Abstract
Interaction between viral proteins is necessary for viral replication and viral particle assembly. We used the yeast two-hybrid assay to identify interactions among all the mature proteins of the hepatitis C virus. The interaction between NS3 and NS3 was one of the strongest viral protein-protein interactions detected. The minimal region required for this interaction was mapped to a specific subdomain of 174 amino acids in the N terminus of the helicase region. Random mutations in the minimal region were generated by PCR, and mutants that failed to interact with a wild-type minimal fragment were isolated using the yeast two-hybrid assay as a screen. Three of these mutations resulted in a reduction or a loss of interaction between helicases. Analytical gel filtration showed that in the presence of an oligonucleotide, wild-type helicases form dimers whereas the mutants remain mostly monomeric. All three mutants were partially or almost inactive when assayed for helicase activity in vitro. Mixing a mutant helicase (Y267S) with wild-type helicase did not dramatically affect helicase activity. These data indicate that dimerization of the helicase is important for helicase activity. The mutations that reduce self-association of the helicase may define the key residues involved in NS3-NS3 dimerization.
Hepatitis C virus (HCV) is a positive-strand RNA virus that belongs to the family Flaviviridae. It is one of the major causes of liver disease, affecting 1 to 2% of the population in most developed countries. Seventy-five percent of HCV infections are chronic; up to 20% of these develop into liver cirrhosis, and another 1 to 5% of cases lead to hepatocellular carcinoma. To date, treatment of this infection using interferon and ribavirin has not been sufficiently effective. The discovery of new drugs to counter HCV-associated liver conditions has been hampered by the lack of reliable cell culture and inexpensive animal model systems.
The HCV positive-strand RNA genome is translated in the host cell into a large polypeptide of about 3,010 amino acids (aa), which is cleaved into 10 individual protein products (6, 20). The three structural proteins—the core (C), two envelope proteins (E1 and E2), and p7, a small peptide of unknown function—are released by host peptidases. The nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B are proteolytic products of NS3, which has a trypsin-like protease domain in the N-terminal one-third of the protein. The assembly of a complex of C with the viral RNA, as well as with E1 and E2, has been described, and this assembly is presumably important for packaging of viral particles. Interaction among the nonstructural proteins has also been shown (18, 22). NS4A acts as an activating cofactor for NS3, and its binding to NS3 modulates the protease activity for NS3-NS4A, NS4A-NS4B, and NS5A-NS5B cleavage sites (14). NS4A binding also serves to uncouple the helicase and ATPase–single-stranded-RNA binding activities of NS3 (7). NS5B, the RNA-dependent RNA polymerase, interacts with NS3 (10), which has a helicase domain in the C-terminal two-thirds of the protein, and this association is probably important for the replication of the viral RNA. NS4A, NS4B, and NS5A have been found to form a complex (16). These data show that nonstructural viral proteins form complexes with one another and possibly also recruit cellular proteins for some process(es) in viral replication and maturation. Long-term sequestration of host proteins for viral functions may indirectly lead to pathogenesis of the liver.
Many antiviral drug screens have been targeted at NS3, the nonstructural protein that has protease, nucleoside triphosphatase (NTPase), and helicase activities. The structures of both protease and helicase domains of NS3 have been elucidated by X-ray crystallography (5, 12, 13, 19). The NS3 helicase belongs to the DEXH family of helicases, with conserved motifs essential for NTP-binding, NTPase, helicase (G207SGKST, D290ECH, T322AT), and RNA-binding (Q460RRGRTGRGRGG) activities. It has been suggested to form a dimer, with a cleft between the components of the dimer through which a single-stranded nucleotide can pass as the helicase unwinds the RNA (5). Other helicases are also known to form and function as dimers or oligomers (1, 3). Dimerization or oligomerization of NS3 helicase has been implicated to be necessary for optimal helicase activity (15), although other studies report that the HCV helicase is monomeric in solution (25) and dimerization is probably not important for helicase activity (17).
Little is known about the role of interactions between HCV proteins in viral replication. We identified viral protein-protein interacting partners using the yeast two-hybrid system and found that NS3 interacts strongly with itself. A fragment of 174 amino acids near the N terminus of the helicase region was defined to be the minimal region necessary for interaction of the region with NS3 and with itself. Under the conditions used for gel filtration, we showed that the presence of a single-stranded oligonucleotide is absolutely required for dimerization. Higher oligomers were not detected. To test the hypothesis that the activity of the HCV helicase is dependent on dimer formation, we generated mutants that showed little or no interaction with another helicase molecule. A reduction in dimerization is correlated with a decrease in helicase activities in three mutants tested. The residues that are involved in dimerization were thus identified, and this could provide useful information for designing compounds that inhibit protein-protein interaction at these positions.
MATERIALS AND METHODS
Construction of plasmids.
cDNA clones encoding NS3 were prepared from HCV-infected serum by reverse transcription-PCR (S. P. Lim et al., unpublished results). For the yeast two-hybrid interaction assays (4), NS3 clones were fused in frame with the Gal4 DNA binding domain (BD) and the Gal4 activating domain in pAS2–1 and pACT2 vectors (Clontech), respectively. Truncations of NS3 are summarized in Table 1 and Fig. 1A. For protein expression, DNA fragments were cloned in frame with the glutathione S-transferase (GST) coding region in a modified pGEX-2TK plasmid (Pharmacia). Proteins were tagged with Flag or myc in pXJ40 (28) for expression in mammalian cells (Table 1).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pAS2-1 | Gal4 DNA-binding domain in a 2μm TRP1 yeast shuttle vector | Clontech Inc. |
| pACT2 | Gal4 activation binding domain in 2μm LEU2 yeast shuttle vector | Clontech Inc. |
| pFG105 | NS3 (aa 1–680) in pAS2-1 | This work |
| pFG106 | NS3 (aa 1–680) in pACT2 | This work |
| pFG109 | myc-NS3 in pXJ40 | This work |
| pFG114 | Flag-NS3 in pXJ40 | This work |
| pFG134 | pAS-CΔ-Stul (NS3 aa 1–538) | This work |
| pFG135 | pAS-CΔ-AflIII (NS3 aa 1–430) | This work |
| pFG136 | pAS-CΔ-BstEII (NS3 aa 1–328) | This work |
| pFG137 | pAS-CΔ-SspI (NS3 aa 1–335) | This work |
| pFG138 | pAS-CΔ-SphI (NS3 aa 1–149) | This work |
| pFG146 | pAS-NΔ-Smal (NS3 aa 162–680) | This work |
| pFG147 | pAS-NΔ-NdeI (NS3 aa 247–680) | This work |
| pFG148 | pAS-NΔ-Sspl (NS3 aa 336–680) | This work |
| pFG150 | pAS-min (SmaI-SspI, NS3 aa 162–335) | This work |
| pFG156 | pAS-NΔ-Ncol (NS3 aa 57–680) | This work |
| pFG157 | pAS-helicase (NS3 aa 182–680) | This work |
| pFG158 | pACT-helicase (NS3 aa 182–680) | This work |
| pFG159 | pAS-Smal-Ndel (NS3 aa 162–246) | This work |
| pFG161 | pAS-Ndel-Sspl (NS3 aa 247–335) | This work |
| pFG162 | pGEX-helicase (NS3 aa 182–680) | This work |
| pFG166 | pFG150 cut with EagI and BstEII and religated | This work |
| pFG171 | pACT-min (SmaI-SacI, NS3 aa 182–375) | This work |
| pFG188 | myc-min (NS3 aa 182–375) in pXJ40 | This work |
| pXJ40 | Mammalian expression vector with cytomegalovirus promoter, Kozak sequence, and simianvirus 40 poly(A) termination signal | 28 |
| pXJ40-myc | Mammalian expression vector for tagging proteins with c-myc at the N terminus | V. Yu lab, IMCBa |
| pXJ40-flag | Mammalian expression vector for tagging proteins with Flag at the N terminus | 21 |
| pGEX-2TX | GST fusion expression vector with modified multiple cloning sites | S. C. Lin lab, IMCBa |
IMCB, Institute of Molecular and Cell Biology, Singapore.
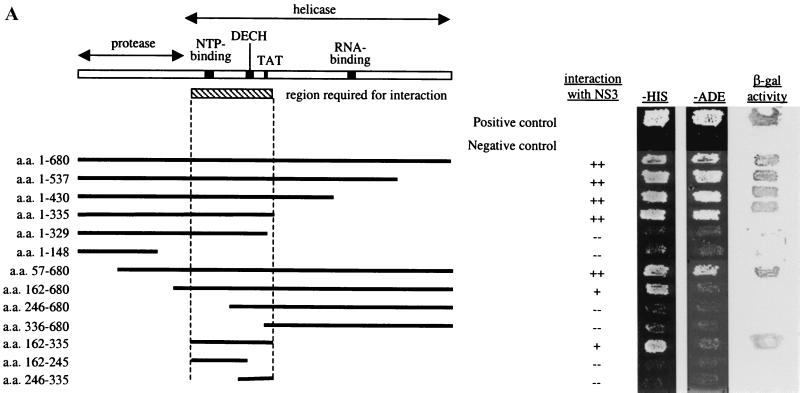
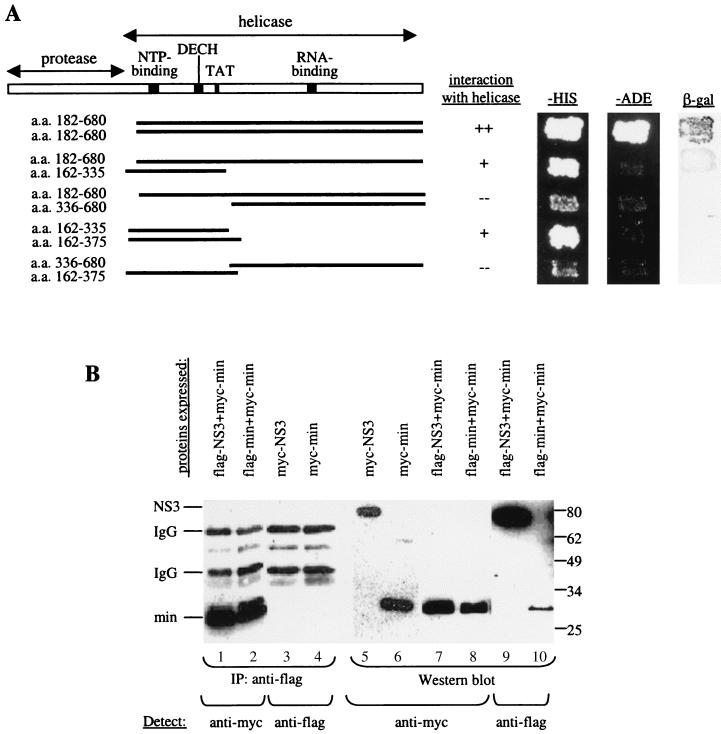
FIG. 1.
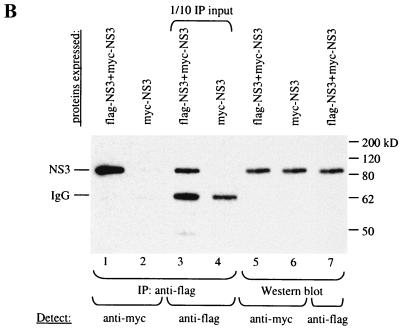
(A) A minimal domain of NS3 required for interaction was defined using the yeast two-hybrid assay. N- and C-terminal truncations in NS3 were made, and the fragments were cloned into pAS2–1. These were transformed into PJ69–2A, and transformants were mated with Y187 carrying pACT-NS3. The resulting diploids were patched onto −Trp −Leu plates, replica plated onto −His and −Ade plates, and also lifted onto a nylon membrane for the β-Gal assay. A fragment encompassing aa 182 to 335 near the N terminus of the helicase domain, which includes conserved residues important for NTP-binding, NTPase, and helicase activities, was defined to be the minimal region required for interaction with NS3. Positive interactions were indicated by growth on −His and −Ade plates and by the presence of β-Gal activity. (B) IP between Flag-NS3 and myc-tagged NS3 proteins. COS cells were transfected with Flag-NS3 and myc-NS3 or with myc-NS3 alone. Total protein (100 μg) from transfected cells was used for IP using an anti-Flag agarose gel. Lane 1, myc-NS3 was detected in the complex precipitated with an anti-Flag gel, demonstrating interaction between myc-NS3 and Flag-NS3. Lane 2, myc-NS3 was not precipitated by the anti-Flag gel in the absence of Flag-NS3, showing the specificity of the anti-Flag gel used for IP. Lanes 3 and 4, 1/10 of the input IP proteins was probed with anti-Flag antibody to show that Flag-NS3 was specifically precipitated by the anti-Flag gel. Lanes 5 through 7, Western blot of total cell lysate (20 μg loaded) to detect the expression of each of the tagged NS3 proteins in transfected cells. IgG, immunoglobulin G.
Manipulation of yeasts and yeast two-hybrid assays.
Manipulation of yeasts for two-hybrid assays was done as described in the Matchmaker user's manual (Clontech). In this system, the presence of an interaction between two viral proteins was indicated by the activation of the reporter genes HIS3 and ADE2, which allow for growth on −His and −Ade media, respectively, and LacZ, a β-galactosidase (β-Gal) that produces a blue color when the substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Sigma) is cleaved. The activation of these three reporters indicates the strength of the interactions, with LacZ+ and Ade+ phenotypes indicating stronger interaction than the His+ phenotype. The pAS2–1 and pACT2 constructs were transformed into PJ69–2A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 ade2-101 gal4Δ gal80Δ LYS2::GALUAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2) and Y187 (MATα trp1-901 leu2-3,112 ura3-52 his3-200 ade2-101 gal4Δ gal80Δ met− URA3::GALUAS-GAL1TATA-LACZ), respectively. To test for interaction, the two strains carrying various fragments of NS3 were mated on yeast extract-peptone-dextrose (YEPD) plates and then transferred onto −Trp −Leu plates to select for diploids that have both plasmids. They were then replica plated onto −Trp −Leu −His and −Trp −Leu −Ade plates and also assayed for β-Gal activity. We confirmed the interactions by retransforming the pairs of constructs into the haploid strains PJ69–2A and Y187 and assaying for the activation of the appropriate reporter genes.
The β-Gal activity assay is done by lifting patches of cells from a plate onto a nylon membrane (Hybond N; Amersham), placing the membrane, with the cells facing up, on a fresh plate to allow the patches of cells to grow for a day, and then performing a “blue” assay on the membrane. The membrane was dipped in liquid nitrogen for a few seconds and then transferred (cells facing up) onto a piece of filter paper (3M) soaked in 1.8 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and 25 μl of X-Gal (25 mg/ml dissolved in dimethylformamide). Membranes were incubated at 30°C to allow the blue color to develop (up to a few hours).
Generation of mutations in NS3 helicase.
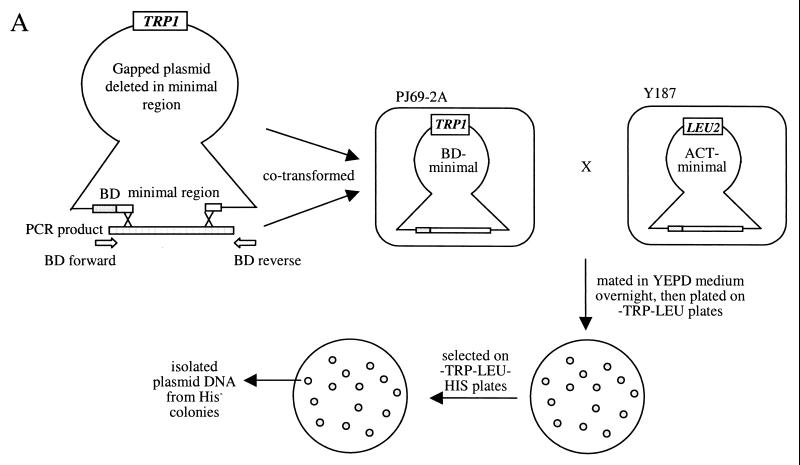
A two-step PCR was used to generate site-directed mutations (9). Random mutations in the minimal region were generated by lowering the nucleotide ratio of each of the four nucleotides to 1:4:4:4 (final concentrations are 25 μM:100 μM) in four separate PCRs using BD forward and BD reverse primers (see Fig. 4A) on template pFG150 (pAS-min). The products of the four reactions were pooled, and the resulting product was cotransformed with a gapped plasmid containing a deletion in the minimal region into PJ69–2A (pFG166 cut with EagI). Plasmids become repaired in yeast to generate a circular plasmid (24). This pool of transformed cells was mated overnight with a Y187 strain containing pACT-min in YEPD liquid medium. An aliquot of mated cells was plated on −Trp −Leu medium, while the rest were kept at 4°C. This gives an estimate of the amount of cells to be plated to give an appropriate density for screening (about 500 to 700 per plate). After about 2 days, the remaining cells were diluted accordingly and then plated on −Trp −Leu plates and incubated at 30°C until colonies appeared. The colonies were replica plated onto −His −Trp −Leu plates to identify colonies that could not grow on −His medium, indicating a lack of interaction. Plasmids from these mutants were extracted and transformed into bacteria. To quickly check that they contained a properly repaired plasmid, PCR was done directly on the bacterial cells with BD forward and BD reverse primers. Those that contained an insert were amplified and retransformed into PJ69–2A carrying pACT-min and tested for interaction on −His −Trp −Leu medium.
FIG. 4.
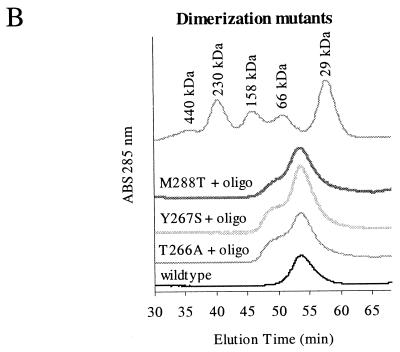
Random mutations that disrupt interaction in the minimal region. (A) Scheme showing how the screen for random mutations that disrupt helicase interaction was done. Random mutations were generated by low-fidelity PCR and recombined with a gapped plasmid in yeasts. Transformed yeasts carrying repaired plasmids were mated with a strain carrying the pACT-min plasmid, and diploids were screened for mutants that did not interact with pACT-min and were therefore His−. (B) Three of these mutants, T266A, Y267S, and M288T (see Fig. 5), were analyzed for their ability to dimerize by gel filtration. All three showed a larger monomer peak and a reduced dimer peak in the presence of an oligonucleotide. The gel filtration profile of the wild-type helicase in the absence of an oligonucleotide was included for comparison. ABS, absorbance.
Protein expression and purification and analytical gel filtration.
To express the helicase in bacteria, the helicase region (aa 182 to 680) was cloned into a derivative of the bacterial expression vector pGEX-2TK (Pharmacia) and transformed into BL21 (Stratagene). The GST-helicase fusion protein expression was induced in the presence of 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) and cultured for 3 h at 37°C. For the dimerization mutants, protein expression was induced at 18°C overnight with 0.5 mM IPTG, and cells were harvested and sonicated once in lysis buffer and a second time in 300 mM NaCl in lysis buffer. These conditions appear to increase the yield and solubility of the mutant proteins. The cells from 1 liter of culture were harvested and disrupted with a Microson ultrasonic homogenizer (model XL2000) in 10 pellet volumes of lysis buffer (50 mM Tris-HCl [pH 7.6], 1 mM β-mercaptoethanol, 1 mM EDTA, 1% Triton X-100, 100 mM NaCl, 5% glycerol). The insoluble materials were pelleted at 18,000 rpm for 45 min in a Sorvall SS34 rotor, and 500 μl of glutathione Sepharose beads (Pharmacia) was added to the clarified supernatant. The beads were allowed to bind for 2 h, and then they were washed four times in lysis buffer and four times in cleavage buffer (50 mM Tris [pH 8], 150 mM NaCl, 0.1% β-mercaptoethanol, 2.5 mM CaCl2, and 1 mM dithiothreitol). The helicase was cleaved from the GST moiety with thrombin (10 U/liter of culture; Sigma) for 45 min. All steps were performed at 4°C unless otherwise stated.
NS3 helicase was then purified by fast-performance liquid chromatography (FPLC) using a 24-ml S200 Sepharose column (Pharmacia) in binding buffer [50 mM Tris-acetate (pH 7.5), 40 mM sodium acetate (NaOAc), 10 mM Mg(OAc)2, 10% glycerol]. NS3 helicase was found to be >95% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining. Dimerization was shown by analytical FPLC on a SMART machine (Pharmacia) in binding buffer in either the absence or presence of a 39-mer oligonucleotide (OLG39: 3′ATAATGTGTGCTGCCTGCCACTCAACTGACTCAACT5′). A 100-μl volume containing helicase at 5 μM and oligonucleotide ranging from 0 to 0.5 μM was used for each run.
Helicase activity assay.
A double-stranded oligodeoxynucleotide was used as a substrate for helicase activity assays, since NS3 helicase was shown to be about as effective in unwinding DNA as RNA duplexes (26). The substrate was made by annealing the high-pressure liquid chromatography-purified oligonucleotides OLG54 (5′GTCAGTTGAGTGGCAGGCGGCACACATTATAGTGTCGTAGGCTTC3′) and OLG55 (GTGTGCCGCCTGCCACTCAACTGACTCAACTACTGTCTTGGGCATCGGCA) (Genset Inc.), which when annealed give a 25-bp 3′ overhang at both ends. OLG54 was end labeled with polynucleotide kinase (New England Biolabs) using [γ-32P]ATP and purified using a nucleotide purification kit (Qiagen). The two overlapping single-stranded DNAs were mixed with a molar excess of the unlabeled oligonucleotide and annealed by cooling the mixture from 75°C to room temperature gradually. Each reaction contained 20 nM substrate and 400 nM helicase in binding buffer made up to a total of 10 μl in reaction buffer (20 mM HEPES-KOH [pH 7.0], 2 mM dithiothreitol, 1.5 mM MnCl2, 2.5 mM ATP, 0.1 mg of bovine serum albumin per ml). The reactions were terminated by adding 6× DNA gel loading dye, and products were run on 10% acrylamide gels (29:1, acryl-bis) in 0.5× Tris-borate-EDTA buffer. The gel was dried, and radioactivity in the single- and double-stranded DNA and protein-bound DNA was detected on an autoradiograph or quantitated on a PhosphorImager (Molecular Dynamics). The percent single-stranded, or unwound, DNA was calculated as the percentage of the total radioactivity that was present in the unwound DNA in each reaction mixture.
Immunoprecipitation (IP).
COS cells were transfected with 2 to 4 μg of DNA using Superfect (Qiagen) according to the supplier's instructions. Twenty-four hours after transfection, cells were washed once in phosphate-buffered saline (PBS) and harvested in lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris [pH 7.4], 1 mM EDTA, 1 mM EGTA, 0.5% NP-40), with protease inhibitors added before use (0.4 mM Na3VO4, 0.4 mM phenylmethylsulfonyl fluoride), at 4°C. Total cell lysate (0.1 mg) was incubated (with rolling) with 20 μl of packed anti-Flag agarose gel (Sigma) overnight at 4°C. The gel was washed five times, each time with 1 ml of lysis buffer, and the proteins bound to the gel were extracted by boiling for 2 min in 2× sample buffer and separated by SDS-PAGE.
Western blot analysis.
To analyze proteins from co-IP experiments and protein expression in transfected cells, protein samples were resolved by SDS-PAGE, transblotted onto Hybond-C membranes (Amersham), and then probed with anti-Flag monoclonal (1:1,000 dilution; Sigma) or anti-myc polyclonal (1:500 dilution; Santa Cruz Biochemicals) antibodies overnight at 4°C or at least 1 h at room temperature. After extensive washes, a secondary antibody conjugated to horseradish peroxidase (1:2,000 dilution; Pierce) was applied to the blots for at least 1 h at room temperature. Washes were done three times for 5 min each in 0.05% Tween 20 in PBS and another three times in PBS. Antibodies were diluted in 3% skimmed milk in PBS. Blots were washed, and reagents for enhanced chemiluminescence (Pierce) were added for 5 min before signal was detected on X-ray film (Hyperfilm).
RESULTS
NS3-NS3 interaction.
We tested protein-protein interaction among all HCV proteins and found that NS3 interacts strongly with itself (Fig. 1A). This interaction was confirmed by coexpressing myc-tagged and Flag-tagged NS3 in COS cells and showing their association by co-IP experiments (Fig. 1B). To delineate the region(s) of NS3 important for interaction with itself, N-terminal and C-terminal deletions of NS3 were constructed in frame with the DNA-binding domain in pAS2–1. These deletions were tested for interaction with a full-length NS3 in the yeast two-hybrid assay (Fig. 1A). The C-terminal region from aa 336 to 680 containing the RNA-binding motif is apparently not required for NS3-NS3 interaction in this assay. The minimal region delineated for interaction is from aa 182 to 335, which includes motifs essential for NTP-binding, NTPase, and helicase activities. Although the minimal region we have defined is sufficient for interaction, other parts of the NS3 protein may also contribute to the stability of the NS3-NS3 interaction. Deletion of the protease domain and the C-terminal half of the NS3 protein containing the RNA-binding motif reduces the strength of the interaction. Since the protease region (aa 1 to 181) is not required for interaction, we focused our studies on the helicase region.
To determine if the interaction occurs in a head-to-head or head-to-tail orientation, we tested for interaction between the N- and C-terminal regions of the helicase using the yeast two-hybrid assay (Fig. 2A). The results show that the minimal region interacts with itself and does not interact with the C-terminal portion containing the RNA-binding region. This interaction between the minimal regions was confirmed by co-IP in mammalian cells (Fig. 2B). However, under gel filtration conditions, the N-terminal minimal region did not bind to a helicase or to itself (data not shown), suggesting that these conditions were not conducive to such interactions and that additional factors in cells or the cellular environment may be required for the minimal region to bind to NS3 and to itself.
FIG. 2.
(A) The NS3 helicase interacts in an N-to-N orientation. The full-length and truncated helicases were tested for interaction with one another using the yeast two-hybrid assay. The minimal region interacts well with the helicase domain and with itself, but not with the C-terminal two-thirds of the helicase domain. The C-terminal two-thirds does not interact with the helicase in this test. (B) IP between the minimal fragments confirms that it physically associates with NS3 and with itself. COS cells were transfected with various constructs as indicated, 0.1 mg of cell lysate was immunoprecipitated with anti-Flag agarose gel, and bound proteins were detected with anti-Flag or anti-myc as indicated. Lane 1, the minimal region (myc-min) was precipitated by the full-length NS3 protein. Lane 2, the minimal region associates with itself, indicating that the NS3 protein can bind in an N-to-N orientation. Lanes 3 and 4, full-length myc-NS3 and myc-min were not precipitated by anti-Flag beads, showing the specificity of the anti-Flag beads. Lanes 5 through 10, Western blots of the transfected cells showing that all the proteins were expressed appropriately. IgG, immunoglobulin G.
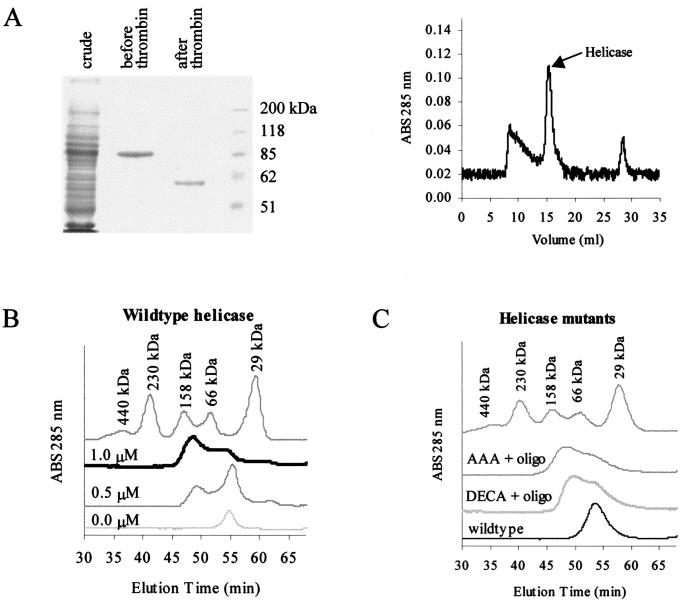
The helicase was bacterially expressed, purified (Fig. 3A), and run through an analytical FPLC column in the presence and absence of an oligonucleotide. This was done to determine if the helicase forms a dimer or higher oligomer, and if the oligomerization is dependent on the binding of an oligonucleotide. We showed that the helicase exists as a monomer in the absence of an oligonucleotide under the conditions used. When an oligonucleotide was added, the protein shifted to a dimer form and no higher oligomer was observed (Fig. 3B). The proportion of dimer versus monomer increased as the molar ratio of protein to oligonucleotide was increased. The monomer and dimer peaks merge, indicating a dynamic equilibrium between the two forms. Under these conditions, the presence of an oligonucleotide is absolutely required for dimerization, and NTP binding and hydrolysis are not necessary, since no NTP was added in these assays. The oligonucleotide we used in these experiments was a 39-mer, which should be long enough to accommodate an active helicase complex, as previous studies showed that a 20-mer 3′ overhang is sufficient for the binding and subsequent unwinding of the double-stranded duplex DNA (26). Our attempts to show the formation of oligomers using chemical cross-linkers, BS3 and glutaraldehyde, demonstrated that these cross-linkers were able to produce higher oligomers even in the absence of an oligonucleotide, suggesting that cross-linking reactions can be nonspecific (data not shown).
FIG. 3.
Gel filtration of the helicase protein shows that it forms a dimer in the presence of an oligonucleotide. (A) Coomassie-stained SDS-PAGE (left) of bacterially expressed GST-helicase proteins before and after thrombin cleavage shows the relative purity of this preparation. This protein thus prepared was still contaminated with other material, as shown by the absorbance graph of proteins detected after separation by FPLC (right). The fractions containing the helicase protein were collected and used for subsequent experiments. (B) In the absence of an oligonucleotide, the helicase runs at around 60 kDa, indicating that it exists as a monomer in the absence of an oligonucleotide. As increasing amounts of oligonucleotide (0, 0.5, and 1 nM) were added, the peak between the 66- and 158-kDa markers increased in height, consistent with the shift from monomer to dimer. (C) Mutants with mutations in the conserved motifs that abolish the helicase activity (DECH→DECA and TAT→AAA) can form dimers in the presence of an oligonucleotide, as shown by gel filtration. The monomeric wild-type protein without an oligonucleotide was included for comparison. ABS, absorbance.
Since oligomerization has been proposed to enhance the helicase activity (15), helicase mutants defective in self-interaction may be inactive for their unwinding activity because of their inability to form dimers or oligomers. To investigate the possibility that mutations in conserved motifs for NTP-binding, NTPase, and helicase activities are inactive because of their failure to form oligomers, we targeted some of these motifs using site-directed mutagenesis. The sequences G207SGKST, D290ECH, and T322AT were mutated to AAGKST, DECA, and AAA, respectively, which have been shown to inactivate the helicase activities (8, 11). These helicase mutants were expressed in yeasts and in bacteria to test for interaction by the yeast two-hybrid method and by gel filtration, respectively. Mutants with changes in these three conserved motifs were able to interact with a wild-type helicase in the yeast two-hybrid test. Gel filtration of these helicase mutants also showed that they were able to form dimers in the presence of an oligonucleotide (Fig. 3C). The loss of helicase activity of these mutants is therefore not due to their inability to dimerize or oligomerize.
Mutations that disrupt dimerization.
Whether the HCV helicase functions as a monomer or a dimer has been in contention for a while. We sought to answer this question by generating mutations that disrupt the interaction between the helicase molecules and to correlate the lack of interaction with the presence or absence of helicase activity of these mutants. To generate random mutants that disrupt the interaction between the minimal interacting domains, the minimal region was amplified under low-fidelity conditions and cotransformed with a gapped plasmid into PJ69–2A (see Materials and Methods) (Fig. 4A). The two fragments were repaired in yeast cells, and colonies were selected and screened for mutants that showed no growth on −His plates. About 500 clones were screened, and 13 His− mutants with mutations that disrupted interaction between a mutant and a wild-type minimal region were isolated. The mutated regions from these mutants were sequenced, and the positions of the mutations found are shown in Fig. 5. The mutations appear to cluster around a few pockets, around aa 200, 268, 290, and 311. These may define the residues that form noncovalent bonds with another helicase.
FIG. 5.
Positions of some of the mutations that disrupted interaction between two minimal regions. Mutations in the minimal fragment resulted in a complete loss of interaction with a wild-type minimal region (min X min). When transferred to a full-length helicase, the mutations resulted in weaker interactions (w) with a wild-type helicase (hel X hel), i.e., His+, Ade−, and β-Gal−, compared to the interaction between two wild-type helicases. The conserved helicase motifs are underlined; mutations selected for in vitro helicase and dimerization assays are in bold.
The mutations were transferred into a full-length helicase, and the mutants were again tested for their ability to interact with a wild-type helicase. None of the dimerization mutants, except S268P, of the full-length helicase showed a complete loss of interaction, as they were His+, in the yeast two-hybrid test. However, the HIS3 gene is a very sensitive reporter gene and allows cells to grow on −His medium even upon weak activation. These mutants showed weaker interaction than the wild-type helicase, which activates all three reporters (Fig. 5). Therefore, single mutations in the helicase context are not sufficient to abolish interaction completely, and additional points of contact outside the minimal region compensate for the binding in vivo. The S268P mutation abolished interaction completely, but the change generated a proline, which is generally disruptive to a protein structure.
To confirm that the mutants show weak interaction with themselves, we expressed three mutants, T266A, Y267S, and M288T, in bacteria and prepared the protein for gel filtration. Using the LOOK program, we chose these mutations for biochemical studies because they were predicted to be within beta-sheets and are less likely to cause major disruption to the structure of the helicase. These mutations show little or no dimerization in the presence of a single-stranded oligonucleotide (Fig. 4B).
Correlation between dimer formation and helicase activity.
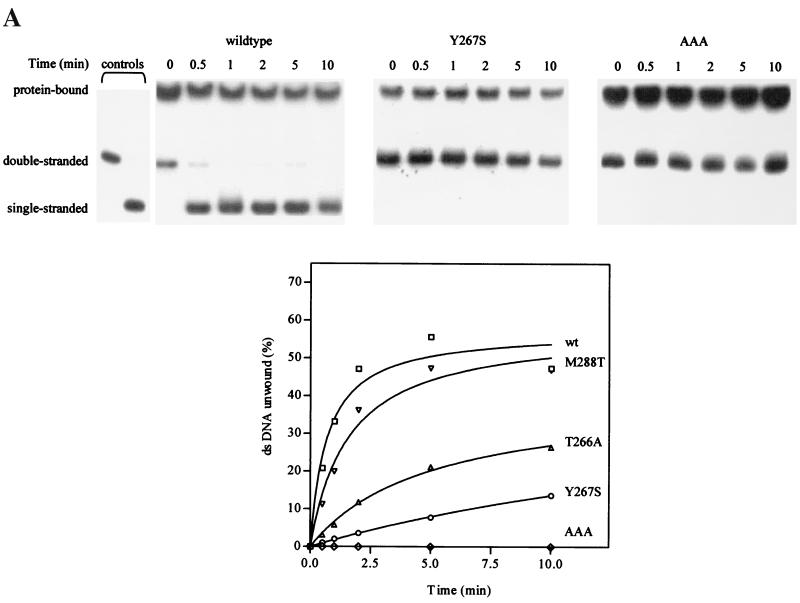
The wild-type helicase and dimerization mutants (T266A, Y267S, and M288T) were assayed for their helicase activities in an in vitro assay (26) using double-stranded DNA oligonucleotides as a substrate. Wild-type helicase unwound the substrate within 30 s, while all the dimerization mutants unwound the substrate at a reduced rate (Fig. 6A). To show a more dramatic difference in helicase activity between the wild type and the dimerization mutants, the substrate-to-enzyme ratio was increased. Under these conditions, the wild-type helicase was very effective in releasing single-stranded oligonucleotide, while mutants T266A and Y267S were relatively inactive (Fig. 6A). The M288T mutant showed a lower initial rate of unwinding but eventually unwound up to 45% of double-stranded DNA. As a negative control, the AAA mutant, which has no helicase activity, was also included. We have therefore identified mutations that abolish the dimer formation and also affect the activities of the HCV helicase to various degrees, indicating that helicase activity is dependent on dimerization.
FIG. 6.
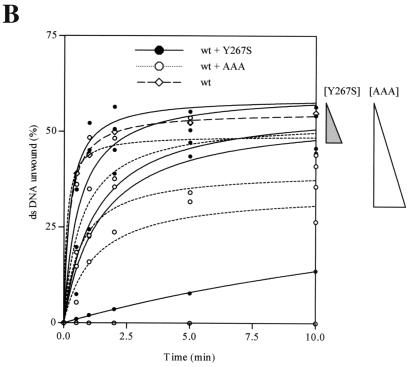
Dimerization mutants show a reduction in helicase activity. (A) Wild-type and mutant (T266A, Y267S, M288T, and AAA) helicases were individually subjected to helicase activity assays, and the radioactive signals were quantitated on a phosphorimager. The autoradiographs of the helicase assays of wild-type helicase, Y267S, and AAA are shown. The percent DNA unwound, expressed as the percentage of total radioactivity per reaction in the single-stranded DNA, was quantitated and plotted on the graph. Each reaction mixture contained 200 nM helicase and 60 nM substrate. The two control lanes on the extreme left showed the positions of single- and double-stranded oligonucleotides (in the absence of helicase). The protein-bound DNA is also indicated. (B) Helicase inhibition by the addition of mutant proteins. Wild-type helicase (400 nM) was mixed with 0, 100, 200, and 400 nM concentrations of mutant helicase Y267S or AAA and 20 nM substrate. The triangles on the right indicate the magnitude of decrease in percent unwinding as the concentration of each of the mutant proteins was increased.
To further confirm this finding, we measured the helicase activity of wild-type helicase when the helicase mutants AAA and Y267S were added to the wild-type helicase in increasing amounts. A previous study showed that addition of an inactive helicase mutant to wild-type helicase was able to inhibit the helicase activity, leading to the conclusion that the HCV helicase acts as a dimer or higher oligomer (15). We have shown that the mutant Y267S is defective both in dimerization and helicase activity. Mutant Y267S is not expected to bind to the wild-type helicase, and the helicase activity of wild-type helicase should not be affected as increasing amounts of mutant protein are added, since inactive wild-type–mutant dimers would not be formed. Indeed, the addition of mutant Y267S to wild-type helicase does not affect the helicase activity significantly. When the ratio of Y267S to wild-type helicase was raised to 1:1, the percent unwound DNA decreased by 10% compared to that obtained with the wild-type helicase (Fig. 6B). As a control, the same experiment was done with the mutant AAA, which is able to dimerize but not able to unwind the double-stranded substrate. We found that the AAA mutant or the AAA–wild-type helicase mixture bound to the substrate and did not release the single-stranded oligonucleotide. As expected, AAA dramatically reduced the helicase activity as the ratio of mutant to wild-type protein increased (Fig. 6). An equimolar ratio of mutant AAA and wild-type helicase reduced the percent unwound DNA by about 30% compared to that obtained with the wild type.
DISCUSSION
We describe the characterization of NS3 homodimerization using a combination of yeast molecular genetic and biochemical techniques. NS3 dimerizes through the N-terminal region of the helicase domain, and several random mutations generated were able to disrupt the interaction between two helicase molecules. This lower affinity correlates with a loss of helicase activity, indicating the importance of dimerization for helicase activity.
The existence and significance of an NS3 helicase dimer have been in contention for a while. From structural (29), mutational (17), and equilibrium and velocity sedimentation centrifugation (25) experiments, the HCV helicase appears to function as a monomer, while others report that it functions as an oligomer (5). This discrepancy may be due to the relative processivity of the various forms of helicase, with monomers and oligomers having different efficiencies in unwinding activity: higher oligomers are more efficient than monomers. DNA helicases have been reported to function as monomers (23) or as oligomers (2). We showed here that the NS3 helicase interacts strongly with itself both in the yeast two-hybrid assay and in co-IP experiments. Gel filtration experiments also demonstrated that the helicase exists as a dimer in solution, and only in the presence of a single-stranded oligonucleotide. The dependence of dimerization on the binding of a single-stranded DNA is reminiscent of the Escherichia coli Rep helicase (27). Others have shown that the HCV helicase forms oligomers up to octamers in cross-linking studies (15). Under gel filtration conditions, we were able to detect only dimers and no higher oligomers. The conditions that we used may not allow for stable formation of higher oligomers, although the buffer we used for gel filtration is similar to those used in cross-linking experiments in which the HCV helicase was shown to form oligomers.
A dimer model for the NS3 helicase has been proposed based on the crystal structure of the NS3 helicase of HCV genotype 1b (5). Those authors suggested a head-to-tail orientation, where the NTPase domain appears to contact the RNA-binding domain. We, however, showed that the interaction between the helicase molecules could occur in a head-to-head orientation through the N terminus of the helicase domain in a yeast two-hybrid test as well as in co-IP experiments. This minimal region is not able to bind to itself under gel filtration conditions, indicating that these conditions may not mimic physiological conditions and/or that additional factors are needed to stimulate the dimerization of the minimal region. From our gel filtration assays, the binding of a single-stranded oligonucleotide plays an important part in inducing dimer formation.
As the helicase shows strong self-association, we wondered if it is important for optimal helicase activity. The minimal region cannot bind to the helicase under gel filtration conditions, presumably because mutual binding of a single-stranded DNA or RNA is necessary for stable dimer formation. We were therefore not able to use the minimal region to inhibit dimerization of the full-length helicase to assay its effect on the helicase activity in an in vitro assay.
To test the hypothesis that dimerization is important for helicase activity, we isolated mutants that disrupted the interaction and used these mutants to assay for helicase activities. One caveat in testing this hypothesis exists: these mutations may disrupt the structure of the helicase, so that both dimerization and helicase activity are affected. We chose mutations (T266A, Y267S, and M288T) that are less likely to perturb the structure of the helicase to assay dimerization and helicase activities. The reduction in helicase activity of the three mutants is not due to their inability to bind the substrate, since all three mutants shifted the substrate to a higher-molecular-weight form (Fig. 6 and our unpublished data). The reduction in helicase activities compared to that of the wild type could therefore be attributed to a reduction in dimer formation, suggesting that dimerization contributes to a more efficient unwinding activity of the NS3 helicase. This conclusion was also confirmed by the reduction of activity of wild-type helicase by the addition of mutant AAA and by a less significant decrease by the addition of a dimerization mutant, Y267S.
The requirement of dimerization of the NS3 protein for optimal helicase activity has interesting implications for HCV replication in an infected cell. Translation of the positive-strand RNA genome and processing of the polypeptide produces a single molecule of NS3. If helicase activity is absolutely dependent on the dimerization of NS3, then multiple rounds of translation must be completed before active NS3 dimers are formed to facilitate RNA replication. The requirement for dimerization may limit the rate of RNA replication and may explain the low rate of replication in the early phase of infection. However, we believe that monomeric NS3 possesses some helicase activity, since two of the dimerization mutants (T266A and M288T) tested showed little dimerization but were able to unwind duplex DNA to some extent (Fig. 4B and 5).
The effect of mutations that disrupt dimerization on the replication of the HCV awaits testing in a cell culture system that expresses the viral genome containing these mutations. The definition of critical residues involved in protein-protein contact could potentially provide useful information for designing compounds that specifically inhibit the dimerization. Such inhibitors will be specific to this helicase, as the dimer structure of the HCV helicase is unique.
ACKNOWLEDGMENTS
We thank A. Tay and her lab members for sequencing DNA clones, A. Ting, Y. J. Tan, and P. Kolatkar for advice, and J. Goh and A. Low for technical assistance.
This work was supported by grants from the National Science and Technology Board, Singapore.
REFERENCES
- 1.Ali J A, Maluf N K, Lohman T M. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- 2.Bujalowski W, Klonowska M M, Jezewska M J. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 3.Chao K L, Lohman T M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991;221:1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- 4.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H-S, Ha N-C, Kang L-W, Chung K M, Back S H, Jang S K, Oh B-H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 6.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of cDNA clone derived from a blood-borne non-A, non-B hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 7.Gallinari P, Paolini C, Brennan D, Nardi C, Steinkuhler C, de Francesco R. Modulation of hepatitis C virus NS3 protease and helicase activities through the interaction with NS4A. Biochemistry. 1999;38:5620–5632. doi: 10.1021/bi982892+. [DOI] [PubMed] [Google Scholar]
- 8.Heilek G M, Peterson M G. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi R. Using PCR to engineer DNA. In: Erlich H A, editor. PCR technology. New York, N.Y: Stockton Press; 1989. pp. 61–70. [Google Scholar]
- 10.Ishido S, Fujita T, Hotta H. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem Biophys Res Commun. 1998;244:35–40. doi: 10.1006/bbrc.1998.8202. [DOI] [PubMed] [Google Scholar]
- 11.Kim D W, Kim J, Gwack Y, Han J H, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O'Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 14.Koch J O, Lohmann V, Herian U, Bartenschlager R. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology. 1996;221:54–66. doi: 10.1006/viro.1996.0352. [DOI] [PubMed] [Google Scholar]
- 15.Levin M K, Patel S S. The helicase from hepatitis C virus is active as an oligomer. J Biol Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 16.Lin C, Wu J W, Hsiao K, Su M S. The hepatitis C virus NS4A protein: interactions with NS4B and NS5A proteins. J Virol. 1997;71:6465–6471. doi: 10.1128/jvi.71.9.6465-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Kim J L. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo S Y, Selby M J, Ou J H. Interaction between hepatitis C virus core protein and E1 envelope protein. J Virol. 1996;70:5177–5182. doi: 10.1128/jvi.70.8.5177-5182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 20.Major M E, Feinstone S M. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 21.Manser E, Huang H Y, Loo T H, Chen X Q, Dong J M, Leung T, Lim L. Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 23.Mechanic L E, Hall M C, Matson S W. Escherichia coli DNA helicase II is active as a monomer. J Biol Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- 24.Mulrad D, Hunter R, Parker R. A rapid method for localised mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 25.Porter D J T, Short S A, Hanlon M H, Preugschat F, Wilson J E, Willard D H, Jr, Consler T G. Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J Biol Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 26.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong I, Chao K L, Bujalowski W, Lohman T M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Biol Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 28.Xiao J H, Davidson I, Matthes H, Garnier J M, Chambon P. Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 29.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]