Abstract
The swarming of self-propelled cytoskeletal filaments has emerged as a new aspect in the field of molecular machines or robotics, as it has overcome one of the major challenges of controlling the mutual interaction of a large number of individuals at a time. Recently, we reported on the photoregulated swarming of kinesin-driven cytoskeletal microtubule filaments in which visible (VIS) and ultraviolet (UV) light triggered the association and dissociation of the swarm, respectively. However, systematic control of this potential system has yet to be achieved to optimize swarming for further applications in molecular machines or robotics. Here, we demonstrate the precise and localized control of a biomolecular motor-based swarm system by varying different parameters related to photoirradiation. We control the reversibility of the swarming by changing the wavelength or intensity of light and the number of azobenzenes in DNA. In addition, we regulate the swarming in local regions by introducing different-sized or shaped patterns in the UV light system. Such a detailed study of the precise control of swarming would provide new perspectives for developing a molecular swarm system for further applications in engineering systems.
Introduction
Living organisms often adopt swarming as a strategy to survive in nature.1,2 Appropriate signal processing is mandatory to trigger swarming in response to signals from the environment.3,4 The signals include chemical and physical cues such as pheromones,5 temperature,6 pH,7 acoustic signals,8 magnetic signals,9 and light.10 Among them, the light signal provides a significant amount of information because of its high propagation directionality and wide range of wavelengths.11 These primary properties are important to initiate communication within swarm units. Inspired by nature, several light-activated artificial swarm systems, including active colloids12 and biological motors,13 have been reported to understand their behavior from fundamental to application aspects. Photoactive colloidal materials exhibit swarming by triggering optical forces14,15 or photochemical reactions16,17 and photophysical effects, i.e., photocatalytic reactions,12,18 photothermal conversion,19 photoisomerization,20−22 and so on. Compared to these externally powered active colloidal systems, self-propelled biomolecular motor-based swarm systems have not yet been well explored. Recently, we reported a light-activated swarm system driven by biological motors (microtubules/kinesins) conjugated with DNA in which visible (VIS) light triggers swarming and ultraviolet (UV) light facilitates the dissociation of swarms.13 In this system, azobenzene was inserted into the DNA backbone as a photoswitch for ON/OFF switching of swarming.23 While it has been shown that this photoregulated swarm system is effective for achieving cooperative and efficient tasks,24 there is still uncharted territory in terms of manipulating swarm behavior by altering the number of photoswitches or adjusting the intensity and wavelength of light exposure. Exploring these possibilities could enable the utilization of the photoactivated swarm system for multifaceted multitasking across a broad spectrum of wavelengths.25−27 In addition, the incorporation of a photosensor in the swarm system has further advantages in regulating the swarm in a spatiotemporal manner. Here, we conducted a thorough investigation of how azobenzene, serving as a photosensors, can be utilized to govern the swarming behavior of kinesin-driven MTs. We have also illustrated how varying the number of photoresponsive azobenzene molecules within DNA sequences and adjusting the intensity of UV light can influence the swarming of MTs. Additionally, we have demonstrated the ability to exert spatial control over the formation and dispersal of swarms in response to light signals. This precise manipulation and optimization of the swarm system have the potential to significantly advance the development of swarm robots, enabling them to tackle intricate tasks in the realms of molecular computation, robotics, and nanotechnology.
Materials and Methods
Purification of Tubulin and Kinesin
Tubulin was purified from the porcine brain through two cycles of polymerization and depolymerization using a high concentration of PIPES buffer (1 M PIPES, 20 mM EGTA, and 10 mM MgCl2) and then preserved in BRB80 buffer (80 mM PIPES, 1 mM EGTA, and 2 mM MgCl2, pH maintained to 6.8 using KOH).28 Recombinant kinesin-1 consisting of the first 573 amino acid residues of human kinesin-1 was prepared as described in the literature.29 Preparation of azide-labeled tubulin was performed using N3–PEG4-NHS following the established protocol of labeling tubulin with a fluorescent dye.30 The tubulin concentration was determined by measuring the absorbance at 280 nm using a UV spectrophotometer (Nanodrop 2000c spectrophotometer). ATTO550- and ATTO488-labeled tubulins (red and green) were also prepared following the same protocol.
Preparation of MTs
MTs were polymerized from a mixture of 70 μM azide-labeled tubulin [80% (v/v)] and dye-labeled tubulin [20% (v/v)] using polymerization buffer (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, and 1 mM polymerizing agent, pH ∼ 6.8). The polymerizing agent for MTs was the nonhydrolyzable guanosine triphosphate analog guanosine-5′-[(α,β)-methyleno]triphosphate (GMPCPP). Then, the mixture was incubated at 37 °C for 30 min to form GMPCPP-stabilized MTs. One microliter of 4× BRB80 buffer and 0.5 μL of 1 mM Taxol were added to the MTs just after polymerization to stabilize the MTs. The copper-free click reaction was initiated by adding 3.5 μL of DBCO-conjugated pDNAs (200 μM) to 5 μL of azide-MTs (56 μM), which allowed the azide–alkyne cycloaddition reaction, and incubated at 37 °C for 6 h.31 One hundred microliters of cushion buffer (BRB80 buffer supplemented with 60% glycerol) was used to separate the MTs by centrifugation at 201,000 g (S55A2–0156 rotor, Hitachi) for 1 h at 37 °C. After removing the supernatant, the pellet of pDNA-conjugated MTs was washed once with 100 μL of BRB80P (BRB80 supplemented with 1 mM Taxol) and dissolved in 15 μL of BRB80P.
Preparation of Swarm of MTs
Five μL of casein buffer [BRB80 buffer supplemented with casein (0.5 mg/mL)] was added to the flow cell. After incubation for 3 min, 800 nM kinesin solution was introduced into the flow cell. After washing the flow cell with 5 μL of wash buffer (BRB80 buffer supplemented with 1 mM dithiothreitol and 10 μM Taxol), 5 μL of green MT (pDNA2-modified ATTO488-labeled MTs) solution was introduced and incubated for 2 min, followed by washing with 10 μL of wash buffer. Subsequently, 5 μL of red MT (pDNA1-modified ATTO550-labeled MTs) solution was introduced and incubated for 2 min, followed by washing with 10 μL of wash buffer. The motility of MTs was initiated by applying 5 μL of ATP buffer [wash buffer supplemented with 5 mM ATP, d-glucose (4.5 mg/mL), glucose oxidase (50 U/ml), catalase (50 U/ml), and 0.2% methylcellulose (w/v)]. The time of ATP addition was set as 0 h. Soon after the addition of ATP buffer, the flow cell was placed in an inert chamber system,32 and pDNA1-MTs and pDNA2-MTs were monitored using an epifluorescence microscope at room temperature.
Results and Discussion
The photoactivated molecular swarm system is illustrated in Figure 1a, in which VIS light irradiation (λ > 480 nm) facilitates the formation of the MT swarm, but UV light (λ = 365 nm) triggers the dissociation of the swarm into single MTs. Swarm units were prepared prior to the swarm demonstration by polymerization of a mixture of azide-labeled tubulins and dye-labeled tubulins to allow modification with DBCO–DNA and visualization under fluorescence microscopy. Swarm units were propelled by surface-adhered recombinant kinesins, consisting of the first 573 amino acid residues of human kinesin-1 in the presence of ATP. To control the swarming of MTs under photoirradiation, two complementary azobenzene-incorporated DNAs (pDNA1 and pDNA2) were employed to modify two different sets of MTs (pDNA1-MT and pDNA2-MT) by an azide–alkyne cycloaddition reaction (DNA sequences are mentioned in Table S1). This DNA conjugation triggers the ON/OFF switching of the MT swarming by VIS-UV light-induced trans–cis isomerization of the azobenzene moiety through the change in melting temperature of DNA hybridization (Figure S1).23
Figure 1.
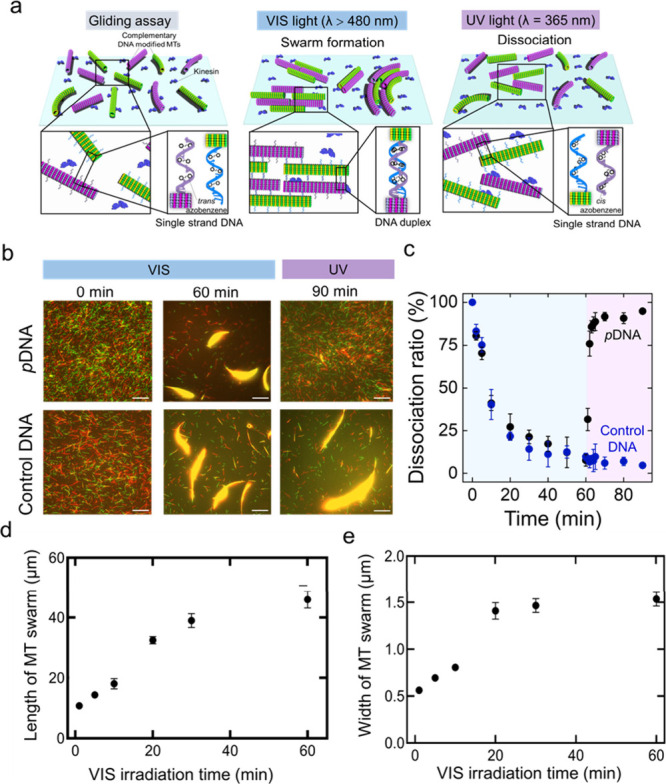
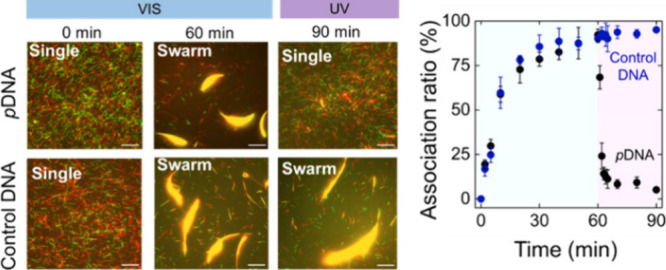
Photocontrol of the swarming of kinesin-driven MTs. a. Schematic illustrations of swarm formation and dissociation of azobenzene-incorporated DNA-conjugated MTs through duplex formation and dissociation of the DNA duplex into single strands under VIS (wavelength, λ>480 nm) and UV light (λ = 365 nm), respectively. b. Time-lapse fluorescence microscopy images show the effect of VIS and UV light on the swarm of pDNA-modified MTs and control DNA (same DNA sequences without azobenzene modification)-modified MTs. Scale bar: 20 μm. c. The time course association ratio of pDNA-modified MTs and control DNA-modified MTs under VIS and UV light irradiation. The blue area shows the VIS irradiation time (60 min), and the purple area shows the UV irradiation time (30 min). d and e. VIS irradiation time changes the length and width of MT swarms respectively upon changing the VIS irradiation time. Error bar: standard deviation. After 30 min of VIS light irradiation, the length and width go to plateau. Length and width of the swarms were measured by Fiji-ImageJ software.
pDNA1-MTs and pDNA2-MTs started to form bundle-like swarms upon collision while moving in the same direction on the kinesin-coated glass surface under VIS light (Figure 1b top). In the absence of VIS light, MTs exhibited movement and collisions, yet fail to organize into swarms due to the absence of conducive interactions. The size of the bundle in terms of length and width changed with VIS irradiation time as cis to trans isomerization of azobenzene in pDNA facilitates the hybridization of the complementary DNA as well as the association of the swarm units. The swarms of MTs exhibited translational motion with a velocity (0.56 ± 0.04 μms–1) slightly lower than that of single MTs (0.58 ± 0.05 μms–1) (Figure S2). Under UV irradiation, the trans-form of azobenzene converts to the cis form, and the nonplanar cis-azobenzene destabilizes the DNA sequence by steric hindrance, which breaks the DNA duplex. To investigate the effect of UV light on swarming, UV light (intensity, I = 0.6 mW/cm2) is irradiated directly on the MT swarm, which triggers the dissociation of the swarm into single MTs. To confirm that only azobenzene in DNA initiates the reversible swarming of MTs, a similar experiment was carried out with the same DNA sequences without azobenzene incorporation (control DNA) (see Table S1). Figure 1b (bottom) confirms the effect of azobenzene-incorporated DNA on the dissociation of the swarm, in which no dissociation was observed under constant irradiation with UV light for 30 min. We characterized the dissociation of the swarm MTs by calculating the association ratio as the fraction of the number of initial MTs incorporated into the swarm at a definite time (See SI for details). Figure 1c shows the time course association ratio of the complementary pDNA-modified MTs and control DNA-modified MTs under VIS-UV irradiation. The dissociation of the swarm into single MTs is quantified by a significant decrease in the association ratio. This result indicates that azobenzene acts as a photoswitch in the swarm system that can sense the wavelength of the light and program the association–dissociation of the MT swarm. Visible light was irradiated on DNA-modified MTs for 60 min, and then UV light was irradiated for 30 min. Under visible light, the association ratio increased and plateaued after 30 min, but after UV light application, the association ratio decreased rapidly within 4 min. Figure 1d and 1e present the increase in MT bundle size in terms of length and width, as the VIS irradiation time progresses.
We then systematically investigated how the number of azobenzenes in DNA influences the dissociation of swarm MTs. The number of azobenzenes (Z) was varied in the pDNA1 sequence from 1 to 7 and then modified to the MTs by an azide–alkyne cycloaddition reaction (DNA sequences and the labeling ratio of DNA to tubulin are mentioned in Table S2). The same intensity of UV light (I = 0.6 mW/cm2) was irradiated directly onto the MT swarms just after swarm formation under 60 min of VIS light irradiation. Figure 2a shows fluorescence microscopy images of the dissociation of the swarm of different numbers of azobenzene-incorporated pDNA1-modified MTs after 5 min of UV irradiation. UV light was irradiated for a longer time, but it was observed that all the MT swarms dissociated at an intensity of 0.6 mW/cm2 within 5 min of UV light irradiation. Figure 2b shows the drastic decrease in the association ratio of MTs with UV irradiation time. Before UV irradiation, the association ratio of the MTs is near 100% in all cases. After 10 min of UV irradiation, the association ratio decreases to almost 10%, which means that almost all the swarm dissociates into single individuals. Figure 2c demonstrates a decrease in the relaxation time of the swarm dissociation with the degree of confinement from 4.4 to 2.8 min of UV light irradiation. The relaxation time or the characteristic time for swarm dissociation is found to depend on the azobenzene number, which means that dissociation of the swarm into single MTs is faster in the case of a higher number of azobenzenes in DNA strands. The relaxation time is calculated from the fitted curve of the association ratio of the MT swarm with the UV irradiation time (Figure 2b, details in the caption). As the trans-to-cis isomerization efficiency increases with the number of incorporated azobenzenes,33 a greater azobenzene presence facilitates the dissociation of the MT swarm within a short UV irradiation time. The kinetics of dissociation of the swarm can be further tuned depending on the optimization of the velocity of the MTs, length of DNA strand, UV light intensity, etc., which will be studied in our future research.
Figure 2.
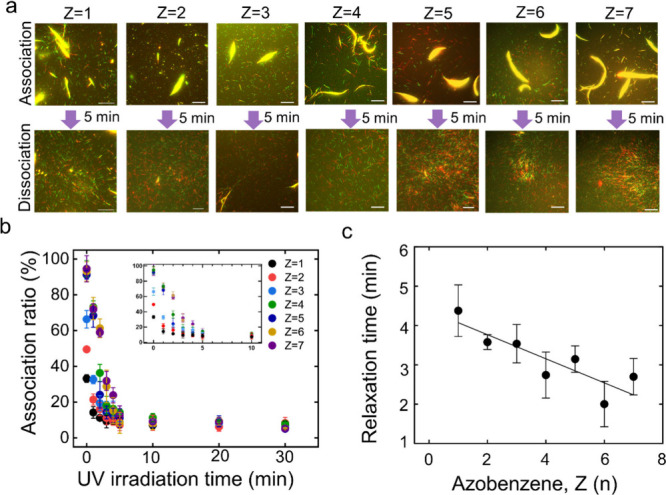
Effect of the number of azobenzenes on the dissociation of the MT swarm. a. Fluorescence microscopy images showing the dissociation of the MT swarms upon varying the number of azobenzene, Z (n) in T12(TTG)(n+1)Z(n) DNA after 5 min of UV irradiation. The intensity of UV light is 0.6 mW/cm2. The association images were captured after 60 min of VIS light irradiation. The labeling ratio of DNA to MT was maintained the same. Scale bar: 20 μm. b. Change in the association ratio of the MTs with the UV irradiation time in which MTs were modified with different numbers of azobenzene-incorporated DNA. Error bar: standard deviation. The inset shows the enlarged association ratio from 0 to 10 min. c. Change in the relaxation time (τ) of MT dissociation from the swarm under UV irradiation with the number of azobenzenes, Z (n) in DNA. Relaxation time was measured from the fitted curve of Figure 2b; the data were fitted with Y = (Y0 - Plateau)*exp(-K*X) + Plateau, where K is the rate constant). Relaxation time is the characteristic time for dissociation, which is computed as the reciprocal of the rate constant. The best fit line has a slope of −0.31 ± 0.08 and a goodness of fit of R2 = 0.82. (Y0 – Plateau), the amplitude of the exponential decay is found to increase with the Azobenzene number of DNA.
As demonstrated earlier, the dissociation of MT swarms is affected by UV light irradiation, and we further carried out a systematic investigation to determine the effect of UV light intensity on the reversible swarming of MTs. Keeping the labeling ratio of DNA to tubulin (Table S3) and the association ratio of MTs almost the same (>80%), we varied the intensity of UV light from 0.1 to 0.6 mW/cm2. Supporting Movie 1 shows the application of different intensities of UV light on the MT swarms formed after 60 min of VIS light irradiation. The fluorescence microscopy image in Figure 3a shows the change in the swarming of MTs after 10 min of UV irradiation at different intensities. At a lower intensity of UV light (0.1–0.2 mW/cm2), the MT swarm did not show complete dissociation, which can be confirmed by the change in the association ratio with UV irradiation time (Figure 3b). At I = 0.1 mW/cm2, the association ratio of the swarm becomes 40% after 10 min of UV irradiation, whereas at I = 0.6 mW/cm2, the association ratio is reduced to almost 10% within 5 min of UV irradiation (Figure S3). At higher intensities of UV light, the association ratio decreases very rapidly with irradiation time. Figure 3c confirms the decrease in the relaxation time to dissociate the MT swarms with the intensity of the UV light. This suggests that an optimum intensity of UV light, which is likely more than 0.2 mW/cm2, is needed to control the reversible swarming of MTs.
Figure 3.
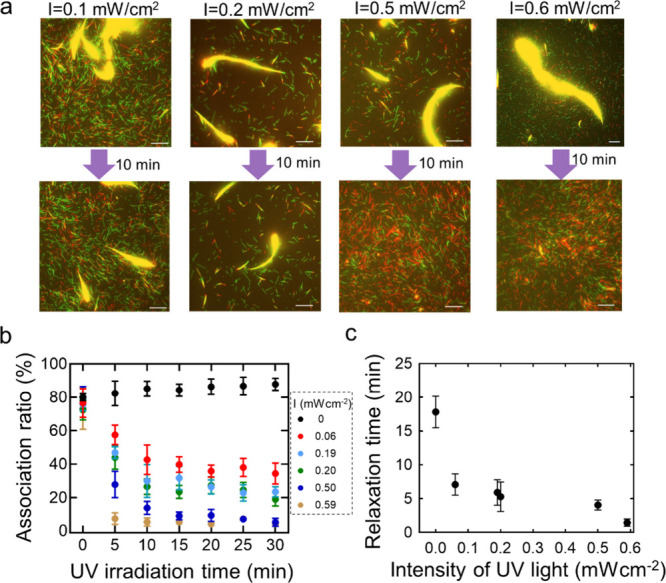
Effect of UV light intensity on the dissociation of the MT swarm. a. Fluorescence microscopy image showing the extent of dissociation of MT swarms after 10 min of UV light irradiation with different intensities (I = 0.59 mW/cm2, 0.50 mW/cm2, 0.20 mW/cm2, and 0.06 mW/cm2). Here, MTs were modified with pDNA1 and pDNA2. Scale bar: 20 μm. b. Change in the association ratio of MTs with the UV irradiation time in which the intensity of UV light irradiation was varied. Error bar: standard deviation. c. c. Change in the relaxation time (τ) of MT swarm dissociation at different intensity of UV irradiation. Relaxation time was measured from the fitted curve of Figure 3b and the data were fitted with Y = (Y0 - Plateau)*exp(-K*X) + Plateau, where K is the rate constant). The best fit line has a slope of 0.17 ± 0.09 and a goodness of fit of R2= 0.94. (Y0 – Plateau), the amplitude of the exponential decay is found to increase with the Azobenzene number of DNA.
We also investigated the localized control of the swarming of MTs in response to photoirradiation (Figure 4a). UV light was applied to the swarming of MTs through a square-shaped photomask (Figure 4b). The intensity of the UV light was kept at 0.6 mW/cm2. The fluorescence microscopy image in Figure 4c shows that MT swarms started to dissociate into single MTs just after entering the square-shaped UV irradiation area due to the dissociation of the conjugated DNAs on MTs (Supporting Movie 2). However, in the visible region, the MT swarms continued gliding with the same gliding motion without showing any dissociation. Moreover, swarms in the VIS region continued to grow into a large bundle with irradiation time. UV irradiation through the differently shaped patterns at different locations indicates the localized remote controllability of our swarm system (Figure S4). UV or visible irradiation through multiple multisized or shaped patterns simultaneously on swarm MTs will facilitate an increase in the controllability of the swarm for further applications as transporters.
Figure 4.
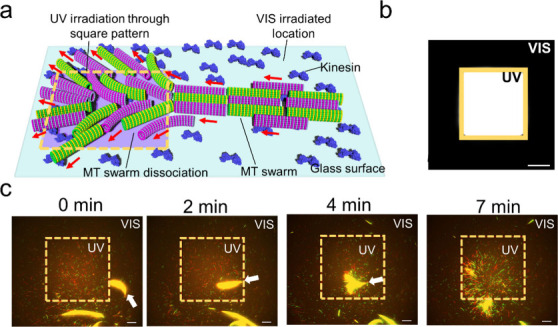
Localized control of the swarming of MTs. a. Schematic illustration of the spatial control of the swarming of MTs through local irradiation of UV light using a square-shaped yellow pattern. b. Fluorescence microscopy image of the square-shaped pattern in which the white area shows the UV light-irradiated region and the black area shows the VIS light-irradiated region. The area of the square region is 120 μm × 120 μm. Scale bar: 40 μm. c. Time-lapse fluorescence microscopy images of spatial control of the reversible swarming of MTs into individuals through local irradiation of UV light via a square pattern. The yellow dotted box shows the UV light irradiated area. Scale bar: 20 μm.
In conclusion, we systematically investigated the effect of wavelength and intensity of light on the DNA-assisted swarming of a self-propelled biomolecular motor system, MT-kinesin. This work demonstrates the advantage of photoirradiation to control the swarm system in a reversible and spatiotemporal manner. Over other external stimuli, light is minimally intrusive, allowing instantaneous manipulation and local triggering of the systems with higher spatiotemporal precision and accuracy.34 This work provides a strategy for precisely and remotely controlling a swarm system by inserting different numbers of appropriate photosensors by means of global or local control.
This study is expected to broaden the application of molecular swarm systems for more intricate sequencing tasks.24 The incorporation of various photosensors, such as different azobenzene derivatives, diarylethenes, spiropyrans, and stilbenes, into DNA can introduce diversity into the molecular swarm system.35 In addition, to understand the effect of DNA melting temperature on the MT swarm dissociation requires more investigation, which constitutes our next area of focus. Moreover, Utilizing light-inducible interactions to regulate MT swarming as well as their gliding assays, opens up new possibilities for reconstructing and comprehending intricate biophysical and biological phenomena.36,37 Given the significance of kinesin-driven microtubule (MT) gliding assays in various nanotechnological applications such as analyte detection, biocomputation, and mechanical sensing, we believe our work will inspire scientists to control the interaction of glided MTs using light, enhancing their practical applications.38,39 Our study will be helpful not only to control the self-assembly of biomolecular motor systems but also to control the self-assembly of synthetic colloidal particles40,41 or inorganic nanomaterials.42 The precise control over swarming in biomolecular motor-based systems demonstrated in this study opens up new possibilities for designing swarming behavior with engineered functionalities in the fields of molecular computation and molecular robotics.43−45
Acknowledgments
This work was financially supported by the Future AI and Robot Technology Research and Development Project (JPNP20006) from the New Energy and Industrial Technology Development Organization (NEDO), Japan; a Grant-in-Aid for Scientific Research on Innovative Areas “Molecular Engine” (JP18H05423); and a Grant-in-Aid for Scientific Research (A) (JP21H04434) to A.K. The Grant-in-Aid for Transformative Research Areas (A) (JP20H05972) and a Grant-in-Aid for Scientific Research (C) (JP21K04846) were awarded to A.M.R.K. H.A. was funded by the Japan Agency for Medical Research and Development 23am0401007 and Grant-in-Aid for Scientific Research (S) JP21H05025.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c03216.
Author Contributions
A.K. conceived the original concept and supervised the project. M.A., A.M.R.K., J.J.K., K.S., and A.K. designed the experiments. M.A. performed the experiments and analyzed the experimental results. H.A. designed the pDNA sequences. M.A. wrote the manuscript. M.A., A.M.R.K. and A.K. edited the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Chakraborty D.; Bhunia S.; De R. Survival chances of a prey swarm: how the cooperative interaction range affects the outcome. Sci. Rep 2020, 10, 1–8. 10.1038/s41598-020-64084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasson B. H.; Haefner J. W.; Bowen M. D. Schooling Increases Risk Exposure for Fish Navigating Past Artificial Barriers. PLoS One 2014, 9, e108220 10.1371/journal.pone.0108220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplimont H.; Georgantzoglou A.; Boulch M.; Walker H. A.; Coombs C.; Papaleonidopoulou F.; Sarris M. Neutrophil Swarming in Damaged Tissue Is Orchestrated by Connexins and Cooperative Calcium Alarm Signals. Curr. Biol. 2020, 30, 2761–2776.e7. 10.1016/j.cub.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reategui E.; Jalali F.; Khankhel A. H.; Wong E.; Cho H.; Lee J.; Serhan C. N.; Dalli J.; Elliott H.; Irimia D. Microscale arrays for the profiling of start and stop signals coordinating human-neutrophil swarming. Nat. Biomed Eng. 2017, 1, 94. 10.1038/s41551-017-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attygalle A. B.; Morgan E. D.. Ant Trail Pheromones. Advances in Insect Physiology; Academic Press, 1985; Vol. 181–30. [Google Scholar]
- Zhu X.; et al. The temperature increase at one position in the colony can predict honey bee swarming (Apis cerana). J. Apic Res. 2019, 58, 489–491. 10.1080/00218839.2019.1632149. [DOI] [Google Scholar]
- Toguchi A.; Siano M.; Burkart M.; Harshey R. M. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: Critical role for lipopolysaccharide. J. Bacteriol. 2000, 182, 6308–6321. 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. M.; Davis K.; Smotherman M. Suppression of emission rates improves sonar performance by flying bats. Sci. Rep 2017, 7, 1–9. 10.1038/srep41641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers C. T.; Hore P. J. Chemical magnetoreception in birds: The radical pair mechanism. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 353–360. 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Arratia P. E.; Patteson A. E.; Gopinath A. Quenching active swarms: effects of light exposure on collective motility in swarming Serratia marcescens. J. R Soc. Interface 2019, 16, 20180960. 10.1098/rsif.2018.0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerle R. Light Propagation in Singly and Doubly Periodic Planar Waveguides. J. Mod. Opt 1987, 34, 1589–1617. 10.1080/09500348714551531. [DOI] [Google Scholar]
- Palacci J.; Sacanna S.; Steinberg A. P.; Pine D. J.; Chaikin P. M. Living crystals of light-activated colloidal surfers. Science (1979) 2013, 339, 936–940. 10.1126/science.1230020. [DOI] [PubMed] [Google Scholar]
- Keya J. J.; Suzuki R.; Kabir A. M. R.; Inoue D.; Asanuma H.; Sada K.; Hess H.; Kuzuya A.; Kakugo A. DNA-assisted swarm control in a biomolecular motor system. Nat. Commun. 2018, 9, 4–11. 10.1038/s41467-017-02778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-F.; Kudo T.; Yuyama K.; Sugiyama T.; Masuhara H. Optically evolved assembly formation in laser trapping of polystyrene nanoparticles at solution surface. Langmuir 2016, 32, 12488–12496. 10.1021/acs.langmuir.6b02433. [DOI] [PubMed] [Google Scholar]
- Jaquay E.; Martínez L. J.; Mejia C. A.; Povinelli M. L. Light-assisted, templated self-assembly using a photonic-crystal slab. Nano Lett. 2013, 13, 2290–2294. 10.1021/nl400918x. [DOI] [PubMed] [Google Scholar]
- Ibele M.; Mallouk T. E.; Sen A. Schooling behavior of light-powered autonomous micromotors in water. Angewandte Chemie - International Edition 2009, 48, 3308–3312. 10.1002/anie.200804704. [DOI] [PubMed] [Google Scholar]
- Yadav V.; Zhang H.; Pavlick R.; Sen A. Triggered “on/off” micropumps and colloidal photodiode. J. Am. Chem. Soc. 2012, 134, 15688–15691. 10.1021/ja307270d. [DOI] [PubMed] [Google Scholar]
- Singh D. P.; Choudhury U.; Fischer P.; Mark A. G. Non-Equilibrium Assembly of Light-Activated Colloidal Mixtures. Adv. Mater. 2017, 29, 1701328. 10.1002/adma.201701328. [DOI] [PubMed] [Google Scholar]
- Weinert F. M.; Braun D. Observation of slip flow in thermophoresis. Phys. Rev. Lett. 2008, 101, 168301. 10.1103/PhysRevLett.101.168301. [DOI] [PubMed] [Google Scholar]
- Klajn R.; Bishop K. J. M.; Grzybowski B. A. Light-controlled self-assembly of reversible and irreversible nanoparticle suprastructures. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 10305–10309. 10.1073/pnas.0611371104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klajn R.; Wesson P. J.; Bishop K. J. M.; Grzybowski B. A. Writing self-erasing images using metastable nanoparticle “inks. Angew. Chem., Int. Ed. 2009, 48, 7035–7039. 10.1002/anie.200901119. [DOI] [PubMed] [Google Scholar]
- Zhang L.; et al. Light-triggered reversible self-assembly of gold nanoparticle oligomers for tunable SERS. Langmuir 2015, 31, 1164–1171. 10.1021/la504365b. [DOI] [PubMed] [Google Scholar]
- Asanuma H.; et al. Synthesis of azobenzene-tethered DNA for reversible photoregulation of DNA functions: hybridization and transcription. Nat. Protoc 2007, 2, 203–212. 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
- Akter M.; Keya J. J.; Kayano K.; Kabir A. M. R.; Inoue D.; Hess H.; Sada K.; Kuzuya A.; Asanuma H.; Kakugo A. Cooperative cargo transportation by a swarm of molecular machines. Sci. Robot 2022, 7, eabm0677 10.1126/scirobotics.abm0677. [DOI] [PubMed] [Google Scholar]
- Forrest N. T.; Vilcapoma J.; Alejos K.; Halvorsen K.; Chandrasekaran A. R. Orthogonal Control of DNA Nanoswitches with Mixed Physical and Biochemical Cues. Biochemistry 2021, 60, 250–253. 10.1021/acs.biochem.0c00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo S.; Sorrentino D.; Ricci F. Orthogonal regulation of DNA nanostructure self-assembly and disassembly using antibodies. Nat. Commun. 2019, 10, 1–9. 10.1038/s41467-019-13104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch M. M.; Hansen M. J.; Velema W. A.; Szymanski W.; Feringa B. L. Orthogonal photoswitching in a multifunctional molecular system. Nat. Commun. 2016, 7, 1–10. 10.1038/ncomms12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S.; Akter M.; Keya J. J.; Rashid M. R.; Afroze F.; Nasrin S. R.; Kakugo A. Purification of Tubulin from Porcine Brain and its Fluorescence Dye Modification. Methods Mol. Biol. 2022, 2430, 3–16. 10.1007/978-1-0716-1983-4_1. [DOI] [PubMed] [Google Scholar]
- Case R. B.; Pierce D. W.; Hom-Booher N.; Hart C. L.; Vale R. D. The Directional Preference of Kinesin Motors Is Specified by an Element Outside of the Motor Catalytic Domain. Cell 1997, 90 (5), 959–966. 10.1016/S0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Akter M.; Keya J. J.; Kabir A. M. R.; Rashid M. R.; Ishii S.; Kakugo A. Functionalization of Tubulin: Approaches to Modify Tubulin with Biotin and DNA. Methods Mol. Biol. 2022, 2430, 47–59. 10.1007/978-1-0716-1983-4_3. [DOI] [PubMed] [Google Scholar]
- Fruh S. M.; Steuerwald D.; Simon U.; Vogel V. Covalent Cargo Loading to Molecular Shuttles via Copper-Free. Click Chemistry 2012, 13, 3908. 10.1021/bm301437c. [DOI] [PubMed] [Google Scholar]
- Kabir A. M. R.; Inoue D.; Kakugo A.; Kamei A.; Gong J. P. Prolongation of the Active Lifetime of a Biomolecular Motor for in Vitro Motility Assay by Using an Inert Atmosphere. Langmuir 2011, 27 (22), 13659–13668. 10.1021/la202467f. [DOI] [PubMed] [Google Scholar]
- Matsunaga D.; Asanuma H.; Komiyama M. Photoregulation of RNA digestion by RNase H with azobenzene-tethered DNA. J. Am. Chem. Soc. 2004, 126, 11452–11453. 10.1021/ja0471976. [DOI] [PubMed] [Google Scholar]
- Tam D. Y.; Zhuang X.; Wong S. W.; Lo P. K. Photoresponsive Self-Assembled DNA Nanomaterials: Design, Working Principles, and Applications. Small 2019, 15, 1805481. 10.1002/smll.201805481. [DOI] [PubMed] [Google Scholar]
- Ishii S.; et al. Kinesin motors driven microtubule swarming triggered by UV light. Polym. J. 2022, 54, 1501–1507. 10.1038/s41428-022-00693-1. [DOI] [Google Scholar]
- Tas R. P.; et al. Guided by Light: Optical Control of Microtubule Gliding Assays. Nano Lett. 2018, 18, 7524–7528. 10.1021/acs.nanolett.8b03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter M.; et al. Photoregulated trajectories of gliding microtubules conjugated with DNA. Chem. Comm 2020, 56 (57), 7953–7957. 10.1039/D0CC03124K. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S.; et al. Label-Free Detection of Microvesicles and Proteins by the Bundling of Gliding Microtubules. Nano Lett. 2018, 18 (1), 117–123. 10.1021/acs.nanolett.7b03619. [DOI] [PubMed] [Google Scholar]
- Kawamata I.; Nishiyama K.; Matsumoto D.; Ichiseki S.; Keya J. J.; Okuyama K.; Ichikawa M.; Kabir A. M. R.; Sato Y.; Inoue D.; et al. Autonomous assembly and disassembly of gliding molecular robots regulated by a DNA-based molecular controller. Sci. Adv. 2024, 10 (22), eadn4490 10.1126/sciadv.adn4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F.; Liebchen B.; Löwen H.; Volpe G. Light-controlled assembly of active colloidal molecules. J. Chem. Phys. 2019, 150, 94905. 10.1063/1.5079861. [DOI] [PubMed] [Google Scholar]
- Borberg E.; et al. Light-Controlled Selective Collection-and-Release of Biomolecules by an On-Chip Nanostructured Device. Nano Lett. 2019, 19, 5868–5878. 10.1021/acs.nanolett.9b01323. [DOI] [PubMed] [Google Scholar]
- Chen H.; et al. Light-activated inorganic CsPbBr2I perovskite for room-temperature self-powered chemical sensing. Phys. Chem. Chem. Phys. 2019, 21, 24187–24193. 10.1039/C9CP03059J. [DOI] [PubMed] [Google Scholar]
- Hess H. Engineering Applications of Biomolecular Motors. Annual Review of Biomedical Engineering 2011, 13, 429–450. 10.1146/annurev-bioeng-071910-124644. [DOI] [PubMed] [Google Scholar]
- Van Den Heuvel M. G. L.; Dekker C. Motor proteins at work for nanotechnology. Science (1979) 2007, 317, 333–336. 10.1126/science.1139570. [DOI] [PubMed] [Google Scholar]
- Hagiya M.; Konagaya A.; Kobayashi S.; Saito H.; Murata S. Molecular robots with sensors and intelligence. Acc. Chem. Res. 2014, 47, 1681–1690. 10.1021/ar400318d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


