Abstract
Background:
With the antibiotic crisis, the topical antibacterial control including chronic wounds gains increasing importance. However, little is known regarding tolerance development when bacteria face repetitive exposure to the identical antiseptics as commonly found in clinical practice.
Materials and Methods:
Clinical isolates foremost of chronic wounds were exposed in vitro to dilutions of two antiseptics used for wound therapy: polyhexanide or octenidine. Adaptive response was determined by growth/kill curves, minimal inhibitory concentration (MIC), and whole genome sequencing. Antiseptic/bacteriophage combinations were studied by liquid-infection assays and bacterial plating.
Results:
Polyhexanide acted stronger against Escherichia coli and Proteus mirabilis while octenidine was more potent against Staphylococcus aureus. Otherwise, the antiseptic efficacy varied across isolates of Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Upon repetitive exposure with constant antiseptic concentrations P. aeruginosa and P. mirabilis adaptation was evident by a reduced lag-phase and a twofold increased MIC. Under increasing octenidine concentrations, P. aeruginosa adapted to an eightfold higher dosage with mutations in smvA, opgH, and kinB affecting an efflux pump, alginate and biofilm formation, respectively. S. aureus adapted to a fourfold increase of polyhexanide with a mutation in the multiple peptide resistance factor MprF, also conferring cross-resistance to daptomycin. Antiseptic/bacteriophage combinations enhanced bacterial inhibition and delayed adaptation.
Conclusion:
Different bacterial species/strains respond unequally to low-level antiseptic concentrations. Bacterial adaptation potential at phenotypic and genotypic levels may indicate the necessity for a more nuanced selection of antiseptics. Bacteriophages represent a promising yet underexplored strategy for supporting antiseptic treatment, which may be particularly beneficial for the management of critical wounds.
Keywords: antiseptic/bacteriophage interactions, chronic wound infection, octenidine, polyhexanide
Introduction
Highlights
Antimicrobial efficacy of polyhexanide and octenidine differs across bacterial species and strains, indicating the need for careful selection of antiseptics.
Bacteria can adapt to low-level amounts of antiseptics at phenotypic and genotypic level, with mutations potentially conferring also cross-resistance to antibiotics.
Suppression of bacteria can be enhanced by combining antiseptics with lytic bacteriophages.
Bacterial colonization and infection are the leading complications in the management of cutaneous wounds. Chronic wounds in particular pose a tremendous structural and financial burden to global healthcare systems1. Beyond mechanical treatment of wounds, foremost surgical debridement, antimicrobial control by either systemic antibiotics or topical antiseptics represent the mainstay in the repertoire of surgeons especially for invasive infections2. In the midst of a rapid surge in resistant bacteria against antibiotics and the continuous rise of pan-drug-resistant bacteria, wound antiseptics gain even more importance while the use of antibiotics needs to be well-considered3–6. Numerous studies report an inappropriate overuse of antibiotics by surgeons for prophylactic or therapeutic wound management7,8. In general, an avoidable antibiotic use in around 30% of cases has been reported across medical specialties9,10 and seems to be particularly true for wound infections11. Hence, a rethinking of existing clinical practice is needed to avert serious repercussions for antimicrobial therapy in the future. This includes also an increasing requirement for prudent use of antiseptics in healthcare settings12,13. In contrast to antibiotics, which are dynamically selected from a large panel of substances according to antibiograms of bacteria isolated from frequently performed wound swabs, antiseptics are unselectively given without any comparable testing. Although various recommendations for the use of wound antiseptics exist14, the choice among the different antiseptics for cutaneous wounds is usually based on individual preference or even availability in the healthcare institution rather than on scientific criteria. The selection of antiseptics may not be critical when bacteria face in-use concentrations. However, penetration capacity in biofilms is impeded15–18 and little is known about antiseptic activity at reduced concentration and the bacterial ability to adapt to lowered antiseptic concentrations.
Polyhexanide and octenidine are among the most frequently used antiseptics, owing to their broad-spectrum antimicrobial efficacy and low number of reported severe adverse reactions19,20. Here, we first compared in vitro the activity of polyhexanide and octenidine dilutions against a panel of clinically isolated bacteria that are relevant in chronic wounds including Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Proteus mirabillis, Escherichia coli, and Staphylococcus aureus. We then examined the bacterial phenotypic and genotypic adaptation potential when bacteria were repetitively exposed to the antiseptics mimicking the recurrent antiseptic wound therapy in the clinical scenario. Finally, we evaluated the feasibility of adding bacteriophages as a novel supplementary strategy to antiseptics.
Material and methods
Bacterial strains
Wounds of all anatomical locations that existed for a minimum of 4 weeks21 were included. All wounds were treated by board-certified plastic surgeons according to standard guidelines of debridement, regular changes of dressings, patient positioning, and multidisciplinary treatment of underlying diseases. Swabs were taken for bacterial sampling using the established Levine technique22 by rotating the wound swab in a 1 cm² area for 5 s. Additional bacteria were acquired from a clinical isolate collection. The patients signed a written informed consent and ethical approval was obtained by the Ethics Committee (Ethics Committee of the RWTH Aachen University Faculty of Medicine, Aachen, Germany; EK 077/18). In total, we included 77 clinical isolates, comprising five gram-negative bacteria, namely P. aeruginosa (16 isolates), K. pneumoniae (13 isolates), P. mirabilis (14 isolates), E. coli (15 isolates), and A. baumannii (10 isolates), along with S. aureus (9 isolates) in this study. Species identity was verified via MALDI-TOF mass spectrometry (Microflex LT, Bruker Daltonik GmbH). Unless otherwise stated, bacterial strains were grown in Mueller Hinton (MA) medium. The automated antibiotic susceptibility testing of clinical isolates was performed using the VITEK2 system (bioMérieux, Marcy-l’Étoile). Selection of isolates was based on their individual antibiotic resistance profiles. Changes in antibiotic sensitivity upon antiseptic adaptation and genomic mutation were verified via determination of MICs using E-tests (bioMérieux, Marcy-l’Étoile) and interpreted according to EUCAST Version 13.1, 2023 (http://www.eucast.org).
Transmission electron microscopy (TEM)
For TEM bacteria were cultivated overnight at 37°C on a shaker at 120 rpm, followed by an OD600nm adjustment to 0.5. Bacteria were then exposed to 0.04% w/v polyhexanide or 0.1% w/v octenidine for 1 h. After subsequent centrifugation, bacteria were resuspended in 3% (vol/vol) glutaraldehyde (Agar Scientific) in PBS. Samples were examined at an acceleration voltage of 60 kV using a TEM Leo 906 (Carl Zeiss) transmission electron microscope.
Assessment of growth dynamics under reduced antiseptic concentrations
The antiseptic products used were polyhexamethylene biguanide hydrochloride (in short: polyhexanide) – 20% aqueous solution (Biosynth) and octenidine dihydrochloride (in short: octenidine) 98% (Thermo Scientific). Initial concentrations were adjusted to those used in the clinical setting, that is 0.04% for polyhexanide and 0.1% for octenidine. Further progressive dilutions were performed until antibacterial efficacy between polyhexanide and octenidine were differentiable. To that end, the antibacterial efficacy of both antiseptics was investigated based on changes in optical density measurements at 600 nm (OD600nm) using the microplate reader SpectraMax (Molecular Devices). Overnight bacterial cultures were adjusted to an OD600nm of 0.5 with fresh MH-medium and 100 µl of the bacterial culture was placed in microtiter plate wells along with 100 µl of either polyhexanide or octenidine, as three biological replicates. Bacterial growth dynamics were recorded every 30 min for 18 h at 37°C, followed by serial dilution and plating for assessment of viable bacteria.
Bacterial adaptation facing constant low-level antiseptic concentrations
Three biological replicates of overnight bacterial cultures were adjusted to OD600nm of 0.2 with fresh MH-medium. Bacterial cultures were placed in cell culture flasks along with polyhexanide or octenidine (at a volume ratio of 1:1) using concentrations allowing time-delayed bacterial growth within 24 h. To this end, the relevant antiseptic concentrations were determined a priori individually for each strain, as described in the aforementioned section. After 24 h (time point T1), bacteria were centrifuged and the pellet resuspended in fresh medium containing the same antiseptic concentration as before, followed by incubation for 24 h (time point T2). This process was repeated two more times leading to the time points T3 and T4. Aliquots from T1 to T4 were stored at −80°C for later joint analysis. In order to determine possible adaptation, bacteria from T1 to T4 were simultaneously challenged with either antiseptic at previous concentrations (and same volume ratios) in microtiter plates. Growth dynamics were measured via optical density for 18 h using the microplate reader SpectraMax (Molecular Devices).
Adaptation experiments with increasing antiseptic concentrations
One P. aeruginosa and one S. aureus strains were grown at initial concentrations of polyhexanide and octenidine of 2 µg/ml in 10 ml medium. After 18 h of incubation 1 ml of each culture was transferred to 9 ml of fresh medium and incubated for 18 h without antiseptic to ensure stability of the phenotype. Thereafter, the cultures were progressively incubated with the double antiseptics’ concentration, whereby between each doubling stage the cultures were allowed to grow overnight without antiseptic. The process was repeated until bacteria failed to further adapt to increasing antiseptic concentrations.
Antiseptic-phage-interactions
The lytic phages used were the P. aeruginosa specific phage NP323, and the S. aureus specific phage Sb-124. Both phages were isolated from the commercially available ‘Pyophage’ and ‘Staphylococcal Bacteriophage’ cocktails25, respectively, obtained from the Eliava Institute of Bacteriophages, Microbiology, and Virology (Tbilisi). Bacteria were exposed to antiseptics with or without lytic phages at an MOI of 0.1 in liquid-infection assays using the microplate reader SpectraMax (Molecular Devices). Lowest antiseptic concentrations where positive interactions with phages occurred were established as 12.5 µg/ml and 1.25 µg/ml of either antiseptic against P. aeruginosa strain PZ and S. aureus strain SA1, respectively. Infection assays were run for 22 h, followed by bacterial plating on antiseptic-free and phage-free solid growth medium. After overnight incubation single colonies were picked for a second exposure to the same antiseptic dilutions in liquid culture with or without phage addition. After incubation for 22 h, bacteria were plated and single colonies were again subjected to a third round of antiseptic/phage challenge.
Following the positive phage/antiseptic interaction observed at lower antiseptic concentrations, the potential of complete bacterial eradication in liquid culture was tested over a broader antiseptic range (i.e. from 0 to 40 µg/ml and from 0 to 400 µg/ml for strains SA1 and PZ, respectively) and verified via plating of treated bacteria on antiseptic-free and phage-free solid media. Single colonies from cultures surviving the highest antiseptic concentrations were picked for a second exposure to all antiseptic dilutions in liquid culture with or without phage addition.
Whole genome sequencing and bioinformatics analysis
Bacterial genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen). Library preparation was performed using the NEBNext Ultra II DNA protocol (New England Biolabs). Samples were sequenced using a high output Sequencing Kit v2.5 (150 cycles) on a NextSeq500 Next-Generation Sequencing System (Illumina). Samples were processed using the nf-core/bacass pipeline version 2.0.0 implemented in Nextflow 22.04.5 with the minimal command. In brief, reads were trimmed via skewer 0.2.2 and evaluated for quality by FastQC 0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were de novo assembled using unicycler 0.4.8 and assembly quality was evaluated by QUAST 5.0.226. The assemblies were verified using kraken2 2.1.1. Genomes were annotated using Prokka 1.14.627.
For variant detection, the QIAGEN CLC Genomics Workbench 23.0.1 was used (https://digitalinsights.qiagen.com/). The function of the mutated genes was investigated using Uniprot (https://www.uniprot.org/) and The Kyoto Encyclopedia of Genes and Genomes database (KEGG; http://www.genome.jp/kegg/).
Statistical analysis
Bacterial growth inhibition was determined from the area under the curve (AUC) calculation of bacterial growth curves relative to the untreated controls. Normality of the data was verified by the Shapiro–Wilk test and significant differences were assessed using Student’s unpaired t-test (two-tailed). Graphics were created using the Microsoft Excel and Graphpad Prism v8, which was also used for AUC calculation and statistical analyses.
Results
Comparison of the antibacterial activity of polyhexanide and octenidine
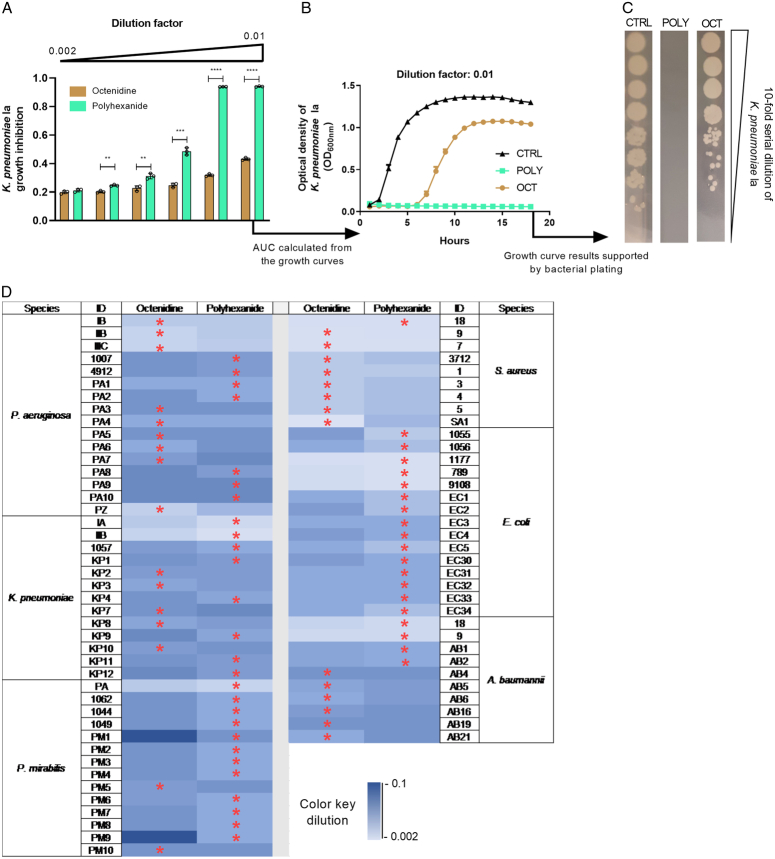
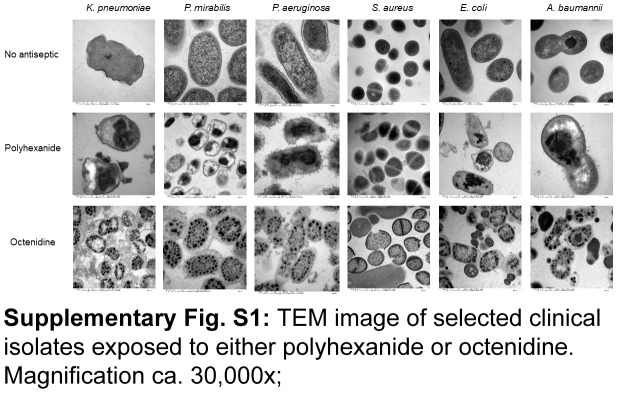
The differential antibacterial mechanisms of polyhexanide and octenidine at cellular level is evident via TEM (Supplementary Figure S1, Supplemental Digital Content 1, http://links.lww.com/JS9/C538) and is due to the fact that polyhexanide causes chromosome condensation28 while octenidine impacts cell membrane and cell wall integrity29. Figure 1A shows that the representative K. pneumoniae isolate Ia was significantly stronger suppressed by various dilutions of polyhexanide compared to equal octenidine dilutions. For instance, in the presence of 1% of the polyhexanide working solution bacterial growth arrest occurred over the whole testing period. In contrast, an equivalent 1% of the octenidine working solution enabled growth after around 6 h of exposure (Fig. 1B). No viable bacteria were recovered on solid growth medium after exposure to polyhexanide as opposed to octenidine (Fig. 1C). Figure 1D depicts the comparative results of both antiseptics for all tested isolates. The range of antiseptic dilutions within which bacteria were still able to grow extended from 0.2 to 10% of clinical in-use formulations. In case of P. aeruginosa, K. pneumoniae and A. baumannii, the superiority of either antiseptic varied from strain to strain. In contrast, for the majority of P. mirabilis isolates and throughout all E. coli isolates, polyhexanide showed superior antibacterial efficacy. Conversely, octenidine was more effective against all but one S. aureus isolates (Fig. 1D). Hence, drug predominance was either species-specific or showed interstrain variability.
Figure 1.
Antibacterial effect of two antiseptics against a panel of clinical isolates. (A) Relative growth inhibition compared to the untreated control (CTRL) of various dilutions of octenidine and polyhexanide on K. pneumoniae strain Ia. Inhibition was determined via area under the curves (AUC) calculated from optical density (OD) measurements of liquid culture assays, as shown in (B), representatively for the dilution factor 0.01. (C) Bacterial plating after 18 h-exposure to the antiseptics, in order to assess retained bacterial viability. (D) Heatmap indicating the growth suppression effect of the antiseptics octenidine and polyhexanide on all tested bacterial isolates. The color scale represents different dilutions of the antiseptics, ranging from deep blue to light blue (i.e. ranging from 10 to 0.2% of the individual working concentrations of both compounds, respectively). The heatmap depicts those dilutions at which bacterial growth arose for the first time. Red stars indicate the more effective antiseptic per tested strain (i.e. bacterial suppression was possible at a higher dilution or the bacteria were suppressed for a longer time period at the same dilution). All experiments were performed as three independent biological replicates. Stars indicate statistical support for an enhanced antibacterial effect of one antiseptic compared to the other at the same dilution. ****P<0.0001, ***P<0.001, **P<0.01.
Bacterial adaptation to constant low-level antiseptic concentrations
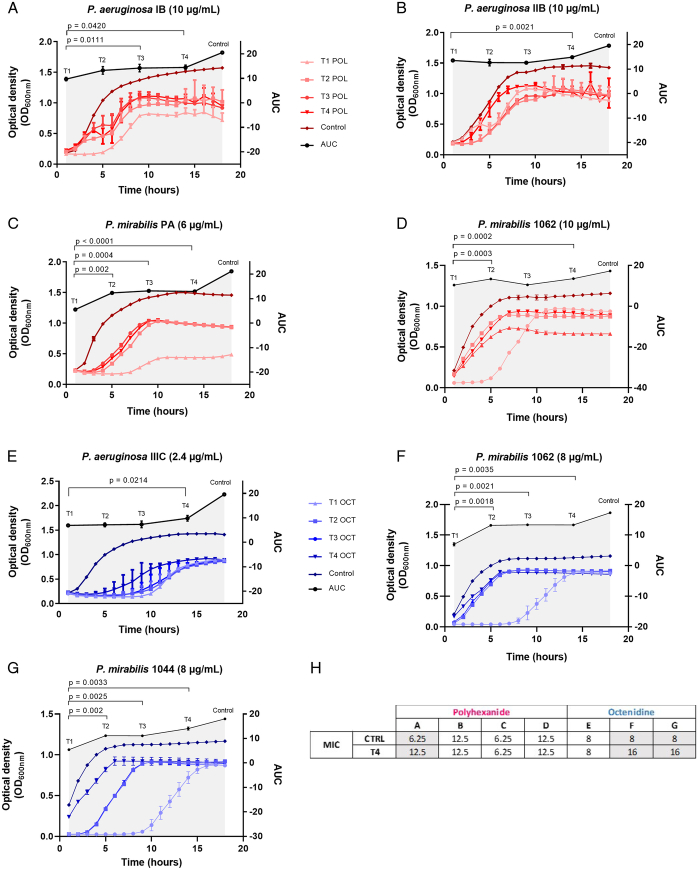
Bacterial adaptation upon four consecutive challenges (T1–T4) with constant low-level concentrations of either polyhexanide or octenidine was observed for three P. aeruginosa and two P. mirabilis strains out of 10 strains tested (Fig. 2, Supplementary Table S1, Supplemental Digital Content 2, http://links.lww.com/JS9/C539). Bacterial adaptation was evident by a reduced lag-phase in the growth curves. Bacterial adaptation emerged either in T4 (Fig. 2B and E), T3 (Fig. 2A), or already from T2 (Fig. 2C, D, F, and G).
Figure 2.
Bacterial adaptation to constant low-level concentrations of polyhexanide and octenidine. (A-G) Different bacterial isolates were challenged with either polyhexanide (POL, red curves) or octenidine (OCT, blue curves) at concentrations indicated in brackets after the strain designation; Shown are liquid culture assays of bacterial isolates that have been challenged with constant low-level concentrations for four times (T1 to T4; T1=first exposure, T2=second exposure, etc.). Bacterial growth was measured via optical density at 600 nm (OD600nm). Adaptation is evident by an earlier re-growth of T2 compared to T1, or T3 compared to T2, and so on. The significance of the acquired tolerance was determined via comparison of the area under the curves (AUC) calculated from the growth curves and is indicated by the gray shade. The control refers to the growth of the parental strain that was not exposed to the antiseptics. All experiments were performed as three independent biological replicates. Given P values indicate statistical support for an enhanced bacterial tolerance between the different generations (T1–T4). (H) Comparison of the MIC values of the adapted strains from T4 with the parental strains (Ctrl=control). The letters A-G refer to the seven bacteria/antiseptic challenges, as displayed in Figure 2.
A doubling of the MIC occurred in P. aeruginosa isolate IB (from 6.25 to 12.5 µg/ml polyhexanide) and P. mirabilis isolates 1062 and 1044 (from 8 to 16 µg/ml octenidine), while in other cases the MIC remained unchanged (Fig. 2H). Whole genome analysis revealed no mutations associated with the MIC increase.
Adaptation to increasing antiseptic concentrations
Upon repeated exposure to increasing polyhexanide concentrations a slight tolerance development was observed for P. aeruginosa strain PZ (i.e. from initial 32 µg/ml to 64 µg/ml). However, the S. aureus strain SA1 survived initially 8 µg/ml of polyhexanide, but was ultimately able to endure a fourfold higher dosage (i.e. 32 µg/ml). Conversely, strain PZ was initially able to survive 32 µg/ml of octenidine but adapted to an eightfold higher dosage (i.e. 256 µg/ml). No tolerance development against octenidine was observed with SA1.
Genomic analysis revealed one de novo non-synonymous mutation in the polyhexanide adapted strain SA1 affecting the gene mprF encoding for the phosphatidylglycerol lysyltransferase. Three mutations were observed in the octenidine adapted strain PZ affecting a MFS efflux pump, as well as glucan and alginate biosynthesis (Supplementary Table S2, Supplemental Digital Content 3, http://links.lww.com/JS9/C540).
The adaptation to polyhexanide conferred strain SA1 cross-resistance to daptomycin (Supplementary Table S2, Supplemental Digital Content 3, http://links.lww.com/JS9/C540). In the octenidine-adapted strain PZ, a 12-fold increase of the MIC for ceftazidime occurred, while the MIC for aztreonam was reduced by around fivefold.
Bacterial growth dynamics upon repeated phage/antiseptic exposures
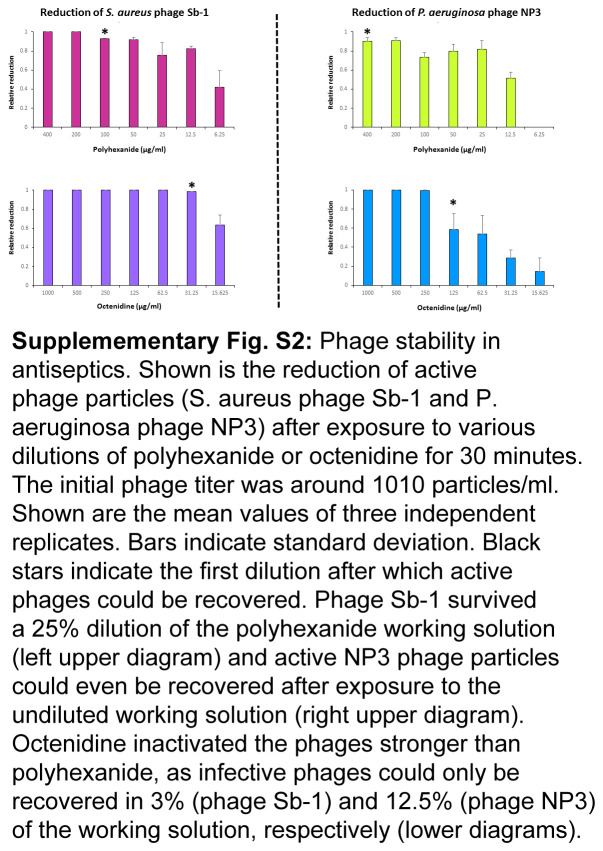
Despite being detrimental at high concentrations, a sufficiently high fraction of infective phages could be recovered from appropriate antiseptic dilutions (Supplementary Figure S2, Supplemental Digital Content 4, http://links.lww.com/JS9/C541).
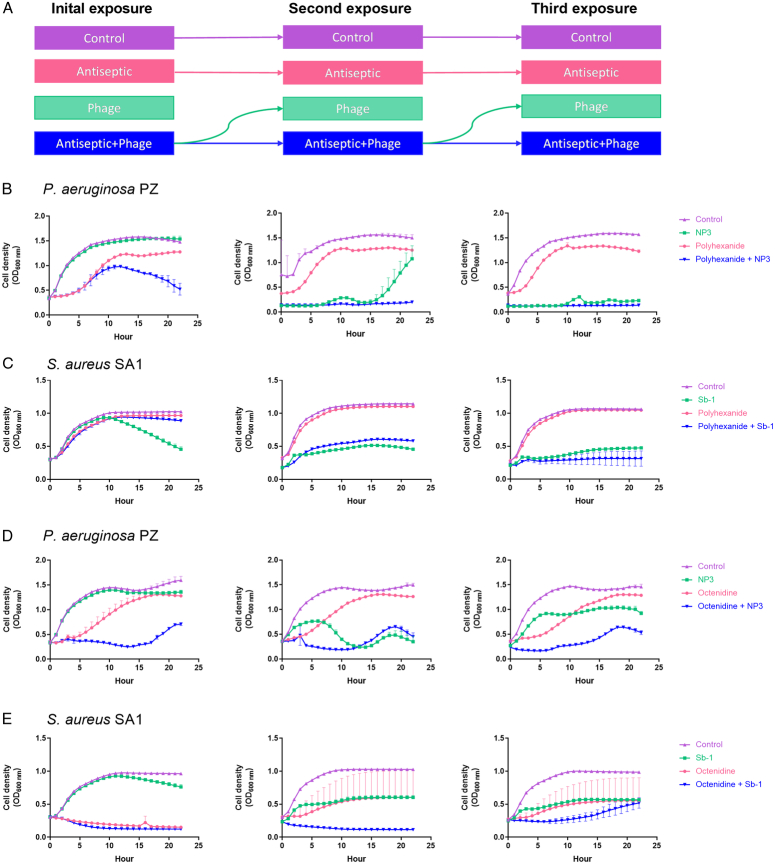
Lowest antiseptic concentrations where positive interactions with phages occurred were established as 12.5 µg/ml and 1.25 µg/ml of either antiseptic against P. aeruginosa strain PZ and S. aureus strain SA1, respectively. Repeated exposure showed that the phage-mono-application and the polyhexanide/phage combinations became more effective after the second and third exposures, in contrast to polyhexanide alone (Fig. 3B and C).
Figure 3.
Bacterial growth dynamics upon repeated phage/antiseptic exposures. (A) Scheme indicating the overall workflow of three subsequent exposures in liquid cultures with either antiseptic or phage alone or both combined (e.g. bacteria that were exposed to the antiseptic/phage combination were subsequently exposed to the phage alone and again with the combination). P. aeruginosa strain PZ (B and D) was challenged with 12.5 µg/ml of polyhexanide or octenidine, respectively, +/- phage NP3. S. aureus strain SA1 (C and E) was challenged with 1.25 µg/ml of polyhexanide or octenidine, respectively, +/- phage Sb-1. The phage MOI was 0.1. All experiments were performed as three independent biological replicates, with the mean bacterial density (OD600nm) displayed. Bars indicate SD.
Likewise, the octenidine-/NP3 combination suppressed strain PZ stronger than octenidine alone and there was an increasing effectiveness of NP3 from the second exposure against strain PZ (Fig. 3D). In addition, while S. aureus adapted to octenidine alone, the co-presence of phage Sb-1 impeded this (Fig. 3E). Furthermore, bacteria became more susceptible to the phage after having been initially exposed to the phage/antiseptic combination (green lines, Fig. 3E).
Bacterial eradication upon antiseptic/phage exposure
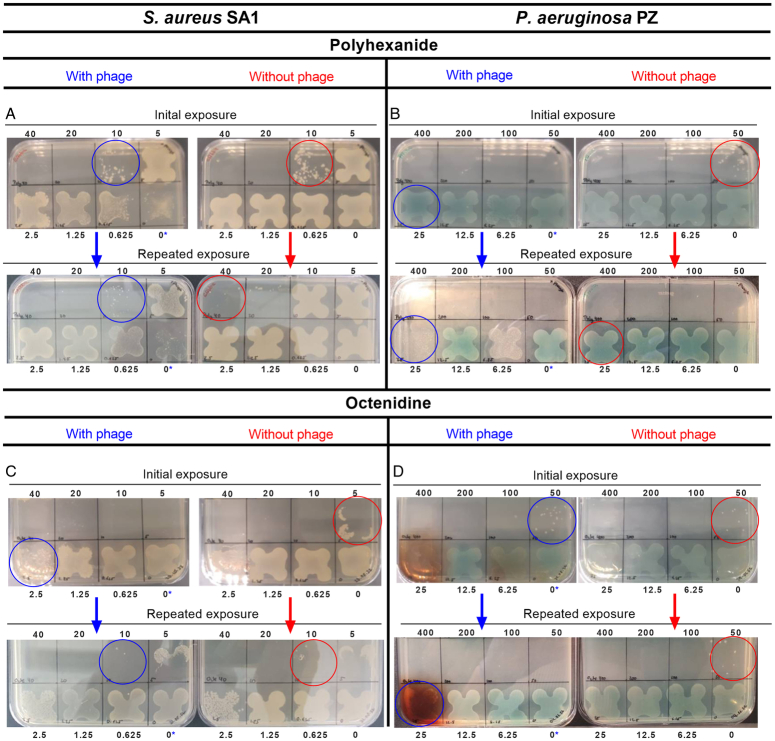
The potential of complete bacterial eradication in liquid culture was tested over a broader antiseptic range (i.e. from 0 to 40 µg/ml and from 0 to 400 µg/ml for strains SA1 and PZ, respectively). A complete eradication of the S. aureus strain SA1 was achieved with 20 µg/ml of polyhexanide with or without phage (Fig. 4A, upper plates). Bacteria surviving the next lower amount of polyhexanide, i.e. 10 µg/ml (Fig. 4A, upper plates, blue, and red circles), were re-exposed to all antiseptic concentrations in liquid culture with and without the phage. Subsequent plating showed that SA1 now survived an antiseptic concentration of 40 µg/ml, without phage (Fig. 4A, lower plates, red circle). However, with the antiseptic/phage combination bacteria remained sensitive and could only survive a concentration of 10 µg/ml (Fig. 4A, lower plates, blue circle).
Figure 4.
Assessment of retained bacterial viability. Bacteria were grown on antiseptic-free and phage-free solid media after two rounds of exposure to polyhexanide (A and B) and octenidine (C and D) with or without the phage Sb-1 (S. aureus strain SA1) or phage NP3 (P. aeruginosa strain PZ) in liquid media for 22 h. Exposure in liquid media was done with twofold serial dilutions of the antiseptics, starting with 40 (S. aureus) or 400 µg/ml (P. aeruginosa), respectively. The phage MOI was 0.1. The upper plates in A to D show the plating results after the first round of exposure. Bacteria surviving the highest concentrations (blue and red circles) were used for the second round of exposure in liquid media, again with all dilutions with and without the phage, as in round 1. The lower plates in A to D show the plating results after the second round of exposure. Bacteria surviving the highest concentrations are again marked by blue and red circles. The numbers above and below the plates indicate the concentrations of the antiseptics in the former liquid culture assays (note, that the zero with a blue star indicates that bacteria were exposed in liquid culture to the phage alone).
P. aeruginosa strain PZ was completely eradicated with phage NP3 combined with polyhexanide at a concentration of 50 µg/ml, while this was achieved with polyhexanide alone at 100 µg/ml (Fig. 4B, upper two plates). With the second exposure both approaches achieved eradication with 50 µg/ml of the antiseptic. However, bacterial density was markedly lower in the phage/antiseptic combination compared to the antiseptic-only approach at 25 µg/ml (Fig. 4B, lower plates red and blue circles).
The use of the octenidine/Sb-1 combination was initially more effective in eradicating SA1, compared to the antiseptic alone (i.e. eradication at 5 µg/ml vs. 10 µg/ml, Fig. 4C, upper plates). During the second exposure, an increase in tolerance with both approaches occurred, as bacteria survived now 10 µg/ml octenidine (Fig. 4C, lower plates, blue and red circles), which means that in this case there was no beneficial effect mediated by phage Sb-1.
The initial challenge of strain PZ by octenidine with or without the phage NP3 was equally effective (i.e. bacteria survived a concentration of 50 µg/ml, Fig. 4D, upper plates, blue, and red circle). Repeated exposure did not show tolerance development with octenidine alone (Fig. 4D, lower plates, red circle). However, with the octenidine/NP3 combination bacteria became more sensitive in the repeated exposure surviving only 25 µg/ml of octenidine (Fig. 4D, lower plates, blue, and red circles).
Discussion
In the present study, various bacteria found in chronic and hard-to-heal wounds were investigated by exposure to low-level concentrations of polyhexanide and octenidine. While in-use concentrations of antiseptics are high enough to exert a bactericidal effect, little is known about the consequences when bacteria get into contact with sub-inhibitory concentrations. This could happen in chronic wound biofilms where drug penetration might be hampered30–33. Other factors, such as dilution of the antiseptics through wound fluids or simple physical factors such as evaporation, drying or dislocation of the wound disinfectant-loaded dressing add to uncertain contact of the antiseptic at the wound34. Bacteria were able to grow at 0.2–10% of clinical in-use formulations but this was characterized by a large interspecies and/or interstrain variability between polyhexanide and octenidine. This fluctuating antimicrobial efficacy underscores the need for a scientifically based antiseptic therapy in wound care. While clear statements in favor of a particular antiseptic may be established for bacteria with a consistent susceptibility against bacterial species (e.g. polyhexanide against E. coli), more sophisticated measures are recommended in bacterial species with high levels of variability such as P. aeruginosa.
There is also an emerging view that bacteria can adapt to antiseptics similar to antibiotics12,35–37. Such adaptation could occur during clinical practice or beyond therapeutic settings, for example, in water sink systems38, an otherwise known reservoir of antibiotic resistance genes39–41. Even worse, adaptation to antiseptics can also lead to cross-resistance to antibiotics, for example, by modification of the membrane or cell wall barrier35,38,42–46 with inconceivable clinical consequences particularly in the severely ill patient. In addition, exposure to low-level concentrations of biocides might promote horizontal transfer of multidrug resistance genes, biofilm formation and the increase of the genetic mutation frequency47 further adding to the importance of a scientifically sound application of antiseptics.
In our study, we exposed bacteria consecutively to either constant or gradually increasing low-level concentrations of antiseptics. Both modes are relevant in the clinical scenario of wound treatment as in common practice the identical wound antiseptic is applied repetitively and often with increasing frequency for the same wound.
Interestingly, adaptation to constant low-level concentrations of antiseptics followed by a subsequent increase of the MIC did occur without mutations. Such phenotypical adaptation may be reversible as soon as the administration of the antiseptic is discontinued. However, repeated antiseptic administrations in wound biofilms could mean the foundation for further adaptation towards stronger antiseptic concentrations linked with stable mutations.
Adaptation to increasing concentrations of antiseptics occur frequently with gene mutations regulating the activity of efflux pumps. For instance, P. aeruginosa adapting to octenidine showed mutations in the gene smvR modulating smvA expression and lipid membrane modification37. Adaptation of P. mirabilis to chlorhexidine and octenidine has also been associated with mutations in smvR 48. In our study, no mutation in smvR were observed in P. aeruginosa, but directly in the gene smvA, which corroborates the importance of this efflux pump system across different species and strains for repelling antiseptic-induced stress49. The additional mutations found in the octenidine adapted strain PZ affected the genes opgH and kinB leading to amino acid changes in the glycosyltransferase H and the alginate biosynthesis sensor protein KinB. Both genes are involved in forming mucoid biofilms, for example, in CF patients, making P. aeruginosa more resistant against antibiotics and the immune system50–52.
The MRSA strain SA1 did not adapt to increasing levels of octenidine aligning with the findings of another in vitro study53. However, after introduction of octenidine for MRSA decolonization in a hospital trust in the UK, S. aureus isolates with reduced susceptibility have emerged36. This indicates that failure to adapt in vitro does not preclude the potential of bacteria to adapt under environmental, ‘real-world’ conditions.
Nevertheless, strain SA1 could adapt to polyhexanide which came along with one de novo mutation in the gene mprF encoding for the phosphatidylglycerol lysyltransferase MprF. Mutations in this enzyme seem to be a universal response of S. aureus to oppose polyhexanide-induced stress, which also leads to cross-resistance against daptomycin54–56, similarly to what has been observed in our study.
One approach to battle most recalcitrant wound infections may be the concomitant use of lytic phages, considering the recognized synergistic antibacterial effects of antibiotics combined with phages57,58. To the best of our knowledge, positive interactions between antiseptics (at low concentration) and phages against wound pathogens have not been described so far. Our study shows an enhanced antibacterial control and a delay in antiseptic adaptation. Further studies involving other bacterial species and phages are therefore warranted. Instead of simultaneous use, phages could be applied first to breakdown the wound biofilm thereby increasing the antibacterial efficacy of the subsequent antiseptic treatment. This order strategy has been described for antibiotics and disinfectants, where bacterial control was stronger, when phages were given beforehand23,59.
A limitation of this study is that it primarily focuses on the antibacterial efficacy of polyhexanide and octenidine in a controlled in vitro setting which does not account for all challenges related to the actual clinical situation found in wounds. For instance, chronic wounds are often polymicrobial60,61 and other confounders need to be considered such as cytotoxicity62, biofilm penetration and breakdown capabilities63 along with individual patients’ conditions, for example, hyperglycemia which promotes biofilm formation and impairs wound healing64,65. However, while those aspects are beyond the scope of the present study, our in vitro results provide basic knowledge needed for designing meaningful in vivo experiments66,67. In order to ensure the overall clinical validity and generalizability, our findings should be corroborated through clinical randomized controlled trials including the investigation of bacterial antiseptic tolerance development in polymicrobial wound infections, potential cross-resistance against antibiotics, and the additional antimicrobial value provided by lytic phages68,69. The use of phages may become particularly important in light of the continuing rise and spread of multidrug resistant bacteria70.
It is also noteworthy, that in our study, bacteria from wounds irrespective of their anatomical area were included as our focus was on the chronicity rather than location. Clearly, in the clinical situation, the anatomical site is a highly relevant factor as bacterial species (e.g. in pressure ulcers, diabetic foot ulcers, and venous leg ulcers) and therapeutic strategies may vary vastly2,71. This means, that future in vivo studies need to account for the features of the individual locations and for the bacterial species typically present in the chronic wounds. Finally, antiseptic solutions are only one element of a multidimensional treatment plan of complex chronic wounds and cannot replace other key components. Wound antiseptics complement a multifaceted network consisting of proper debridement, which can range from less invasive solutions (e.g. enzymatic, biological, and mechanical) to surgical debridement72, regular wound assessment and application of various dressings, advanced therapies such as hyperbaric oxygen therapy, patient positioning, nutrition, and suitable surgical wound coverage2.
Concluding remarks
Our results underline the possible emergence of bacterial tolerance/resistance in the clinical setting in response to recurrent utilization of antiseptics. Therefore, a more nuanced choice of available antiseptics based on results of wounds swabs in close collaboration with microbiologists represents a facile solution surgeons may embark on.
The change of antiseptics may be a potential solution to counteract bacterial adaptation and may also account for the fact that different bacterial species and/or strains respond unequally to low-level antiseptic concentrations.
Notably, bacteria can change phenotype or genotype in order to withstand repeated exposures to antiseptics. This adaptation can induce cross-resistance against clinically relevant antibiotics, the consequences of which should be addressed by in vivo investigations. In addition, research should be intensified to device strategies for combined approaches employing lytic phages and antiseptics for increasing our antibacterial power against bacterial wound pathogens.
Ethical approval
Ethical approval was obtained by the Ethics Committee of the RWTH Aachen University Faculty of Medicine, Aachen, Germany; (EK 077/18).
Consent
Not applicable.
Sources of funding
This work was supported by a grant of the Volkswagenstiftung (grant number 93726, recipient Bong-Sung Kim). The study sponsor had no involvement in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Author contribution
B.S.K., H.P.H., C.S., J.P.B., and T.R.: conceptualization; B.S.K.: funding acquisition; T.L.T., P.M., K.S., A.K., and S.R.: experiments; T.L.T and H.P.H: writing – original draft preparation; H.P.H., T.L.T., B.S.K, J.P.B., C.S., and T.R.: writing – review and editing.
Conflicts of interest disclosure
The authors declare no conflict of interest.
Research registration unique identifying number (UIN)
Name of the registry: not applicable.
Unique identifying number or registration ID: not applicable.
Hyperlink to your specific registration (must be publicly accessible and will be checked): not applicable.
Guarantor
Bong-Sung Kim and Hans-Peter Horz.
Data availability statement
Datasets generated during this study are available on reasonable request.
Provenance and peer review
This paper was not invited.
Supplementary Material
Footnotes
Hans-Peter Horz and Bong-Sung Kim shared last authorship.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.lww.com/international-journal-of-surgery.
Published online 13 May 2024
Contributor Information
Thaysa Leite Tagliaferri, Email: ttagliaferri@ukaachen.de.
Sophie Rhode, Email: s.rhode@uke.de.
Priscila Muñoz, Email: priscila.munoz@rwth-aachen.de.
Kevin Simon, Email: k.simon@simontech.de.
Alex Krüttgen, Email: akruettgen@ukaachen.de.
Christian Stoppe, Email: christian.stoppe@gmail.com.
Tim Ruhl, Email: truhl@ukaachen.de.
Justus P. Beier, Email: jbeier@ukaachen.de.
Hans-Peter Horz, Email: jhorz@ukaachen.de.
Bong-Sung Kim, Email: Bong-Sung.Kim@usz.ch.
References
- 1. Carter MJ, DaVanzo J, Haught R, et al. Chronic wound prevalence and the associated cost of treatment in Medicare beneficiaries: changes between 2014 and 2019. J Med Econ 2023;26:894–901. [DOI] [PubMed] [Google Scholar]
- 2. Eriksson E, Liu PY, Schultz GS, et al. Chronic wounds: treatment consensus. Wound Repair Regen 2022;30:156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nobel FA, Islam S, Babu G, et al. Isolation of multidrug resistance bacteria from the patients with wound infection and their antibiotics susceptibility patterns: a cross-sectional study. Ann Med Surg (Lond) 2022;84:104895. doi: 10.1016/j.amsu.2022.104895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ruegsegger L, Xiao J, Naziripour A, et al. Multidrug-resistant gram-negative bacteria in burn patients. Antimicrob Agents Chemother 2022;66:e00688-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lachiewicz AM, Hauck CG, Weber DJ, et al. Bacterial infections after burn injuries: impact of multidrug resistance. Clin Infect Dis 2017;65:2130–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karakonstantis S, Kritsotakis EI, Gikas A. Pandrug-resistant gram-negative bacteria: a systematic review of current epidemiology, prognosis and treatment options. J Antimicrob Chemother 2020;75:271–282. [DOI] [PubMed] [Google Scholar]
- 7. Salas A, Williams MC, Van Etten E, et al. Prolonged perioperative antibiotics: a hidden problem. Hosp Pediatr 2022;12:125–132. [DOI] [PubMed] [Google Scholar]
- 8. Lim MK, Lai PSM, Ponnampalavanar SSLS, et al. Antibiotics in surgical wards: use or misuse? A newly industrialized country’s perspective. J Infect Dev Ctries 2015;9:1264–1271. [DOI] [PubMed] [Google Scholar]
- 9. Hersh AL, King LM, Shapiro DJ, et al. Unnecessary antibiotic prescribing in US ambulatory care settings, 2010–2015. Clin Infect Dis 2021;72:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caputo WJ, Monterosa P, Beggs D. Antibiotic misuse in wound care: can bacterial localization through fluorescence imaging help? Diagnostics 2022;12:3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serena TE, Gould L, Ousey K, et al. Reliance on clinical signs and symptoms assessment leads to misuse of antimicrobials: Post hoc analysis of 350 chronic wounds. Adv Wound Care 2022;11:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Dijk HFG, Verbrugh HA, Ad hoc advisory committee on disinfectants of the Health Council of the Netherlands . Resisting disinfectants. Commun Med 2022;2:6; Abee T, Andriessen JW, van Dijk HFG, u. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts CD, Leaper DJ, Assadian O. The role of topical antiseptic agents within antimicrobial stewardship strategies for prevention and treatment of surgical site and chronic open wound infection. Adv Wound Care 2017;6:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kramer A, Dissemond J, Kim S, et al. Consensus on wound antisepsis: update 2018. Skin Pharmacol Physiol 2018;31:28–58. [DOI] [PubMed] [Google Scholar]
- 15. Albayaty YN, Thomas N, Hasan S, et al. Penetration of topically used antimicrobials through Staphylococcus aureus biofilms: a comparative study using different models. J Drug Deliv Sci Technol 2018;48:429–436. [Google Scholar]
- 16. Bartoszewicz M, Rygiel A, Krzemiński M, et al. Penetration of a selected antibiotic and antiseptic into a biofilm formed on orthopedic steel implants. Ortop Traumatol Rehabil 2007;9:310–318. [PubMed] [Google Scholar]
- 17. Bridier A, Briandet R, Thomas V, et al. Resistance of bacterial biofilms to disinfectants: a review. Biofouling 2011;27:1017–1032. [DOI] [PubMed] [Google Scholar]
- 18. Stuermer EK, Besser M, Brill F, et al. Comparative analysis of biofilm models to determine the efficacy of antimicrobials. Int J Hyg Environ Health 2021;234:113744. [DOI] [PubMed] [Google Scholar]
- 19. Hübner NO, Kramer A. Review on the efficacy, safety and clinical applications of polihexanide, a modern wound antiseptic. Skin Pharmacol Physiol 2010;23:17–27. [DOI] [PubMed] [Google Scholar]
- 20. Hübner NO, Siebert J, Kramer A. Octenidine dihydrochloride, a modern antiseptic for skin, mucous membranes and wounds. Skin Pharmacol Physiol 2010;23:244–258. [DOI] [PubMed] [Google Scholar]
- 21. Spear M. Acute or chronic? What’s the difference? Plast Surg Nurs 2013;33:98–100. [DOI] [PubMed] [Google Scholar]
- 22. Levine NS, Lindberg RB, Mason AD, et al. The quantitative swab culture and smear: a quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma 1976;16:89–94. [PubMed] [Google Scholar]
- 23. Chaudhry WN, Concepción-Acevedo J, Park T, et al. Synergy and order effects of antibiotics and phages in killing pseudomonas aeruginosa biofilms. PLOS ONE 2017;12:e0168615; Rozen DE, Herausgeber. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simon K, Pier W, Krüttgen A, et al. Synergy between Phage Sb-1 and oxacillin against methicillin-resistant staphylococcus aureus. Antibiotics 2021;10:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCallin S, Sarker SA, Sultana S, et al. Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo‐controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ Microbiol 2018;20:3278–3293. [DOI] [PubMed] [Google Scholar]
- 26. Gurevich A, Saveliev V, Vyahhi N, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 28. Chindera K, Mahato M, Kumar Sharma A, et al. The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes. Sci Rep 2016;6:23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malanovic N, Ön A, Pabst G, et al. Octenidine: novel insights into the detailed killing mechanism of gram-negative bacteria at a cellular and molecular level. Int J Antimicrob Agents 2020;56:106146. [DOI] [PubMed] [Google Scholar]
- 30. Alves PJ, Barreto RT, Barrois BM, et al. Update on the role of antiseptics in the management of chronic wounds with critical colonisation and/or biofilm. Int Wound J 2021;18:342–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999;284:1318–1322. [DOI] [PubMed] [Google Scholar]
- 32. Fazli M, Bjarnsholt T, Kirketerp-Møller K, et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 2009;47:4084–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Donnell JA, Wu M, Cochrane NH, et al. Efficacy of common antiseptic solutions against clinically relevant microorganisms in biofilm. Bone Jt J 2021;103-B:908–915. [DOI] [PubMed] [Google Scholar]
- 34. Augustin M, Herberger K, Wille A, et al. Impact of human wound exudate on the bactericidal efficacy of commercial antiseptic products. J Wound Care 2023;32:422–427. [DOI] [PubMed] [Google Scholar]
- 35. Wand ME, Bock LJ, Bonney LC, et al. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother 2017;61:e01162–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hardy K, Sunnucks K, Gil H, et al. Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus . mBio 2018;9:e00894–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bock LJ, Ferguson PM, Clarke M, et al. Pseudomonas aeruginosa adapts to octenidine via a combination of efflux and membrane remodelling. Commun Biol 2021;4:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shepherd MJ, Moore G, Wand ME, et al. Pseudomonas aeruginosa adapts to octenidine in the laboratory and a simulated clinical setting, leading to increased tolerance to chlorhexidine and other biocides. J Hosp Infect 2018;100:e23–e29. [DOI] [PubMed] [Google Scholar]
- 39. Muzslay M, Moore G, Alhussaini N, et al. ESBL-producing gram-negative organisms in the healthcare environment as a source of genetic material for resistance in human infections. J Hosp Infect 2017;95:59–64. [DOI] [PubMed] [Google Scholar]
- 40. Amoureux L, Riedweg K, Chapuis A, et al. Nosocomial Infections with IMP-19−Producing Pseudomonas aeruginosa Linked to Contaminated Sinks, France. Emerg Infect Dis 2017;23:304–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. VanderElzen K, Zhen H, Shuman E, et al. The hidden truth in the faucets: a qualityimprovement project and splash study of hospital sinks. Am J Infect Control 2019;47:S26. [Google Scholar]
- 42. Sheldon AT. Antiseptic resistance: real or perceived threat? Clin Infect Dis 2005;40:1650–1656. [DOI] [PubMed] [Google Scholar]
- 43. Adkin P, Hitchcock A, Smith LJ, et al. Priming with biocides: a pathway to antibiotic resistance? J Appl Microbiol 2022;133:830–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davin-Regli A, Pagès JM. Cross-resistance between biocides and antimicrobials: an emerging question. Rev Sci Tech Int Off Epizoot 2012;31:89–104. [PubMed] [Google Scholar]
- 45. Russell A. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect Dis 2003;3:794–803. [DOI] [PubMed] [Google Scholar]
- 46. Kampf G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 2018;7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hemati S, Kouhsari E, Sadeghifard N, et al. Sub-minimum inhibitory concentrations of biocides induced biofilm formation in Pseudomonas aeruginosa. New Microbes New Infect 2020;38:100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pelling H, Bock LJ, Nzakizwanayo J, et al. Derepression of the smvA efflux system arises in clinical isolates of proteus mirabilis and reduces susceptibility to chlorhexidine and other biocides. Antimicrob Agents Chemother 2019;63:e01535–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wand ME, Jamshidi S, Bock LJ, et al. SmvA is an important efflux pump for cationic biocides in Klebsiella pneumoniae and other Enterobacteriaceae. Sci Rep 2019;9:1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hentzer M, Teitzel GM, Balzer GJ, et al. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 2001;183:5395–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mah TF, Pitts B, Pellock B, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003;426:306–310. [DOI] [PubMed] [Google Scholar]
- 52. Lequette Y, Rollet E, Delangle A, et al. Linear osmoregulated periplasmic glucans are encoded by the opgGH locus of Pseudomonas aeruginosa. Microbiology 2007;153:3255–3263. [DOI] [PubMed] [Google Scholar]
- 53. Al-Doori Z, Goroncy-Bermes P, Gemmell CG, et al. Low-level exposure of MRSA to octenidine dihydrochloride does not select for resistance. J Antimicrob Chemother 2007;59:1280–1281. [DOI] [PubMed] [Google Scholar]
- 54. Renzoni A, Von Dach E, Landelle C, et al. Impact of exposure of methicillin-resistant staphylococcus aureus to polyhexanide In Vitro and In Vivo . Antimicrob Agents Chemother 2017;61:e00272–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wendel AF, Otchwemah R, Layer-Nicolaou F, et al. Investigating a possible link between antiseptic treatment and the increased occurrence of daptomycin-resistant Staphylococcus aureus. Clin Microbiol Infect 2023;29:1334.e1–1334.e6. [DOI] [PubMed] [Google Scholar]
- 56. Ernst CM, Slavetinsky CJ, Kuhn S, et al. Gain-of-function mutations in the phospholipid flippase MprF confer specific daptomycin resistance. mBio 2018;9:e01659–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Torres-Barceló C, Hochberg ME. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol 2016;24:249–256. [DOI] [PubMed] [Google Scholar]
- 58. Tagliaferri TL, Jansen M, Horz HP. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol [Internet] 2019;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stachler E, Kull A, Julian TR. Bacteriophage treatment before chemical disinfection can enhance removal of plastic-surface-associated pseudomonas aeruginosa. Appl Environ Microbiol 2021;87:e00980–21; McBain AJ, Herausgeber. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14:244–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dalton T, Dowd SE, Wolcott RD, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PloS One 2011;6:e27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim BS, Ott V, Boecker AH, et al. The effect of antiseptics on adipose-derived stem cells. Plast Reconstr Surg 2017;139:625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Percival SL, McCarty SM, Lipsky B. Biofilms and wounds: an overview of the evidence. Adv Wound Care 2015;4:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burgess JL, Wyant WA, Abdo Abujamra B, et al. Diabetic Wound-Healing Science. Medicina (Mex) 2021;57:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dhall S, Do DC, Garcia M, et al. Generating and reversing chronic wounds in diabetic mice by manipulating wound redox parameters. J Diabetes Res 2014;2014:562625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. MacGowan A, Rogers C, Bowker K. In vitro models, in vivo models, and pharmacokinetics: what can we learn from in vitro models? Clin Infect Dis 2001;33:S214–S220. [DOI] [PubMed] [Google Scholar]
- 67. Ding X, Tang Q, Xu Z, et al. Challenges and innovations in treating chronic and acute wound infections: from basic science to clinical practice. Burns Trauma 2022;10:tkac014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Casey E, van Sinderen D, Mahony J. In vitro characteristics of phages to guide ‘real life’ phage therapy suitability. Viruses 2018;10:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pinto AM, Cerqueira MA, Bañobre-Lópes M, et al. Bacteriophages for chronic wound treatment: from traditional to novel delivery systems. Viruses 2020;12:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brives C, Pourraz J. Phage therapy as a potential solution in the fight against AMR: obstacles and possible futures. Palgrave Commun 2020;6:100. [Google Scholar]
- 71. Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 2008;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thomas DC, Tsu CL, Nain RA, et al. The role of debridement in wound bed preparation in chronic wound: a narrative review. Ann Med Surg 2021;71:102876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets generated during this study are available on reasonable request.