Abstract
We developed a recombinant virus technique to determine the coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from plasma samples, the source expected to represent the most actively replicating virus population in infected subjects. This method is not subject to selective bias associated with virus isolation in culture, a step required for conventional tropism determination procedures. The addition of a simple subcloning step allowed semiquantitative evaluation of virus populations with a different coreceptor (CCR5 or CXCR4) usage specificity present in each plasma sample. This procedure detected mixtures of CCR5- and CXCR4-exclusive virus populations as well as dualtropic viral variants, in variable proportions. Sequence analysis of dualtropic clones indicated that changes in the V3 loop are necessary for the use of CXCR4 as a coreceptor, but the overall context of the V1-V3 region is important to preserve the capacity to use CCR5. This convenient technique can greatly assist the study of virus evolution and compartmentalization in infected individuals.
Human immunodeficiency virus type 1 (HIV-1) entry into target cells relies on a complex interaction between viral and cellular proteins, eventually leading to viral and plasma membrane lipid mixing. The binding of the viral gp120 envelope glycoprotein to the cellular CD4 molecule induces a conformational change in the gp120 protein, contributing to the exposure of the binding site for a coreceptor (reviewed in reference 77). Several chemokine receptors can serve as coreceptors for HIV-1 entry (1, 11, 20–23, 28, 46, 58, 60) (see references 3, 5, 12, 13, and 50 for reviews), but the relevant coreceptors used in vivo seem limited to CCR5 and CXCR4 (78, 79) and viral isolates that use alternative chemokine receptors also use either CCR5 or CXCR4. The initial phases of HIV-1 infection in vivo involve viral strains characterized by the ability to grow both in stimulated peripheral blood mononuclear cells (PBMCs) and in macrophages, but not in established T-cell lines (63, 64, 80). These strains are now known to use CCR5 as a coreceptor and are hence named R5 viruses (4). Later on in the course of disease progression, some of the HIV-1-infected population harbor virus strains capable of productive infection of established T-cell lines as well as stimulated PBMCs (17, 64). These viral strains grow more rapidly than R5 viruses (16, 71) and use CXCR4 as a coreceptor; therefore, they are called X4 strains (4). A fraction of the CXCR4-using viruses can also use CCR5 and are called dualtropic or R5X4 viruses (15, 22, 68).
R5 strains are most commonly transmitted in vivo (29, 64, 72, 75, 80), suggesting that a poorly understood selective force acts against X4 strains at the level of virus transmission independently of the route of transmission. Examples of X4 virus suppression after blood-borne infection also supports this hypothesis (18, 44). Accordingly, despite the fact that few (two or three) mutations in the third variable loop (V3 loop) of the envelope gene can confer the ability to use CXCR4 (14, 30, 37), X4 viruses appear only late after infection, indicating that these strains are selected against by a competent immune system (9, 53). On the other hand, the association of the emergence of X4 viruses with the marked decline in CD4 T-cell counts (39, 41, 59), together with the observation of increased cytopathogenicity of these viruses in experimental systems (31, 38, 54, 57), may support the alternative hypothesis that disease progression could be the consequence, rather than the cause, of the development of X4 strains. The pathogenic potential of R5 viruses, however, is proven, given that most individuals who die from AIDS-related complications appear to harbor only R5 viruses (59).
Current understanding of the precise kinetics of the appearance of X4 and dualtropic viruses is hampered by technical difficulties and by the selective bias that is associated with virus isolation, a step required for the determination of coreceptor usage. Virus isolation in culture (47) typically relies on cultivating patient PBMCs in the presence of interleukin-2 after stimulation by phytohemagglutinin or anti-CD3 antibodies. Fresh donor PBMCs are added to provide new target cells for the production of a virus culture with a sufficient titer for testing on indicator cells. Extensive virus culturing may lead to the selection of viruses adapted to culturing conditions that misrepresent the original virus source. In vitro culturing of PBMCs may alter the level of chemokine receptor expression on the surface of the cells (6, 8), generally favoring the expression of CXCR4 with respect to CCR5. In addition, virus isolation from patient PBMCs may lead to the production of virus that does not necessarily reflect the actively replicating population, because activation of infected PBMCs can induce the expression of archived integrated proviral genomes.
We describe here a new method to determine primary HIV-1 coreceptor usage that does not require virus propagation in tissue culture. This method allows the study of plasma virus, which best represents the actively replicating virus population in the patient (25, 55, 73). We compared and validated our method by analyzing sequential series of plasma samples and viral isolates from pediatric patients that had been previously characterized by conventional phenotypic procedures. We found that R5 viruses persisted in the plasma of patients throughout the observation period and were paralleled by, but not replaced by, the emergence of X4 viruses. Using a subcloning procedure, we could determine that viral populations found after the acquisition of CXCR4 usage capacity consisted of R5 virus mixed with X4-exclusive and/or authentic dualtropic viruses. Sequence analysis of the primary dualtropic viruses showed that CXCR4 usage is associated with an increase in the positive charge of the V3 loop, as previously described for X4 (exclusive) viruses. In addition, the finding that some X4 and dualtropic primary viruses share an identical V3 loop sequence suggests that domains within V1-V2 and the second conserved region (C2), located immediately upstream of V3, contain the determinants for the expanded tropism of dualtropic viruses.
MATERIALS AND METHODS
Vector construction.
The SalI-BamHI fragment from pNL4.3 (from nucleotide 5785 to 8465) containing most of the envelope gene sequence was subcloned in pBluescript II SK(+) (Stratagene) to provide a convenient vector (SK-SB) for site-directed mutagenesis. The sequence spanning the V1-V2 loops, the C2 domain, and V3 loop (nucleotides 6610 to 7250) was excised by oligonucleotide-directed mutagenesis using the Quick change mutagenesis kit (Stratagene) with the following oligonucleotides: Delta V+, 5′-CCCCACTCTGTGTTAGTTTTAAGTGCTAGCAAATTAAGAGAAC-3′, and Delta V−, 5′-GTTCTCTTAATTTGCTAGCACTTTAAACTAACACAGAGTGGGG-3′. The natural NheI site just downstream of the V3 loop was preserved and used in combination with SalI to transfer the V1-V3 deleted fragment of the envelope gene back into a variant of pNL4.3 (43XCS [49]), creating the final construct 43-ΔV. The same NheI site can be used to linearize the 43-ΔV vector between the envelope domains C1 and C3.
Virus RNA amplification.
Viral RNA was isolated from frozen patient plasma samples or (where indicated) from PBMC culture supernatants, using the Roche Amplicor kit (Roche Diagnostics). An initial reverse transcription-PCR (RT-PCR) amplification was carried out using the following primers: E00, 5′-TAGAAAGAGCAGAAGACAGTGGCAATGA-3′ (nucleotides 6196 to 6224 of pNL4.3), and ES8B, 5′-CACTTCTCCAATTGTCCCTCA-3′ (nucleotides 7638 to 7662 of pNL4.3). An aliquot of the RT-PCR product was then used in a nested PCR with the following primers: E20, 5′-GGGCCACACATGCCTGTGTACCCACAG-3′ (nucleotides 6426 to 6452 of pNL4.3), and E115, 5′-AGAAAAATTCCCCTCCACAATTAA-3′ (nucleotides 7341 to 7364 of pNL4.3). By this approach, 900-bp-long products that span the V1-V3 region deleted from the 43-ΔV vector were obtained, with approximately 150-bp extensions on each side to allow homologous recombination during transfection. PCR products were verified by agarose gel electrophoresis and were column purified (Qiagen) prior to use in transfection.
Cell culture and tropism recombinant test (TRT).
293-T cells and U373MG-CD4 cells (34) were cultivated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS) and antibiotics. U373MG-CD4 cells stably transfected with an expression vector for the chemokine receptor CCR5 or CXCR4 (43) were cultured in the presence of 10 μg of puromycine/ml and 100 μg of hygromycin-B. The three U373MG-CD4-derived cell lines (373, 373-CCR5, and 373-CXCR4) contain an HIV-1 long terminal repeat (LTR)-LacZ cassette that allows the detection of single cycle infection by a colorimetric assay based on Tat-induced expression of β-galactosidase (β-Gal) (43).
Subconfluent 293-T cells in 25-cm2 flasks were transfected with 8 μg of NheI-linearized 43-ΔV vector DNA and 1 μg of PCR-amplified DNA from patient samples, using the calcium phosphate precipitation method. Virus-containing supernatant (3 ml) was collected 36 h after transfection and clarified by centrifugation. Triplicates of 100 μl were used to infect subconfluent target cells cultured in 96-well plates in the presence and in the absence of 2 μg of DEAE-dextran/ml. Parallel cultures of 373, 373-CCR5, and 373-CXCR4 cells were used as target cells in all assays. At 24 to 36 h after infection, virus infectivity was determined in target cell cultures by measurement of β-Gal activity in cell lysates, using a colorimetric assay based on cleavage of chlorophenolred-β-d-galactopyranoside (CPRG) by β-Gal (termed here CPRG assay), adapted from a study by Eustice et al. (24). Briefly, following elimination of the supernatant, target cells were lysed in 100 μl of lysis buffer (5 mM MgCl2, 0.1% NP-40 in phosphate-buffered saline). After incubation for 5 min at room temperature, 100 μl of reaction buffer (6 mM CPRG in lysis buffer) was added to the cell lysates and incubated for between 5 min and 2 h at 37°C. Optical densities in the reaction wells were read at 570 nm with a reference filter set at 690 nm. The CCR5 or CXCR4 coreceptor usage of recombinant viruses was determined by measurement of CPRG in the different target cells and comparison with cells in wells exposed to supernatant produced by transfection in the absence of PCR products. Optical density values greater than twice the background value were considered positive. Values between 2 and 10 times the background value were confirmed in at least two independent experiments.
PCR product subcloning and sequence analysis.
RT-PCR-amplified material from patient plasma samples and from PBMC culture supernatants were cloned using the Topo-TA Cloning kit (Invitrogen) according to the manufacturer's instructions. Low-cycle PCR was conducted using the primers E20 and E115 directly on single colonies from Luria-Bertani agar plates, and 500 ng of PCR product was used in the recombinant virus assay to determine the tropism associated with single envelope sequences. Nucleotide sequencing of the V3 loop of the env gene was performed from the subcloned V1-V3 RT-PCR product. Samples of 4 μl of PCR product were used for cycle sequencing (Thermo sequenase fluorescent labelling primer cycle sequencing kit; Amersham Pharmacia Biotech, Little Chalfont Buckinghamshire, United Kingdom) with CY5-labeled J53Y primer (5′-AATTTCTGGGTCCCCTCCTG-3′) according to the manufacturer's protocol. The generated fragments were analyzed in a 6% polyacrylamide gel with an automated laser fluorescent sequencing apparatus (Pharmacia Biotech, Uppsala, Sweden).
Phylogenetic analysis was conducted with Phylogeny Inference Package (PHYLIP) version 3.57c programs (26, 27) as previously described (61). Briefly, nucleotide distances were estimated by means of the maximum-likelihood model. Phylogenies were reconstructed by both the neighbor-joining method and the Fitch-Margoliash distance method in order to increase confidence in the reconstructed phylogenies. Bootstrap resampling (200 replicates) was applied to the neighbor-joining trees to assess the strength of support for each branch.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in the GenBank database under the following accession numbers: for patient 136 samples, no. AF284505 to AF284526, and for patient 145 samples, no. AF284527 to AF284552.
RESULTS
Vector construction and experimental procedure.
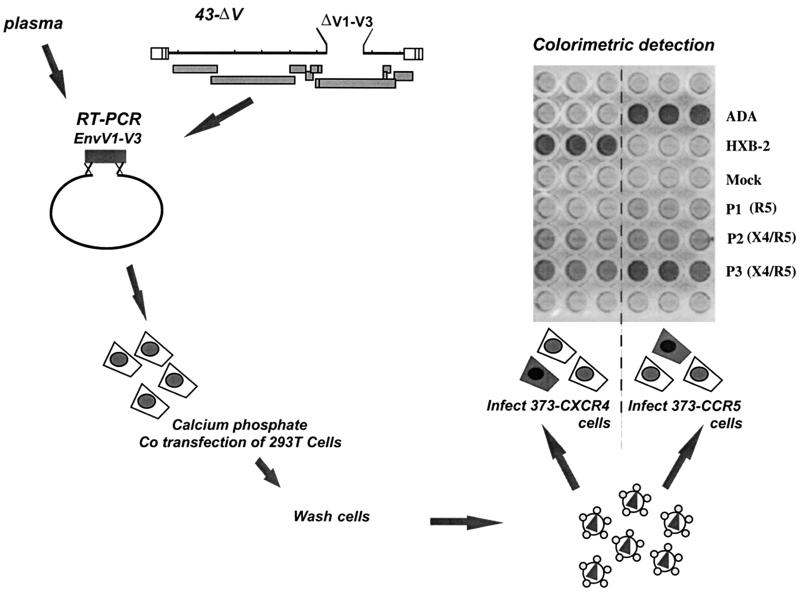
The region spanning the three N-terminal variable loops of the gp120 surface glycoprotein contain the main determinants for HIV-1 coreceptor usage (2, 7, 14, 32, 33, 35, 36, 48, 51, 52, 66, 69, 74, 76). To generate a vector for the phenotypic determination of cellular tropism of primary viruses, we deleted the entire V1-V3 region (600 bp) of the envelope from a previously described pNL4-3 derivative, thus obtaining the vector 43-ΔV. The natural and unique NheI restriction site just downstream of the deletion was used to linearize the vector. Recombinant viruses were produced by complementation of the 43-ΔV-linearized vector with PCR products from patients' samples encompassing the deleted region and short (100 to 150 nucleotides) overlaps that allowed homologous recombination. In our experimental system, 900-bp-long amplification products were produced by RT-PCR from plasma virus. As controls, PCR products from plasmids encoding HXB-2 or ADA envelope, an X4 viral clone and an R5 viral clone, respectively, were also used (Fig. 1).
FIG. 1.
Schematic representation of the TRT.
The recombinant virus released in the transfected 293-T-cell supernatant was used to infect indicator cells, expressing CD4 and either CXCR4 (373-CXCR4) or CCR5 (373-CCR5) coreceptor molecules on their surface (Fig. 1). These indicator cell lines also carry an inducible LTR-LacZ cassette, which allows colorimetric assessment of virus infection by HIV-1 Tat-induced β-Gal expression. As shown in Fig. 1, complementation of the 43-ΔV vector with PCR product from the HXB-2 envelope sequence resulted in a strong and specific signal in target cells expressing CXCR4 but not CCR5, consistent with the known coreceptor specificity of this virus. Conversely, vector complementation with a PCR product from the ADA envelope tested positive only in cells expressing CCR5. This finding indicates that the V1-V3 region of the envelope glycoprotein is sufficient for the prediction of virus tropism using the recombinant virus assay described here, in agreement with several observations pointing to this region as the major tropism determinant. As expected, supernatant from cells transfected with the 43-ΔV-linearized vector only, in the absence of a PCR product, did not induce β-Gal expression in target cells (Fig. 1, mock wells). As a control, 373-CD4 cells that carried the LTR-LacZ cassette but that did not express a coreceptor molecule always tested negative when exposed to virus-containing supernatant (data not shown). Figure 1 also shows the result obtained when RT-PCR products from representative plasma samples were used to complement the 43-ΔV vector. Some samples tested positive exclusively for CCR5 usage (samples P1) while others tested positive both on CCR5- and on CXCR4-expressing 373 cells (samples P2 and P3). These data show that the TRT described here allows the determination of coreceptor usage of viruses from plasma samples, without the need for virus isolation in culture.
Comparison of TRT with current phenotyping procedures.
We compared our method with conventional virus tropism assays, testing consecutive plasma samples from three selected perinatally infected patients (Table 1) whose viruses underwent a phenotypic change from R5 to X4 during disease progression (62). Emergence of X4 plasma virus was detected by the TRT in all three patients at the same time points as conventional phenotyping procedures based on MT-2 syncytium scoring and on virus replication in U87-CD4+ cells expressing CXCR4 (Table 1). Interestingly, TRT analysis indicated that the ability of plasma viruses to use CCR5 was preserved throughout the follow-up (Table 1). In contrast, the R5 virus population was not detected using the CCR5-expressing U87-CD4+ indicator cells at months 48 and 67 for patient 145 (Table 1). This discrepancy between the two detection methods may be due to the different virus sources or may reflect the replicative advantage of X4 strains in the PBMC cultures used for the U87-CD4+ assay (see below).
TABLE 1.
Comparison of TRT results with those of other phenotyping procedures
| Patient no. | Age (months) | Plasma virus on 373 CD4+ cellsa
|
PBMC isolate on MT2 cellsbc | PBMC isolate on U87 CD4+ cellsbd
|
||
|---|---|---|---|---|---|---|
| CCR5 | CXCR4 | CCR5 | CXCR4 | |||
| 3 | 18 | + | − | − | +++ | − |
| 40 | + | − | − | +++ | − | |
| 54 | + | + | + | +/− | +++ | |
| 65 | + | + | + | +/− | +++ | |
| 136 | 3 | + | − | − | +++ | − |
| 34 | + | − | − | +++ | − | |
| 60 | + | + | + | + | +++ | |
| 64 | + | + | + | + | +++ | |
| 67 | + | + | + | +/− | +++ | |
| 145 | 5 | + | − | − | +++ | − |
| 48 | + | + | + | − | +++ | |
| 67 | + | + | + | − | +++ | |
Infection of 373-CD4 cells expressing one coreceptor was performed with virus supernatant produced by the recombinant virus technique TRT as described in Materials and Methods.
Data were published previously (62).
Viral isolates capable of infecting MT2 cells were defined as positive (+) cultures, and those unable to infect were defined as negative (−).
Infection of U87-CD4 cells expressing one coreceptor was achieved by culturing PBMCs infected with each viral isolate, and cultures were evaluated at day 7 after infection. −, no syncytia; +/−, no syncytia but increasing p24 production; +, rare syncytia; ++, medium-size syncytia; +++, abundant large syncytia.
Coreceptor usage of single virus clones from a mixed plasma virus population.
The ability to use both CCR5 and CXCR4 could be the result of two nonexclusive phenomena: (i) the expanded coreceptor usage capacity conferred by a single envelope sequence or (ii) the presence of a mixed virus population in which different envelope sequences allow the use of different coreceptors. To analyze the composition of plasma virus samples from patient 145 at different time points, we subcloned the V1-V3 RT-PCR product. Individual clones were then screened for coreceptor specificity by TRT, using plasmid DNA as a PCR substrate. Clones obtained from an authentic dualtropic virus are expected to test positive on both 373-CCR5 and 373-CXCR4 cells, while clones from R5 and X4 viruses can infect only one of the target cell lines. As shown in Table 2, for the sample obtained at month 5 from patient 145, which was determined to be R5-exclusive both by our method and by current procedures, all tested clones (11 out of 11) harbored R5-specific envelope sequences when analyzed by TRT. At month 48, the first available time point for which X4 viruses were detected for patient 145, an analysis of the clones showed that the plasma virus was composed of a mixed population, with an equivalent proportion of X4 and dualtropic isolates (4 out of 9 clones, each) and a minority of R5 isolates (1 out of 9 clones). The capacity of the mixed plasma virus population to infect target cells using both coreceptors was thus due to the coexistence of all three possible virus populations in this plasma sample: R5-exclusive, X4-exclusive, and dualtropic viruses. At 67 months, plasma virus from patient 145 again tested positive on indicator cells expressing either coreceptor (Table 1). This sample harbored similar proportions of R5-exclusive and dualtropic viruses (5 and 6 clones, respectively, out of 11 tested), while the X4-exclusive virus population was no longer detected (Table 2).
TABLE 2.
Comparison of coreceptor usage by clones from plasma and culture supernatanta
| Patient no. | Age (months) | No. of clones from plasma
|
No. of clones from PBMC supernatant
|
||||
|---|---|---|---|---|---|---|---|
| R5 | X4 | R5X4 | R5 | X4 | R5X4 | ||
| 145 | 5 | 11 | 0 | 0 | 11 | 0 | 0 |
| 48 | 1 | 4 | 4 | 0 | 2 | 6 | |
| 67 | 5 | 0 | 6 | 0 | 1 | 8 | |
| 136 | 3 | 11 | 0 | 0 | NA | NA | NA |
| 34 | 12 | 0 | 0 | NA | NA | NA | |
| 60 | 11 | 1 | 0 | 1 | 6 | 4 | |
| 64 | 6 | 3 | 1 | 2 | 3 | 5 | |
Comparison is of the number of clones carrying an envelope sequence from plasma samples and from PBMC culture supernatant, characterized by specific coreceptor usage. NA, sample not available.
Clones from all available plasma samples were obtained also for patient 136 (Table 2). Clones from early time points (month 3 and 34) confirmed that the virus population was homogeneous with respect to coreceptor usage, since all clones tested positive only on cells expressing the CCR5 coreceptor. At month 60, the first time point at which X4 viruses were detected, only 1 of the 12 clones was indeed able to use CXCR4, while the vast majority of the clones were still R5 exclusive (Table 2). At month 64, however, plasma from patient 136 harbored all three virus populations, mostly R5-exclusive virus (6 out of 10) with some X4-exclusive virus (3 out of 10) and a single dualtropic component (Table 2). Thus, patient isolates able to use both CCR5 and CXCR4 coreceptors consist of mixtures of R5-exclusive viruses with X4-exclusive and/or authentic dualtropic variants.
Virus culture biases the coreceptor usage determination of virus replicating in patients.
To determine why the R5 virus population detected by our recombinant virus technique (Tables 1 and 2) was not recognized in the phenotypic assay using cultured virus (Table 1), we further analyzed the case of patient 145. Viral RNA was extracted from the supernatants of the patient PBMC cultures used to infect U87-CD4 cells, and PCR amplification products were cloned as described above. Clones were tested for coreceptor usage by TRT and compared to clones from plasma samples obtained at the same time points (Table 2). Month 5 clones from plasma and culture supernatants were all R5 exclusive, in agreement with previous characterization on MT2 cells and U87-CD4 cells (62). Interestingly, for subsequent time points (months 48 and 67), no R5-exclusive virus was detected in the culture supernatant samples, in contrast to plasma samples obtained at the same time points (Table 2). Dualtropic and X4-exclusive viruses were instead found in both supernatants (Table 2). A comparison of virus population with different tropisms in plasma and PBMC culture supernatants was performed also for the available samples from patient 136 (months 60 and 64; Table 2). The majority of clones from PBMC supernatant consisted of X4 and dualtropic clones (Table 2), while in the corresponding plasma samples, the large majority of the clones were R5 (Table 2). These results suggest that either the source of the virus (PBMCs versus plasma) or selection against R5-exclusive variants in culture may have caused the misrepresentation of plasma virus population by tropism assays that rely on virus propagation in culture.
Genetic determinants of virus tropism.
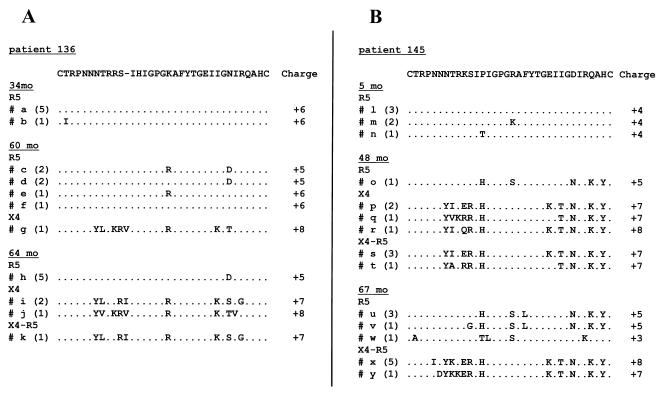
An additional advantage offered by the subcloning approach used here consists of the ability to correlate virus tropism with the amino acid sequences of the cloned fragments. The V3 loop has been shown to contain signature sequences typical of either R5 or X4 viruses, consisting both of specific amino acid substitutions and of a change in the overall charge of the domain (10, 19, 30, 36, 42, 67, 70). We analyzed the amino acid sequence of the V3 loop for clones obtained from selected plasma samples from patients 136 and 145 (Fig. 2A and B, respectively). Six pre-switch sequences were analyzed for both patients 136 and 145 and were found to be quite homogeneous within each patient, differing at most by one residue. For patient 136, after the phenotypic switch, the majority of the clones corresponded to R5 viruses (Table 2), and their sequences were still highly conserved (Fig. 2A). The single X4-exclusive clone detected displayed a marked increase of the positive charge of the V3 domain and a characteristic positively charged residue at position 11 (Fig. 2A). Four months later, all R5-exclusive clones were still identical, while several differences were observed in the sequences of the clones able to use CXCR4, consistent with active replication and diversification of X4 viruses. Interestingly, the single dualtropic clone obtained from this sample displayed a V3 loop amino acid sequence identical to that of some of the X4-exclusive clones from the same patient (Fig. 2A). Concerning patient 145, only one clone from the month 48 plasma sample was R5 exclusive, and it harbored several amino acid differences with respect to pre-switch R5 clones (Fig. 2B), a likely consequence of the relatively long time elapsed between the collection of these samples. Clones shown to use CXCR4 that were from the month 48 plasma sample displayed a net increase in the positive charge of the V3 domain associated with the replacement of a positively charged residue at position 11, among other changes. Some variability was observed between single clones, and again some of the dualtropic clones had a V3 loop sequence identical to that of some X4-exclusive clones. At month 67, R5-exclusive clones displayed increasing variability, while minor changes were detected in the dualtropic clones at this time point.
FIG. 2.
Multiple alignment of deduced amino acid sequences of the V3 domain from plasma samples from patients 136 (A) and 145 (B) obtained at different time points. Sequences of the clones are aligned against the prevalent clone from the first time point. Letters on the left of each sequence correspond to letters on the phylogenetic tree in Fig. 3. The frequency of the clones with identical amino acid sequences is given in parentheses, and the net positive charge of the domain is given. Dots indicate identity and dashes represent gaps introduced to maximize alignment.
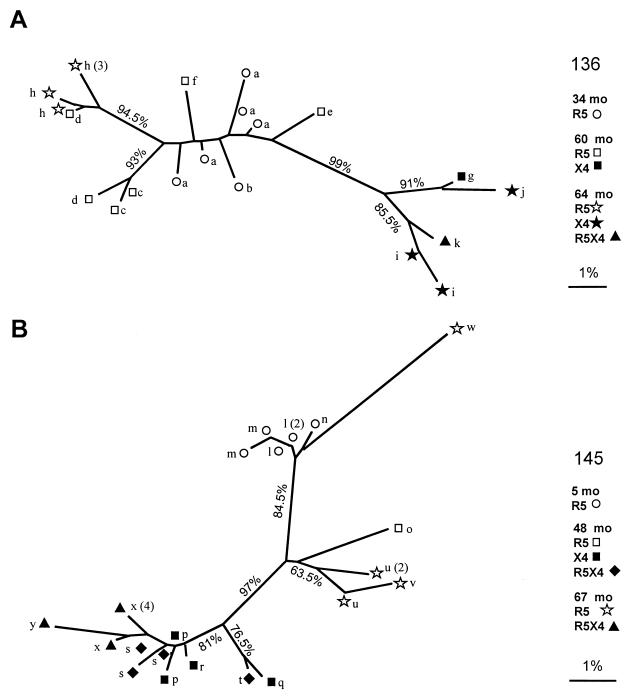
To further analyze the relationship between the viral variants, a phylogenetic tree was reconstructed by the neighbor-joining method using 200-nucleotide-long sequences encompassing the V3 domain from all clones obtained from plasma samples. The phylogenetic analysis for patient 136 (Fig. 3A) showed that all sequences of R5 clones clustered together, separately from the sequences of the X4 and dualtropic clones. This separation was supported by a high bootstrap value (99%). The separation of R5 sequences obtained at month 64 from all other R5 sequences indicates an increase in the evolutionary distance over time, despite similar V3 loop amino acid sequences. The dualtropic sequences clustered with X4 sequences, with a relatively high bootstrap value (85.5%), thus confirming the genetic relatedness of these variants. For patient 145 (Fig. 3B), several sequences corresponding to those of X4 and dualtropic clones spread on different branches, indicating the presence of multiple distinct lineages. Of note, as for patient 136, we found that sequences of X4 and dualtropic clones clustered together separately from the R5 cluster at a distance that was well supported by bootstrap resampling (97%).
FIG. 3.
Unrooted neighbor-joining tree of the V3 nucleotide sequences of virus clones from plasma samples. Branch lengths are drawn to scale. Bar, 1% nucleotide sequence distance. The number at the node indicates the proportion of support in 200 bootstrap replicates. The single-letter code corresponds to letters in Fig. 2 to allow the identification of each sequence on the tree. A number in parentheses following the letter indicates the number of clones with an identical nucleotide sequence.
DISCUSSION
Determination of the coreceptor usage of HIV-1 in vivo is relevant both to the prediction of disease progression and to the precise understanding of the dynamic process of virus evolution in infected subjects. Faster disease progression rates (39, 41, 59) as well as increased replication kinetics and pathogenicity in vitro (31, 38, 54, 57) are associated with the capacity of HIV-1 strains to use CXCR4 as a coreceptor. Although phenotypic characterization of the cellular tropism of patient virus populations has led to major advances in the understanding of the infectious process in vivo, it remains unclear whether the appearance of X4 viruses in patients is a cause or a consequence of immunodeficiency. Arguments can be found to support both views, and it is only by frequently sampling large patient populations by using a sensitive technique that the temporal relationship between these two events will be elucidated. Extensive studies in this field have been hampered by the fact that conventional procedures are time-consuming and involve complex sample manipulation. Most of the viruses characterized by current phenotypic procedures originate from PBMC cocultivation followed by in vitro culture to achieve a sufficient infectious titer for testing on selective target cells (47). Phenotyping techniques based on virus culture suffer from some drawbacks: they are difficult to standardize due to primary cell variability, HIV-1 genomes integrated in host DNA can be reactivated in stimulated PBMCs, and the virus produced can differ from the population replicating in the patient, which is best approximated by plasma virus (25, 55, 73). In addition, culture conditions exert a selective pressure that may also lead to misrepresentation of the virus population, even after a limited number of passages.
To overcome some of these technical difficulties, we have developed an assay that determines the coreceptor usage of primary HIV-1 viruses which does not rely on virus isolation in culture, thus providing an additional tool for the study of the kinetics and equilibrium of virus populations in infected subjects. This approach eliminates the selective bias associated with virus culture, allows easy monitoring of large patient cohorts, and was used here to study the tropism of virus populations found in plasma. We evaluated the tropism of virus from three infected children for which sequential samples preceding and following the emergence of X4 strains were available. These viruses had been previously characterized with tropism assays using both MT-2 syncytium scoring and U87-CD4+ cells expressing a single coreceptor (62). For all three patients, our recombinant method detected viruses able to use CXCR4 (X4 or dualtropic) at the same time points they were first detected by assays based on MT-2 and U87 cells. Besides, the addition of a simple step, consisting of the subcloning of PCR products, allowed semiquantitative analysis of the complexity of the patient virus population. We could thus identify different proportions of R5- and X4-exclusive viral components as well as authentic primary dualtropic viruses present in plasma samples. A more precise quantification of viruses with different tropisms that are present in a population would require the screening of very large number of clones. Even then, the quantification could suffer from the possibility that some primary sequences may not be functional in the context of the NL4-3 vector used here.
Primary dualtropic viruses have been previously obtained by limiting dilution of PBMC culture supernatants from two patients (68). In that study, no X4-exclusive viruses were found, suggesting that the syncytium-inducing phenotype of the overall isolate reflected CXCR4 usage by dualtropic clones (68). Accordingly, some of the virus populations analyzed here by TRT were a mixture of R5-specific and dualtropic viruses (Table 2, patient 145, month 67). Nevertheless, we unambiguously showed that X4-exclusive viruses can be found in some plasma samples (Table 2, patient 136, month 64, and patient 145, month 48). Our finding that X4 viruses are present in plasma samples shows that evolution towards the exclusive usage of CXCR4 is a relevant phenomenon in patients and not only the consequence of in vitro virus culture on established T-cell lines. Dualtropic isolates were proposed to represent a transitional state in the evolution from R5 to X4 viruses (15, 22). The finding that dualtropic viruses displayed reduced replication kinetics, with respect to X4 viruses, in primary cells expressing only the CXCR4 coreceptor (56) also supports the hypothesis that they precede X4 viruses. Improved CXCR4 usage would then confer to X4 viruses an increased colonization potential, given that this molecule is more widely expressed in primary cells than CCR5 (see reference 77 and references within). An alternative hypothesis is that R5 viruses could switch to an X4 phenotype by just a few changes in their envelope sequence, and dualtropism may represent a subsequent evolutionary adaptation with peculiar biological properties. The understanding of the temporal relationship between these forms would provide valuable information on the selective forces at play and on the viral replication strategy.
Our study also establishes that primary dualtropic viruses have V3 loop sequence characteristics similar to those of X4 viruses, with comparable values of net positive charge, the same substitutions at specific positions with respect to R5 viruses, and, in some cases, complete amino acid sequence identity with X4 viruses from the same patient. V3 loop sequences from dualtropic and X4 plasma viruses from the same patient clustered together, separate from all R5 sequences analyzed here. These findings suggest that modification of the V3 loop sequence is required for the use of CXCR4 as a coreceptor, while the possibility to maintain the capacity to use CCR5, in addition to CXCR4, depends on the overall context of the V1-V3 region. The molecular determinants responsible for the expanded tropism of dualtropic primary viruses are not well established, and the study of a larger cohort is required before we can generalize on the implication of specific residues or domains. As mentioned above, losing the capacity to use CCR5 could represent the evolutionary cost for specialized CXCR4-mediated entry.
Interestingly, we showed here that the R5 viral population persisted after the emergence of X4 and/or dualtropic viruses in all three patients. The lack of detection of the R5 component in one of the three patients (patient 145) using U87 cells, was due to the differences in the virus source and technical approach. Persistent CCR5 utilization was previously shown for both blood- and tissue-derived isolates, with the additional observation that isolates obtained from late stages of disease displayed increased replication ability in macrophages (45). Similarly, R5 viruses isolated from patients with AIDS also displayed faster replication kinetics and increased T-cell depletion in SCID-hu mice with respect to viruses isolated from the same patients at earlier time points (65). These reports suggest that for HIV, as shown for simian immunodeficiency virus (SIV) (40), virus pathogenicity increases with time in an infected individual independently of coreceptor usage change. The analysis of sequential R5 viruses with a recombinant virus technique similar to the one described here could determine whether changes in the envelope sequence are solely responsible for the different biological characteristics of late R5 viruses. These could be the consequence of increased affinity for or increased ability to use CCR5. Alternatively, faster virus replication kinetics and pathogenicity could result from the slower process of coevolution of several viral genes. Such a study would provide important information on the relationship between HIV and the host.
Persistence of the R5 virus population indicates that the environmental conditions that favor X4 and dualtropic virus replication in vivo are not necessarily more restrictive for R5 viruses. Accordingly, the emergence of X4 viruses would represent the colonization of new target cells in an expanding infectious process, rather than the appearance of virus population directly competing with R5 viruses. The development of X4 and dualtropic strains could reflect the colonization of a tissue compartment in which the usage of CXCR4 confers an advantage either by allowing increased replication kinetics or because the local immune surveillance is decreased. Although the present study does not analyze HIV tropism in specific tissue compartments, the molecular approach used might provide important insights on this issue. We envisage the use of samples from different compartments, feasible as long as a PCR product can be generated, to compare the kinetics of appearance of X4 viruses and the complexity of the virus isolates.
Virus tropism can be expected to evolve also under the selective pressure of antiviral molecules aimed at inhibiting virus entry by competing for coreceptor binding. Natural and synthetic molecules that block coreceptors and exert strong inhibition of virus infection in culture are being evaluated for their potential use in the treatment of HIV-1 infection, and a few have progressed to clinical trials (5). Blocking one coreceptor may select for variants that use alternative coreceptors, leading to a shift in the virus population. Monitoring the coreceptor usage of viruses from patients treated with entry inhibitors can be facilitated by our assay. Overall, we believe that the study of the kinetics and of the conditions under which viruses with different coreceptor specificity evolve in patients can be greatly assisted by the recombinant virus approach described here.
ACKNOWLEDGMENTS
We thank Allan Hance and Esther Race for critically reading the manuscript.
This work was supported in part by a grant from the Agence Nationale de Recherche sur le SIDA (ANRS). F.S. was the recipient of a fellowship from Istituto Superiore di Sanità, Rome, Italy.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1998. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Andeweg A C, Leeflang P, Osterhaus A D, Bosch M L. Both the V2 and V3 regions of the human immunodeficiency virus type 1 surface glycoprotein functionally interact with other envelope regions in syncytium formation. J Virol. 1993;67:3232–3239. doi: 10.1128/jvi.67.6.3232-3239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 4.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 5.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 6.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll R G, Riley J L, Levine B L, Feng Y, Kaushal S, Ritchey D W, Bernstein W, Weislow O S, Brown C R, Berger E A, June C H, St. Louis D C. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 9.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 12.Clapham P R. HIV and chemokines: ligands sharing cell-surface receptors. Trends Cell Biol. 1997;7:264–268. doi: 10.1016/S0962-8924(97)01075-1. [DOI] [PubMed] [Google Scholar]
- 13.Clapham P R, Reeves J D, Simmons G, Dejucq N, Hibbitts S, McKnight A. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol Membr Biol. 1999;16:49–55. doi: 10.1080/096876899294751. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 15.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelissen M, Mulder-Kampinga G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, Goudsmit J. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Jong J J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 22.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 23.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 24.Eustice D, Feldman P, Colberg-Poley A, Buckery R, Neubauer R. A sensitive method for the detection of β-galactosidase in transfected mammalian cells. BioTechniques. 1991;11:739–743. [PubMed] [Google Scholar]
- 25.Feinberg M B. Changing the natural history of HIV disease. Lancet. 1996;348:239–246. doi: 10.1016/s0140-6736(96)06231-9. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. PHYLIP manual version 3.25c. Berkeley University Herbarium, University of California, Berkeley; 1993. [Google Scholar]
- 28.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 29.Fenyo E M, Fiore J, Karlsson A, Albert J, Scarlatti G. Biological phenotypes of HIV-1 in pathogenesis and transmission. Antibiot Chemother. 1994;46:18–24. doi: 10.1159/000423630. [DOI] [PubMed] [Google Scholar]
- 30.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glushakova S, Grivel J C, Fitzgerald W, Sylwester A, Zimmerberg J, Margolis L B. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- 32.Groenink M, Andeweg A C, Fouchier R A, Broersen S, van der Jagt R C, Schuitemaker H, de Goede R E, Bosch M L, Huisman H G, Tersmette M. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J Virol. 1992;66:6175–6180. doi: 10.1128/jvi.66.10.6175-6180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van't Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 34.Harrington R D, Geballe A P. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman T L, Doms R W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol Membr Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 36.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science. 1992;257:535–537. doi: 10.1126/science.1636088. [DOI] [PubMed] [Google Scholar]
- 37.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 38.Kaneshima H, Su L, Bonyhadi M L, Connor R I, Ho D D, McCune J M. Rapid-high, syncytium-inducing isolates of human immunodeficiency virus type 1 induce cytopathicity in the human thymus of the SCID-hu mouse. J Virol. 1994;68:8188–8192. doi: 10.1128/jvi.68.12.8188-8192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keet I P, Krijnen P, Koot M, Lange J M, Miedema F, Goudsmit J, Coutinho R A. Predictors of rapid progression to AIDS in HIV-1 seroconverters. AIDS. 1993;7:51–57. doi: 10.1097/00002030-199301000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 41.Koot M, Keet I P, Vos A H, de Goede R E, Roos M T, Coutinho R A, Miedema F, Schellekens P T, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 42.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retrovir. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 43.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lathey J L, Pratt R D, Spector S A. Appearance of autologous neutralizing antibody correlates with reduction in virus load and phenotype switch during primary infection with human immunodeficiency virus type 1. J Infect Dis. 1997;175:231–232. doi: 10.1093/infdis/175.1.231. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif H M. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73:9741–9755. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liesnard C, Delforge M L, Tchetcheroff M, De Maertelaer V, Farber C M, Van Vooren J P. Importance of method in the determination of syncytium-inducing phenotype of human immunodeficiency virus type 1 clinical isolates. J Virol Methods. 1997;64:137–145. doi: 10.1016/s0166-0934(96)02152-0. [DOI] [PubMed] [Google Scholar]
- 48.Malykh A, Reitz M S, Jr, Louie A, Hall L, Lori F. Multiple determinants for growth of human immunodeficiency virus type 1 in monocyte-macrophages. Virology. 1995;206:646–650. doi: 10.1016/s0042-6822(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 49.Mammano F, Trouplin V, Zennou V, Clavel F. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J Virol. 2000;74:8524–8531. doi: 10.1128/jvi.74.18.8524-8531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 51.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 52.Palmer C, Balfe P, Fox D, May J C, Frederiksson R, Fenyo E M, McKeating J A. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology. 1996;220:436–449. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 53.Pantaleo G, Cohen O J, Schacker T, Vaccarezza M, Graziosi C, Rizzardi G P, Kahn J, Fox C H, Schnittman S M, Schwartz D H, Corey L, Fauci A S. Evolutionary pattern of human immunodeficiency virus (HIV) replication and distribution in lymph nodes following primary infection: implications for antiviral therapy. Nat Med. 1998;4:341–345. doi: 10.1038/nm0398-341. [DOI] [PubMed] [Google Scholar]
- 54.Penn M L, Grivel J C, Schramm B, Goldsmith M A, Margolis L. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc Natl Acad Sci USA. 1999;96:663–668. doi: 10.1073/pnas.96.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 56.Picchio G R, Gulizia R J, Mosier D E. Chemokine receptor CCR5 genotype influences the kinetics of human immunodeficiency virus type 1 infection in human PBL-SCID mice. J Virol. 1997;71:7124–7127. doi: 10.1128/jvi.71.9.7124-7127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picchio G R, Gulizia R J, Wehrly K, Chesebro B, Mosier D E. The cell tropism of human immunodeficiency virus type 1 determines the kinetics of plasma viremia in SCID mice reconstituted with human peripheral blood leukocytes. J Virol. 1998;72:2002–2009. doi: 10.1128/jvi.72.3.2002-2009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 59.Richman D D, Bozzette S A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 60.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salvatori F, Masiero S, Giaquinto C, Wade C M, Brown A J, Chieco-Bianchi L, De Rossi A. Evolution of human immunodeficiency virus type 1 in perinatally infected infants with rapid and slow progression to disease. J Virol. 1997;71:4694–4706. doi: 10.1128/jvi.71.6.4694-4706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 63.Schuitemaker H. Macrophage-tropic HIV-1 variants: initiators of infection and AIDS pathogenesis? J Leukoc Biol. 1994;56:218–224. doi: 10.1002/jlb.56.3.218. [DOI] [PubMed] [Google Scholar]
- 64.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scoggins R M, Taylor J R, Jr, Patrie J, van't Wout A B, Schuitemaker H, Camerini D. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J Virol. 2000;74:3205–3216. doi: 10.1128/jvi.74.7.3205-3216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 67.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van't Wout A B, Blaak H, Ran L J, Brouwer M, Kuiken C, Schuitemaker H. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J Virol. 1998;72:5099–5107. doi: 10.1128/jvi.72.6.5099-5107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van't Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Investig. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 74.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolinsky S M, Wike C M, Korber B T, Hutto C, Parks W P, Rosenblum L L, Kunstman K J, Furtado M R, Munoz J L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 76.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 77.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 78.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, Ho D D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, KewalRamani V N, Moore J P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]