Abstract
The major structural protein of the retroviral core (CA) contains a conserved sequence motif shared with the CA-like proteins of distantly related transposable elements. The function of this major region of homology (MHR) has not been defined, in part due to the baffling array of phenotypes in mutants of several viruses and the yeast TY3. This report describes new mutations in the CA protein of Rous sarcoma virus (RSV) that were designed to test whether these different phenotypes might indicate distinct functional subdomains in the MHR. A comparison of 25 substitutions at 10 positions in the RSV conserved motif argues against this possibility. Most of the replacements destroyed virus infectivity, although either of two lethal phenotypes was obtained depending on the residue introduced. At most of the positions, one or more replacements (generally the more conservative substitutions) caused a severe replication defect without having any obvious effects on virus assembly, budding, Gag-Pol and genome incorporation, or protein processing. The mutant particles exhibited a defect in endogenous viral DNA synthesis and showed increased sensitivity of the core proteins to detergent, indicating that the mutations interfere with the formation and/or activity of the virion core. The distribution of these mutations across the MHR, with no evidence of clustering, suggests that the entire region is important for a critical postbudding function. In contrast, a second class of lethal substitutions (those that destroyed virus assembly and release) consists of alterations that are expected to cause severe effects on protein structure by disruption either of the hydrophobic core of the CA carboxyl-terminal domain or of the hydrogen bond network that stabilizes the domain. We suggest that this duality of phenotypes is consistent with a role for the MHR in the maturation process that links the two parts of the life cycle.
CA is the major structural component of retroviruses, forming a protein shell that encases the ribonucleoprotein complex in the virion core. The ribonucleoprotein complex is in turn composed of viral RNA (vRNA), tRNA, and the associated nucleocapsid (NC), reverse transcriptase (RT), and integrase (IN) proteins in an enzymatic complex that is capable of vDNA synthesis (4, 14, 41). Structural and sequence similarities have been noted among CA proteins of retroviruses and CA-like proteins of other reverse-transcribing elements (20, 21, 25, 27, 34, 47). The most highly conserved region of CA is a motif of about 20 amino acids that lies within the carboxy-terminal domain of the protein (termed the major homology region [MHR]). The MHR has been identified in all retroviruses with the exception of spumaretroviruses (12). Related sequences are present in hepatitis B virus (47), the retrotransposon Ty3 of Saccharomyces cerevisiae (32, 36), and certain non-long terminal repeat (LTR) transposable elements (33), suggesting that the motif is very probably involved in an aspect of replication common to all these reverse-transcribing elements. Nevertheless, the functions in the viral replication cycle of the MHR domain and of CA itself remain unknown.
The CA protein influences both viral assembly and postassembly replication activities. CA exists first as a domain of the Gag precursor, the protein that directs virus assembly and budding, and is released as a mature structural protein by the action of the viral protease during subsequent maturation. In Rous sarcoma virus (RSV), the CA sequence within Gag is not essential for the budding process, since none of the numerous deletions that span the CA region interfere with the formation of budding-competent particles in a Gag expression system (13, 29). However, CA clearly determines the internal organization of the assembled and budded material, as indicated by the effects of deletions on particle size (29) and core integrity (11, 12; N. K. Krishna, T. M. Cairns, and R. C. Craven, unpublished data).
The strongest case for the involvement of CA in postentry replication events comes from experiments with murine leukemia virus (MuLV). Certain mutations in the CA domain of MuLV Gag cripple the ability of the virus to synthesize vDNA upon entry (1). The CA protein is the target of a restriction event mediated by the Fv-1 gene product in the target cell that results in a block to replication prior to viral integration (3, 5, 15, 26, 38). Furthermore, MuLV CA has been found in association with core-derived complexes that are in the process of DNA synthesis in the cytosol of the newly infected cell (18) and with complexes containing completed and integration-competent vDNA molecules (6). A number of mutations in HIV-1 CA also interfere with DNA synthesis in the cell (8).
In hopes of better defining the function of the CA protein, the MHR has been targeted by mutagenesis in several retroviruses and the yeast Ty3 element. The result has been a perplexing array of phenotypes. In Ty3, human immunodeficiency virus type 1 (HIV-1), RSV, MuLV, Mason-Pfizer monkey virus (M-PMV), and bovine leukemia virus, many substitutions completely abolish particle assembly or cause the release of particles of aberrant morphology (1, 12, 31, 36, 40, 44). Also, certain mutations that alter or delete the MHR sequence of HIV-1 Gag have been reported to cause defects in Gag-membrane binding (17) and in Gag-Pol packaging into particles (24, 39). A number of additional MHR mutations have been described in RSV, MuLV, HIV-1, M-PMV, and bovine leukemia virus that have no discernible defect in assembly yet cause a severe loss of infectivity (1, 12, 31, 40, 44). Likewise, certain Ty3 mutations also result in the formation of particles that have severe defects in reverse transcription and transposition (36). The existence of this latter class of mutants (those with apparently normal assembly) argues strongly that the MHR motif influences one or more events of replication that are distinct from the assembly of budding-competent particles.
The principal goal of the work presented below was to define the reason for the failure of such RSV mutants to establish persistent infections. The mutations present in the five assembly-competent lethal MHR mutants previously described (12) all mapped to the second half of the motif, raising the possibility that the two structural elements within the MHR (a strand-loop element followed by an α-helix) recently defined in the CA proteins of RSV, HIV, equine infectious anemia virus, and human T-cell leukemia virus (9, 20, 25, 27) might possess distinguishable functions. The study of new mutations presented here supports the contrary conclusion. Mutations whose lethal phenotype cannot be explained by effects on particle assembly and release map to numerous locations throughout the MHR. In all cases, the lethality is due to a failure to accomplish a very early stage of reverse transcription, suggesting that the mutations interfere with either the formation or the activity of the reverse transcription complex in the viral core.
MATERIALS AND METHODS
Plasmid DNAs and cell lines.
The infectious RSV genome, carried in plasmid pRC.V8, was derived from pBH.RCAN.HiSV (16) by replacement of the Schmidt-Ruppin A gag gene with that of the Prague C strain (12). The viral mutants F167Y, L171I, and R170Q were previously described (12). New substitutions (see Fig. 1) were created by oligonucleotide-directed mutagenesis as described previously (45) with the following primers (nucleotide changes are underlined): D155N (GGCGCACAT), D155Y (GGCGAACAT), I156V (GGACGTGATGC), Q158E (CATGGAAGGAC), Q158N (CATGAACGGAC), E162D (CTGATTCCT), E162Q (ATCTCAATCCT), F164L (GTCCCTTGT), F164V (GTCCGTTGT), F164Y (TCCTACGTTG), and L171A (TCGGGCTATA). Cell lines stably expressing the F167Y, L171A, L171I and R170Q mutant genomes were created by calcium phosphate transfection of QT6 cells (35) with mutant proviral DNAs and selection of hygromycin-resistant colonies. Colonies were screened for virus production by Western blotting for RSV antigens. Released particles were analyzed for normal patterns of Gag cleavage products, envelope glycoproteins, and RT activity on an exogenous template. The phenotypes of the released particles remained stable over at least 2 to 3 months of culturing of the cell lines.
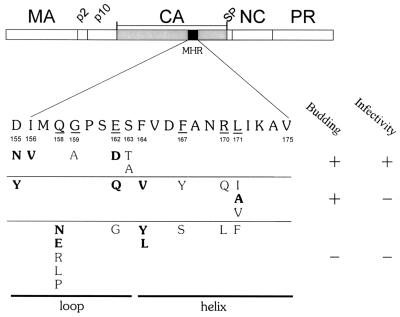
FIG. 1.
Amino acid substitutions in the major homology region of the RSV Gag protein. The Gag protein is shown with CA shaded and the MHR in black. The wild-type MHR sequence is positioned below Gag, with the most highly conserved amino acids underlined and numbered according to their position in CA. Amino acid substitutions listed below the MHR are grouped by phenotype. Substitutions resulting in >50% budding efficiency were scored as budding positive. New substitutions created in this study are in bold type; the others were previously described (12).
Particle release and infectivity.
The expression of Gag protein from the mutant proviral vectors and the ability of the mutant proteins to assemble and release virus-like particles were evaluated by transfection of QT6 cells followed by radiolabeling and immunoprecipitation as described previously by our laboratory (2, 12, 29, 45). To determine the efficiency of release, QT6 cells were transfected in duplicate and 24 h later were radiolabeled with l-[35S]methionine (50 μCi, 1,000 Ci/mmol) using two different methods. The cells of one plate were lysed after only 5 min to evaluate the levels of Gag expression; from the second plate, extracellular particles were collected after 3 h of labeling. The Gag and CA proteins were subsequently immunoprecipitated with an anti-RSV serum and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and phosphorimaging. A ratio of the CA released into the medium to the Gag expression level in the lysates was obtained as an estimate of budding efficiency.
Infections of QT6 cells were initiated by transfection of duplicate plates with proviral DNAs (12). One set of plates was used to confirm Gag expression by radioimmunoprecipitation with an anti-RSV serum 24 h later. The second set was passaged over a period of 30 days, at which point the mutants that continued to produce extracellular particles (detected by the accumulation of RT activity and Gag antigens in the medium) were scored as infectious.
Detection of viral DNA in cell lysates.
QT6 cells were infected with particle suspensions produced from stable cell lines, concentrated 50-fold using a Centriprep-50 spin column (Amicon), and standardized by an exogenous RT assay (10). After 18 h, low-molecular-weight viral DNA was isolated from the cells (23) and used as a template in the Taq PCR with primers derived from the RSV LTR and the 5′ and 3′ untranslated regions: primer 3 (CTTCATGCAGGTGCTCGTAGTCG) and primer 5 (GCCATTTTACCATTCACCACA) for minus-strand strong stop DNA; primer 3 and primer 8 (GGATTGGACGAACCACTGAA) for first strand switch; primer 7 (CAACGACTCTCTGAGTTCTC) and primer 5 for second-strand switch; and primer 8 and primer 9 (CAGGAGTATTGCATAAGACTAC) for 2LTR circles. Additional primers (TTTTAACCTAACTCCCCTACTTA and GCCTGAAGCTAGTCACGGAAT) were used to amplify quail mitochondrial DNA. Each primer pair was tested with plasmid DNA over a range of annealing temperatures, MgCl2 concentrations, and DNA concentrations to ensure high sensitivity (≤10 copies of DNA) and responsiveness of the PCR signal to template dilution. One primer of each pair was end labeled with [γ-33P]ATP using T4 polynucleotide kinase and included with template DNA in a 100-μl reaction mixture containing 2.5 U of Taq polymerase, 200 μM each deoxynucleoside triphosphate, and 15 mM MgCl2. Standard cycle parameters and product analysis were described previously (37).
Endogenous reverse transcription.
Particles produced from cell lines were prepared as described above. Particles produced by transfection were pelleted through 25% sucrose (400 μl) in a TLA100.4 rotor at 126,000 × g for 40 min, resuspended in 200 μl of Tris-buffered saline containing 10 mM MgCl2, and incubated with 600 U of DNase I for 1 h at 37°C. Destruction of the plasmid DNA was confirmed by PCR amplification of the ampicillinase gene in the vector. Virions were incubated in an endogenous RT reaction buffer (37) with 125 μg of melittin per ml for 3 h at 42°C, and then viral DNA was extracted and analyzed by PCR as above.
Detergent treatment of viral particles.
Resistance of the CA protein to extraction with 1% Triton X-100 was evaluated as previously described (12), using a rabbit anti-CA serum to detect radiolabeled protein in supernatant and pellet fractions. The RT protein was analyzed similarly, except that ≥4 × 106 transfected QT6 cells were labeled with l-[35S]methionine (250 μCi, >1,000 Ci/mmol) for 24 h, after which the particles in the medium were treated with detergent and centrifuged through a 25% sucrose cushion. Labeled RTα, RTβ, and IN proteins in the pellet and supernatant fractions were analyzed by immunoprecipitation with a goat anti-RT (α/β) antibody (no. 765-168; National Institutes of Health).
RESULTS
In our previous study of RSV, all point mutations that allowed the release of noninfectious particles mapped within second half of the MHR (specifically, F167Y, L171I, L171V, and R170Q) whereas the mutations that blocked assembly were scattered throughout the MHR (Fig. 1) (12). This uneven distribution may simply reflect the limited number of mutations tested or, alternatively, could be indicative of different functional roles for the two halves of the domain. Therefore, before focusing on the reasons for the replication defect in the assembly-competent lethal mutants, we created several additional mutations targeted primarily at the conserved sites in the first half of the RSV MHR domain.
Substitutions in the MHR strand-loop subdomain.
A total of 11 new substitutions were created (Fig. 1), including substitutions at three conserved positions in the strand-loop (I156, Q158, and E162) and at the nonconserved D155, which lies at the boundary of the conserved motif and the upstream interdomain linker (9). Additional substitutions were also constructed at the first amino acid (F164) of the helical subdomain. The L171A mutation was created for a separate study but is included in many of the experiments presented below.
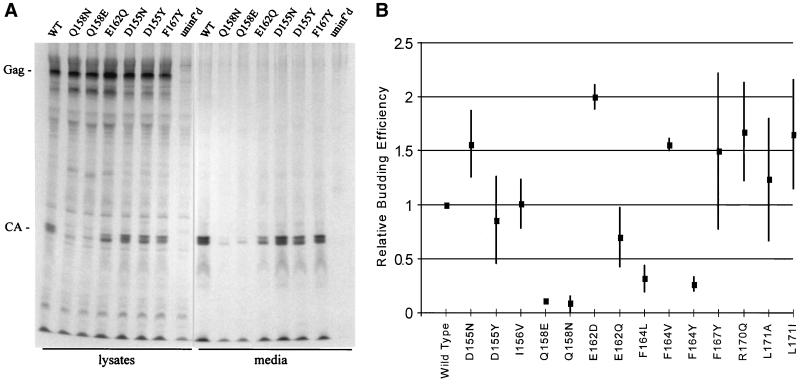
All of the new mutant genomes were capable of expressing Gag protein, detected by radioimmunoprecipitation with an anti-RSV antibody as described in previous studies (2, 12, 29, 45). The protein profiles of a representative set of mutants are shown in Fig. 2A. In a quantitative experiment (Fig. 2B), the particle release ability of each mutant (a ratio of particle release to Gag expression as described in Materials and Methods) was compared with that of the wild-type parent. The mutants could be separated easily into two groups. The D155N, I156V, E162D, and L171A mutants released particles at levels equal to or greater than those of the wild-type parent, and the D155Y mutant averaged 86% of the wild-type level. The E162Q mutant was slightly less efficient, showing release levels ranging from 50 to 100% with an average of 70% (Fig. 2). In each case, the particles that were released contained properly processed CA protein (Fig. 2A); no uncleaved Gag precursor or unusual cleavage intermediates were present in the particles. In contrast, the remaining mutants showed clear defects in particle release, with budding efficiencies of <32% of the wild-type level (Q158N, 9%; Q158E, 11%; F164Y, 27%; and F164L, 32%) (Fig. 2B).
FIG. 2.
Evaluation of mutant particle release in QT6 cells. (A) Virions released into the media were visualized by radioimmunoprecipitation. (B) Budding efficiencies were calculated as a ratio of the amount of Gag produced to the CA released into the medium, as discussed in Materials and Methods. All numbers were normalized to the wild-type value (WT), which was set at 1.00 efficiency. Average budding efficiencies (center box) and standard deviations (lines) are shown. uninf'd, uninfected.
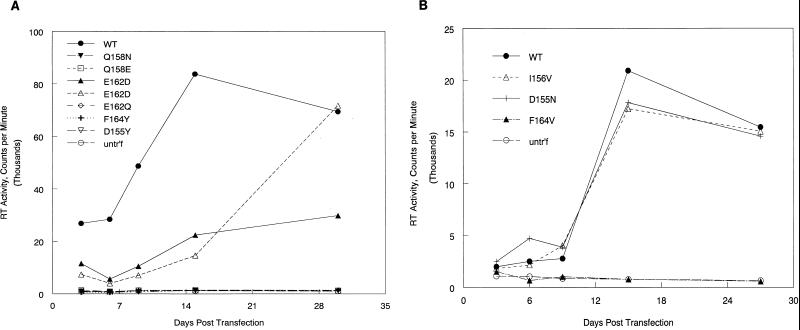
The potential for infectivity was analyzed for each of the new mutants by monitoring the appearance of RT activity in the medium after transfection of QT6 cells with proviral plasmid DNA. Not surprisingly, all four of the budding-defective mutants were completely noninfectious in this assay, as illustrated by Q158E, Q158N, and F164Y (Fig. 3A). Two of the particle-producing mutants (D155N and I56V) were indistinguishable from the wild-type parent in their ability to establish infection following transfection (Fig. 3B). A third mutant (E162D) was invariably successful in establishing infection (Fig. 3A); however, the appearance of RT in the medium during the 4-week assay period was slower than seen with the wild-type parent or the other two infectious mutants. Whether genetic reversion or second-site suppressor mutations occurred during this lag time has not yet been determined but seems likely, given that the glutamic acid at position 162 is absolutely conserved in all 20 retroviruses compared in an earlier study (12). Interestingly, three more recently described fish retroviruses lack this glutamic acid (30).
FIG. 3.
Establishment of infection followed by monitoring RT activity in the media of transfected QT6 cells. (A and B) Medium samples were collected at intervals over a span of 1 month following transfection. Viruses with RT activity at background (i.e., equal to the untransfected) levels after this period were scored as noninfectious. untr'f, untransfected.
Most important for the remainder of this study is a group of four mutants (D155Y, E162Q, F164V, and L171A) which failed to infect (Fig. 3), even though their ability to assemble and release virus particles was near normal. These new mutants join four mutants with a similar phenotype (F167Y, R170Q, L171I, and L171V), which were described in our earlier study (12). Quite striking is the fact that the combined members of this mutant class map across the MHR sequence with no obvious clustering (Fig. 1). The substitutions that cause the assembly-defective phenotype are similarly distributed. Only at one position (Q158) did every substitution destroy assembly. This greater sensitivity may be indicative of the unusual role of the conserved glutamine in the CA structure (see Discussion). Other than this, there is no evidence that the two structural elements that compose the MHR (the strand-loop and α-helical elements) are functionally distinct.
Detection of viral DNA in infected cells.
Having obtained a sizeable collection of mutations that destroyed infectivity but not particle assembly, we investigated the reasons for this replication defect. The L171I mutant, which has been the most carefully characterized RSV mutant, served as the focus of the experiments below. Additional mutants with substitutions in the loop (D155Y and E162Q) and in the α-helical subdomain (F167Y, L171A, and R170Q) were included in most experiments as well.
Initially, low-molecular-weight DNA extracted from QT6 cells infected with the mutant viruses for 18 h was analyzed for full-length viral DNA by Southern blotting and by PCR amplification of the 2LTR circle junction DNA (formed in the cell as a result of circularization of full-length linear viral DNA). In both tests, no completed viral DNA molecules could be detected in samples from cells infected with the L171A, L171I, and F167Y mutants (data not shown). The R170Q, D155Y, and E162Q mutants also failed to produce detectable 2LTR DNA in infected cells (data not shown).
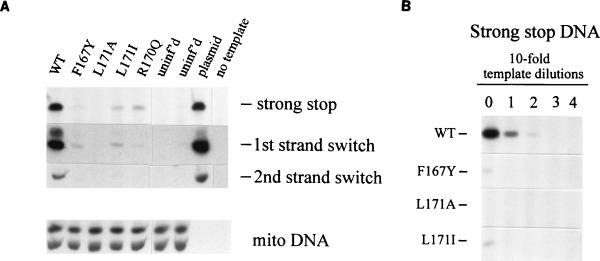
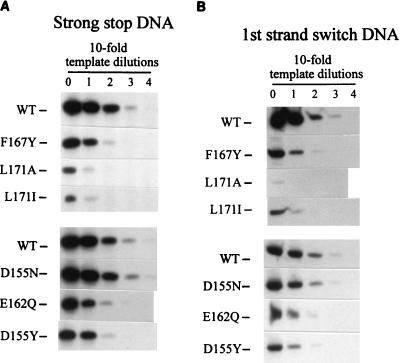
The DNA samples were next analyzed for the presence of incomplete DNA products by using PCR primer pairs that amplify viral DNAs containing the minus-strand strong-stop sequence (the first 101 bases) and molecules whose synthesis proceeded at least through the point of the first strand switch (169 bases) or the second strand switch (586 bases). All three viral DNA sequences were easily detected in samples from cells infected with wild-type virus (Fig. 4A), whereas little or no viral DNA could be detected in samples from mutant-infected cells. Quail mitochondrial DNA was present in all samples, however, confirming the consistent performance of the DNA extraction and PCR steps. To estimate the degree of the defect, template DNAs were serially diluted prior to amplification (Fig. 4B). The level of strong-stop DNA in L171A-infected cells proved to be reduced >1,000-fold compared to that in wild-type-infected cells. The F167Y and L171I mutations caused a reduction of 100-fold. Finally, detection of viral DNA was no better when the infected cells were lysed and DNA was extracted at earlier times after infection (2 to 8 h postinfection [data not shown]), giving no indication that the viral DNA was synthesized and then degraded. Thus, it appears that even at the earliest stages of infection tested (synthesis of the first 101 nucleotides of DNA), these mutants are severely deficient in viral DNA synthesis.
FIG. 4.
Viral DNAs in infected QT6 cells. Cells were infected with RSV particles collected from stable cell lines. Low-molecular-weight DNA was extracted and analyzed by amplification using the LTR primers described in Materials and Methods and with the quail mitochondrial DNA primers. (A) Viral DNA detected in lysates of cells infected for 18 h. (B) Template DNAs analyzed in panel A were diluted prior to amplification in order to compare the relative amounts of minus-strand strong-stop DNA in cells infected with wild-type (WT) and MHR mutant virus. uninf'd, uninfected; mito, quail mitochondrial DNA.
Endogenous reverse transcription.
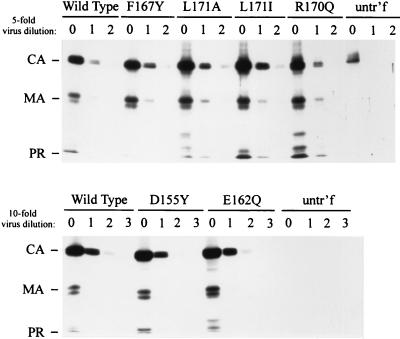
The next step was to determine whether the defect that caused the severe loss of DNA synthesis in infected cells was also evident in extracellular particles. Although our earlier report showed the presence of some endogenous reverse transcription ability in mutant particles isolated from cell lines (12), this was reevaluated by applying the more quantitative PCR-based assay to the broader set of mutants now available. Minus-strand strong-stop DNA was clearly detectable in all samples (Fig. 5A), as previously reported (12). However, dilution of the template DNA showed that the level of viral DNA in the mutant particles was actually lower than that in wild-type particles by a factor ranging from 10-fold (D155Y, E162Q, and F167Y) to as much as 100-fold (L171A and L171I). The infectious mutant D155N was indistinguishable from the wild-type parent in the endogenous reverse transcription assay.
FIG. 5.
Efficiency of reverse transcription of the endogenous viral RNA. Particles were collected from stable cell lines (wild type [WT], F167Y, L171A, and L171I) or from transfected cells (WT, D155N, D155Y, and E162Q), permeabilized with melittin, and incubated with dNTPs for three hours at 42°C. The resulting DNA was extracted, diluted, and amplified as before. The relative amounts of minus-strand strong stop (A) or first strand switch (B) DNA products in wild-type and MHR mutant virions were compared. F167Y particles collected from transfected cells showed the same 10-fold deficit in endogenous reverse transcription activity as those collected from the stable cell line (data not shown).
The failure of the previous study (12) to find this defect in the F167Y, R170Q, and L171I mutant particles was probably due to the use of a crude and nonquantitative assay method that was dependent on observation of the strong-stop DNA band among a smear of DNA products on a sequencing gel. It is also likely that the batch-selected cultures used to produce the R170Q and F167Y mutant particles may have contained revertant viruses (J. B. Bowzard, unpublished data). In the present study, the use of the more quantitative and highly reproducible PCR-based method allowed the detection of a DNA synthesis defect in each of the assembly-competent but noninfectious mutants, regardless of whether the particles were produced by cloned cell lines (F167Y, L171I, and L171A) or by transfection (F167Y, E162Q, and D155Y).
The presence of more extended DNA products was also evaluated in the endogenous reverse transcription reactions. The noninfectious mutants were deficient in the synthesis of the first strand-switch product, but with the exception of the L171A mutant, the defect appeared no more severe than at the strong-stop stage (Fig. 5B); in other words, the response to template dilution was identical with the two sets of primers. Finally, the sensitivity of detection of the second strand-switch product was too low even in the wild-type samples to allow a quantitative comparison with the strong-stop and first strand- switch products.
RT content.
The major defect at an early stage of the endogenous reverse transcriptase reaction implies that something is very wrong with the protein-RNA complex that makes up the reverse transcription machinery. For this reason, the mutant viruses were examined for abnormalities that might explain the defect in viral DNA synthesis.
The mutant particles were not deficient in enzymatically active RT. The CA protein in mutant and wild-type particles was evaluated by Western blotting of particle suspensions that had been standardized for their RT content by using an exogenous poly(A) template and an oligo(dT) primer (Fig. 6). No variation in the enzyme activity-to-CA ratio that could explain the DNA synthesis defect was found in any of the mutants. Furthermore, L171I mutant particles contain normal ratios of RTα, RTβ, and IN protein (see below), as do the F167Y, R170Q, and L171A mutants (data not shown). Thus, the failure of mutants to synthesize DNA is not explained by defects in the packaging or processing of the Gag-Pol precursor protein.
FIG. 6.
The ratio of CA to RT per particle is equivalent between wild-type and mutant viruses. Particles were normalized by RT activity on an exogenous template/primer, serially diluted, and separated by SDS-PAGE. Viral proteins were transferred to nitrocellulose by Western blotting and incubated with anti-RSV serum. The relative amount of CA protein per sample was examined. untr'f, untransfected.
Viral RNA content.
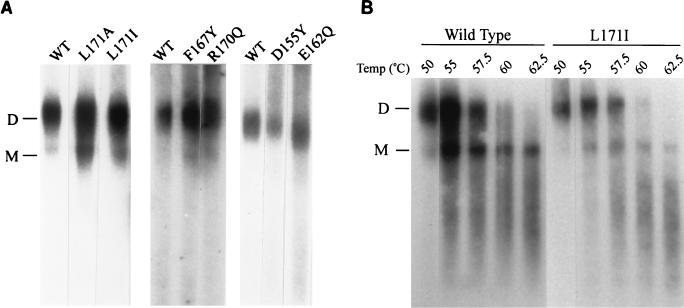
Although the F167Y and L171I particles were shown previously by slot blot analysis to contain viral RNA (12), the failure of DNA synthesis raised the possibility that viral RNA might be degraded inside the particle. Therefore, the viral RNA was extracted and analyzed by standard methods (19, 37) (Fig. 7). Genomic RNA from the D155Y, E162Q, F167Y, L171A, L171I, and R170Q mutants proved to be intact and was present primarily in the form of dimers (Fig. 7A), as was that from the mutant F164V (data not shown). No degradation of the viral RNA template and no deficit in the amount recovered were detected in any case. The thermostability of the dimeric RNAs of the L171I mutant was tested by exposing the nucleic acid to temperatures as high as 65°C prior to electrophoresis. The mutant RNA showed the same melting profile as that from wild-type particles (Fig. 7B); the same result was obtained with F167Y and L171A particles as well (data not shown). Thus, we detected no gross distortion of the viral genomic RNA that could explain the defect in reverse transcription.
FIG. 7.
Analysis of viral RNA in wild-type and mutant particles. (A) Nondenaturing Northern blot analysis of RNA extracted from particles. (B) RNA melting profiles were generated by incubating the viral RNAs at the indicated temperatures for 10 min prior to loading on a nondenaturing agarose gel. WT, wild type; D, dimer; M, monomer.
Association of CA and RT with detergent-resistant core material.
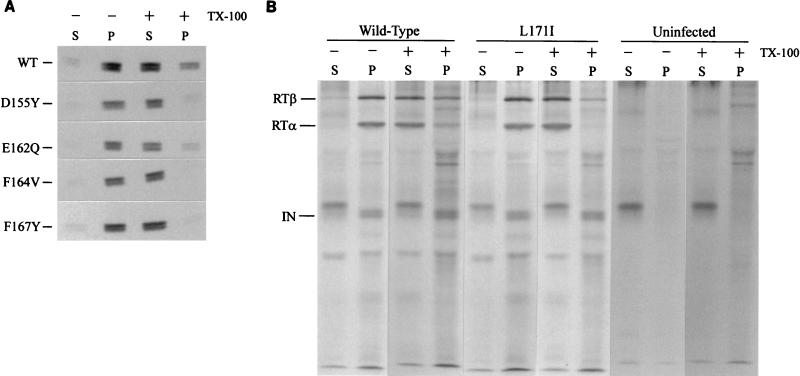
A number of laboratories have isolated material derived from the RSV core, including CA, by centrifugation of detergent-treated virions through sucrose (4, 12, 41). In our laboratory, such a method has allowed the reproducible recovery of approximately 25 to 30% of the total CA protein in particulate form from infectious wild-type virions (12) (Fig. 8A). However, with the F164V or F167Y mutants, the amount of CA protein recovered in the detergent-resistant pellet was greatly reduced, ranging between 0 and 3% of the total amount of CA (Fig. 8A). This was similar to our previous findings with the L171I mutant (12). For the D155Y and E162Q mutants, a somewhat less severe defect was seen; approximately 10% of CA was recovered in the pellet, still reproducibly lower than the 30% recovered from the wild-type particles.
FIG. 8.
CA and RT are displaced from detergent-resistant complexes by mutations in the MHR. Particles produced by transfection were incubated with or without detergent prior to ultracentrifugation through a sucrose cushion. Pellet and supernatant fractions were collected and immunoprecipitated with a monoclonal antibody against CA (A) or RT (B). A background band recovered by the RT immunoprecipitation migrated slightly above the IN protein and appeared in both infected and uninfected controls. WT, wild type; TX-100, Triton X-100; P, pellet; S, supernatant.
The distribution of RT between the particulate and pellet fractions was also altered by the L171I mutation. For wild-type particles, approximately 30% of RTα and 40% of RTβ remained with the pellet after detergent treatment (Fig. 8B). In contrast, detergent treatment of the L171I mutant particles solubilized all the detectable RTα and left only a trace amount of RTβ (4 to 6%, as quantitated by phosphorimaging analysis of the results of two independent experiments) associated with the pellet. The IN protein behaved differently in that the protein from the wild-type virus was entirely associated with the particulate fraction after detergent treatment and this distribution was not altered by the L171I mutation. Although the exact structure of the detergent-resistant material isolated from both infectious and noninfectious virions remains to be determined, these results imply that the intermolecular interactions that hold CA and RT in place in the core have been altered in some way by the mutations. This finding is consistent with the DNA synthesis defect identified in both infected cells and cell-free virions.
DISCUSSION
With the addition of new mutations in this study, we present the most comprehensive collection of such mutations, consisting of a total of 25 substitutions placed at 10 positions (Fig. 1). A few substitutions in this sequence, primarily alterations at the less highly conserved positions in the motif, caused no demonstrable effects on virus infectivity; these were not studied further. Two classes of lethal mutations could be distinguished—those that caused a severe assembly and release defect and others that allowed near-normal particle release. Strikingly, the distributions of these two phenotypes across this region of CA are almost identical. At five of seven positions, replacement with two or more different amino acids yielded both types of noninfectious mutants. Only at Q158 did all five replacements destroy assembly (see section below). From this analysis, there is no evidence for functionally distinct subregions within this region of CA; rather, the entire domain probably functions as a whole. This conclusion is quite significant, given the wide variety of phenotypes that have been seen in HIV-1 and M-PMV MHR mutants (31, 40).
Implications of the budding-defective phenotype.
The block to release from avian cells in certain MHR mutants seems contradictory to the published findings that the CA region of Gag is not part of the minimal protein segments needed to drive budding, a conclusion drawn from Gag expression studies in mammalian cells (12, 29). However, the pattern of intracellular Gag proteins observed in avian cells with these mutants, i.e., minimal Gag cleavage and very little CA protein in cell lysates (Fig. 2A) (12), resembles that seen with Gag mutants that are deficient in Gag-membrane binding (a function of Gag MA) or Gag-Gag interaction (the NC region) (2, 13, 42). The budding defect in the MHR mutants could conceivably result from interference with either activity via long-range structural effects. However, our experience with the modularity of these functions and the plasticity of the RSV Gag to mutation (2, 13, 29, 43, 45) make this seem unlikely. An alternate scenario is that a localized structural perturbation within the CA carboxy-terminal domain (CTD) of Gag leads to inappropriate aggregation of the precursors in the avian cell. Such a dead-end pathway could also result from inappropriate interactions of Gag with a cellular factor(s). This could explain the cell type dependence of certain RSV MHR mutants, i.e., Gag proteins that allow budding from mammalian cells but not avian cells (12).
Of the mutations that interfered with assembly, R170L and E162G and all five replacements at the Q158 position alter the conserved hydrophilic residues in the MHR. The homologous residues in the HIV CA protein form an unusual network of hydrogen and ionic bonds that links the two halves of the MHR to one another and to the α-helix immediately following the MHR. Recently obtained nuclear magnetic resonance spectroscopy data on the RSV CA CTD are consistent with the presence of a similar network (28). The central role of the glutamine in this network (20) suggests that it is especially important for the folding of the CTD, and this may explain why all substitutions at Q158 destroyed RSV assembly. The R170L and E162G substitutions that also interfere with assembly (12) should likewise have dramatic effects on this bonding network. The more conservative substitutions (R170Q and E162D), on the other hand, may be compatible with particle assembly and release because they are able to maintain at least some of the hydrogen bond network. Thus, these findings suggest that a similar bonding network forms in the Gag precursor and is important for maintaining its assembly function. The remaining budding-defective mutations are alterations that create rather substantial changes in the hydrophobic core of the CTD. For example, introduction of a polar amino acid (F167S) or of a larger, bulkier hydrophobic residue (L171F) had dramatic effects on the assembly of the Gag protein, supporting the idea that these changes are destabilizing the hydrophobic core of the CTD.
Budding-competent, noninfectious mutants.
When the lethal mutations that destroyed the budding capacity of Gag are removed from consideration, all those that remain have a common feature—a defect in reverse transcription. Many of these are the more conservative substitutions placed at the same sites where more radical changes blocked virus assembly. The L171I mutant is the most thoroughly characterized. The disability in the other MHR mutants of this class, although less severe in some cases, is qualitatively the same, as is that of the D155Y mutant, whose substitution lies at the boundary of the MHR and the interdomain linker.
The L171I particles contain properly processed Gag proteins and have physical properties (size and density) identical to those of the wild-type parent (12, 29). The Gag-Pol precursor is packaged and processed to form enzymatically active RT. The packaging and integrity of the viral genome also appear normal. What is not known is whether the placement of tRNA on the primer binding site of the viral RNA is qualitatively and quantitatively normal, but we have been able to detect the presence of the primer on the viral RNA by end labeling with [32P]dATP (data not shown). The ability of the mutant virus to bind and enter cells has not been tested, although failure of virus entry seems an unlikely explanation since Env glycoproteins have been detected on the particles (12) (data not shown) and there is no reason a priori to guess that CA mutations would interfere with this step.
Defects consistent with the in vivo results were found in extracellular mutant particles—a serious deficit in either the initiation of DNA synthesis or the elongation of the nascent viral DNA and the disruption of core integrity indicated by the ease of detergent extraction of CA and RT. The first strand-switching step does not appear to be affected by the mutations (with the possible exception of the L171A mutant); however, it is not known whether extension of the DNA molecules after this step occurs with normal efficiency. Finally, the defect in DNA synthesis in infected cells exceeds that observed in the endogenous reverse transcription reaction by a factor of 10, suggesting that the instability of the mutant cores becomes even more critical in the environment of the cytosol. It is possible that other, unidentified factors are limiting the efficiency of strong-stop DNA formation in the cell. Regardless, the reduced endogenous reverse transcription activity suggests that the phenotype of these RSV mutants may truly be different from that of Fv-1-restricted MuLV (26, 38), HIV CA mutants that fail to incorporate cyclophilin A (7), and other MuLV CA mutants (1) that fail to make viral DNA in the cell although they are able to do so in extracellular particles.
Clearly, the primary function of the MHR is not one of packaging the viral genome or the Gag-Pol precursor nor of controlling the PR-mediated processing of Gag or Gag-Pol. The viral DNA synthesis deficit and the unusual detergent sensitivity of the cores in extracellular mutant particles (in spite of apparently normal incorporation and processing of the component parts) argue instead that the defect is due to inappropriate interactions between the core components. In other words, the parts are not put together properly. It is tempting to conclude that this results from a failure of the MHR residues to perform a normal activity that is needed for putting the parts together. It must also be recognized, however, that the phenotype could be an indirect consequence of structural abnormalities in Gag or CA which interfere with the formation or activity of the viral core by some means unrelated to the normal function of the MHR. Thus, although characterization of these RSV mutants has eliminated a number of possibilities from consideration, the question of the function of the MHR residues in the wild-type virus remains open.
Structural studies of the CA proteins may yet yield some useful clues to CA function. Work by Gamble et al. has described a CA-CA interface in the CTD of HIV CA that appears to explain the dimerization activity of both the isolated domain and the intact CA protein in solution (20). This interface is expected to exist in the assembled CA shell as well. In the HIV protein, the MHR does not appear to participate directly in the CA-CA interface. However, this region is integral to the CA CTD structure and is in intimate contact with the α-helix that forms the dimer interface (20, 46). In vitro assembly studies with the HIV CA protein indicate that different conformations correlate with different modes of protein packing (22). Thus, it is quite conceivable that certain MHR substitutions in the HIV protein could perturb the domain structure and thereby alter CA-CA interactions. In the RSV mutants, a similar scenario might explain the loss of detergent resistance in the virion core as well as the lethal phenotype. However, the RSV protein, unlike the one from HIV, fails to dimerize in solution (9, 28); therefore, the possible consequences of the lethal MHR substitutions on CA multimerization cannot be evaluated as yet.
The possibility that the MHR is critical to the structure of the Gag precursor and the mature CA protein does not appear sufficient to explain its unusually strong conservation. It may be significant that the MHR immediately follows the flexible interdomain linker. It seems quite possible that the MHR could be a conformational switch that controls the interactions between CA subunits or heterotypic interactions that need to occur in an ordered fashion during maturation. Such a dynamic role in the progression of events that connects Gag assembly to the final formation of an active core might be consistent with the duality of mutant phenotypes observed. Finally, the DNA synthesis defect in the mutants raises the interesting possibilities that the MHR is a site of interaction with other viral or cellular factors necessary for vDNA synthesis and that CA could be a direct contributor to the DNA synthesis machinery. Although we cannot distinguish between these possibilities at this time, the field has been narrowed considerably as a result of these studies, and the contribution of CA to the DNA synthesis pathway will continue to be explored.
ACKNOWLEDGMENTS
Many thanks are due to John Wills for support, encouragement, and constructive criticisms during this work. We also thank Volker Vogt, Rich Kingston, and Michael Rossmann for sharing their structural models of the RSV CA protein prior to publication. We appreciate the critical review of the manuscript by Leslie Parent and Brad Bowzard. Brad Bowzard also assisted this project by constructing the L171A proviral vector.
This work was supported by funds from the Four Diamonds Research Fund (to R.C.C.) and by NIH grant R01 CA47482 (to J. W. Wills).
REFERENCES
- 1.Alin K, Goff S P. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 1996;222:339–351. doi: 10.1006/viro.1996.0431. [DOI] [PubMed] [Google Scholar]
- 2.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best S, Tissier P L, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 4.Bolognesi D P, Luftig R, Shaper J H. Localization of RNA tumor virus polypeptides. I. Isolation of further virus substructures. Virology. 1973;56:549–564. doi: 10.1016/0042-6822(73)90057-3. [DOI] [PubMed] [Google Scholar]
- 5.Boone L R, Myer F E, Yang D M, Ou C-Y, Koh C K, Roberson L E, Tennant R W, Yang W K. Reversal of Fv-1 host range by in vitro restriction endonuclease fragment exchange between molecular clones of N-tropic and B-tropic murine leukemia virus genomes. J Virol. 1983;48:110–119. doi: 10.1128/jvi.48.1.110-119.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 7.Braaten D, Aberham C, Franke E K, Yin L, Phares W, Luban J. Cyclosporin A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J Virol. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos-Olivas R, Newman J L, Summers M F. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 10.Craven R C, Bennett R P, Wills J W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991;65:6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven R C, Leure-duPree A E, Weldon R A, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 14.Davis N L, Rueckert R R. Properties of a ribonucleoprotein particle isolated from Nonidet P-40-treated Rous sarcoma virus. J Virol. 1972;10:1010–1020. doi: 10.1128/jvi.10.5.1010-1020.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DesGroseillers L, Jolicoeur P. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J Virol. 1983;48:685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, Dubay J W, Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 18.Fassati A, Goff S P. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sunquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 21.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 22.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 24.Huang M, Martin M A. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J Virol. 1997;71:4472–4478. doi: 10.1128/jvi.71.6.4472-4478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z, Jin L, Peterson D L, Lawson C L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 26.Jolicoeur P, Rassart E. Effect of FV-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J Virol. 1980;33:183–195. doi: 10.1128/jvi.33.1.183-195.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khorasanizadeh S, Campos-Olivas R, Summers M F. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 28.Kingston R L, Fitzon-Ostendorp T, Eisenmesser E Z, Schatz G W, Vogt V M, Post C B, Rossmann M G. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 29.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaPierre L A, Holzschu D L, Bowser P R, Casey J W. Sequence and transcriptional analysis of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: evidence for a gene duplication. J Virol. 1999;73:9393–9403. doi: 10.1128/jvi.73.11.9393-9403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer B, Bannert H, Darai G, Flugel R M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClure M A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991;8:835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- 34.McClure M A, Johnson M S, Feng D F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moscovici C, Moscovici M G, Jimenez H, Lai M M C, Hayman M J, Vogt P K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 36.Orlinsky K J, Gu J, Hoyt M, Sandmeyer S, Menees T M. Mutations in the TY3 major homology region affect multiple steps in TY3 retrotransposition. J Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parent L J, Cairns T M, Albert J A, Wilson C B, Wills J W, Craven R C. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J Virol. 2000;74:164–172. doi: 10.1128/jvi.74.1.164-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryciak P M, Varmus H E. Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J Virol. 1992;66:5959–5966. doi: 10.1128/jvi.66.10.5959-5966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasakumar N, Hammarskjold M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strambio de Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1974;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willems L, Kerkhofs P, Attenelle L, Burny A, Portetelle D, Kettmann R. The major homology region of bovine leukemia virus p24gag is required for virus infectivity in vivo. J Gen Virol. 1997;78:637–640. doi: 10.1099/0022-1317-78-3-637. [DOI] [PubMed] [Google Scholar]
- 45.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worthylake D K, Wang H, Soo S, Sundquist W I, Hill C P. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr Ser D. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 47.Zlotnick A, Stahl S J, Wingfield P T, Conway J F, Cheng N, Steven A C. Shared motifs of the capsid proteins of hepadnaviruses and retroviruses suggest a common evolutionary origin. FEBS Lett. 1998;431:301–304. doi: 10.1016/s0014-5793(98)00755-8. [DOI] [PubMed] [Google Scholar]