Abstract
Cilia are dynamic subcellular systems, with core structural and functional components operating in a highly coordinated manner. Since many environmental stimuli sensed by cilia are circadian in nature, it is reasonable to speculate that genes encoding cilia structural and functional components follow rhythmic circadian patterns of expression. Using computational methods and the largest spatiotemporal gene expression atlas of primates, we identified and analyzed the circadian rhythmic expression of cilia genes across 22 primate brain areas. We found that around 73% of cilia transcripts exhibited circadian rhythmicity across at least one of 22 brain regions. In 12 brain regions, cilia transcriptomes were significantly enriched with circadian oscillating transcripts, as compared to the rest of the transcriptome. The phase of the cilia circadian transcripts deviated from the phase of the majority of the background circadian transcripts, and transcripts coding for cilia basal body components accounted for the majority of cilia circadian transcripts. In addition, adjacent or functionally connected brain nuclei had large overlapping complements of circadian cilia genes. Most remarkably, cilia circadian transcripts shared across the basal ganglia nuclei and the prefrontal cortex peaked in these structures in sequential fashion that is similar to the sequential order of activation of the basal ganglia-cortical circuitry in connection with movement coordination, albeit on completely different timescales. These findings support a role for the circadian spatiotemporal orchestration of cilia gene expression in the normal physiology of the basal ganglia-cortical circuit and motor control. Studying orchestrated cilia rhythmicity in the basal ganglia-cortical circuits and other brain circuits may help develop better functional models, and shed light on the causal effects cilia functions have on these circuits and on the regulation of movement and other behaviors.
Keywords: brain, cilia, circadian, rhythm, transcriptome
1 |. INTRODUCTION
Cilia are evolutionarily conserved organelles that protrude from the membranes of almost all cell types. The distinct designs of cilia, which served swimming through fluids in ancient unicellular organisms, were repurposed by evolution to drive a wide range of completely different functions from their original ones. These functions range from guiding a worm to explore its environment to regulating complex cognitive functions in primates. With recent discoveries highlighting the importance of primary cilia in brain function (Breunig et al., 2008; Guo et al., 2017; Han et al., 2008; Liu et al., 2019; Nechipurenko et al., 2013; Oishi et al., 2006), cilia have emerged as an essential component of sensory perception and transduction signaling pathways and a crucial center for non-synaptic neuronal signaling (Nachury & Mick, 2019). Cilia, however, have preserved two features throughout their long evolutionary history and across all cell types—highly dynamic physical structures (length and assembly/disassembly) and the capability of sensing and transducing a variety of environmental stimuli. In this context, cilia act as cell “antennas” to sense environmental signals and transduce them into biochemical responses that regulate a wide range of cellular activities (see Berbari et al., 2009; Wheatley, 2008 for reviews).
Numerous extracellular and environmental stimuli can be detected and transduced by cilia, including light (Insinna & Besharse, 2008) odorant (Kang et al., 2010), mechanical stimuli and fluid flow (Nauli et al., 2013; Nonaka et al., 1998; Yoshiba et al., 2012), pH (Atkinson et al., 2019; Bargmann, 2006), osmolarity (Choi et al., 2019; Christensen et al., 2005), temperature (Clary-Meinesz et al., 1992; Humphries, 2013; Kuhara et al., 2008; O’Callaghan et al., 1995), gravity (Moorman & Shorr, 2008; Shi et al., 2017), and chemical signals (signaling molecules, neurotransmitter, hormones, growth factors) (Bargmann, 2006). Besides performing these highly specialized sensory and transducing functions, cilia also act as signaling centers that mediate cell-to-cell communication through extracellular vesicles (O’Hagan et al., 2017; Wang et al., 2015). The signal transduction properties of cilia are mediated by specific molecular receptors associated with cilia membranes, including ion channels, receptor tyrosine kinases, and G protein–coupled receptors (GPCRs) (Berbari et al., 2008; Brailov et al., 2000; Domire et al., 2011; Handel et al., 1999; Omori et al., 2015; Varela & Horvath, 2018).
Cilia are highly dynamic systems in terms of length, ultrastructural morphology, subcompartments, and protein composition and trafficking, with the core structural and functional components of cilia functioning in a highly coordinated manner (Domire et al., 2011; Orbach & Howard, 2019; Phua et al., 2017; Silverman & Leroux, 2009). Many environmental stimuli that are sensed by cilia are oscillatory in nature and follow rhythmic temporal patterns of stimulation such as light–dark cycles (Dewan et al., 2011; Wright et al., 2013), temperature (Buhr et al., 2010), nutrient availability (Damiola et al., 2000), and gravity (Casey et al., 2015; Fuller, 1994; Robinson & Fuller, 2000). The rhythmic nature of external signals sensed by cilia alongside their highly dynamic nature lead us to speculate that genes encoding cilia substructural and functional components are expressed in rhythmic patterns.
More precisely many functions, in which ciliary signaling is implicated, oscillate in circadian fashion, such as sleep-wake, feeding (Gu et al., 2004; Nagata et al., 2013; Omori et al., 2015), energy homeostasis (Hughes et al., 2020), metabolism (Lee et al., 2015), body temperature (Berbari et al., 2013), sexual/reproductive behaviors (Diniz et al., 2020; Koemeter-Cox et al., 2014), and even higher brain functions related to memory, and mood and social behavior (Bae & Barr, 2008; Benton et al., 2007; Wang et al., 2011). We speculate that genes encoding cilia structural and functional components, responsible for driving cilia oscillatory regulation of metabolic, physiological, and behavioral processes, follow rhythmic circadian patterns of expression. To test this hypothesis, we acquired transcriptomic data from the largest spatiotemporal gene expression atlas of primates to profile rhythmic brain cilia transcriptome and to define the patterns of cilia gene expression in the brain (Mure et al., 2018).
2 |. METHODS AND EXPERIMENTAL DESIGNS
2.1 |. Cilia genes’ list
We used a list of 281 genes (Table S1) that were identified and verified to be cilia genes expressed in the human brain using cilia databases (Arnaiz et al., 2009, 2014; van Dam et al., 2013), as described in our recent report (Alhassen et al., 2021). Briefly, we used SysCilia Gold Standard version 1 and CiliaCarta database to compile a list of gold standard cilia genes as well as CilDB database to further confirm the cilia genes. We then screened the genes to be sure that they were expressed in the brain using GTEX. It is important to note that cilia gene databases are constantly being revised, and no list can truly be considered “complete” because new cilia genes are discovered all the time. As a result, our list of cilia genes is a working list that will be revised as needed.
2.2 |. Cilia circadian genes data
We extracted the transcriptomic time series associated with the 281 cilia genes from the full transcriptomic time series performed in 22 baboon brain regions tissue collection was performed at the Institute of Primate Research (IPR, National Museums of Kenya, Nairobi; see the original study for full details on the protocols, ethics, sample collection, and processing; Mure et al., 2018). Brain regions included the following: amygdala (AMY), arcuate nucleus (ARC), cerebellum (CER), dorsomedial hypothalamus (DMH), habenula (HAB), hippocampus (HIP), prefrontal cortex (PRC), putamen (PUT), lateral globus pallidus (LGP), lateral hypothalamus (LH), mammillary body (MMB), medial globus pallidus (MGP), olfactory bulb (OLB), paraventricular nucleus (PVN), preoptic area (PRA), pons (PON), substantia nigra (SUN), suprachiasmatic nucleus (SCN), supraoptic nucleus (SON), thalamus (THA), ventromedial hypothalamus (VMH), and visual cortex (VIC).
2.3 |. Circadian analysis
For the circadian analyses, the transcriptomic time series associated with the cilia genes was analyzed through CircadiOmics (Ceglia et al., 2018; Patel et al., 2012), the largest repository of circadian omic time series data sets. We then identified which transcripts were oscillating in a circadian manner under control and experimental conditions. BIO_CYCLE (Agostinelli et al., 2016a, 2016b), a deep learning-based model developed to analyze periodicity in transcriptomic time series data, was used to identify statistically significant circadian transcripts, as well as the amplitude and the phase of their oscillations. BIO_CYCLE is trained on both synthetic and real-world biological time series data sets containing labels for periodic and aperiodic signals. A classification deep neural network (DNN) is trained to classify signals as periodic or not, and a regression DNN is trained to estimate the period, phase, and amplitude of the signal. Whether a gene is oscillating or not is determined by the p value (cutoff at 0.05) provided by BIO_CYCLE. BIO_CYCLE calculates a p value as follows: N aperiodic signals are first generated from the synthetic time series data sets, and the N output values V(i) (i = 1, …, N) of the classification DNN on these aperiodic signals are calculated. These values are used to establish the distribution for the null hypothesis. Then, the output value V of the new signal s is compared to V(i)(i = 1, …, N), producing the estimate for the probability of obtaining an output of size V or greater (p value), assuming that the signal s comes from the null distribution (the distribution of aperiodic signals). Therefore, the smaller the p value, the more likely that s is periodic. The corresponding q-values are obtained through the Benjamini and Hochberg procedure. BIO_CYCLE is publicly available from the CircadiOmics web portal at: http://circadiomics.igb.uci.edu.
Cilia circadian transcripts were grouped within four time phases of 6-hr intervals (quarter-phases), associated with their peak in gene expression: first quarter-phase (ZT0–ZT5), second quarter-phase (ZT6–ZT11), third quarter-phase (ZT12–ZT17), and fourth quarter-phase (ZT18–ZT23). A Fisher’s exact test was then used to compare the proportions of circadian cilia genes in the 22 brain regions with the brain area background circadian transcriptome. The overlap of cilia rhythmic genes across regions was determined as gene-view and region-view intersections. The distribution of cilia circadian genes across main structural and functional components of the cilia was examined. Thus, cilia genes were grouped based on their localization to the cilium, including basal body, axoneme, kinesin, dynein, IFT-A, IFT-B, transition zone, BBsome, Golgi, nucleus, cytosol, and the ciliary membrane. When localization is unknown, the genes were placed in a group labeled as “other.” The percentages of substructural cilia transcripts that exhibited circadian rhythm were calculated.
3 |. RESULTS
3.1 |. Circadian oscillation of cilia transcriptome in the brain is region specific
All 22 brain regions reported in the primate diurnal transcriptome atlas (Mure et al., 2018) had circadian cilia transcripts (Figure 1a,b, Table S2) (www.brainmaps.org) (Mikula et al., 2007), and 206 cilia genes (72.8% of all cilia genes) were determined to have circadian oscillations in at least one brain region. Prefrontal cortex (PRC) exhibited the highest number of cilia circadian cycling genes (66 cilia cycling genes), followed by PUT, LGP and MMB (58 cilia cycling genes in each), and MGP (56 cilia cycling genes), accounting for 23%, 21%, 21%, and 21% of the cilia genes in PRC, PUT, LGP, and MGP, respectively. Amygdala and lateral hypothalamus exhibited the lowest number of cilia circadian genes (four cilia cycling genes in each). The overwhelming majority (greater than 99%) of the cilia rhythmic transcripts oscillate with the period of 22–24 hr (Figure 1c). The rhythmic expression was region specific and, in general, the number of cilia circadian genes in a given brain region was correlated with the number of background circadian genes in the same region (Pearson’s correlation coefficient was high, r2 = 0.966, p < 0.0001, Figure 1d). We computed the enrichment of circadian oscillating genes in these 22 brain regions, and found that cilia circadian rhythmicity was overrepresented in 12 brain regions (Fisher’s exact test, p < 0.05). Brain regions, in which cilia transcripts exhibited higher rhythmicity than the non-cilia transcripts, included the PRC (Odds ratio (OR) = 1.8), PUT (OR = 1.6), LGP (OR = 1.67), MMB (OR = 1.64), MGP (OR = 1.5), DMH (OR = 2.3), PVN (OR = 2.2), VIC (OR = 1.74), SCN (OR = 1.71), ARC (OR = 2.38), PON (OR = 1.49), and PRA (OR = 1.85) (Figure 1e).
FIGURE 1.
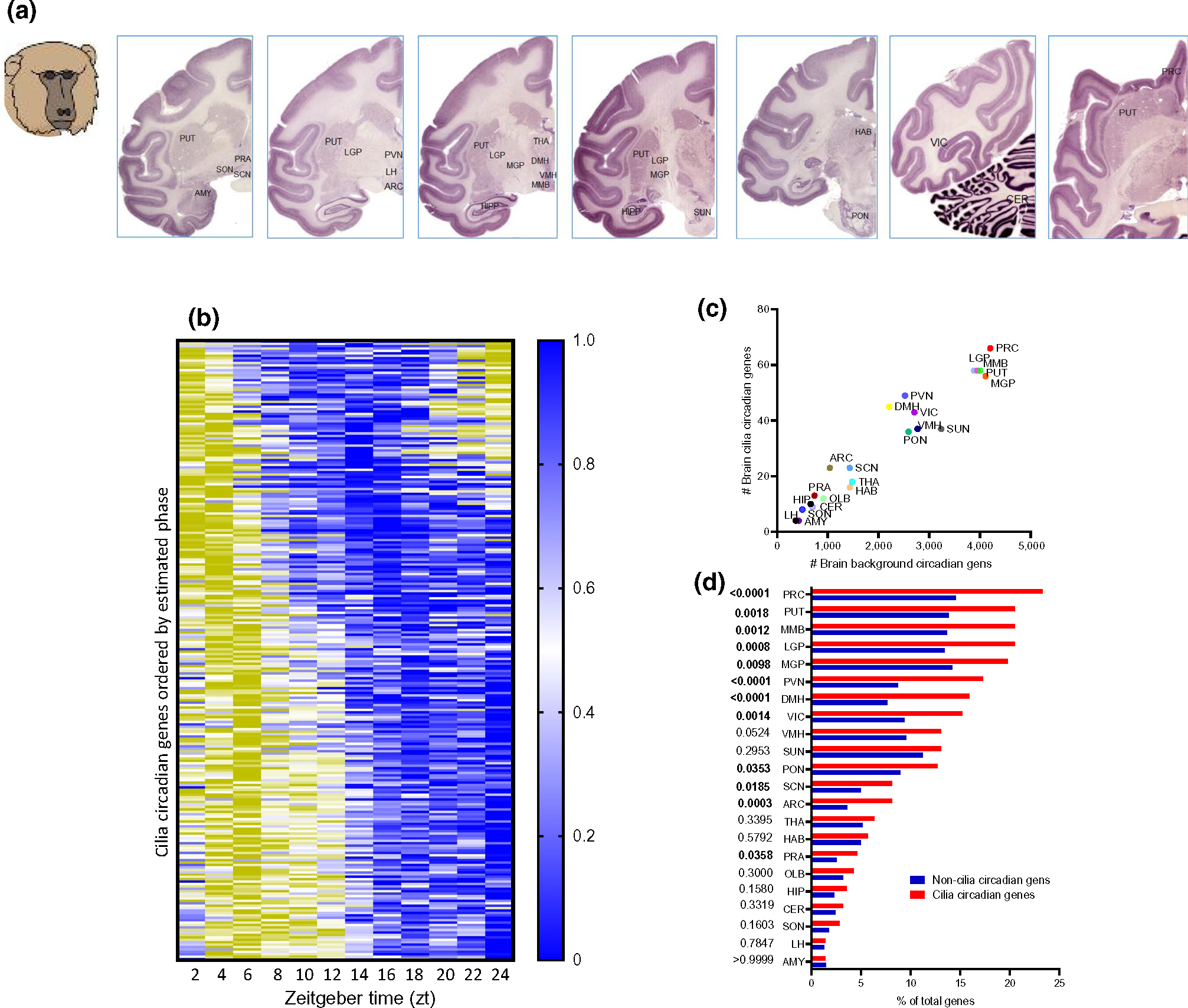
Twenty-four-hour circadian rhythms of cilia genes abundance in the brain of primates. (a) Representative brain sections from Macaca mulatta brains (Brain maps, Primates: www.brainmaps.org (Mikula et al., 2007)), showing the regions and nuclei from which tissues were collected (Mure et al., 2018). AMY, Amygdala; ARC, arcuate nucleus; CER, cerebellum; DMH, dorsomedial hypothalamus; HAB, habenula; HIP, hippocampus; LGP, lateral globus pallidus; LH, lateral hypothalamus; MGP, medial globus pallidus; MMB, mammillary body; OLB, olfactory bulb; PRA, preoptic area; PRC, prefrontal cortex; PON, pons; PUT, putamen; PVN, paraventricular nucleus; SCN, suprachiasmatic nucleus; SON, supraoptic nucleus; SUN, substantia nigra; THA, thalamus; VIC, visual cortex; VMH, ventromedial hypothalamus. (b) Heatmap representation of 24-hr oscillation of 206 circadian cilia genes in primate brain, with mean expressions ordered by phase (p < 0.05). Gene expressions are normalized between 0 and 1, yellow (1) indicates a peak of expression and blue (0) indicates a trough of expression. (c) Linear correlation between number of cilia circadian genes in a given brain region and the number of general circadian genes in the same region (Pearson’s r, r2 = 0.966, p < 0.0001). (d) Histogram showing the enrichment analysis (Fisher’s exact test) of cilia genes that are circadian in the 22 brain regions; 12 regions exhibited significant enrichment (i.e., with overrepresented circadian cilia transcripts including: ARC, arcuate nucleus; DMH, dorsomedial hypothalamus; LGP, lateral globus pallidus; LH, lateral hypothalamus; MGP, medial globus pallidus; MMB, mammillary body; PON, pons; PRA, preoptic area; PRC, prefrontal cortex; PUT, putamen; PVN, paraventricular nucleus; SCN, suprachiasmatic nucleus; VIC, visual cortex)
We grouped the circadian ciliary genes into four 6-hr phases (quarter-phases) (Figure 2a). Across all cilia circadian genes and the 22 brain regions, the combination of peak phases of expression revealed three main peaks, early afternoon (44 and 83 cilia genes peaked at ZT6 and ZT7, respectively), midnight (41 genes peaked at ZT14), and late night (39 genes peaked at ZT20) (Figure 2a,b). With a few exceptions, the peak phases in each brain region were mostly bundled in one or two narrow (<6 hr) temporal intervals (Figure 2c,d). In regions such as ARC, DMH, LGP, MGP, PRC, PUT, PVN, VIC, SCN, and PRA, the majority of circadian genes peaked within a narrow window, whereas some regions such as PON, MMB, and SCN had transcripts that peaked within two distinct time windows (Figure 2d). However, in anatomically and functionally connected regions, these phase clusters were temporally close. For example, in the basal ganglia, the main phase cluster was at ZT4 for the PUT, and in the LGP and MGP, it appeared at ZT6 and ZT7, respectively. Similarly, except for the PRA, the phase clusters of the hypothalamic nuclei (ARC, DMH, SCN, and PVN) were similar (Figure 2c,d).
FIGURE 2.
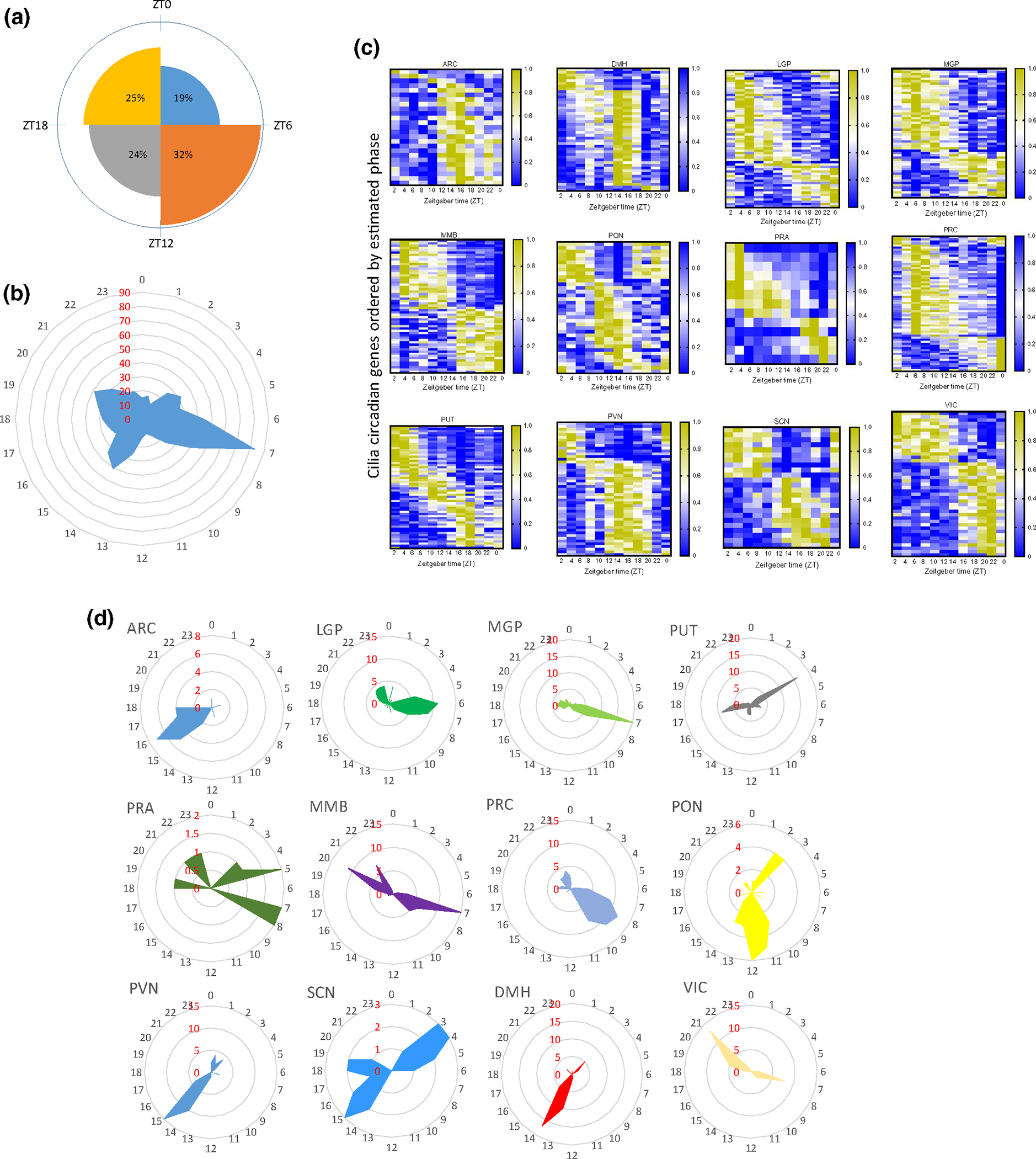
Cilia circadian gene display region-specific expression. (a) Rose diagram showing the percentage of cilia circadian genes in each of the four time phases. (b) Radial diagram of the distribution of the peak phase of expression of the circadian cilia genes in the whole brain. The radial plot displays phases (hr) on the circumference and the number of gene peaks of expression on the radius (red). (c) Heatmap representation of 24-hr oscillation of circadian cilia genes in the 12 brain regions that exhibited overrepresented circadian cilia genes in primate brain (p < 0.05), with mean expressions ordered by phase (p < 0.05). Gene expressions are normalized between 0 and 1, yellow (1) indicates a peak of expression and blue (0) indicates a trough of expression. (d) Radial plot of the distribution of the peak phase of expression of the cilia circadian genes in each of the 12 brain regions that exhibited overrepresentation (in Figure 1d). Phases (hours) are displayed on the circumference and the numbers of gene peaks of expression are displayed on the radius (red)
3.2 |. Overlap of expression of cilia circadian genes across brain regions
3.2.1 |. Gene view overlap
Confined expression overlap of cilia circadian genes between brain regions was observed, with no cycling genes being shared across all brain regions (Figure 3a). The maximal number of regions that shared one circadian gene was 14 regions, whereas 54 cilia genes exhibited circadian oscillation only in a single brain region (Figure 3a). We surveyed genes that were detected as being circadian in multiple brain regions. The gene shared in the largest number of brain regions was FOPNL (14 regions), followed by RILPL1, which was circadian in 12 brain regions, and TUBGCP2, TUBGCP6, and RAB8A, which were circadian in nine regions (Figure 3b). Out of these five genes, only FOPNL did peak within a distinct narrow interval (ZT17–ZT22) in all the 14 shared brain regions (Figure 3b).
FIGURE 3.
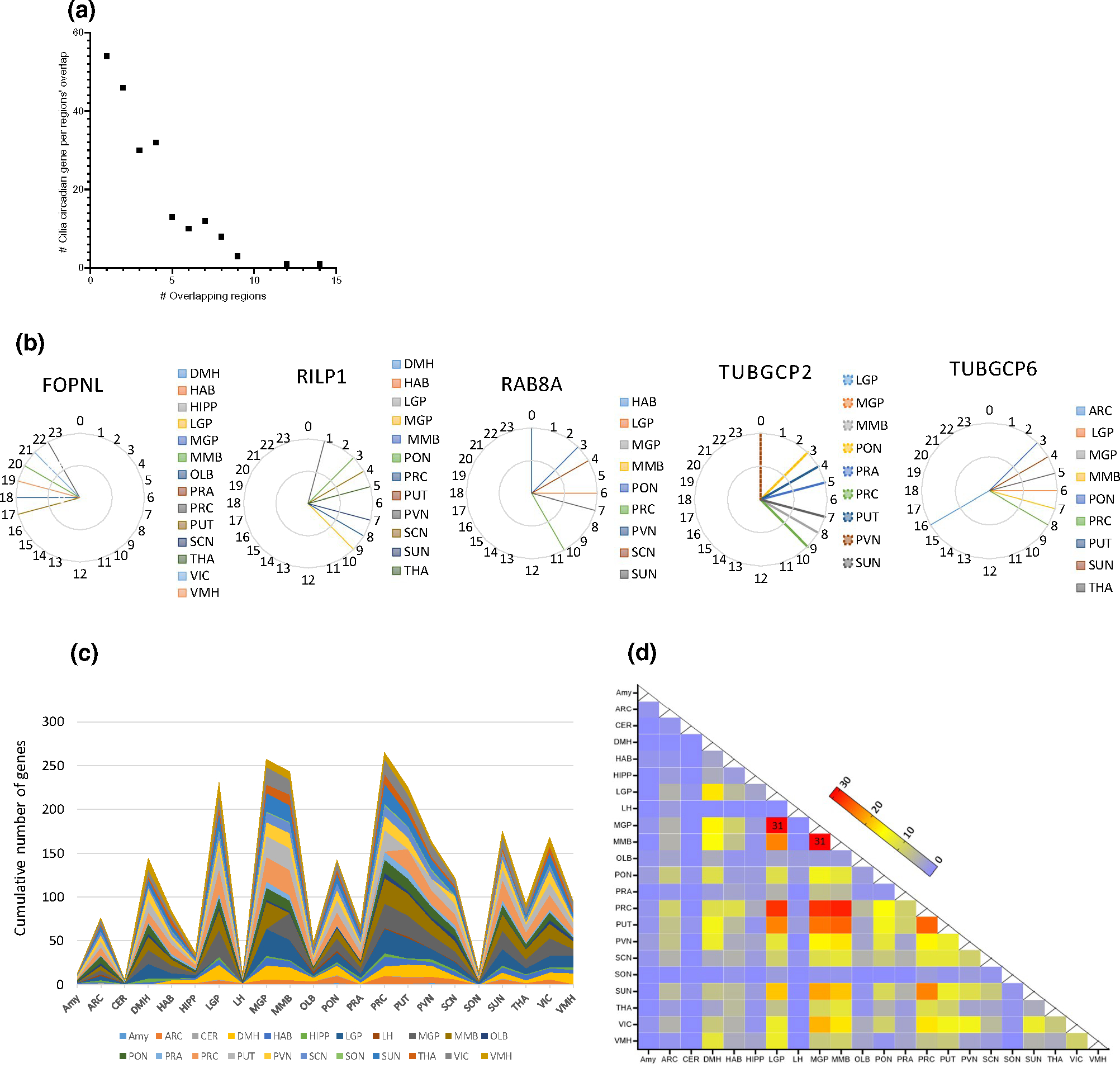
Overlap of cilia circadian transcriptome in brain nuclei/regions. (a) Number of cilia circadian transcripts (y axis), whose expressions exhibit overlap in corresponding number of brain regions’ intersections (x axis). (b) Peak expression phase of top overlapping cilia circadian genes (genes showing highest overlap among brain regions). (c) Stacked area plot showing the cumulative distribution of the overlapped genes in different brain regions. (d) Heatmap of nucleus-by-nucleus intersections of the cilia circadian genes. The size of the intersections ranged from one gene (blue) to 31 transcripts (red), which occurred in two intersections
3.2.2 |. Region view overlap
Region pairwise intersection analysis revealed 184 (out of 231 possible) non-empty intersections. The size of these intersections ranged from one gene for 37 intersections to 31 genes which occurred in two intersections (Figure 3c,d). PUT exhibited an overlap of its cilia circadian transcriptome with all other brain regions (21 regions), and PON, PRC, and SCN exhibited overlaps with 20 brain regions. As per the number of shared circadian genes, MGP exhibited the highest overlap in the transcripts shared (31 genes with LGP and MMB), followed by PRC which overlapped with MMB (29 genes), and with MGP and LGP (28 genes each). In turn, PUT overlapped with MMB (25 shared genes), MGP (24 genes), and LGP (23 genes) (Figure 3d).
3.3 |. Structural and functional organization of cilia circadian genes
We then examined the distribution of cilia circadian genes’ expressions across main structural and functional components of the cilia (Figure 4a). We found that genes encoding the components of the basal body exhibited the highest number (61) of circadian genes (78% of total basal body genes) (Figure 4a). Interestingly, 100% of genes encoding the components of the Golgi were circadian, whereas 87%, 87.5%, 67%, 79%, 62%, and 47.6% of IFT-B, kinesin, transition zone, GPCRs, axoneme, and dynein, respectively, exhibited circadian rhythms in at least one brain region (Figure 4a,b). This strongly suggests that the regulation of cilia assembly and functions have a strong circadian component.
FIGURE 4.
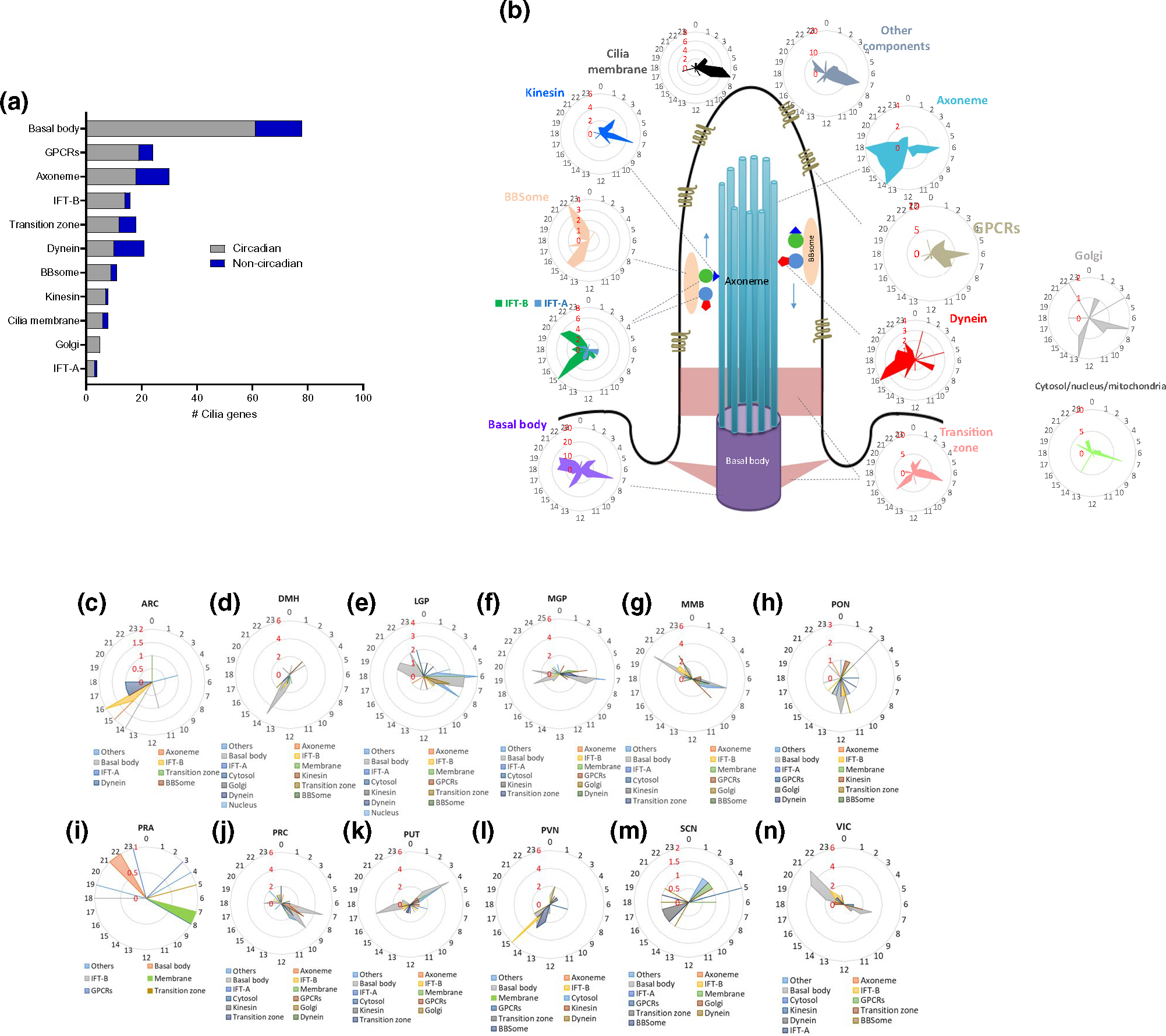
Substructural organization of cilia circadian genes. (a) Number of circadian and non-circadian genes in each of cilia substructural compartments. (b) Schematic of cilia structure and radial diagrams of the distribution of the peak phase of expression of the cilia substructural components’ transcripts in the whole brain. The radial plot displays phases (hours) on the circumference and the number of gene peaks of expression on the radius (in red). (c–n) Radial diagram of the distribution of the peak phase of expression of the circadian cilia genes in the 12 brain regions that showed significant enrichment. The radial plot displays phases (hours) on the circumference and the number of gene peaks of expression on the radius
We further analyzed the phase distributions of the genes encoding most cilia structural and functional components in the entire brain. We found that most genes were clustered in one or two narrow windows (Figure 4b–n), whereas some components were widely distributed within 14–18 hr, such as the genes of the basal body and transition zone. Notably, the circadian transcripts associated with the axoneme, which gives rise to the cilia cytoskeletal structure, peaked within wide intervals. Apparently, the majority of cilia circadian transcripts that peaked at ZT7, as noted above, encode components of the basal body, transition zone, kinesin, cilia membrane, and Golgi apparatus. Of particular interest are the temporal patterns of the transport machinery genes (kinesin, dynein, BBsomes, and IFT-A and IFT-B). The circadian genes associated with kinesin motors, which are primarily involved in anterograde trafficking, peaked during the 12-hr light phase, whereas the circadian genes associated with dynein motors, which are responsible for the retrograde transport, peaked during the 12-hr dark phase. The circadian transcripts of BBsomes and IFT-B particles peaked in the same phase as dynein in the dark phase (ZT13–ZT22). Most interestingly, most circadian genes encoding cilia GPCRs, which are transported through cilia by the kinesin-dependent trafficking machinery, peaked in a way similar to kinesin, during the light phase.
Although the abundance of rhythmic gene expression of cilia substructures components varied across brain regions, the peak phases of expression of the cilia substructural components were mostly consistent with their rhythmic patterns in the whole brain (Figures 4b–n and S1a–l). Circadian transcripts encoding specific cilia substructures, however, exhibited dispersed temporal phases in different brain regions. Unlike their multiphasic oscillating patterns in the whole brain, genes of the basal body and the transition zone tended to peak in one or two phases in different brain regions. These results support the idea that specific cilia genes oscillate in circadian manner distinctively across brain regions (Figure 4b–n and S1a–l).
3.4 |. Rhythmicity of brain circuits’ functions
Given the high level of circadian rhythmicity and confinement of shared oscillatory patterns in multiple brain nuclei, we speculated that cilia components display orchestrated oscillatory patterns in anatomically and functionally defined brain circuits. We focused on a brain circuit involved in movement control because its nuclei and structures, PUT, SUN, MGP, LGP, and PFC, share the highest number of circadian cilia transcripts. Notably, PUT, LGP, MGP, and SUN shared 10 cilia circadian genes (Figures 3d and 5a). These four nuclei collectively form the basal ganglia, a set of subcortical nuclei that are mainly responsible for motor control and motor learning, in addition to other functions such as executive functions and emotions. Circadian genes in the PUT, the input region of the basal ganglia, peaked mostly at two sharp phases: ZT4 (first half of the morning) and ZT17 (around midnight), whereas LGP and MGP genes peaked within 1–2 hr of PUT cilia genes peaking (ZT6–ZT7 and ZT17–ZT20). Interestingly, nine out of the 10 shared circadian transcripts among the basal ganglia structures (B9D1, C21ORF2, CCDC28B, FUZ, PARD6A, PKD1, RILPL1, TUBGCP2, and TUBGCP6) peaked at ZT4 in the PUT and ZT6–ZT8 in the other regions. All these nine genes were also rhythmic in the PRC and peaked there at ZT9–ZT10. The tenth common gene (PCM1) in the basal ganglia nuclei peaked at ZT17 in the PUT and at ZT18–ZT20 in the three other nuclei, maintaining a 12-hr phase shift with respect to the other nine genes in the four basal ganglia structures.
FIGURE 5.
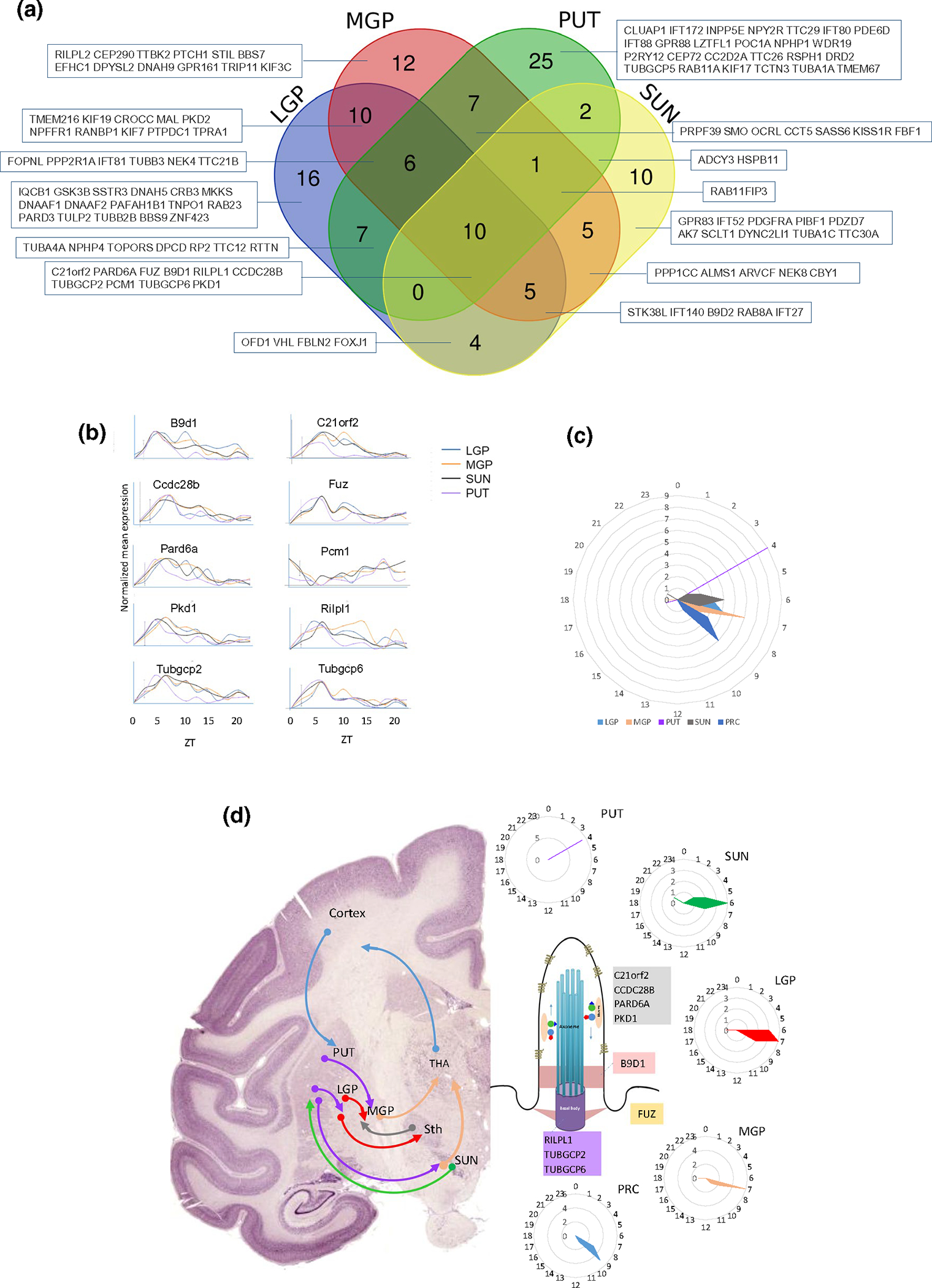
Rhythmicity of the nuclei/regions of the basal ganglia-cortex circuit. (a) Venn diagram showing the overlap between the cilia circadian genes identified in the motor control circuit (nuclei of the basal ganglia and PRC). The overlapping transcripts are shown in boxes. (b) Twenty-four-hour oscillating cilia genes in the basal ganglia nuclei, displayed as normalized expression levels. (c) Radial plot of the distribution of the peak phase of expression of the cilia circadian genes in each of the five brain regions of the motor control circuit. Phases (hours) are displayed on the circumference and the numbers of gene peaks of expression are displayed on the radius. (d) Schematic diagram illustrating the arrangement of information flow through the cortical-basal ganglia-cortical circuit in primate brain (left), and the peak phase of the nine overlapping genes in the nuclei/regions of the circuit (right), with their localized expression in the cilia substructures (middle)
4 |. DISCUSSION
This study presents a systematic evaluation of rhythmicity and spatiotemporal expression patterns of cilia genes in 22 baboon brain regions. To examine whether cilia-associated genes oscillate in a circadian manner in various brain areas, we acquired publicly available transcriptomic data from the largest spatiotemporal gene expression atlas of a primate (Mure et al., 2018).
Around 73% of cilia transcripts exhibited circadian rhythmicity across at least one of the 22 brain regions reported in the primate diurnal transcriptome atlas (Mure et al., 2018); and in 12 brain regions, cilia circadian transcripts were statistically overrepresented compared to the brain circadian background transcriptome, supporting the notion that cilia are highly dynamic systems.
4.1 |. Cilia circadian genes follow distinct rhythmic patterns
To our surprise, cilia circadian transcripts did not strictly follow the rhythmic fashion of the majority of the background circadian transcriptomes. For example, in contrast to many circadian transcripts which tend to peak in the early morning (Mure et al., 2018), cilia rhythmic transcripts tend to peak during the early afternoon at ZT7 (83 transcripts). Furthermore, there were a few major differences in the rhythmic patterns between cilia genes and the background of all circadian transcripts in specific brain regions (Mure et al., 2018). For example, contrary to the expression patterns of the majority of background circadian genes in the PVN, which peaked in the early morning (first quarter-phase), cilia circadian genes peaked at ZT14–ZT15 (third quarter-phase) in this region. In addition, unlike the majority of ARC background rhythmic transcriptomes, which peaked within three phases (first, third, and fourth quarter-phases), cilia rhythmic genes in this region peaked only within a 4-hr interval (ZT14–18) (Mure et al., 2018). On the other hand, cilia circadian transcripts in the SCN (master clock) had peaks distributed across three phases (the first, third, and fourth quarter-phases), compared to the majority of background circadian transcripts, which peaked mainly in the first quarter-phase (Mure et al., 2018). These discrepancies raise the question of whether the gene expression of cilia circadian components is regulated in a specific way that is different from the background oscillating transcriptomes.
4.2 |. Rhythmicity of cilia structural and functional components
The primary core structure of the cilia comprises 9+0 axoneme that consists of doublet microtubules nucleating and extending from the basal body through the ciliary transition zone (Christensen et al., 2007; Satir & Christensen, 2007). The basal body is a centriolar structure composed of a radial array of nine triplet microtubules and comprised of a mother and daughter centrioles (Garcia & Reiter, 2016; Thomas et al., 2010). Genes coding for the basal body components produced the largest fraction of circadian transcripts in each of the 22 brain regions. The peak phases of circadian genes encoding components of axoneme, basal body, and transition zones were dispersed, with the majority of genes peaking at ZT4, ZT7, ZT14, and ZT16–ZT18. This strongly suggests that cilia assembly/disassembly and length, which are governed by axoneme and basal body components, might also oscillate following similar rhythmic patterns to that of axoneme, basal body components.
Ciliogenesis, elongation, and maintenance are contingent on the proper function of the intraflagellar transport (IFT) machinery (Pedersen & Rosenbaum, 2008). IFT is associated with a bidirectional transport process that relies on kinesin and dynein molecular motors, IFT particle subcomplexes (A and B), and IFT-associated proteins (e.g., Bardet–Biedl syndrome (BBS) proteins) (Mourao et al., 2016; Taschner & Lorentzen, 2016). Kinesin-dependent motors, associated with IFT-B particles, traffic cargo (e.g., axonemal components and receptors) toward the tip of the cilia. In contrast, dynein-mediated retrograde motor, associated with IF-A particles, recycles components back to the base (Mourao et al., 2016; Nakayama & Katoh, 2018; Taschner & Lorentzen, 2016). Circadian genes encoding components of kinesin peaked in the light phase (ZT0–ZT11), whereas those encoding dynein components peaked in the dark phase (ZT12–ZT23). Unexpectedly, genes encoding IFT-B particles peaked, unlike kinesin, in the dark phase, along with other components of the IFT system (BBsomes). Although the number of rhythmic genes encoding IFT-B particles seem to be higher than that of IFT-A, rhythmic IFT-A genes accounted for 100% of the entire brain IFT-A genes, whereas rhythmic IFT-B genes accounted for 82% of brain IFT-B genes.
Ciliary GPCRs play essential roles in the signal transduction mediated by neuronal cilia. Interestingly, cilia rhythmic GPCRs included receptors that bind to known neurotransmitters/ neuropeptides (DRD2, KISSR1, SMO, MCHR1, SSTR3, GALR3, GALR2, DRD5, and HTR6) as well as orphan receptors (GPR88, GPR161, and GPR83) (Green et al., 2012; Hirano et al., 2017; Mykytyn & Askwith, 2017; Omori et al., 2015; Schou et al., 2015). Signaling through these cilia GPCRs has been implicated in numerous physiological functions such as movement control, feeding behavior, cognitive processes, wake/sleep, and reproduction (Berbari et al., 2008, 2014; Engle et al., 2020; Mykytyn & Askwith, 2017; Omori et al., 2015). Cilia receptors recruit kinesin/IFT-B transport machinery for their trafficking to the cilia membrane (Kobayashi et al., 2020; McIntyre et al., 2016). Therefore, peaking of the brain cilia primarily in the light phase (ZT2–ZT3, ZT7, and ZT8–ZT9) and the similar rhythmic patterns of GPCRs and IFT-B particles are, thus, not surprising.
4.3 |. Cilia circadian transcripts exhibit higher overlap in adjacent and functionally connected nuclei
Despite the limited overlap of cilia circadian genes between regions, nuclei with anatomical and/or functional connectivity exhibited higher overlap than non-connected regions. For example, cilia rhythmic genes in the hypothalamic nuclei including SCN, PVN, ARC, and DMH shared peaking in the third quarter-phase (early evening (ZT14–ZT16)). On the other hand, cilia cycling genes in the anatomically and/or functionally connected nuclei of basal ganglia (PUT, LGP, MGP, and SUN) and the PFC peaked in the light phase when the animal is awake. Interestingly, the amygdala, which is involved in emotional behaviors and fear response, and the cerebellum showed the lowest overlap of cilia rhythmic genes with any other brain regions.
The fact that physiological functions that are regulated by cilia follow circadian patterns raises the questions of whether the circadian rhythm of cilia gene expressions regulates circadian rhythmicity of these functions or that cilia gene expressions follow the physiological needs associated with the different brain regions. For example, the expression levels of specific cilia genes such as MC4R, MCHR1, and ADCY3 directly alter feeding behavior (Kobayashi et al., 2021; Siljee et al., 2018). On the other hand, cilia length is controlled by the feeding status (they are shorter in fasted mice than fed mice) (Hamamoto et al., 2016; Kobayashi et al., 2021). Given that feeding behavior is regulated by a number of hypothalamic orexigenic and anorexic neuropeptides, the circadian rhythm of cilia gene expressions in different brain regions may simply follow the circadian expressions of these hypothalamic neuropeptides.
4.4 |. Basal ganglia as a model of brain rhythmic functional circuit
The dynamic organization and spatiotemporal coordination of activities within and across different parts of the basal ganglia-cortex loop are at the basis of motor initiation, coordination, and learning (Graybiel et al., 1994). Remarkably, the nine shared cilia rhythmic genes in the basal ganglia nuclei and the PRC peaked in these structures in a sequential fashion, as in a wave: PUT (ZT4) → SUN (ZT6) → LGP/MGP (ZT7) → PRC (ZT9). This order is interesting, as it is the same order of activation, albeit on a completely different timescale, of the basal ganglia-cortical circuitry connected with movement coordination.
The PUT, with caudate (CA), is the main input to the basal ganglia, whereas the MGP and pars reticulata of the SUN (SUNr) represent the output stations of the basal ganglia. Interestingly, the PUT was the only nucleus that shared rhythmic cilia genes with all the other brain regions, whereas MGP shared the highest number of cilia rhythmic genes with other brain regions (31 genes with each of LGP and MMB, 28 with PRC, 27 with PUT, 21 with SUN, and 20 with VIC).
The basal ganglia control the movement by regulating motor planning, sequencing, feedback processing and learning; and by acting as a coincidence detector of cortical and thalamic input (Bednark et al., 2015; Jin et al., 2009; Li et al., 2011; Talakoub et al., 2016). According to the functional circuit model of the basal ganglia, information about movement is collected from the cortex, processed in the PUT/CA, and then transmitted to the output structures of the basal ganglia (SUNr/MGP), through two pathways, the direct and indirect pathways (for review (Foley & Riederer, 2000; Ikemoto et al., 2015; Kreitzer & Malenka, 2008; Morita & Hikida, 2015)). The direct pathway projects monosynaptically to the output nuclei of the basal ganglia, whereas the indirect pathway projects to the output regions, bisynaptically (via LGP) and trisynaptically (via LGP and the subthalamic nucleus (STN)). When the animal is not moving, PUT/CA neurons are mute, while neurons in the output regions of the basal ganglia are tonically active. The firings of the SUNr/MGP drive an inhibitory (GABAergic) tone on the ventral thalamic nucleus, which in turn project back to the cortex. Apparently, the spatiotemporal expressions of circadian cilia genes in the basal ganglia neurons follow the sequential order of this circuitry while controlling movement, though on different timescales. The highly coordinated rhythmicity of cilia genes in the basal ganglia and the peaking of these genes during the light phase suggest an essential role for cilia genes in the control of motor sequencing and activity. In support of this notion, many GPCRs in the basal ganglia that are involved in motor control (including D2 and D1 dopamine receptors) are localized on the cilia membrane (Avasthi et al., 2012; Domire et al., 2011; Omori et al., 2015). The pars compacta of the SUN (SUNc) sends dopaminergic projections to the PUT/CA, which stimulates the direct pathway via D1 receptors and inhibits the indirect pathway via D2 receptors. Interestingly, D2 but not D1 receptors displayed circadian rhythms in the PUT, suggesting a key role for cilia rhythmicity in the function of the indirect pathway of the basal ganglia (for review, see Foley and Riederer (2000); Haber (2014)).
The basal ganglia circuit is believed to perform central clock functions in the brain (Macar et al., 2006; Wiener et al., 2010), and its role in timing is thought to result from the dopaminergic projection from the SUNc to the PUT/CA. SUNc and PUT/CA are the two basal ganglia regions that are necessary for interval timing (Matell & Meck, 2004). It is accepted now that the pacemaker pulses are action potentials of dopaminergic neurons, and that dopamine controls the clock speed (Bussi et al., 2014; Jones et al., 2008; Yang et al., 2004). Thus, the increases in synaptic dopamine in the PUT/CA result in a faster internal clock process, whereas the decreases in PUT/CA synaptic dopamine slow down the clock speed (Bussi et al., 2014; Jones et al., 2008; Yang et al., 2004).
The dysfunctions of the basal ganglia underlie a spectrum of movement disorders including Parkinson’s disease, which results from the degeneration of dopaminergic neurons in the SUNc, and Huntington’s disease, which results from the degeneration of projection neurons in the PUT/CA. Of great interest, patients in both disorders exhibit profound difficulty in performing rhythmic movements and show decreased ability to calculate the timing of the initiation and termination of voluntary actions, especially for sequential movements (Avanzino et al., 2016; Boyd et al., 2009; Nagasaki et al., 1978). The rhythmicity of cilia genes encoding therapeutic targets (Dopamine receptors) for Parkinson’s disease and Huntington’s disease suggests that the therapeutic efficacies might be influenced by the time-of-day of administration of the corresponding drugs.
We speculate that spatiotemporal orchestration of cilia gene expression in the basal ganglia, particularly in the SUN, PUT, LGP, and MGP, is essential to maintain normal physiology of the basal ganglia-cortical circuit and proper motor control, and that abnormal orchestration of cilia genes might contribute to the pathophysiology of several neurological and psychiatric disorders. These speculations are supported by our recent study, which provided evidence for the association between cilia gene expressions and psychiatric disorders such as schizophrenia, autism, bipolar and major depressive disorders (Alhassen et al., 2021).
Further mechanistic studies are warranted to better characterize and understand cilia rhythmicity in the basal ganglia-cortical circuits and other brain circuits, which in turn may help develop better functional models, and shed light on the causal effects cilia functions have on these circuits and on the regulation of movement and other behaviors.
Supplementary Material
TABLE S1 List of cilia genes with their substructure localizations
FIGURE S1 Subcilia localization of the circadian genes in brain regions, in which cilia transcripts exhibited higher rhythmicity than the non-cilia transcripts. Subcilia localization of genes that exhibit circadian expression in (a) ARC, (b) DMH, (c) LGP, (d) MGP, (e) MMB, (f) PON, (g) (PRA), (h) PRC, (i) PUT, (j) PVN, (k) SCN, and (l) (VIC)
TABLE S2 Cilia gene transcripts that oscillate in a circadian manner in 22 baboon brain regions together with the oscillating signal’s period, lag, and amplitude
Significance.
Understanding orchestrated cilia rhythmicity in brain neurocircuits will help develop better functional models, and shed light on the causal effects cilia functions have on the regulation of oscillatory metabolic, physiological, and behavioral processes.
ACKNOWLEDGMENTS
The work of AA was supported by the department of pharmaceutical sciences, UCI. The work of SC and PB was supported in part by NIH grant GM123558.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jnr.24919.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
Transparent Science Questionnaire for Authors
Transparent Peer Review Report
DATA AVAILABILITY STATEMENT
All the circadian data sets and the deep learning model BIO_CYCLE are publically available on the CircadiOmics web portal (circadiomics.igb.uci.edu). The raw data of cilia circadian gene expressions are available in Table S2.
REFERENCES
- Agostinelli F, Ceglia N, Shahbaba B, Sassone-Corsi P, & Baldi P (2016a). What time is it? Deep learning approaches for circadian rhythms. Bioinformatics, 32(19), 3051. 10.1093/bioinformatics/btw504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinelli F, Ceglia N, Shahbaba B, Sassone-Corsi P, & Baldi P (2016b). What time is it? Deep learning approaches for circadian rhythms. Bioinformatics, 32(12), i8–i17. 10.1093/bioinformatics/btw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhassen W, Chen S, Vawter M, Robbins BK, Nguyen H, Myint TN, Saito Y, Schulmann A, Nauli SM, Civelli O, Baldi P, & Alachkar A (2021). Patterns of cilia gene dysregulations in major psychiatric disorders. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 109, 110255. 10.1016/j.pnpbp.2021.110255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Cohen J, Tassin AM, & Koll F (2014). Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia, 3, 9. 10.1186/2046-2530-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Malinowska A, Klotz C, Sperling L, Dadlez M, Koll F, & Cohen J (2009). Cildb: A knowledgebase for centrosomes and cilia. Database, 2009, bap022. 10.1093/database/bap022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson KF, Sherpa RT, & Nauli SM (2019). The role of the primary cilium in sensing extracellular pH. Cells, 8(7), 704. 10.3390/cells8070704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzino L, Pelosin E, Vicario CM, Lagravinese G, Abbruzzese G, & Martino D (2016). Time processing and motor control in movement disorders. Frontiers in Human Neuroscience, 10, 631. 10.3389/fnhum.2016.00631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Marley A, Lin H, Gregori-Puigjane E, Shoichet BK, von Zastrow M, & Marshall WF (2012). A chemical screen identifies class a g-protein coupled receptors as regulators of cilia. ACS Chemical Biology, 7(5), 911–919. 10.1021/cb200349v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, & Barr MM (2008). Sensory roles of neuronal cilia: Cilia development, morphogenesis, and function in C. elegans. Frontiers in Bioscience, 13, 5959–5974. 10.2741/3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI (2006). Chemosensation in C. elegans. In WormBook; (pp. 1–29). http://www.wormbook.org/chapters/www_chemosensation/chemosensation.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednark JG, Campbell ME, & Cunnington R (2015). Basal ganglia and cortical networks for sequential ordering and rhythm of complex movements. Frontiers in Human Neuroscience, 9, 421. 10.3389/fnhum.2015.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, & Vosshall LB (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature, 450(7167), 289–293. 10.1038/nature06328 [DOI] [PubMed] [Google Scholar]
- Berbari NF, Lewis JS, Bishop GA, Askwith CC, & Mykytyn K (2008). Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proceedings of the National Academy of Sciences of the United States of America, 105(11), 4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Malarkey EB, Yazdi SMZR, McNair AD, Kippe JM, Croyle MJ, Kraft TW, & Yoder BK (2014). Hippocampal and cortical primary cilia are required for aversive memory in mice. PLoS ONE, 9(9), e106576. 10.1371/journal.pone.0106576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, & Yoder BK (2009). The primary cilium as a complex signaling center. Current Biology, 19(13), R526–R535. 10.1016/j.cub.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Pasek RC, Malarkey EB, Yazdi SMZ, McNair AD, Lewis WR, Nagy TR, Kesterson RA, & Yoder BK (2013). Leptin resistance is a secondary consequence of the obesity in ciliopathy mutant mice. Proceedings of the National Academy of Sciences of the United States of America, 110(19), 7796–7801. 10.1073/pnas.1210192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd LA, Edwards JD, Siengsukon CS, Vidoni ED, Wessel BD, & Linsdell MA (2009). Motor sequence chunking is impaired by basal ganglia stroke. Neurobiology of Learning and Memory, 92(1), 35–44. 10.1016/j.nlm.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Brailov I, Bancila M, Brisorgueil MJ, Miquel MC, Hamon M, & Verge D (2000). Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Research, 872(1–2), 271–275. 10.1016/s0006-8993(00)02519-1 [DOI] [PubMed] [Google Scholar]
- Breunig JJ, Sarkisian MR, Arellano JI, Morozov YM, Ayoub AE, Sojitra S, Wang B, Flavell RA, Rakic P, & Town T (2008). Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proceedings of the National Academy of Sciences of the United States of America, 105(35), 13127–13132. 10.1073/pnas.0804558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, & Takahashi JS (2010). Temperature as a universal resetting cue for mammalian circadian oscillators. Science, 330(6002), 379–385. 10.1126/science.1195262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi IL, Levin G, Golombek DA, & Agostino PV (2014). Involvement of dopamine signaling in the circadian modulation of interval timing. European Journal of Neuroscience, 40(1), 2299–2310. 10.1111/ejn.12569 [DOI] [PubMed] [Google Scholar]
- Casey T, Patel OV, & Plaut K (2015). Transcriptomes reveal alterations in gravity impact circadian clocks and activate mechanotransduction pathways with adaptation through epigenetic change. Physiological Genomics, 47(4), 113–128. 10.1152/physiolgenomics.00117.2014 [DOI] [PubMed] [Google Scholar]
- Ceglia N, Liu Y, Chen S, Agostinelli F, Eckel-Mahan K, Sassone-Corsi P, & Baldi P (2018). CircadiOmics: Circadian omic web portal. Nucleic Acids Research, 46(W1), W157–W162. 10.1093/nar/gky441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Madhu V, Shapiro IM, & Risbud MV (2019). Nucleus pulposus primary cilia alter their length in response to changes in extracellular osmolarity but do not control TonEBP-mediated osmoregulation. Scientific Reports, 9(1), 15469. 10.1038/s41598-019-51939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ST, Pedersen LB, Schneider L, & Satir P (2007). Sensory cilia and integration of signal transduction in human health and disease. Traffic, 8(2), 97–109. 10.1111/j.1600-0854.2006.00516.x [DOI] [PubMed] [Google Scholar]
- Christensen ST, Voss JW, Teilmann SC, & Lambert IH (2005). High expression of the taurine transporter TauT in primary cilia of NIH3T3 fibroblasts. Cell Biology International, 29(5), 347–351. 10.1016/j.cellbi.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Clary-Meinesz CF, Cosson J, Huitorel P, & Blaive B (1992). Temperature effect on the ciliary beat frequency of human nasal and tracheal ciliated cells. Biology of the Cell, 76(3), 335–338. 10.1016/0248-4900(92)9043-5 [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, & Schibler U (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development, 14(23), 2950–2961. 10.1101/gad.183500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan K, Benloucif S, Reid K, Wolfe LF, & Zee PC (2011). Light-induced changes of the circadian clock of humans: Increasing duration is more effective than increasing light intensity. Sleep, 34(5), 593–599. 10.1093/sleep/34.5.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz GB, Battagello DS, Klein MO, Bono BSM, Ferreira JGP, Motta-Teixeira LC, Duarte JCG, Presse F, Nahon J-L, Adamantidis A, Chee MJ, Sita LV, & Bittencourt JC (2020). Ciliary melanin-concentrating hormone receptor 1 (MCHR1) is widely distributed in the murine CNS in a sex-independent manner. Journal of Neuroscience Research, 98(10), 2045–2071. 10.1002/jnr.24651 [DOI] [PubMed] [Google Scholar]
- Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, & Mykytyn K (2011). Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cellular and Molecular Life Sciences, 68(17), 2951–2960. 10.1007/s00018-010-0603-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle SE, Bansal R, Antonellis PJ, & Berbari NF (2020). Cilia signaling and obesity. Seminars in Cell & Developmental Biology, 110, 43–50. 10.1016/j.semcdb.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley P, & Riederer P (2000). The motor circuit of the human basal ganglia reconsidered. Journal of Neural Transmission, 58, 97–110. 10.1007/978-3-7091-6284-2_8 [DOI] [PubMed] [Google Scholar]
- Fuller CA (1994). The effects of gravity on the circadian timing system. Journal of Gravitational Physiology, 1(1), P1–P4. [PubMed] [Google Scholar]
- Garcia G 3rd, & Reiter JF (2016). A primer on the mouse basal body. Cilia, 5, 17. 10.1186/s13630-016-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM, Aosaki T, Flaherty AW, & Kimura M (1994). The basal ganglia and adaptive motor control. Science, 265(5180), 1826–1831. 10.1126/science.8091209 [DOI] [PubMed] [Google Scholar]
- Green JA, Gu C, & Mykytyn K (2012). Heteromerization of ciliary G protein-coupled receptors in the mouse brain. PLoS ONE, 7(9), e46304. 10.1371/journal.pone.0046304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Geddes BJ, Zhang C, Foley KP, & Stricker-Krongrad A (2004). The prolactin-releasing peptide receptor (GPR10) regulates body weight homeostasis in mice. Journal of Molecular Neuroscience, 22(1–2), 93–103. 10.1385/JMN:22:1-2:93 [DOI] [PubMed] [Google Scholar]
- Guo J, Otis JM, Higginbotham H, Monckton C, Cheng JG, Asokan A, Mykytyn K, Caspary T, Stuber GD, & Anton ES (2017). Primary cilia signaling shapes the development of interneuronal connectivity. Developmental Cell, 42(3), 286–300.e284. 10.1016/j.devcel.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN (2014). The place of dopamine in the cortico-basal ganglia circuit. Neuroscience, 282, 248–257. 10.1016/j.neuroscience.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto A, Yamato S, Katoh Y, Nakayama K, Yoshimura K, Takeda S, Kobayashi Y, & Saito Y (2016). Modulation of primary cilia length by melanin-concentrating hormone receptor 1. Cellular Signalling, 28(6), 572–584. 10.1016/j.cellsig.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, & Alvarez-Buylla A (2008). Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nature Neuroscience, 11(3), 277–284. 10.1038/nn2059 [DOI] [PubMed] [Google Scholar]
- Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, & Höllt V (1999). Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience, 89(3), 909–926. 10.1016/s0306-4522(98)00354-6 [DOI] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, & Nakayama K (2017). Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Molecular Biology of the Cell, 28(3), 429–439. 10.1091/mbc.E16-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JW, Cho JH, Conway HE, DiGruccio MR, Ng XW, Roseman HF, Abreu D, Urano F, & Piston DW (2020). Primary cilia control glucose homeostasis via islet paracrine interactions. Proceedings of the National Academy of Sciences of the United States of America, 117(16), 8912–8923. 10.1073/pnas.2001936117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries S (2013). A physical explanation of the temperature dependence of physiological processes mediated by cilia and flagella. Proceedings of the National Academy of Sciences of the United States of America, 110(36), 14693–14698. 10.1073/pnas.1300891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S, Yang C, & Tan A (2015). Basal ganglia circuit loops, dopamine and motivation: A review and enquiry. Behavioral Brain Research, 290, 17–31. 10.1016/j.bbr.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, & Besharse JC (2008). Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Developmental Dynamics, 237(8), 1982–1992. 10.1002/dvdy.21554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DZ, Fujii N, & Graybiel AM (2009). Neural representation of time in cortico-basal ganglia circuits. Proceedings of the National Academy of Sciences of the United States of America, 106(45), 19156–19161. 10.1073/pnas.0909881106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CR, Malone TJ, Dirnberger G, Edwards M, & Jahanshahi M (2008). Basal ganglia, dopamine and temporal processing: Performance on three timing tasks on and off medication in Parkinson’s disease. Brain and Cognition, 68(1), 30–41. 10.1016/j.bandc.2008.02.121 [DOI] [PubMed] [Google Scholar]
- Kang N, Ro H, Park Y, Kim HT, Huh TL, & Rhee M (2010). Seson, a novel zinc finger protein, controls cilia integrity for the LR patterning during zebrafish embryogenesis. Biochemical and Biophysical Research Communications, 401(2), 169–174. 10.1016/j.bbrc.2010.08.124 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ishida Y, Hirano T, Katoh Y, & Nakayama K (2020). Cooperation of the IFT-A complex with the IFT-B complex is required for ciliary retrograde protein trafficking and GPCR import. Molecular Biology of the Cell, 32(1), 45–56. 10.1091/mbc.E20-08-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Okada T, Miki D, Sekino Y, Koganezawa N, Shirao T, Diniz GB, & Saito Y (2021). Properties of primary cilia in melanin-concentrating hormone receptor 1-bearing hippocampal neurons in vivo and in vitro. Neurochemistry International, 142, 104902. 10.1016/j.neuint.2020.104902 [DOI] [PubMed] [Google Scholar]
- Koemeter-Cox AI, Sherwood TW, Green JA, Steiner RA, Berbari NF, Yoder BK, Kauffman AS, Monsma PC, Brown A, Askwith CC, & Mykytyn K (2014). Primary cilia enhance kisspeptin receptor signaling on gonadotropin-releasing hormone neurons. Proceedings of the National Academy of Sciences of the United States of America, 111(28), 10335–10340. 10.1073/pnas.1403286111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer AC, & Malenka RC (2008). Striatal plasticity and basal ganglia circuit function. Neuron, 60(4), 543–554. 10.1016/j.neuron.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A, Okumura M, Kimata T, Tanizawa Y, Takano R, Kimura KD, Inada H, Matsumoto K, & Mori I (2008). Temperature sensing by an olfactory neuron in a circuit controlling behavior of C. elegans. Science, 320(5877), 803–807. 10.1126/science.1148922 [DOI] [PubMed] [Google Scholar]
- Lee H, Song J, Jung JH, & Ko HW (2015). Primary cilia in energy balance signaling and metabolic disorder. BMB Reports, 48(12), 647–654. 10.5483/bmbrep.2015.48.12.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Luo F, Shi L, Woodward DJ, & Chang J (2011). Ensemble neural activity of the frontal cortical basal ganglia system predicts reaction time task performance in rats. Neuroscience Research, 71(2), 149–160. 10.1016/j.neures.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tu H, Kang Y, Xue Y, Ma D, Zhao C, Li H, Wang LU, & Liu F (2019). Primary cilia regulate hematopoietic stem and progenitor cell specification through Notch signaling in zebrafish. Nature Communications, 10(1), 1839. 10.1038/s41467-019-09403-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macar F, Coull J, & Vidal F (2006). The supplementary motor area in motor and perceptual time processing: fMRI studies. Cognitive Processing, 7(2), 89–94. 10.1007/s10339-005-0025-7 [DOI] [PubMed] [Google Scholar]
- Matell MS, & Meck WH (2004). Cortico-striatal circuits and interval timing: Coincidence detection of oscillatory processes. Cognitive Brain Research, 21(2), 139–170. 10.1016/j.cogbrainres.2004.06.012 [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Hege MM, & Berbari NF (2016). Trafficking of ciliary G protein-coupled receptors. Methods in Cell Biology, 132, 35–54. 10.1016/bs.mcb.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Mikula S, Trotts I, Stone JM, & Jones EG (2007). Internet-enabled high-resolution brain mapping and virtual microscopy. Neuroimage, 35(1), 9–15. 10.1016/j.neuroimage.2006.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman SJ, & Shorr AZ (2008). The primary cilium as a gravitational force transducer and a regulator of transcriptional noise. Developmental Dynamics, 237(8), 1955–1959. 10.1002/dvdy.21493 [DOI] [PubMed] [Google Scholar]
- Morita M, & Hikida T (2015). Distinct roles of the direct and indirect pathways in the basal ganglia circuit mechanism. Nihon Shinkei Seishin Yakurigaku Zasshi, 35(5–6), 107–111. [PubMed] [Google Scholar]
- Mourao A, Christensen ST, & Lorentzen E (2016). The intraflagellar transport machinery in ciliary signaling. Current Opinion in Structural Biology, 41, 98–108. 10.1016/j.sbi.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, & Panda S (2018). Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science, 359(6381), eaao0318. 10.1126/science.aao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, & Askwith C (2017). G-protein-coupled receptor signaling in Cilia. Cold Spring Harbor Perspectives in Biology, 9(9), a028183. 10.1101/cshperspect.a028183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, & Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nature Reviews Molecular Cell Biology, 20(7), 389–405. 10.1038/s41580-019-0116-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Nakamura R, & Taniguchi R (1978). Disturbances of rhythm formation in patients with Parkinson’s disease: Part II. a forced oscillation model. Perceptual and Motor Skills, 46(1), 79–87. 10.2466/pms.1978.46.1.79 [DOI] [PubMed] [Google Scholar]
- Nagata A, Hamamoto A, Horikawa M, Yoshimura K, Takeda S, & Saito Y (2013). Characterization of ciliary targeting sequence of rat melanin-concentrating hormone receptor 1. General and Comparative Endocrinology, 188, 159–165. 10.1016/j.ygcen.2013.02.020 [DOI] [PubMed] [Google Scholar]
- Nakayama K, & Katoh Y (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. Journal of Biochemistry, 163(3), 155–164. 10.1093/jb/mvx087 [DOI] [PubMed] [Google Scholar]
- Nauli SM, Jin X, AbouAlaiwi WA, El-Jouni W, Su X, & Zhou J (2013). Non-motile primary cilia as fluid shear stress mechanosensors. Methods in Enzymology, 525, 1–20. 10.1016/B978-0-12-397944-5.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko IV, Doroquez DB, & Sengupta P (2013). Primary cilia and dendritic spines: Different but similar signaling compartments. Molecules and Cells, 36(4), 288–303. 10.1007/s10059-013-0246-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, & Hirokawa N (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell, 95(6), 829–837. 10.1016/s0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Silva M, Nguyen KCQ, Zhang W, Bellotti S, Ramadan YH, Hall DH, & Barr MM (2017). Glutamylation regulates transport, specializes function, and sculpts the structure of cilia. Current Biology, 27(22), 3430–3441.e3436. 10.1016/j.cub.2017.09.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan C, Achaval M, Forsythe I, & Barry PW (1995). Brain and respiratory cilia: The effect of temperature. Biology of the Neonate, 68(6), 394–397. 10.1159/000244261 [DOI] [PubMed] [Google Scholar]
- Oishi I, Kawakami Y, Raya A, Callol-Massot C, & Izpisua Belmonte JC (2006). Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nature Genetics, 38(11), 1316–1322. 10.1038/ng1892 [DOI] [PubMed] [Google Scholar]
- Omori Y, Chaya T, Yoshida S, Irie S, Tsujii T, & Furukawa T (2015). Identification of G protein-coupled receptors (GPCRs) in primary cilia and their possible involvement in body weight control. PLoS ONE, 10(6), e0128422. 10.1371/journal.pone.0128422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach R, & Howard J (2019). The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nature Communications, 10(1), 1838. 10.1038/s41467-019-09779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VR, Eckel-Mahan K, Sassone-Corsi P, & Baldi P (2012). CircadiOmics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nature Methods, 9(8), 772–773. 10.1038/nmeth.2111 [DOI] [PubMed] [Google Scholar]
- Pedersen LB, & Rosenbaum JL (2008). Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Current Topics in Developmental Biology, 85, 23–61. 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Setou M, Rohatgi R, Reiter JF, Ikegami K, & Inoue T (2017). Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell, 168(1–2), 264–279.e215. 10.1016/j.cell.2016.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EL, & Fuller CA (2000). Gravity and thermoregulation: Metabolic changes and circadian rhythms. Pflügers Archiv, 441(Suppl. 2–3), R32–R38. 10.1007/s004240000329 [DOI] [PubMed] [Google Scholar]
- Satir P, & Christensen ST (2007). Overview of structure and function of mammalian cilia. Annual Review of Physiology, 69, 377–400. 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- Schou KB, Pedersen LB, & Christensen ST (2015). Ins and outs of GPCR signaling in primary cilia. EMBO Reports, 16(9), 1099–1113. 10.15252/embr.201540530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Xie Y, He J, Zhou J, Gao Y, Wei W, Ding N, Ma H, Xian CJ, Chen K, & Wang J (2017). Microgravity induces inhibition of osteoblastic differentiation and mineralization through abrogating primary cilia. Scientific Reports, 7(1), 1866. 10.1038/s41598-017-02049-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siljee JE, Wang YI, Bernard AA, Ersoy BA, Zhang S, Marley A, Von Zastrow M, Reiter JF, & Vaisse C (2018). Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nature Genetics, 50(2), 180–185. 10.1038/s41588-017-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman MA, & Leroux MR (2009). Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends in Cell Biology, 19(7), 306–316. 10.1016/j.tcb.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Talakoub O, Neagu B, Udupa K, Tsang E, Chen R, Popovic MR, & Wong W (2016). Time-course of coherence in the human basal ganglia during voluntary movements. Scientific Reports, 6, 34930. 10.1038/srep34930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, & Lorentzen E (2016). The intraflagellar transport machinery. Cold Spring Harbor Perspectives in Biology, 8(10), a028092. 10.1101/cshperspect.a028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, & Durand B (2010). Transcriptional control of genes involved in ciliogenesis: A first step in making cilia. Biology of the Cell, 102(9), 499–513. 10.1042/BC20100035 [DOI] [PubMed] [Google Scholar]
- van Dam TJP, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, & Huynen MA (2013). Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proceedings of the National Academy of Sciences of the United States of America, 110(17), 6943–6948. 10.1073/pnas.1221011110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela L, & Horvath TL (2018). Neuronal cilia: Another player in the melanocortin system. Trends in Molecular Medicine, 24(4), 333–334. 10.1016/j.molmed.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, Gravato-Nobre M, Hodgkin J, Hall DH, Murphy CT, & Barr MM (2015). Cell-specific transcriptional profiling of ciliated sensory neurons reveals regulators of behavior and extracellular vesicle biogenesis. Current Biology, 25(24), 3232–3238. 10.1016/j.cub.2015.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Phan T, & Storm DR (2011). The type 3 adenylyl cyclase is required for novel object learning and extinction of contextual memory: Role of cAMP signaling in primary cilia. Journal of Neuroscience, 31(15), 5557–5561. 10.1523/JNEUROSCI.6561-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN (2008). Nanobiology of the primary cilium–paradigm of a multifunctional nanomachine complex. Methods in Cell Biology, 90, 139–156. 10.1016/S0091-679X(08)00807-8 [DOI] [PubMed] [Google Scholar]
- Wiener M, Turkeltaub P, & Coslett HB (2010). The image of time: A voxel-wise meta-analysis. Neuroimage, 49(2), 1728–1740. 10.1016/j.neuroimage.2009.09.064 [DOI] [PubMed] [Google Scholar]
- Wright KP Jr., McHill AW, Birks BR, Griffin BR, Rusterholz T, & Chinoy ED (2013). Entrainment of the human circadian clock to the natural light-dark cycle. Current Biology, 23(16), 1554–1558. 10.1016/j.cub.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YK, Yeh TL, Chiu NT, Lee IH, Chen PS, Lee LC, & Jeffries KJ (2004). Association between cognitive performance and striatal dopamine binding is higher in timing and motor tasks in patients with schizophrenia. Psychiatry Research, 131(3), 209–216. 10.1016/j.pscychresns.2003.07.002 [DOI] [PubMed] [Google Scholar]
- Yoshiba S, Shiratori H, Kuo IY, Kawasumi A, Shinohara K, Nonaka S, Asai Y, Sasaki G, Belo JA, Sasaki H, Nakai J, Dworniczak B, Ehrlich BE, Pennekamp P, & Hamada H (2012). Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science, 338(6104), 226–231. 10.1126/science.1222538 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 List of cilia genes with their substructure localizations
FIGURE S1 Subcilia localization of the circadian genes in brain regions, in which cilia transcripts exhibited higher rhythmicity than the non-cilia transcripts. Subcilia localization of genes that exhibit circadian expression in (a) ARC, (b) DMH, (c) LGP, (d) MGP, (e) MMB, (f) PON, (g) (PRA), (h) PRC, (i) PUT, (j) PVN, (k) SCN, and (l) (VIC)
TABLE S2 Cilia gene transcripts that oscillate in a circadian manner in 22 baboon brain regions together with the oscillating signal’s period, lag, and amplitude
Data Availability Statement
All the circadian data sets and the deep learning model BIO_CYCLE are publically available on the CircadiOmics web portal (circadiomics.igb.uci.edu). The raw data of cilia circadian gene expressions are available in Table S2.


