired growth kinetics in cultured cells (Fig. 6). The replication defect of vATC can be partially overcome by the expression of p40sac or p145sac-pol in trans, suggesting that the defect stems, at least in part, from the mutant's inability to generate these novel proteins. This observation is consistent with the concept that p40sac and/or p145sac-pol play an important role in viral replication in culture.
It has been shown that sequences within the native gag 5′ UTR can inhibit cap-dependent translation of reporter proteins. The Tat-responsive element (TAR) is an RNA secondary structural element that is responsible for binding HIV-1 Tat protein and may play a role in the inhibition of cellular interferon-induced double-stranded RNA-activated kinase (12, 22). The TAR is located at the 5′ end of all HIV-1 RNA transcripts. Because the presence of secondary structure at or near the 5′ end of RNAs reduces the accessibility of the 5′ cap to eIF4F (49), it is thought that this feature of HIV-1 mRNAs can inhibit their cap-dependent translation (64, 78). Other stable HIV-1 RNA structural elements involved in regulating the splicing, polyadenylation, dimerization, packaging, primer binding, and reverse transcription of viral RNAs are thought to inhibit translation by blocking the scanning of 40S ribosomes (and their associated cofactors) through the gag 5′ UTR (29, 57). Thus, a possible function of the HIV-1 gag IRES might be to serve as a mechanism to bypass the structural barriers to cap-dependent translation by recruiting ribosomes directly to the gag ORFs.
It has been demonstrated that infection with HIV-1 can arrest cells in the G2 phase of the cell cycle (reviewed in reference 13). The fact that cap-dependent translation is inhibited during some phases of the cell cycle (8) led Vagner and colleagues to speculate that an HIV-1 IRES might allow for enhanced HIV-1 gene expression under growth-arrest conditions where the cap-dependent translation of host cell mRNAs is inhibited (91). Recent reports have found that the IRESs of hepatitis C virus (HCV) and host mRNAs encoding ornithine decarboxylase and p58PITSLRE are significantly more active during the G2/M phase of the cell cycle than in the G1 or S phases (11, 34, 69). The possible interplay between HIV-1 cell cycle arrest and IRES utilization deserves future investigation.
In addition to the IRESs found in the 5′ UTRs of a variety of plus-stranded RNA viruses (3, 9, 31, 42, 43, 66, 88) and a number of eukaryotic cellular mRNAs (6, 27, 37, 38, 61, 62, 84, 90), IRESs have recently been described in several type C retroviruses. These include MMLV (91), Friend murine leukemia virus (FMLV) (4, 19), the Rat VL30 region of the Harvey murine sarcoma virus (5), and avian reticuloendotheliosis virus type A (53). A variety of other retroid family members, including human T-cell leukemia virus type 1 (2), human foamy virus (51), LINE-1 and VL30 (two groups of endogenous vertebrate retro-elements) (39, 52, 56, 87), and Drosophila R2 retrotransposable elements (30), have also been shown to exhibit possible IRES-like activities. These data, when taken together with our observation in this report that HIV-1 encodes an IRES, suggest the possibility that IRES-mediated translation is a typical feature of the retroviridae.
Murine leukemia viruses encode two different Gag proteins. Glyco-Gag, a nonstructural transmembrane glycoprotein important for viral infectivity, is initiated from a CUG codon located upstream of and in frame with the AUG-initiated gag gene (67, 68). The IRES of MMLV has been found to be situated entirely within the viral glyco-gag ORF in a 126-bp sequence just upstream of the gag AUG. The MMLV IRES was shown to initiate translation only at the gag AUG, whereas the FMLV IRES, which has not been fully localized, was shown to be capable of directing translation of both Gag and Glyco-Gag.
Localization of the MMLV gag IRES to a region entirely contained within an ORF was striking in that it (and perhaps the IRES of FMLV) was the first naturally occurring (10) IRES entirely contained within a translated ORF. More recently, a similar ORF-localized IRES has been described for the host mRNA encoding p110PITSLRE and p58PITSLRE (11). The HIV-1 Pr55gag IRES activity described here is thus conceptually similar to the MMLV and PITSLRE IRESs in that it is located entirely within an ORF and is similar to the FMLV IRES in that it can direct translation of two different gag gene products.
Miele and colleagues have argued that the HIV-1 packaging signal does not encode an IRES (57). Using a series of bicistronic constructs in which the entire gag 5′ UTR and as much as the first 200 nt of the gag ORF were inserted between two reporter cistrons, these investigators found no evidence for potentiation of translation of the downstream reporter cistron. In contrast to Miele and colleagues' findings, a recent report from Ohlmann and colleagues has found that sequences in the 5′ UTR of the gag gene of another primate lentivirus, SIVmac, do exhibit IRES activity (63). Thus, it may be that IRES activity is typical of the lentiviridae, while the exact location of sequences required for the activity varies.
Perhaps the most surprising feature of the HIV-1 gag IRES is that it directs translational initiation at (or, conceivably, beyond) its 5′ border. Precedents for translational enhancement by sequence elements located 3′ of the ORF they control include the mRNAs of certain viruses (14, 85, 94), and the 3′ UTR of the mRNA for the β subunit of mitochondrial H+-ATP synthase (40). Important future work on this topic will include identification of the minimal sequence elements required for HIV-1 gag ORF IRES activity and the potential mechanisms underlying its surprising, seemingly backward, arrangement.
Das et al. have demonstrated that constitutive expression of a small inhibitor RNA (IRNA) molecule isolated from yeast (15) can effectively protect cultured cells from productive poliovirus infection (17) without significantly impacting cell growth or morphology. The suppression of poliovirus replication in cells expressing this IRNA is likely the result of competition between the IRNA and the poliovirus IRES for access to host proteins required for viral IRES-mediated translation (16, 93). It is thus formally possible that similar antagonism of the HIV-1 gag IRES could disrupt the replication of HIV-1 or other IRES-bearing retroviruses.
ACKNOWLEDGMENTS
We thank Mark J. Selby, Lucy Carruth, Tim Tobery, Monika Hermankova, and Xiao Fang Yu for valuable advice. We also thank Elizabeth K. Flynn for field trials of the Benchmark Protein Ladder which facilitated the discovery of p40sac. We are especially grateful to Ellie Ehrenfeld and Richard Lloyd for patiently providing advice and materials which were crucial for the completion of this research.
This work was supported by NIH grants AI28108 and AI37924 and by a grant from the Markey Trust.
REFERENCES
- 1.Arfin S M, Bradshaw R A. Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry. 1988;27:7979–7984. doi: 10.1021/bi00421a001. [DOI] [PubMed] [Google Scholar]
- 2.Attal J, Theron M C, Taboit F, Cajero-Juarez M, Kann G, Bolifraud P, Houdebine L M. The RU5 (‘R’) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 1996;392:220–224. doi: 10.1016/0014-5793(96)00815-0. [DOI] [PubMed] [Google Scholar]
- 3.Belsham G J, Brangwyn J K. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlioz C, Darlix J L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus Gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlioz C, Torrent C, Darlix J L. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 7.Blum H E, Galun E, Liang T J, von Weizsacker F, Wands J R. Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. J Virol. 1991;65:1836–1842. doi: 10.1128/jvi.65.4.1836-1842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonneau A M, Sonenberg N. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J Biol Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- 9.Borman A, Jackson R J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 12.Cullen B R. Does HIV-1 Tat induce a change in viral initiation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- 13.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 14.Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S, Coward P, Dasgupta A. A small yeast RNA selectively inhibits internal initiation of translation programmed by poliovirus RNA: specific interaction with cellular proteins that bind to the viral 5′-untranslated region. J Virol. 1994;68:7200–7211. doi: 10.1128/jvi.68.11.7200-7211.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, Kenan D J, Bocskai D, Keene J D, Dasgupta A. Sequences within a small yeast RNA required for inhibition of internal initiation of translation: interaction with La and other cellular proteins influences its inhibitory activity. J Virol. 1996;70:1624–1632. doi: 10.1128/jvi.70.3.1624-1632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, Ott M, Yamane A, Tsai W, Gromeier M, Lahser F, Gupta S, Dasgupta A. A small yeast RNA blocks hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation and inhibits replication of a chimeric poliovirus under translational control of the HCV IRES element. J Virol. 1998;72:5638–5647. doi: 10.1128/jvi.72.7.5638-5647.1998. . (Erratum, 72:9419.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dasso M C, Milburn S C, Hershey J W, Jackson R J. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur J Biochem. 1990;187:361–371. doi: 10.1111/j.1432-1033.1990.tb15313.x. [DOI] [PubMed] [Google Scholar]
- 19.Deffaud C, Darlix J L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J Virol. 2000;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl P L, Hugin A W, Moss B. Removal of cryptic poxvirus transcription termination signals from the human immunodeficiency virus type 1 envelope gene enhances expression and immunogenicity of a recombinant vaccinia virus. J Virol. 1990;64:2448–2451. doi: 10.1128/jvi.64.5.2448-2451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl P L, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. Boston, Mass: John Wiley and Sons; 1993. pp. 16.17.1–16.17.12. [Google Scholar]
- 22.Edery I, Petryshyn R, Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 23.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 549–573. [Google Scholar]
- 24.Ellerbrok H, Serpente N, Pancino G, Vanhee C, D'Auriol L, Sitbon M, Vaquero C. Sequences in the rev-responsive element responsible for premature translational arrest in the human-immunodeficiency-virus-type-1 envelope. Eur J Biochem. 1993;216:459–467. doi: 10.1111/j.1432-1033.1993.tb18164.x. [DOI] [PubMed] [Google Scholar]
- 25.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison C A D, Loeb D D, Swanstrom R. Cleavage of HIV-1 Gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retrovir. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 26.Evans L A, Homsy J M, Morrow W J, Gaston I, Sooy C D, Levy J A. Human monoclonal antibody directed against gag gene products of the human immunodeficiency virus. J Immunol. 1988;140:941–943. [PubMed] [Google Scholar]
- 27.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 28.Geballe A P. Regulation of translation by upstream AUG codons. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 173–197. [Google Scholar]
- 29.Geballe A P, Gray M K. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 1992;20:4291–4297. doi: 10.1093/nar/20.16.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.George J A, Eickbush T H. Conserved features at the 5′ end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol Biol. 1999;8:3–10. doi: 10.1046/j.1365-2583.1999.810003.x. [DOI] [PubMed] [Google Scholar]
- 31.Glass M J, Jia X Y, Summers D F. Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology. 1993;193:842–852. doi: 10.1006/viro.1993.1193. [DOI] [PubMed] [Google Scholar]
- 32.Hammond S A, Johnson R P, Kalams S A, Walker B D, Takiguchi M, Safrit J T, Koup R A, Siliciano R F. An epitope-selective, transporter associated with antigen presentation (TAP)-1/2-independent pathway and a more general TAP-1/2-dependent antigen-processing pathway allow recognition of the HIV-1 envelope glycoprotein by CD8+ CTL. J Immunol. 1995;154:6140–6156. [PubMed] [Google Scholar]
- 33.Hatfield D, Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell G A, Lemon S M. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iizuka N, Chen C, Yang Q, Johannes G, Sarnow P. Cap-independent translation and internal initiation of translation in eukaryotic cellular mRNA molecules. Curr Top Microbiol Immunol. 1995;203:155–177. doi: 10.1007/978-3-642-79663-0_8. [DOI] [PubMed] [Google Scholar]
- 38.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and Cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilves H, Kahre O, Speek M. Translation of the rat LINE bicistronic RNAs in vitro involves ribosomal reinitiation instead of frameshifting. Mol Cell Biol. 1992;12:4242–4248. doi: 10.1128/mcb.12.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izquierdo J M, Cuezva J M. Internal-ribosome-entry-site functional activity of the 3′-untranslated region of the mRNA for the beta subunit of mitochondrial H+-ATP synthase. Biochem J. 2000;346:849–855. [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 42.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joachims M, Van Breugel P C, Lloyd R E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986;47:481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- 48.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 50.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lochelt M, Flugel R M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol. 1996;70:1033–1040. doi: 10.1128/jvi.70.2.1033-1040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Lastra M, Ulrici S, Gabus C, Darlix J L. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J Virol. 1999;73:8393–8402. doi: 10.1128/jvi.73.10.8393-8402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopezlastra M, Gabus C, Darlix J L. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type a RNA and development of novel Mlv-Rev-based retroviral vectors. Hum Gene Ther. 1997;8:1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 54.Luukkonen B G, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 56.McMillan J P, Singer M F. Translation of the human LINE-1 element, L1Hs. Proc Natl Acad Sci USA. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miele G, Mouland A, Harrison G P, Cohen E, Lever A M. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morley S J, Curtis P S, Pain V M. eIF4G: translation's mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 59.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1171–1176. [Google Scholar]
- 60.Myers G, Korber B, Wain-Hobson S, Jeang K T, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory Theoretical Biology and Biophysics; 1994. [Google Scholar]
- 61.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 62.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 63.Ohlmann T, Lopez-Lastra M, Darlix J L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J Biol Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 64.Parkin N T, Cohen E A, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavlakis G N, Schwartz S, D'Agostino D M, Felber B K. Structure, splicing, and regulation of expression of HIV-1: a model for the general organization of lentiviruses and other complex retroviruses. In: Kennedy R, Wong-Staal F, Koff W C, editors. Annual review of AIDS research. New York, N.Y: Marcel Dekker, Inc.; 1991. pp. 41–63. [Google Scholar]
- 66.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 67.Pillemer E A, Kooistra D A, Witte O N, Weissman I L. Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated Gag polyproteins demonstrates their unusual orientation in the cell membrane. J Virol. 1986;57:413–421. doi: 10.1128/jvi.57.2.413-421.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prats A C, De Billy G, Wang P, Darlix J L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 69.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 70.Reil H, Kollmus H, Weidle U H, Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz S, Felber B K, Benko D M, Fenyo E M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwartz S, Felber B K, Fenyo E M, Pavlakis G N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartz S, Felber B K, Pavlakis G N. Expression of human immunodeficiency virus type 1 Vif and Vpr mRNAs is Rev-dependent and regulated by splicing. Virology. 1991;183:677–686. doi: 10.1016/0042-6822(91)90996-o. [DOI] [PubMed] [Google Scholar]
- 77.Schwartz S, Felber B K, Pavlakis G N. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol Cell Biol. 1992;12:207–219. doi: 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SenGupta D N, Berkhout B, Gatignol A, Zhou A M, Silverman R H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shastri N, Nguyen V, Gonzalez F. Major histocompatibility class I molecules can present cryptic translation products to T-cells. J Biol Chem. 1995;270:1088–1091. doi: 10.1074/jbc.270.3.1088. [DOI] [PubMed] [Google Scholar]
- 80.Shatkin A J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 81.Shatkin A J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 82.Sherman F, Stewart J W, Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985;3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- 83.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Teerink H, Voorma H O, Thomas A A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 85.Timmer R T, Benkowski L A, Schodin D, Lax S R, Metz A M, Ravel J M, Browning K S. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J Biol Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- 86.Tobery T W, Siliciano R F. Targeting of HIV-1 antigens for rapid intracellular degradation enhances cytotoxic T lymphocyte (CTL) recognition and the induction of de novo CTL responses in vivo after immunization. J Exp Med. 1997;185:909–920. doi: 10.1084/jem.185.5.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Torrent C, Berlioz C, Darlix J L. Stable MLV-VL30 dicistronic retroviral vectors with a VL30 or MoMLV sequence promoting both packaging of genomic RNA and expression of the 3′ cistron. Hum Gene Ther. 1996;7:603–612. doi: 10.1089/hum.1996.7.5-603. [DOI] [PubMed] [Google Scholar]
- 88.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vacca J P, Dorsey B D, Schleif W A, Levin R B, McDaniel S L, Darke P L, Zugay J, Quintero J C, Blahy O M, Roth E, et al. L-735,524: an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc Natl Acad Sci USA. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 92.Venaud S, Yahi N, Fehrentz J L, Guettari N, Nisato D, Hirsch I, Chermann J C. Inhibition of HIV by an anti-HIV protease synthetic peptide blocks an early step of viral replication. Res Virol. 1992;143:311–319. doi: 10.1016/s0923-2516(06)80119-6. [DOI] [PubMed] [Google Scholar]
- 93.Venkatesan A, Das S, Dasgupta A. Structure and function of a small RNA that selectively inhibits internal ribosome entry site-mediated translation. Nucleic Acids Res. 1999;27:562–572. doi: 10.1093/nar/27.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang S, Browning K S, Miller W A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Williams M A, Lamb R A. Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J Virol. 1989;63:28–35. doi: 10.1128/jvi.63.1.28-35.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]