Abstract
Normal mRNA polyadenylation signals are composed of an AAUAAA motif and G/U box spaced 20 to 30 bp apart. If this spacing is increased further, then polyadenylation is disrupted. Previously it has been demonstrated that insertion of an intron will similarly disrupt this signal even though such introns are removed during a nuclear splicing reaction (X. Liu and J. Mertz, Nucleic Acids Res. 21:5256–5263, 1993). This observation has led to the suggestion that polyadenylation site selection is undertaken prior to intron excision. We now present results that both support and extend these observations and in doing so create a novel class of retroviral expression vector with improved qualities. We found that when an intron-disrupted polyadenylation signal is inserted within a retroviral expression vector, such a signal, although reformed in the producer cell, remains benign until transduction, where it is then preferentially used. Thus, we demonstrate that upon transduction these vectors now produce a majority of shortened subgenomic species and as a consequence have a reduced tendency for subsequent mobilization from transduced cells. In addition, we demonstrate that the use of this internal signal leads to enhanced expression from such vectors and that this is achieved without any loss in titer. Therefore, split polyadenylation signals confer enhanced performance and improved safety upon retroviral expression vectors into which they are inserted. Such split signals may prove useful for the future optimization of retroviral vectors in gene therapy.
Almost all RNA transcripts synthesized by RNA polymerase II contain a tail of between 20 and 250 adenosine residues at their 3′ termini. These residues are added to the RNA by a cleavage and polyadenylation reaction of the pre-mRNA, which is catalyzed by a multicomponent protein complex in the nucleus of the cell (31). The position at which this occurs is determined by the location of a polyadenylation signal found in the 3′ untranslated region of the RNA to be polyadenylated. This signal consists of two elements: the highly conserved AAUAAA hexanucleotide and the more poorly conserved G/U-rich element (the G/U box) normally located 20 to 30 residues downstream (24). The spacing between the two elements is important, as it has been demonstrated that if it is increased beyond 40 nucleotides, the polyadenylation signal becomes disabled (10).
Like polyadenylation, splicing is another posttranscriptional modification of polymerase II-synthesized pre-mRNA transcripts. Although the precise relationship between polyadenylation and splicing of transcripts is not resolved, it is now believed that the former does not actually precede the latter but is instead only seen to precede it because of faster reaction kinetics (16, 22). This is supported by observations that in longer mRNA transcripts in which there is a significant time lag between the synthesis of 5′ and 3′ ends, 5′ splicing reactions can be completed prior to synthesis of the 3′ polyadenylation signal and thus prior to polyadenylation (4, 18, 23). Consequently, most studies in this field are instead concerned only with the relationship between the polyadenylation signal and the 3′ terminal intron, and it is now thought that the 3′ splice site of this intron and polyadenylation signals can in some way cooperate (3, 26).
Previously, Liu and Mertz (20) chose to investigate not the order in which splicing and polyadenylation occur but rather the order in which splicing and polyadenylation site selection occur. This was undertaken by disrupting the optimal spacing of the AAUAAA hexanucleotide and G/U box of a polyadenylation signal by intron insertion. They demonstrated that when such an intron-containing signal is placed within an RNA it is never used in vivo, even though the optimal AAUAAA and G/U box spacing is restored by splicing of the transcript. Consequently they showed that unless a second, functional polyadenylation signal is present downstream, transcripts that harbor such intron-disrupted polyadenylation signals (IDPAs) are never polyadenylated. Because of these findings, they tentatively concluded that polyadenylation site selection must occur at an early step in mRNA processing and prior to 3′ intron excision.
In a retrovirus, the R-U5 border defines the precise point at which the genomic transcript is cleaved and polyadenylated. Consequently, for most such viruses the G/U box is located in U5 while the AAUAAA motif is just upstream in R. Exceptions to this, however, include the signal found in human T-cell lymphotropic virus type 1 (HTLV-1) in which the AAUAAA box is located farther away, 276 residues upstream in U3 (1, 5). By such positioning, HTLV-1 thus ensures that only one copy of its complete polyadenylation signal is present per viral transcript, as the 5′ U3 of a provirus is never transcribed. Despite this arrangement, normal spacing between the two elements is still a prerequirement for such viruses for efficient polyadenylation, and thus a highly ordered R region is used to physically draw the two elements to the equivalent of 20 to 30 bases apart (1, 5).
Unlike the polyadenylation reaction of most host cell mRNAs, the full-length genomic viral transcript is polyadenylated but unspliced, and thus cooperation between the 3′ intron and the polyadenylation signal is not observed. Perhaps partly due to this, transcript read-through in an integrated provirus can rise to as high as 15% (11, 13). As a consequence, viral transcripts can sometimes fuse with the host cell sequence located downstream of the site of integration and eventually utilize host cell elements as polyadenylation signals. However, as is the case with low-fidelity reverse transcriptase, such read-through may not simply be viral inefficiency but instead be evolutionarily advantageous. This is because by such read-through a retrovirus is occasionally supplied with new heterologous RNA sequences of host cell origin, which it might utilize by recombination events within the diploid virion (11, 29, 33).
For retrovirus-derived expression vectors, such transcript read-through is, however, undesirable, as it will lead to both lowered expression levels and transcript-genome RNA fusions. In spite of this fact, most of the commonly used lentivirus- and oncoretrovirus-derived expression vectors still tend to use such viral polyadenylation signals for desired cDNA expression in transduced cells. For this reason we have attempted to improve the signal used by such vectors in transduced cells. Presented here is a prototypic murine leukemia virus-derived vector in which this is achieved by exploiting the single-cycle infection profile of expression vectors and the use of IDPAs.
MATERIALS AND METHODS
Cell culture.
293T (9) and HT1080 (25) cell lines were maintained in Dulbecco's modified Eagle medium containing 10% (vol/vol) fetal calf serum supplemented with penicillin, streptomycin, and l-glutamine.
Transient three-plasmid expression system and viral titration.
Retroviral vector stocks were produced according to the three-plasmid transient-transfection protocol described by Soneoka et al. (28). Briefly, 10 μg each of pHIT456 (4070A envelope), pHIT60 (gag-pol), and the appropriate vector was transfected by overnight calcium phosphate treatment into 293T cells at 70 to 80% confluence in 10-cm-diameter dishes. The following morning, the cell medium was replaced with medium containing sodium butyrate (10 mM final concentration) for 12 h. Subsequently this medium was replaced with 5 ml of fresh medium, and the viral supernatants were harvested 12 h later (therefore 48 h posttransfection).
Target cells were subcultured into six-well plates 24 h prior to virus harvest so that they were 50% confluent on the day of transduction (usually about 1 × 105 to 2 × 105 cells per well). Just before transduction, the virus supernatants were diluted up to 106-fold in fresh medium containing Polybrene (8 μg/ml). One milliliter of each different viral dilution was added to each well. After 2 h, an additional 1 ml of fresh medium was added to each well. Transduced cells were incubated for 48 h at 37°C in a 5% CO2 atmosphere before viral titers were established.
DNA analysis.
Genomic DNA was prepared from HT1080 stable cell lines with the DNeasy Tissue Kit (Qiagen). The HT1080 cell lines were transduced by the different viral supernatants produced by transient transfection and then selected on neomycin (1 mg/ml) for 3 weeks.
PCR amplification was used to analyze the IDPA fragment in the integrated vectors using a forward primer (Fp: AAGCGCGATCACATGGTCC) and a reverse primer (Rp: AAGACGGCAATATGGTGGA) that flank this fragment. The PCR product was then cloned into pGEM-Teasy (Promega, Madison, Wis.) and sequenced.
RNA analysis.
Total RNA was prepared with the RNeasy Mini Kit (Qiagen). RNA was extracted from transduced HT1080 stable cell lines that had been selected on neomycin (1 mg/ml) for 3 weeks. Selection started 48 h after transduction with viral supernatants produced by three-plasmid transient transfection of 293T cells (28). A 10-μg amount of each RNA sample was separated on a 1% denaturing agarose gel and then blotted onto a Hybond-N+ filter (Amersham). After UV cross-linking, the membranes were hybridized to a [32P]CTP-labeled enhanced green fluorescent protein (EGFP) probe overnight. The filters were washed three times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 37°C and three times with 0.1× SSC–0.1% sodium dodecyl sulfate at 50°C. The membrane was exposed for 12 h to a phosphorimager screen, and the RNA bands were analyzed with a Storm 860 Imager (Molecular Dynamics).
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis of EGFP was performed using the FACSCaliber System (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). Samples were prepared from trypsinized cells cultured on 6-cm-diameter dishes and resuspended in 0.5 ml of phosphate-buffered saline.
RESULTS
Vector design and construction.
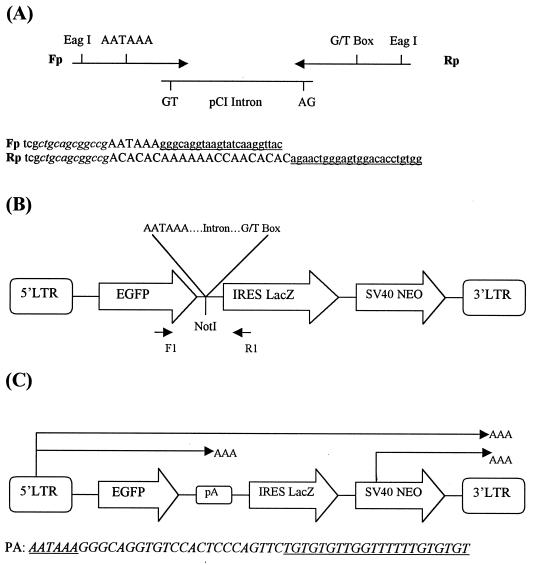
To investigate the potential for using IDPAs within a retroviral expression cassette, we placed such a signal downstream of EGFP and upstream of the internal ribosome entry site (IRES) in a long terminal repeat (LTR)-driven EGFP-IRES-LacZ reporter cassette contained within a derivative of the Moloney murine leukemia virus-based expression vector pHIT111, named pGiresZN (12). Also contained within this vector is a separate downstream simian virus 40 promoter-neomycin phosphotransferase cassette. As a control, the same IDPA was also inserted at the same location but in the reverse orientation to make the reverse complement sequence such that intron excision will not occur. The resulting vectors, named pAINT [for poly(A) intron] and pAINT-R, respectively, are shown in Fig. 1. Also shown is the consensus polyadenylation sequence generated upon intron removal. The intron used in this study was taken from the expression vector pCI (Promega).
FIG. 1.
Design and function of the pAINT vectors. (A) Design of the IDPA. The forward (Fp) and reverse (Rp) primers shown were used to PCR amplify the intron from the pCI expression vector (Promega). The primers include 3′ sequences complementary to the pCI intron (underlined), the consensus AAUAAA and G/U box sequence elements that comprise a polyadenylation signal (shown in uppercase letters), and useful unique restriction sites (italic) (PstI and EagI). The resulting PCR product consists of a polyadenylation signal separated by the pCI-derived intron. (B) Schematic outline of the pAINT vector. This vector was constructed by insertion of the EagI-EagI PCR-amplified IDPA fragment into a unique NotI site located downstream of the EGFP and upstream of the IRES sequence of the expression vector pGiresZN (12). Also constructed was the control vector pAINT-R, in which the IDPA is inserted in the reverse orientation to include the reverse complement sequence of the IDPA within the vector transcript. The reverse complement sequence contains no functional splice sites or polyadenylation elements. (C) Schematic representation of potential genomic and subgenomic transcripts made by the pAINT vector. Also shown is the DNA sequence of the intron-excised reconstituted polyadenylation site (PA).
Comparative primary and secondary titer analysis.
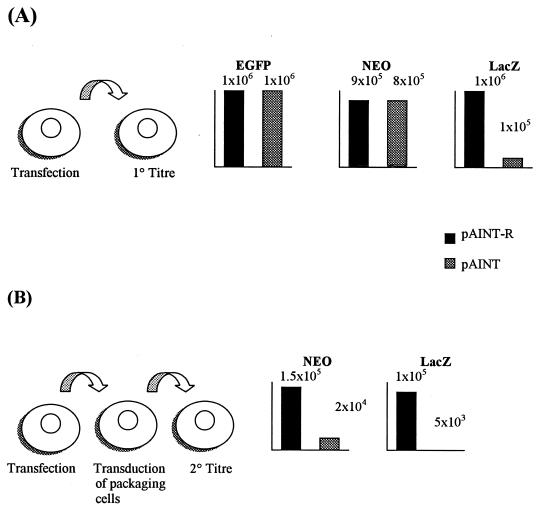
It has been shown that within a short mRNA transcript, an intron-disabled polyadenylation signal is not selected for cleavage and polyadenylation, even after the intron is excised (20). For this reason it might be expected that inclusion of an IDPA within a retroviral expression vector would have little effect on full-length viral transcript production and thus little effect on the primary titers produced. However, because of this intron excision, it might also be expected that after one round of transduction from such vectors, the now reconstituted polyadenylation signal would be selected for in preference to that of the downstream LTR signal in transduced cells. Utilization of this internal polyadenylation site would result in the production of only short EGFP mRNA transcripts. As a consequence, it would be expected that any subsequent, secondary titer produced from transduced cells would be lower because essential viral elements required for reverse transcription and integration (the 3′-polypurine tract, U3, and R) would no longer be present in the shorter subgenomic transcript. For this reason, the pAINT and pAINT-R viral titers from both a primary harvest (taken from transfected 293T producer cells) and secondary harvest (taken from a transduced packaging cell line) were compared. The results of this comparison are presented in Fig. 2 and demonstrate that while inclusion of a sense-oriented IDPA in pAINT has no effect on Neo or EGFP primary titers, subsequent secondary titers drop by 90% compared to those with the control vector pAINT-R. This therefore supports the model in which the initial intron-excised signal is available only for site selection in subsequent transduced cells. Further support for this model stems from LacZ titer analysis. This is because even the primary titers conferred by LacZ in the pAINT vector drop by 90% (and significantly more if X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] staining is shortened from 12 to 1 h [data not shown]). This apparent drop in even primary titers when using LacZ would also be expected because LacZ expression in the pAINT vector is from an IRES and thus expressed only if the upstream polyadenylation signal is not utilized.
FIG. 2.
Analysis of pAINT primary and secondary titers. (A) Primary titers of pAINT and pAINT-R expression vectors. pAINT and pAINT-R vector stocks were made by the three-plasmid transient-transfection method, and neomycin resistance (NEO; CFU per milliliter), EGFP (GFP forming units per milliliter), and LacZ (LacZ forming units per milliliter) titers were then subsequently established. Data show that upon transduction and relative to the control, LacZ titers from pAINT drop by 90% while EGFP and NEO titers remain unaffected. This suggests that there is a reduction in full-length IRES-LacZ-containing transcript in the pAINT vector, which would signify the use of the subgenomic polyadenylation signal in the transduced cell. (B) Secondary titers of the pAINT and pAINT-R vectors. One milliliter of the viral stocks used for panel A was added to the HT1080-derived packaging line FLY-RD18 (8). After 24 h, the viral supernatant on the FLY-RD18 cells was replaced with fresh medium. Forty-eight hours later, a viral harvest was undertaken and subsequent secondary titers were established. Data show that relative to the control the neomycin resistance and LacZ secondary titers are reduced in the pAINT vector. This would support a model in which upon transduction into the FLY-RD18 packaging line, the intron-excised polyadenylation signal is utilized such that fewer full-length genomic transcripts are produced and thus the subsequent titer is reduced.
pAINT transcript analysis.
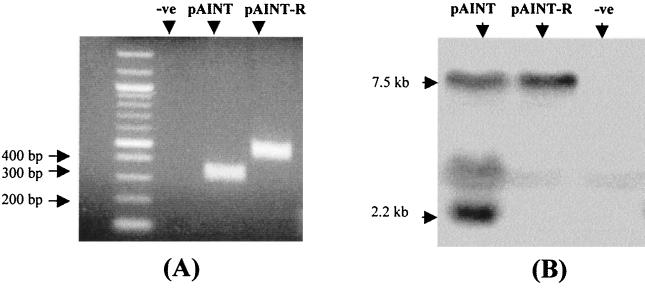
Although the primary LacZ titers and secondary Neo titers are reduced, this reduction is not absolute, which therefore suggests that upon transduction the internal polyadenylation signal is not always used. This might be either because intron excision in the producer cells is not complete and thus a reconstituted polyadenylation signal is not always present in transduced cells or because although intron excision is complete, the subsequent signal is not always used in a transduced cell. The most sensitive method to investigate if intron excision is completed by the time of transduction is PCR. Consequently, primers were designed at either side of the IDPA and PCR was performed on genomic DNA extracts taken from stable cells lines transduced with pAINT and pAINT-R. If intron excision was completed in the nucleus of the producer cell, then only a shorter intron-minus form of the integrated provirus would be detected. The results of this analysis are presented in Fig. 3A and reveal that while no intron-excised provirus can be detected in the pAINT-R control vector, only intron-excised provirus can be detected in the pAINT vectors. The identity of these PCR products was confirmed by sequencing. This therefore demonstrates that intron excision is completed during viral production and thus suggests that any remaining full-length transcripts produced in transduced cells are because the internal, reconstituted polyadenylation signal is not always utilized. To investigate if this is indeed the case, Northern analysis of cells transduced with either pAINT or pAINT-R was undertaken. The results of this analysis are shown in Fig. 3B and reveal that while there exists an abundance of shortened subgenomic transcripts that terminate at the internal reconstituted polyadenylation signal found in pAINT, there is still a significant level of full-length transcript synthesized from such vectors. Consequently it can be concluded that within the context of a retrovirus, an internal, consensus, optimal polyadenylation signal is not always used. This contrasts with the results of a number of previous studies using far shorter nonviral mRNA transcripts which demonstrate that such a signal is always used in preference of downstream equivalents (5, 20).
FIG. 3.
Analysis of pAINT poly(A) site utilization. (A) PCR-based analysis of the integrated proviral vector in stable HT1080 cell lines. Forward (F1) and reverse (R1) primers were so designed to amplify the IDPA sequence from genomic DNA extracted from either pAINT- or pAINT-R-transduced cells (Fig. 1B). While the amplified product derived from control vector (pAINT-R)-transduced cells is still of full length, the product from cells transduced with the pAINT vector is smaller. Subsequent cloning and sequence analysis confirmed that the smaller product lacked the intronic sequence observed in the control vector-derived amplicon. Intron excision means that the AAUAAA and G/U elements are now correctly spaced to generate a functional polyadenylation signal (Fig. 1C). Genomic DNA from HT1080 cells was used as a negative PCR control. (B) Northern analysis of vector transcripts in transduced HT1080 stable cell lines. An EGFP cDNA probe was used to discern the genomic to subgenomic ratios of vector transcripts present in transduced cells. While control (pAINT-R) and transduced cells contain only full-length genomic transcripts, those cells transduced with pAINT contain a significant fraction of smaller subgenomic species of a size expected if the internal intron-excised polyadenylation site is utilized. Total RNA from HT1080 cells was used as a negative control.
Expression levels.
During EGFP titer analysis it was observed that relative to the control vector, the EGFP expression levels from the pAINT vector appeared visually higher. This suggests that the presence of an internal polyadenylation signal enhances the expression of the upstream gene. This would be in agreement with previous observations regarding the use of strong polyadenylation signals (19, 30, 32). To confirm this observation, we undertook FACS analysis on cells both transfected and transduced with both pAINT and pAINT-R. The results of this analysis can be seen in Table 1 and reveal that while EGFP expression levels are similar in transfected cells, in transduced cells the pAINT vector produces 40% higher fluorescence. Therefore, only once it is reconstituted does the polyadenylation signal confer higher expression levels on the upstream EGFP marker.
TABLE 1.
EGFP expression analysisa
| Vector | EGFP fluorescence in:
|
|
|---|---|---|
| Transfected cells | Transduced cells | |
| pAINT | 3,322.2 ± 400.1 | 587.5 ± 37.6 |
| pAINT-R | 3,272.7 ± 77.4 | 305.5 ± 14.2 |
FACS-based EGFP analysis was performed on cells transfected and transduced with pAINT and the control vector pAINT-R. EGFP levels in 293T cells transfected by the pAINT vector are identical to levels from cells transfected by the control vector pAINT-R. Upon transduction, however, there is a significant increase (40 to 50%) in EGFP expression levels only in cells transduced by pAINT vector. This enhanced expression correlates with the observed use of the internal polyadenylation signal by the pAINT vector as described in the legend to Fig. 3. The results are means ± standard deviations (n = 3).
DISCUSSION
Efficient polyadenylation has been suggested to influence virtually all aspects of mRNA metabolism. Its proposed functions include conferring mRNA stability, promoting mRNA translational efficiency, and playing a role in transport of processed mRNA from the nucleus to the cytoplasm (7, 19, 27, 32).
In some retroviruses, polyadenylation at the correct site is not always accomplished (6, 11, 13). For example, in the case of avian leukosis virus (ALV), it has been demonstrated that a significant number (15%) of viral transcripts retain cellular sequences (11). The major biological significance of these read-through transcripts is likely to be their role in the acquisition of cellular proto-oncogenes, as read-through transcripts can still be packaged into virions (29). However, unlike ALV, in most other retroviruses (e.g., human immunodeficiency virus [HIV] and spleen necrosis virus [SNV]) the proportion of read-through transcripts generated is much lower, suggesting more efficient utilization of their 3′ LTR polyadenylation site. Nevertheless, in expression vectors derived from both HIV and SNV, the inclusion of nonretroviral polyadenylation signals downstream of the 3′ LTR still resulted in increases in steady-state RNA levels in the producer and improved vector titer (13, 14). This demonstrates that most if not all retrovirus-derived expression vectors would still benefit from improved polyadenylation signals. Presently, however, and as described above for HIV- and SNV-based vectors, most polyadenylation signal improvements are restricted to transfected producer cells because upon transduction the less efficient 5′ U5 supplies the polyadenylation signal for the 3′ LTR. Consequently there remains no simple manner by which polyadenylation signals within a retroviral vector can be optimized in a transduced cell, and it is for this reason that we have investigated the potential use of IDPAs within retroviral vectors. The results presented show that the inclusion of such intron-disabled polyadenylation signals led to significant improvements in expression levels of a gene upstream of the IDPA, reduced marker levels from the downstream LacZ gene, and reduced risk of subsequent vector mobilization within transduced cells. Such traits are all desirable in retroviral expression vectors, be they oncoretroviral or lentiviral in origin. Regarding their use in lentiviral vectors, however, it must be noted that REV/RRE requirements of such vectors might impede efficient intron excision from IDPAs in some circumstances. Therefore, minimal lentiviral vectors in which REV/RRE elements are no longer included, such as those recently described by our laboratory (15), might be the preferred choice of lentiviral vector for IDPA inclusion.
Upon transduction, polyadenylation at the internal, reconstituted polyadenylation signal in the vector described here is not 100% efficient. Precisely why this is the case remains unclear, but the result does support previous observations regarding the inefficient use of polyadenylation signals inserted within a retrovirus (2, 21). Such observed inefficiency might be due to the influence of neighboring viral sequence elements such as the major splice donor and/or the proximity of the LTRs. Consequently, to increase the observed subgenomic-to-genomic transcript ratio still further, extra modifications such as using multiple IDPAs might be required in future.
The work presented here confirms and extends the observations made by Liu and Mertz (20), who demonstrated that short transcripts containing IDPAs upstream of a conventional signal failed to be polyadenylated at the IDPA even though the intron had been properly excised to regenerate a functional site. Unlike the Liu and Mertz findings, however, the reconstituted yet benign polyadenylation signal in this study is located 5.0 kb upstream of the 3′ LTR signal. Therefore, the distance between the IDPA and downstream polyadenylation signal is far greater than that in the previous study and would suggest a sufficient time lag to allow intron excision even prior to the synthesis of the 3′ LTR signal. There are a number of possibilities why an intron-excised, reconstituted signal is simply never used in transfected cells. First, it might be due to an artifact of the IDPA design, that the splicing machinery masks the intron-excised signal from site selection. For example, a recent report shows evidence that pre-mRNA splicing leads to stable association of proteins at exon-exon junctions (17). This may interfere with selection of an overlapping poly(A) signal. An alternative possibility is that polyadenylation site selection and splicing truly do occur in a stepwise manner, with site selection occurring prior to splicing machinery engagement and never after. A third and related possibility is that polyadenylation site selection occurs at a very early stage close to the point of synthesis or, and somewhat unimaginably, even earlier. Such discussions aside and on a more pragmatic note, the results presented here suggest that IDPA insertion may have universal appeal in future vector design because although not 100% efficient, inclusion is relatively simple and can still confer significant advantages on retroviral expression vectors in which they are contained.
ACKNOWLEDGMENTS
S. I. Ismail was supported by the Karim Rida Said Foundation and has also been awarded an Overseas Research Studentship by the University of Oxford.
We gratefully acknowledge Melvin Yap, Kyriacos Mitrophanous, Ekaterini Kotsopoulou, and Rachel Harrison for their excellent help and advice.
REFERENCES
- 1.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 2.Bender A M, Miller A D, Gelinas R E. Expression of the human beta-globin gene after retroviral transfer into murine erythroleukemia cells and human BFU-E cells. Mol Cell Biol. 1988;8:1725–1735. doi: 10.1128/mcb.8.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 4.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 5.Brown P H, Tiley L S, Cullen B R. Effect of RNA secondary structure on polyadenylation site selection. Genes Dev. 1991;5:1277–1284. doi: 10.1101/gad.5.7.1277. [DOI] [PubMed] [Google Scholar]
- 6.Coffin J M, Hughs S H, Varmus H E. Synthesis and processing of viral RNA. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 7.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 8.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuBridge R B, Tang P, Hsia H C, Leong P-M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil A, Proudfoot N J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 11.Herman S A, Coffin J M. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science. 1987;236:845–848. doi: 10.1126/science.3033828. [DOI] [PubMed] [Google Scholar]
- 12.Ismail S I, Kingsman S M, Kingsman A J, Uden M. Split-intron retroviral vectors: enhanced expression with improved safety. J Virol. 2000;74:2365–2371. doi: 10.1128/jvi.74.5.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwakuma T, Cui Y, Chang L J. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology. 1999;261:120–132. doi: 10.1006/viro.1999.9850. [DOI] [PubMed] [Google Scholar]
- 14.Iwasaki K, Temin H M. The U3 region is not necessary for 3′ end formation of spleen necrosis virus RNA. J Virol. 1990;64:6329–6334. doi: 10.1128/jvi.64.12.6329-6334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotsopoulou E, Kim V N, Kingsman A J, Kingsman S M, Mitrophanous K A. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leff S E, Rosenfeld M G, Evans R M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- 17.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 18.LeMaire M F, Thummel C S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis J D, Gunderson S I, Mattaj I W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J Cell Sci Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Mertz J E. Polyadenylation site selection cannot occur in vivo after excision of the 3′-terminal intron. Nucleic Acids Res. 1993;21:5256–5263. doi: 10.1093/nar/21.22.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller A D, Ong E S, Rosenfeld M G, Verma I M, Evans R M. Infectious and selectable retrovirus containing an inducible rat growth hormone minigene. Science. 1984;225:993–998. doi: 10.1126/science.6089340. [DOI] [PubMed] [Google Scholar]
- 22.Nevins J R, Darnell J E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978;15:1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- 23.Ohno M, Sakamoto H, Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc Natl Acad Sci USA. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 25.Rasheed S, Nelson-Rees W A, Toth E M, Arnstein P, Gardner M B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs A B, Buratowski S. Common themes in translational and transcriptional regulation. Trends Biochem Sci. 1997;22:189–192. doi: 10.1016/s0968-0004(97)01051-7. [DOI] [PubMed] [Google Scholar]
- 28.Soneoka Y, Cannon P M, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:623–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swain A, Coffin J M. Polyadenylation at correct sites in genome RNA is not required for retrovirus replication or genome encapsidation. J Virol. 1989;63:3301–3306. doi: 10.1128/jvi.63.8.3301-3306.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitelaw E, Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 32.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q Y, Clausen P A, Yatsula B A, Calothy G, Blair D G. Mutation of polyadenylation signals generates murine retroviruses that produce fused virus-cell RNA transcripts at high frequency. Virology. 1998;241:80–93. doi: 10.1006/viro.1997.8947. [DOI] [PubMed] [Google Scholar]