Abstract
Attenuated simian immunodeficiency viruses (SIVs) have been described that produce low levels of plasma virion RNA and exhibit a reduced capacity to cause disease. These viruses are particularly useful in identifying viral determinants of pathogenesis. In the present study, we show that mutation of a highly conserved tyrosine (Tyr)-containing motif (Yxxφ) in the envelope glycoprotein (Env) cytoplasmic tail (amino acids YRPV at positions 721 to 724) can profoundly reduce the in vivo pathogenicity of SIVmac239. This domain constitutes both a potent endocytosis signal that reduces Env expression on infected cells and a sorting signal that directs Env expression to the basolateral surface of polarized cells. Rhesus macaques were inoculated with SIVmac239 control or SIVmac239 containing either a Tyr-721-to-Ile mutation (SIVmac239Y/I) or a deletion of Tyr-721 and the preceding glycine (ΔGY). To assess the in vivo replication competence, all viruses contained a stop codon in nef that has been shown to revert during in vivo but not in vitro replication. All three control animals developed high viral loads and disease. One of two animals that received SIVmac239Y/I and two of three animals that received SIVmac239ΔGY remained healthy for up to 140 weeks with low to undetectable plasma viral RNA levels and normal CD4+ T-cell percentages. These animals exhibited ongoing viral replication as determined by detection of viral sequences and culturing of mutant viruses from peripheral blood mononuclear cells and persistent anti-SIV antibody titers. In one animal that received SIVmac239Y/I, the Ile reverted to a Tyr and was associated with a high plasma RNA level and disease, while one animal that received SIVmac239ΔGY also developed a high viral load that was associated with novel and possibly compensatory mutations in the TM cytoplasmic domain. In all control and experimental animals, the nef stop codon reverted to an open reading frame within the first 2 months of inoculation, indicating that the mutant viruses had replicated well enough to repair this mutation. These findings indicate that the Yxxφ signal plays an important role in SIV pathogenesis. Moreover, because mutations in this motif may attenuate SIV through mechanisms that are distinct from those caused by mutations in nef, this Tyr-based sorting signal represents a novel target for future models of SIV and human immunodeficiency virus attenuation that could be useful in new vaccine strategies.
Infection by human immunodeficiency viruses (HIVs) is characterized by persistent viral replication that in most individuals in the absence of treatment leads to a progressive and irreversible decline in the number of CD4+ lymphocytes and eventually to AIDS (33). Although host immune responses and genetic factors can influence the level of virus replication (10, 12, 13, 77, 81), viral determinants undoubtedly play key roles in affecting the development of disease (2, 25, 48, 60). The related simian immunodeficiency viruses (SIVs) have provided powerful animal models to determine the contribution of viral and host factors to disease progression (53, 92). Interestingly, a number of SIVs that are markedly attenuated in vivo have been derived. These viruses characteristically establish low to undetectable plasma viral RNA levels and cause limited depletion of CD4+ lymphocytes (3, 42). Although some attenuated viruses have been reported to cause disease in neonatal animals (4, 5) and in some adults after prolonged infection (4, 5, 28, 89), they have been extremely useful in identifying viral determinants for virulence in vivo. Moreover, animals infected with attenuated SIVs have, in some cases, been protected from infection and/or disease when challenged intravenously (24) or mucosally (22, 44, 66, 72) with more pathogenic homologous (24, 56, 79) or heterologous (15, 103) strains. Thus, these viruses may provide important clues for determining the nature of protective immunity for SIV and HIV.
Mechanisms responsible for HIV and SIV attenuation in vivo could be due to (i) intrinsic defects in virus infectivity and/or replicative capacity, (ii) reduced cytopathic effects of viral infection on target cells, and/or (iii) increased susceptibility of the virus to a host immune response. Among the attenuated viruses studied to date, the best characterized are those containing mutations in the viral nef gene (42, 46). This gene encodes a 27-kDa myristoylated protein that has been associated with a number of biological effects, which include (i) downregulating CD4 and major histocompatibility complex class I molecules (1, 17, 19, 21, 37, 39, 59, 75, 85), (ii) augmenting virus infectivity (14, 26, 58, 67, 90), and (iii) altering T-cell signaling pathways and/or development (6, 11, 37, 40, 83, 87, 100, 104). Consequently, Nef could affect viral pathogenesis through multiple mechanisms (29). In addition, other studies have demonstrated that Nef-deleted viruses can be rendered more attenuated by deletions of other accessory genes, such as vpr, vpx, and vif (30, 44). Another well-characterized SIV variant, SIVmac1A11, exhibits a markedly attenuated phenotype that appears to involve determinants in multiple regions of the genome (63). Interestingly, given the importance of the viral structural genes for infectivity, it is remarkable that few examples exist in which specific mutations in these genes are associated with reduced pathogenicity. This observation could result from the high viral mutation rate, creating reversions or compensatory mutations that can be selected over time to generate progressively more fit viruses (16, 47, 94). Alternatively, mutations in structural genes may be poorly tolerated due to their deleterious effects on viral replication. Nonetheless, it is reasonable to predict that mutations in important functional domains of SIV and HIV structural proteins would have significant consequences for pathogenesis and could, depending on their mechanism of action, be combined with mutations in accessory genes in strategies to produce viruses with reduced virulence.
In the present study we evaluated the in vivo effects of mutations in a highly conserved tyrosine (Tyr)-dependent sorting motif in the SIV Env cytoplasmic tail. For SIV and HIV, this Yxxφ motif (where Y is a Tyr, x is any amino acid, and φ is an amino acid with a bulky hydrophobic side chain) (61, 69, 84) has been shown in vitro to constitute both a potent endocytosis signal (8, 74, 78, 82) and a basolateral sorting signal (27, 54, 55). These signals are analogous to those in cellular proteins that are constitutively endocytosed from the plasma membrane, where binding of the Yxxφ to μ2 chains of AP2 adapter complexes recruits cell surface proteins into clathrin-coated pits (7, 8, 74). Interactions with other adapter proteins, AP1 in particular, probably underlie the ability of this motif to direct Env expression to the basolateral surface of polarized cells (27, 54, 55). We (82) and others (7, 27, 78) have demonstrated that this membrane-proximal motif can modulate the surface expression of Env on infected cells by recruiting Env glycoproteins that are not incorporated into virions into clathrin-coated pits. We have proposed that this motif could function in vivo to reduce the susceptibility of infected cells to host immune responses (62). Alternatively, the basolateral function of this signal could influence pathogenesis by facilitating cell-to-cell infection and/or directing the spread of virus to key anatomic areas (27). Here we report that disruption of the membrane-proximal Yxxφ motif in the pathogenic molecular clone SIVmac239 results in viruses that are able to replicate in vitro but are markedly attenuated in their ability to sustain high viral loads in vivo. These novel findings indicate the importance of this highly conserved Env sorting signal in SIV (and probably HIV) pathogenesis. Mutations in this signal could be useful in complementary strategies to attenuate SIV and HIV through mechanisms independent of those mediated by viral accessory genes.
MATERIALS AND METHODS
Construction of recombinant viruses.
SIVmac239/nef-stop containing mutations in the Yxxφ motif, where Y is at amino acid position 721 of the SIVmac239 sequence, were produced using pVP-1 and pVP-2 plasmids as previously described (50). The mutagenesis primers were 5′-TTAAGGCAGGGGATTAGGCCAGTGTTCC-3′ for the Y721I mutation and 5′-TTAAGGCAGAGGCCAGTGTTCTC-3′ for the ΔGY mutation (Fig. 1). To produce viruses, pVP-1 and pVP-2 were codigested with SphI, ligated with T4 ligase, linearized with ApaI, and electroporated into CEMx174 cells. Virus was harvested when cytopathic effects were observed, and the stocks were quantified by measuring reverse transcriptase activity (50).
FIG. 1.
Partial amino acid sequences of the membrane-spanning and cytoplasmic domains of SIVmac239 and Tyr-721 mutants. Tyr-721 is indicated by the arrow. Residues that are critical to the formation of the endocytosis signal include Gly-720, Tyr-721, and Val-724 (8, 9).
Infection of macaques.
Eight juvenile rhesus macaques (Macaca macaca), seronegative for antibodies to SIV, simian T-cell leukemia virus, simian type D retroviruses, and herpesvirus type B, were used in this study. Macaques were housed in isolation facilities at the University of Alabama at Birmingham in accordance with institutional and Animal Welfare Act guidelines. Before inoculation of virus or collection of blood samples, the macaques were anesthetized by intramuscular injection of ketamine hydrochloride (10 mg/kg), weighed, and examined for lymphadenopathy or other signs of disease. The macaques were randomly placed in three groups and inoculated intravenously with approximately 1,000 50% tissue culture infective doses of either SIVmac239/nef-stop (three macaques) or one of two SIVmac239/nef-stop env mutant viruses: two macaques were inoculated with the Y721I cytoplasmic tail mutant, and three macaques were inoculated with the ΔGY mutant. After virus inoculation, blood samples were collected from each animal at regular intervals for hematologic analysis, to isolate and quantify virus, and for serologic assays. The macaques were euthanized when moribund, and complete necropsies were performed.
In vitro replication of virus.
Macaque peripheral blood mononuclear cells (PBMC) were separated from whole blood by centrifugation through lymphocyte separation medium and then stimulated with concanavalin A at 106 cells/ml in RPMI 1640 medium containing antibiotics and 10% fetal bovine serum (10% medium). After 3 days, the PBMC were washed and aliquots of 107 cells were infected for 2 h with 1,000 to 3,000 50% tissue culture infective doses of the different virus strains before being washed and placed in 10% medium containing 8 U of interleukin-2/ml. Viral stocks were titrated by limiting dilution and infection of CEMx174 cells. Every 4 to 5 days, approximately 30% of the culture medium was removed, assayed for reverse transcriptase activity, and replaced with fresh medium.
Detection and quantification of virus.
To isolate virus, macaque PBMC were cultured with phytohemagglutinin-stimulated normal human PBMC; culture supernatants were tested every 4 to 5 days for reverse transcriptase activity using standard procedures. The cultures were monitored 6 to 7 weeks before being identified as virus negative. Two assays were used to quantify plasma virion RNA: a branched-DNA (bDNA) signal amplification assay specific for SIV (performed at Chiron, now Bayer Corp.) was used with plasma containing heparin, and a subset of EDTA-treated plasma samples were assayed using a real-time reverse transcription-PCR (RT-PCR) assay (93). The results of the two assays were comparable.
Serum antibodies.
Titers of antibodies specific for SIVmac239 were determined on serum samples after twofold serial dilutions using a highly cross-reactive HIV-2 enzyme immunoassay kit (Sanofi Pasteur Diagnostics, Seattle, Wash.). A CEMx174 cell-killing assay was used to determine neutralizing-antibody titers against either H9 cell line-adapted SIVmac251 or SIVmac239/nef-open grown in rhesus macaque PBMC (68).
Flow cytometry.
To monitor changes in lymphocyte subsets, EDTA-treated blood samples were stained by a whole-blood lysis procedure with fluorochrome-labeled mouse anti-human monoclonal antibodies with high cross-reactivity to macaque cell surface antigens. All antibodies, reagents, and the FACS-STAR flow cytometer on which the analyses were performed were purchased from Becton Dickinson (Mountain View, Calif.). Lymphocytes were gated according to forward-scatter–versus–side-scatter characteristics, with negative cell populations excluded after identification with isotype-matched control antibodies.
PCR amplification and sequence analyses.
Nested PCR of portions of the env TM and nef genes was performed with 1 μg of DNA from macaque PBMC obtained at various times after infection. Fragments of 393 and 310 bp, respectively, of the env TM were amplified with the following sets of primers: outer, 239env-I (5′-TAGAGGAGG-CACAAATTCAACAAG-3′) (bases 8823 to 8846) and 239env-II (5′-TGCTGAATAGCCAA-GTCAAGAG-3′) (bases 9215 to 9194); and inner, 239env-III (5′-GAATTACAAAAGTTG-AATAGCTGGG-3′) (bases 8861 to 8885) and 239env-IV (5′-AATGAATATATTCTATCTG-CCAAGG-3′) (bases 9170 to 9146). For the nef gene, fragments of 505 and 389 bp were amplified with the following sets of primers: outer, 239nef-I (5′-CTCTTAGGAGAGGT-GGAAGATGG-3′) (bases 9423 to 9445) and 239nef-IV (5′-GAGCTGGATGCATTAAATAA-TGCTC-3′) (bases 9927 to 9903); and inner, 239nef-II (5′-TTGAGCTCACTCTCTTGTGA-GGG-3′) (bases 9480 to 9502) and 239nef-III (5′-GGGACTAATTTCCATAGCCAGCC-3′) (bases 9868 to 9846). The positions of all primers are those of SIVmac239. PCR products were purified, cloned into the PCR vector (TA cloning kit; Invitrogen, San Diego, Calif.), and transfected into competent Escherichia coli cells. DNA from selected clones was prepared and sequenced using the Sequenase version 2 kit (U.S. Biochemicals, Cleveland, Ohio). PCR of the env TM cytoplasmic tail was also performed using the outside primers listed above on genomic DNA from cultures in which cell-free viruses had been serially passaged six times in CEMx174 cells.
Competition studies of viral fitness. (i) Preparation of infected-cell and virus stocks.
Infectious virions were produced by calcium phosphate transfection of HEK293T cells. The 5′ and 3′ hemigenomes of SIVmac239 (or specific mutant derivatives) were linearized with SphI, purified by phenol-chloroform extraction, and resuspended in water prior to transfection (50). At 24 h posttransfection, the supernatant was clarified by centrifugation, filtered through a 0.2-μm-pore-size filter, and incubated with 4 × 106 CEMx174 cells in the presence of Polybrene (8 μg/ml) for 6 h at 37°C. The cultures were washed once with phosphate-buffered saline and maintained by adding fresh cells and/or medium as appropriate.
(ii) Fitness assay.
For the competitions between SIVmac239 and SIVmac239ΔGY, infected CEMx174 cultures were established with the respective virus strains. Different ratios of infected cells from each culture were mixed with uninfected CEMx174 cells, and uininfected cells were added daily. For analysis of DNA, 106 cells were lysed in 500 μl of PCR lysis buffer (0.001% sodium dodecyl sulfate, 0.001% Triton X-100, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) with proteinase K (1 mg/ml), incubated at 58°C for 1 h, and heat inactivated at 95°C for 10 min. A 25-μl sample was for PCR amplification of env regions with primers oJD11 and oJD13 (sequences are given in Table 1). A 5-μl volume of reaction products was analyzed on a 3.5% MetaPhor agarose gel. For fluorescence resonance energy transfer (FRET) analysis, the cell lysates were further cleaned by phenol-chloroform extraction and resuspended in 500 μl of distilled water. Fluorescent PCR was performed with the LightCycler (Roche Molecular Biochemicals) as specified by the manufacturer, with FRET probes purchased from GenSet Oligos. Briefly, the 20-μl PCR mixes contained 2 μl of cell lysate, 0.2 μM M1 probe, 0.4 μM A1 probe, 0.3 μM oJD32 primer, 0.3 μM oJD33 primer, 3 mM MgCl2, and 2 μl of DNA-Hybridization Probe Master Mix (Roche Molecular Biochemicals). The lysates were subjected to 50 cycles of amplification (95°C for 0 s, 52°C for 10 s, 72°C for 21 s) followed by melting-curve analysis. To generate the melting curves, fluorescence was monitored continuously as the reaction temperature was increased from 43 to 95°C at a rate of 0.1°C/s.
TABLE 1.
Primers and probes used in this study
| Primer or probe | Sequencea |
|---|---|
| oJD11 primer | 5′-ttggtttgaccttgcttcttgg-3′ |
| oJD13 primer | 5′-ggaaataagagggtggggaagag-3′ |
| oJD32 primer | 5′-cttggggatgtgcgtttag-3′ |
| oJD33 primer | 5′-agaggcgtatcagttggcg-3′ |
| A1 probe | 5′-(RED 640)-cttccccaccctcttatttccagca-PO4-3′ |
| M1 probe | 5′-caggggtataggccagtgttc-(fluorescein)-3′ |
| A2 probe | 5′-(RED 640)-tatttccagcagacccatatccaacag-PO4-3′ |
| M2 probe | 5′-gtgttctcttccccaccctc-(fluorescein)-3′ |
Bold type indicates the site of mutation.
For competitions between SIVmac239ΔGY and SIVmac239Y/I and between SIVmac239ΔGY and SIV-7-14, infected cells, mixed cultures, and lysates were prepared as described above. The same primers, probes, and reaction mix were used for the SIVmac239ΔGY-versus-SIVmac239Y/I competition. For SIVmac239ΔGY versus SIV-7-14, the M1 and A1 probes were replaced with A2 and M2 probes, respectively, and 0.32 μl of the TaqStart anti-Taq polymerase antibody (Sigma) was added. For this competition the amplification parameters were altered (95°C for 0 s, 56°C for 5 s, and 72°C for 21 s).
RESULTS
Derivation and characterization of SIVs with mutations in the Yxxφ sorting motif.
To evaluate the effects of the conserved Yxxφ motif in an in vivo model, we constructed two mutants using a pathogenic molecular clone SIVmac239 (46) (Fig. 1). In one mutant, designated SIVmac239Y/I, the critical Tyr in the motif (YRPV) was changed to an isoleucine (Ile) by mutating two nucleotides in env codon 721 (TAT→ATT). In a second mutant, designated SIVmac239ΔGY, the Tyr and the preceding glycine (Gly) were removed by deleting the six nucleotides coding for these amino acids. Previous studies from our laboratory (9) and by Boge et al. (8) have shown that both Gly-720 and Tyr-721 are critical for the formation of a functional endocytosis signal. To monitor the replication competence of these viruses in vivo, we elected to use as the parental virus a SIVmac239 clone that contained a stop codon in nef at position 92. As shown previously, this premature termination codon characteristically reverts to an open reading frame within 2 to 4 weeks of inoculation of rhesus macaques, and within 1 year the majority of infected animals develop high viral loads, decreased numbers of CD4+ cells, and clinical AIDS (46). As a result of the rapid reversion of this nef point mutation, the clinical course of animals infected with this clone and the course of animals infected with SIVmac239 with an intact nef open reading frame are identical (46).
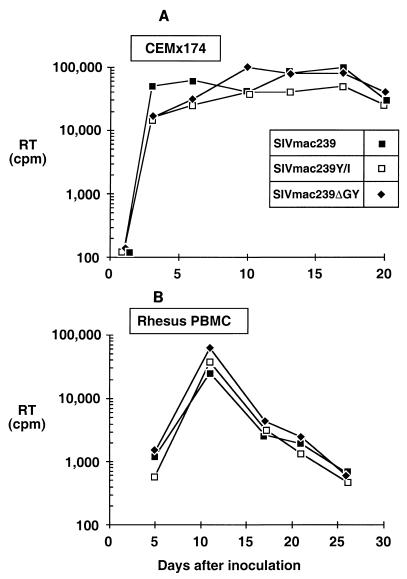
Using virus stocks generated by electroporation of CEMx174 cells, the replication competence of SIVmac239Y/I and SIVmac239ΔGY was compared to that of wild-type SIVmac239 in CEMx174 cells and PBMC from rhesus macaques (Fig. 2). Both mutants were infectious and yielded growth curves similar to that of wild-type SIVmac239. The stability of the mutations during in vitro propagation was also determined by multiple serial passages of these viruses in CEMx174 cells, followed by amplification and sequencing of env genes from genomic DNA. After as many as six passages in CEMx174 cells, the mutations introduced into SIVmac239Y/I and SIVmac239ΔGY remained intact and no new mutations were observed in the membrane-spanning domain or elsewhere in the TM cytoplasmic tail (data not shown). Thus, mutations that abrogated this Tyr-based sorting signal were stable in vitro and resulted in viruses that were replication competent in macaque PBMC and a human lymphoid cell line.
FIG. 2.
Growth curves of SIVmac239 and Tyr-721 mutants. Stocks of SIVmac239, SIVmac239Y/I, and SIVmac239ΔGY were normalized for RT activity and used to infect CEMx174 cells (A) or phytohemagglutinin-stimulated PBMC from rhesus macaques (B). Viral replication was quantified by serial determination of RT activity in culture supernatants. The experiments were performed twice for CEMx174 and three times for PBMC.
Inoculation of rhesus macaques with Tyr mutants.
The virus stocks of the wild-type and mutant SIVs generated by electroporation of CEMx174 cells were used to inoculate rhesus macaques intravenously. Two animals received SIVmac239Y/I, three received SIVmac239ΔGY, and three received wild-type SIVmac239. After inoculation, plasma viral RNA levels, percentages of CD4+ T cells, platelet counts, and antibody responses to viral antigens were monitored every 2 to 4 weeks. Among the three animals (K7K, N7K, and N2V) that received wild-type SIVmac239 containing the Nef point mutation, viral RNA levels in plasma peaked between 1.7 × 105 and 2.7 × 107 copies/ml during the first month of infection (Fig. 3). This acute phase was followed by establishment of viral “set points” that ranged between approximately 5 × 104 and 1 × 106 RNA copies/ml. All three animals exhibited declines in CD4+ T-cell percentages and platelet counts and were euthanized at weeks 70, 73, and 81 with weight loss and clinical features of immunodeficiency (Table 2).
FIG. 3.
Changes in viral burdens and hematologic parameters in macaques inoculated with SIVmac239 and Tyr-721 mutants. Animals designated by a dagger in the plasma RNA panels either died or were euthanized due to SIV-induced immunodeficiency. Plasma RNA levels were determined by the bDNA assay, which had levels of sensitivity of either 104 or 1,500 copies/ml, depending on the time at which the assay was performed. Some samples were tested by QC RT-PCR, for which the sensitivity was 300 copies/ml. All points below the dotted lines were negative by both assays, with the exception of two values marked by an asterisk. These values were 350 and 800 copies/ml, respectively, for AGP at 4 weeks and N8G at 20 weeks after infection.
TABLE 2.
Status of macaques infected with SIVmac239 and Tyr-721 mutants
| Virus | Animal | Outcome (wk)a | Clinical courseb |
|---|---|---|---|
| SIVmac239 | N2V | Died (81) | Weight loss, lymphadenopathy, B-cell leukemia, splenomegaly |
| K7K | Died (70) | Weight loss, pneumonia | |
| N7K | Died (73) | Weight loss, diarrhea | |
| SIVmac239Y/I | VF3 | Died (89) | Weight loss, pneumonia, glomerulonephritis |
| N8G | Alive (139) | No disease | |
| SIVmac239ΔGY | WD1 | Alive (120) | No disease |
| AGP | Alive (120) | No disease | |
| 2AJ | Died (77) | Weight loss, lymphadenopathy, pneumonia, pulmonary arteriopathy |
Outcome is shown as the number of weeks after inoculation when an animal died or at the end of the study.
The clinical course is shown for animals inoculated with wild-type SIVmac239 and SIVmac239 containing Y721I and the ΔGY mutation in the Env cytoplasmic domain. Weight loss indicates a loss of >10% of body weight.
Markedly different outcomes were observed for the two animals (VF3 and N8G) that received SIVmac239Y/I. Both exhibited peak viral loads of approximately 105 RNA copies/ml followed by a decline. In animal VF3, viral RNA levels subsequently rose to nearly 107 copies/ml and the percentages of CD4+ cells and platelet numbers progressively decreased (Fig. 3). This animal was euthanized at week 89 with wasting, pneumonia, and glomerulonephritis, which are all characteristic of severe immunodeficiency (Table 2). In contrast, after the initial peak of viremia, N8G showed a progressive decline in plasma viral RNA levels to below the limits of detection by the bDNA assay (<104 copies/ml) or a more sensitive quantitative competitive RT-PCR assay (<300 copies/ml) performed on selected samples (93). N8G also exhibited stable percentages of CD4+ T cells and numbers of platelets.
Differences in pathogenicity were noted among the three animals (WD1, AGP, and 2AJ) that received SIVmac239ΔGY. In WD1 and AGP, peak viral levels of 5.5 × 104 and 1.2 × 104 RNA copies/ml, respectively, were observed. Both of these animals have remained healthy with low or undetectable levels of RNA by the bDNA or the QC RT-PCR assay (Fig. 3) and have maintained normal hematologic values. Interestingly, the remaining animal (2AJ) exhibited a high peak level of 4.8 × 105 RNA copies/ml and an elevated viral set point of approximately 2 × 105 RNA copies/ml of plasma. This animal subsequently experienced a progressive increase in viral load to 2.8 × 106 RNA copies/ml and a progressive decline in CD4+ T cells and platelets and was euthanized at week 77 due to AIDS-defining conditions. In addition to levels of cell-free virus in plasma, the frequency with which virus is isolated from PBMC of SIV-infected macaques can be a direct reflection of the viral burden (41). Virus was isolated on 100% of attempts from the three macaques infected with the wild-type SIVmac239 as well as from macaque VF3, infected with the SIVmac239Y/I mutant. Likewise, 95.7% of virus isolation attempts were successful for macaque 2AJ, infected with the SIVmac239ΔGY mutant. That the last two animals were the only two mutant-virus recipients that maintained high plasma viral RNA levels and died of AIDS is consistent with the frequencies of virus isolation. In contrast, virus was successfully isolated from PBMC from the three surviving macaques in only 24, 33, and 40% of attempts.
Thus, of the five animals that received viruses with mutations in the Yxxφ motif, one of two animals infected with SIVmac239Y/I and two of three animals infected with SIVmac239ΔGY had clinical courses characterized by low to undetectable viral loads, stable CD4+ T-cell percentages, and no symptoms of disease. Similar to other published reports for SIV and HIV, the viral load was predictive of the clinical outcome (41, 47, 65, 73, 88, 101). After the initial peak of viremia, all five animals in which viral loads ultimately stabilized at >105 viral RNA copies/ml later developed immunodeficiency and other characteristics of SIV-related disease.
Antibody responses in infected macaques.
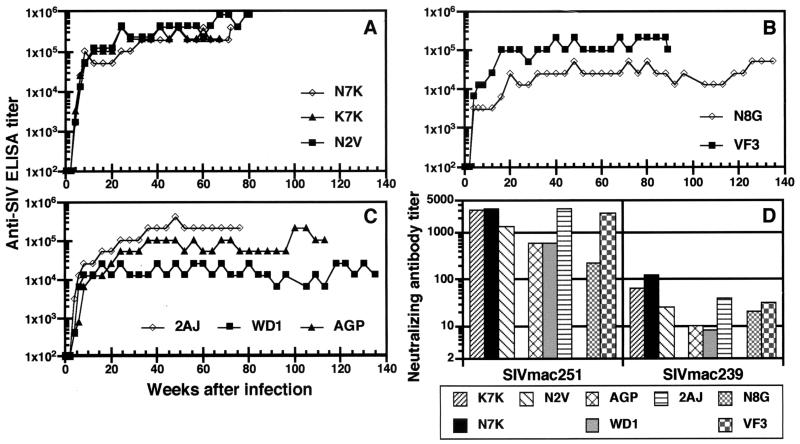
It was possible that the attenuated course of infection in the three animals that received mutant viruses but survived resulted from differences in host humoral immune responses. To test this hypothesis, serum antibody titers and the ability of antibodies to neutralize SIV were measured by enzyme-linked immunosorbent assay (ELISA) and a CEMx174 cell-killing assay, respectively. Within 4 weeks after inoculation, antibodies to SIV were detected in sera from all animals (Fig. 4A to C). Also within 4 weeks, all eight macaques had antibodies capable of neutralizing H9-adapted SIVmac251; however, until week 16 none had antibodies that neutralized the parental SIVmac239 grown in rhesus PBMC (Fig. 4D). Without exception, the highest titers in both the ELISA and neutralization assays were generated in the animals that had the highest viral loads and succumbed to disease. Thus, these results show that the failure of animals to develop disease was not due to a more efficient overall humoral immune response. However, despite the low to undetectable levels of plasma viral RNA in the surviving macaques, N8G, WD1, and AGP, these animals maintained antibody titers ranging from 1:25,600 to 1:102,400 for up to 139 weeks after inoculation, indicating ongoing viral replication (Fig. 4).
FIG. 4.
Humoral immune responses of macaques infected with SIVmac239 and Tyr-721 mutants. (A to C) Serum antibody titers after infection of macaques with SIVmac239 (A), SIVmac239Y/I (B), or SIVmac239ΔGY (C) are shown. The titers are expressed as the reciprocal of the highest dilution of serum yielding a value above the cutoff for the ELISA. (D) Neutralizing-antibody titers against H9-adapted SIVmac251 and SIVmac239 grown in rhesus PBMC are shown for serum samples collected 16 weeks after infection.
Molecular analysis of Env.
To determine the stability of the Y721I and ΔGY mutations in the inoculated animals, partial env sequences encompassing the TM membrane-spanning domain and the entire cytoplasmic domain were amplified by nested PCR from peripheral blood cellular DNA. In animals that received wild-type SIVmac239, only sporadic mutations were observed in clones from PBMC obtained 12 and 26 weeks after inoculation (Fig. 5A); no mutations involved the YRPV signal. Mutation of an Arg to a Gly at codon 751 in the cytoplasmic tail was observed consistently, however, indicating selection pressure at this position. The mutation also encodes a Lys-to-Arg substitution at codon 81 in Rev. Although the significance of this mutation is unclear, an identical change has been reported in other studies of SIVmac239 in rhesus macaques (34). Nonetheless, the lack of any mutations in the YRPV signal is consistent with the highly conserved nature of this domain (50, 62).
FIG. 5.
Molecular evolution of SIVs recovered from animals infected with the SIVmac239 and Tyr-721 mutants. Peripheral blood DNA from rhesus macaques infected with SIVmac239 (A), SIVmac239Y/I (B), or SIVmac239ΔGY (C) was extracted, amplified by PCR, cloned, and sequenced. Partial amino acid sequences of Env in cells from individual animals infected with each virus are shown. For panels B and C, the sequence of the mutant virus used for inoculation is shown relative to SIVmac239. Wk, weeks following inoculation; C, code designation for individual clones. Amino acid identity to SIVmac239 is indicated by a dash, deletions are indicated by a dot, and a stop codon (found only in week 14, clone 4, from AGP) is shown by an asterisk. In each panel, the YRPV sorting signal is underlined.
Of the two animals that received SIVmac239Y/I, clones from animal N8G, which remained healthy with a low viral load, showed persistence of the Y721I mutation in env sequences for up to 80 weeks (Fig. 5B). No other consistent changes in these clones were seen, although the R751G mutation, noted in the SIVmac239-inoculated animals, was present sporadically in clones from early time points. Interestingly, in animal VF3, which developed a high viral load and immunodeficiency, the mutation to Ile at position 721 had reverted to a Tyr in some clones obtained at 12 weeks after inoculation (Fig. 5B). Among 12 clones analyzed at this time, an Ile was present in 5 while a Tyr was present in 4. Remarkably, three clones exhibited a Phe at this position (codon TTT), probably reflecting a transitional intermediate as the Ile codon (ATT) reverted to a Tyr (TAT). We have showed previously that a Phe at this position can constitute a weak endocytosis signal in a reporter construct containing an SIVmac TM tail (82). As seen in the SIVmac239-infected animals, the R751G mutation was also found in the majority of clones from VF3. By 36 weeks and until its death at week 89, all sequences from VF3 contained a Tyr reversion at codon 721.
Among animals receiving SIVmac239ΔGY, viral sequences from both WD1 and AGP continued to show the GY deletion without other mutations for up to 56 and 52 weeks, respectively. However, although the ΔGY mutation persisted in all clones from animal 2AJ, which developed a high viral load and immunodeficiency, other mutations were noted, including Y696H in the membrane-spanning domain and a S727P and R746K in the cytoplasmic domain (Fig. 5C). Additional sporadic mutations seen in more than three clones included P723S, T789N, and H831L (data not shown), as well as the R751G mutation observed in other animals.
Therefore, in the two animals that received Y/I viruses, reversion of the mutation at position 721 was associated with a high viral load and disease while persistence of the Ile was associated with a markedly attenuated clinical course. In the three animals that received the ΔGY mutant virus, all maintained the GY deletion, with two exhibiting low viral loads. The remaining animal developed a high viral load and disease in association with novel mutations in the TM tail, raising the possibility that one or more of these changes could have compensated for the ΔGY mutation.
Evaluation of the Nef termination codon.
As noted above, to monitor the replication competence of viruses in vivo, animals were inoculated with SIVs that contained a premature termination codon in nef. We reasoned that if viruses could replicate in vivo, this codon would be repaired rapidly. Indeed, in all three control animals that received wild-type SIVmac239, the nef stop codon (TAA) reverted to an open reading frame by 4 to 8 weeks after inoculation (Table 3). Codons for a variety of amino acids were seen in this position with Glu (GAA or GAG) or Gln (CAA) predominating and Lys (AAA), Ser (TCA), Tyr (TAC), Leu (TTA), and Asp (GAC) observed less frequently. A stop codon in one of eight clones from animal N2V was still detectable at 8 weeks after inoculation. In one (VF3) of the two animals that received SIVmac239Y/I, the nef stop codon reverted to Glu, Tyr, or Gln in 11 of 11 clones by 2 weeks. In N8G, reversion was observed in 4 of 10 clones at 4 weeks, 6 of 6 clones at 12 weeks, and 7 of 10 clones at 80 weeks. Among the animals that received the ΔGY mutant, 11 of 11 clones from 2AJ had reverted to an open reading frame by 2 weeks whereas reversions were seen in clones from both WD1 and AGP at 2 and 8 weeks, respectively. By 44 and 25 weeks, all the nef clones had reverted in these animals. Thus, although stop codons in nef were still detected in clones from N8G at late time points, by 2 to 4 weeks after inoculation all the animals that remained healthy harbored viruses with clear evidence of correction of the nef termination codon. This finding indicated that the mutant viruses replicated sufficiently in vivo to repair nef and, more importantly, that a loss of Nef function could not account for the attenuated disease course in these animals.
TABLE 3.
Evaluation of the nef open reading frame in animals inoculated with Tyr-721 mutantsa
| Virus | Animal | Time (wk) | Total no. of clones | No. of clones with following amino acid encoded by nef codon 92:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stop (TAA) | Glu (GAA) | Ser (TCA) | Tyr (TAC) | Gln (CAA) | Lys (AAA) | Leu (TTA) | Glu (GAG) | Asp (GAC) | ||||
| SIVmac239 | N2V | 8 | 8 | 1 | 1 | 2 | 3 | 1 | ||||
| K7K | 8 | 9 | 3 | 1 | 2 | 1 | 2 | |||||
| N7K | 4 | 7 | 4 | 1 | 2 | |||||||
| SIVmac239Y/I | VF3 | 2 | 11 | 6 | 3 | 2 | ||||||
| N8G | 4 | 10 | 6 | 1 | 3 | |||||||
| N8G | 12 | 6 | 1 | 3 | 2 | |||||||
| N8G | 44 | 6 | 5 | 1 | ||||||||
| N8G | 56 | 7 | 3 | 2 | 1 | 1 | ||||||
| N8G | 80 | 10 | 3 | 3 | 4 | |||||||
| SIVmac239ΔGY | WD1 | 2 | 11 | 7 | 2 | 2 | ||||||
| WD1 | 44 | 11 | 9 | 1 | 1 | |||||||
| WD1 | 56 | 2 | 1 | 1 | ||||||||
| AGP | 4 | 9 | 9 | |||||||||
| AGP | 8 | 10 | 3 | 3 | 4 | |||||||
| AGP | 25 | 5 | 2 | 3 | ||||||||
| AGP | 52 | 5 | 1 | 4 | ||||||||
| 2AJ | 2 | 11 | 10 | 1 | ||||||||
DNA from peripheral blood lymphocytes of the animals indicated was amplified using nested primers that flanked the premature termination codon in nef at position 92 that was present in all viruses during the initial infection. Fragments were cloned into pCDNA and sequenced. Codon 92 and the predicted amino acids are shown at the indicated weeks postinoculation. nef reverted to an open reading frame in wild-type SIVmac239 and the Y/I and ΔGY mutants, although persistence of the stop codon was seen in some clones for animal N8G at 56 and 80 weeks.
Evaluation of replication competence in vitro using virus fitness assays.
Given the reduced viral RNA load in three of five animals that received mutant viruses, it was possible that the mutations had introduced subtle defects in viral replicative capacity that were not detectable in the initial growth curve experiments on PBMCs or CEMx174 cells (Fig. 1). To evaluate this possibility, viruses were compared in a series of fitness assays. Wild-type SIVmac239 and viruses containing the Y/I or ΔGY mutations were combined in different ratios to establish starting mixtures for each competition and added to CEMx174 cells. Cultures were replenished with uninfected cells (usually every 2 to 3 days) and maintained for 4 to 5 weeks. Total cellular DNA was isolated at various time points for analysis.
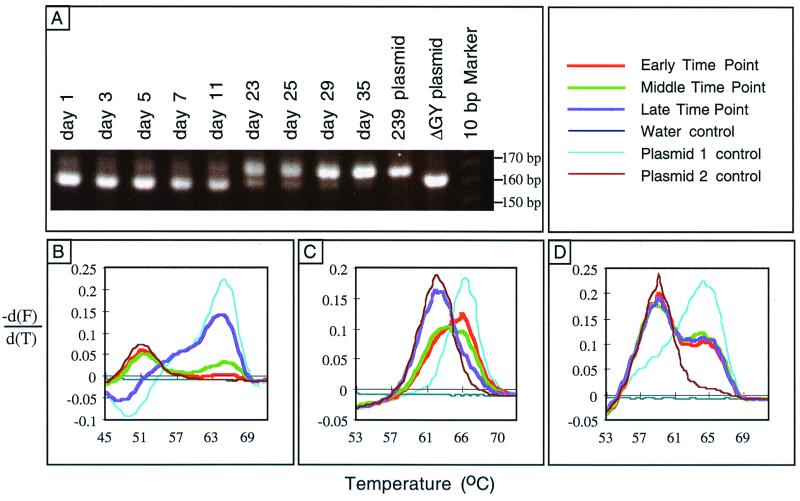
Initially, wild-type SIVmac239 was mixed with SIVmac239ΔGY at a starting ratio of ∼1:10. Serial DNA samples were evaluated by PCR amplification of the region of env that included the ΔGY mutation and analyzed by agarose gel electrophoresis. The 6-nucleotide deletion present in SIVmac239ΔGY yielded a fragment with higher electrophoretic mobility than that from the wild-type virus. As shown in Fig. 6A, although SIVmac239ΔGY was the predominant virus at early time points, by day 23 it was substantially replaced by wild-type viruses and by day 35 only SIVmac239 could be detected. No reversion of the ΔGY mutation was observed when SIVmac239ΔGY was passaged alone (data not shown). Therefore, in the context of this fitness assay, the ΔGY mutation did introduce a replication defect in SIVmac239.
FIG. 6.
Comparative evaluation of virus replication in competition assays in CEMx174 cells. (A) PCR analysis of SIVmac239 versus SIVmac239ΔGY competition. The env region was PCR amplified from cell lysates harvested on the indicated days. The larger band (158 bp) corresponds to the wild-type virus, and the smaller band (152 bp) corresponds to the mutant virus. (B) FRET analysis of SIVmac239 versus SIVmac239ΔGY competition. The samples from panel A were subjected to melting-curve analysis. Plasmid controls 1 and 2 correspond to SIVmac239 and SIVmac239ΔGY, respectively. The early, middle, and late time points are days 1, 11, and 35, respectively. (C) FRET analysis of SIVmac239ΔGY versus SIV-7-14 competition. Plasmid controls 1 and 2 correspond to SIVmac239ΔGY and SIV-7-14, respectively. The early, middle, and late time points are days 4, 7, and 17, respectively. (D) FRET analysis of SIVmac239 versus SIVmac239Y/I competition. Plasmid control 1 and 2 correspond to SIVmac239 and SIVmac239Y/I, respectively. The early, middle, and late time points are days 7, 17, and 30, respectively.
For subsequent experiments, we used an alternative PCR-based assay in which the products were amplified by primers spanning the mutation site and then subjected to FRET over a range of temperatures by using pairs of fluorescently labeled probes, one of which overlapped the mutation site (see Materials and Methods). In this analysis, the differential fluorescence, shown as a melting curve, reflected the proportional amounts of each amplified DNA species in the sample. When the same samples used in Fig. 6A were analyzed, the relative proportion of wild-type SIVmac239 (exhibiting a higher Tm) to SIVmac239ΔGY (exhibiting a lower Tm) increased over time until only SIVmac239 was detected on day 35 (Fig. 6B). Thus, this result corresponded precisely to that determined by gel analysis and validated the use of this technique for subsequent fitness assays.
Experiments were next performed on a derivative of SIVmac239ΔGY, designated 7-14, that contained the Y696H, P723S, S727P, R746K, R751G, T789N, and H831L mutations. These mutations had been noted in several env clones from 2AJ, the SIVmac239ΔGY-inoculated animal that developed a high virus load and disease (Fig. 5 and data not shown). Interestingly, in fitness assays, 7-14 possessed a clear replication advantage over SIVmac239ΔGY when these viruses were mixed at an initial ratio of ∼1:3 (Fig. 6C). Therefore, some or all of the env mutations that were acquired in vivo in 2AJ were able to improve the replication competence of SIVmac239ΔGY in vitro.
In contrast to SIVmac239ΔGY, no differences were observed in fitness assays when SIVmac239Y/I was mixed with wild-type SIVmac239. When approximately a 3:1 ratio of SIVmac239Y/I to SIVmac239 was added, the same proportion was present after 28 days (Fig. 6D). Similar results were seen over a range of input ratios (data not shown). Thus, although striking in vivo differences were observed between animals N8G, in which the Y/I mutation was maintained, and VF3, in which the Ile reverted to a Tyr, no differences in replication between SIVmac239Y/I and wild-type SIVmac239 were apparent in this in vitro fitness assay.
DISCUSSION
In this study we show that mutation of a highly conserved sorting motif in the cytoplasmic tail of the SIV Env protein results in marked in vivo attenuation of a pathogenic molecular clone. Among five animals inoculated with viruses containing mutations that abrogated the function of this motif, three of them exhibited an initial peak of viremia followed by a profound reduction in viral load and have remained healthy for up to 140 weeks. In one (N8G) of two animals that received SIVmac239Y/I, the Y721I mutation was maintained and was associated with an attenuated phenotype, while in the other animal (VF3) the Ile reverted to a Tyr and was associated with a high viral load, loss of CD4+ cells, and disease. Similarly, two (AGP and WD1) of the three animals that received SIVmac239ΔGY remained healthy, with low to undetectable plasma viral RNA levels. The third animal (2AJ) developed high viral RNA levels and disease and exhibited novel and possibly compensatory mutations that flanked the ΔGY mutation. The replication competence of SIVmac239Y/I and SIVmac239ΔGY was demonstrated in cultured PBMC and CEMx174 cells. Moreover, these viruses produced initial peaks of viremia in all animals and corrected a premature stop codon in nef that has been shown to revert in the setting of active in vivo but not in vitro replication (49). Although virus fitness experiments demonstrated a replication defect for SIVmac239ΔGY compared to SIVmac239, no defect was demonstrated for viruses with a Y/I mutation. In addition, although viral RNA levels remained low or undetectable in the three animals showing an attenuated clinical course, all have maintained antibody titers of >1:10,000, indicating ongoing replication of the mutant viruses. These findings demonstrate that the Tyr-dependent Env sorting motif, while dispensable for SIV replication in vitro, has a marked effect on viral pathogenesis.
Several attenuated SIVs that establish low viral loads and exhibit reduced pathogenicity in infected animals have been described (30, 44, 46, 57, 63, 102). However, even when the determinants for this phenotype were mapped to particular genes, such as nef, the underlying mechanisms remained unclear (23, 75, 91). In most SIV models, the risk of developing disease can be predicted by plasma viral RNA levels (41, 47, 88, 97, 101). Therefore, any intrinsic defect in virus replication could result in lower viral loads and decreased pathogenicity. Alternatively, attenuation in vivo could result from reduced cytopathicity of virus for infected cells, an inability of the virus to infect critical target sites, and/or an increased susceptibility of the virus to host immune responses. For the nef-deleted viruses, Nef downregulates major histocompatibility complex class I molecules, thereby reducing the susceptibility of infected cells to killing by virus-specific cytotoxic T lymphocytes (CTL) (17–19). Given that animals infected with nef-deleted viruses exhibit potent antiviral CTL activity, it is tempting to speculate that these viruses are less fit to survive a host cellular immune response (38, 43). However, Nef also binds cellular kinases (11, 58, 83) and increases viral infectivity (14, 26, 58, 67, 90), and as a result, nef-deleted viruses could exhibit multiple defects to account for their reduced virulence.
Although the viral env gene mediates functions that are crucial for infectivity, including binding to CD4 and a chemokine receptor and fusion with the cell membrane, there are few reports of mutations in distinct functional domains of Env that reduce pathogenicity in vivo. Reitter et al. have recently shown that SIVmac239 mutants lacking glycosylation sites in the V1/V2 loops of gp120 were markedly attenuated in vivo, possibly as a result of their ability to elicit novel humoral immune responses (76). Interestingly, in SIVs with prematurely truncated cytoplasmic tails, strong selection pressures in vivo restore a full-length TM (49). For one well-characterized attenuated isolate, SIVmac1A11, reversion of a truncated to a full-length cytoplasmic tail correlated with increases in viral load (57). A recent study by Shacklett et al. showed that multiple stop codons in the TM cytoplasmic tail of SIVmac239 reduced the replicative capacity in vitro and produced a highly attenuated phenotype in vivo (86). In addition to the Tyr-based motif evaluated in our study, the TM tail of SIV and HIV contains other domains that have been implicated in mediating cytopathic effects (95, 96) and in interacting with the viral matrix protein (20, 70). Thus, the cytoplasmic domain is likely to play an important role in virus assembly, replication, and pathogenesis (7, 31, 35, 36, 71), and in this regard our findings draw particular attention to the importance of this conserved Tyr-sorting signal.
A number of mechanisms could account for the striking in vivo effects of mutations in this TM sorting motif. As noted previously, this domain functions as a potent endocytosis signal that mediates clathrin-dependent internalization of Env molecules (7, 8, 50, 74, 78, 82). As a result, Env glycoproteins that are not incorporated into budding virions are rapidly internalized and cleared from the plasma membrane (32). SIVs and HIVs lacking this motif characteristically exhibit a higher steady-state level of Env on the cell surface (27, 50, 82) and could be more susceptible to humoral immune responses (7, 27, 62). Indeed, we have shown that cells infected by SIVmac239Y/I characteristically show three to four times more Env on the cell surface than do cells infected by wild-type SIVmac239 (50), and we have seen a similar increase in Env production in cells infected by SIVmac239ΔGY (J. A. Hoxie, unpublished data). Although this latter effect is modest, it is possible that even a slight increase in the susceptibility of virus-producing cells to host immune responses could have significant consequences on net virus production since this defect is amplified through subsequent rounds of infection. Alternatively, the ability of this domain to also function as a basolateral sorting signal could be important in directing virus infection to crucial target sites, such as gut-associated lymphoid tissue, where viral replication is particularly active in early stages of infection (99). Finally, it is also possible that the loss of a determinant for directional virus spread could impact on the efficiency of cell-to-cell transmission in tissues. Indeed, the loss of the corresponding Yxxφ signal in an HIV-1 TM recently was shown to be associated with a defect in cell-to-cell spread in vitro (27).
It remains possible that the subtle defect in the replicative capacity of SIVmac239ΔGY contributed to its attenuated in vivo phenotype. Although intrinsic differences in infection kinetics compared to those of wild-type SIVmac239 were not seen in growth curves generated with PBMC or CEMx174 cells, SIVmac239ΔGY was less fit in competition assays. The slightly lower levels of plasma RNA reached during the acute phase of infection in two of three animals infected with SIVmac239ΔGY are consistent with this observation. However, for SIVmac239Y/I, no differences were detectable with respect to SIVmac239 in fitness assays, although viruses in the one animal that maintained this mutation exhibited an attenuated phenotype. Although a selection pressure to restore the Tyr was evident in animal VF3, the finding that this mutation did not revert in vitro during multiple passages (50; Hoxie, unpublished) indicates that elimination of this sorting motif has little or no effect on virus replication, at least under these conditions. Finally, as noted above, reversion of the nef premature termination codon was observed in all animals, indicating that there was a sufficient level of replication to restore this open reading frame. Although it is possible that the attenuating effect of mutations in the Yxxφ motif is context dependent and applicable only for viruses containing a stop codon in nef, we consider this possibility unlikely, given that reversion to a nef open reading frame was observed in all animals within 8 weeks after inoculation. Our data are consistent with the view that the Yxxφ sorting motif in SIV is a crucial determinant of virus replication in vivo and/or in modulating susceptibility of virus-producing cells to host immune responses.
It is interesting that animal 2AJ developed a high viral load despite the continued presence of the ΔGY mutation. Although it is possible that additional mutations in the 2AJ cytoplasmic domain compensated for this effect, none of the mutations in clones from this animal created an alternate Yxxφ motif. However, it remains possible that one or more of these new mutations facilitated an interaction with cellular adapter proteins or other molecules to generate a novel endocytosis and/or basolateral sorting signal. Alternatively these changes could have affected other parts of the viral life cycle, a finding suggested by the improved fitness of a virus that contained the ΔGY mutation and the compensatory mutations in a 2AJ clone (Fig. 6C). Additional in vivo studies are required to determine if one or more of these mutations is sufficient to account for the aggressive disease course that was observed in this animal.
Attenuated SIVs have enabled determinants for pathogenesis to be identified that could not have been inferred from in vitro studies. Moreover, as shown by the occurrence of nef-deleted HIV-1s that are associated with either absent or delayed progression to disease, these animal models will probably have relevance for HIV infection (25, 48, 51). Importantly, attenuated SIV models have provided the best evidence to date that protective immune responses can be generated, and intensive efforts are being undertaken to understand the basis for this effect (3, 28, 42, 52). The use of an attenuated SIV or HIV vaccine poses obvious safety concerns and has prompted the development of second- and third-generation viruses with multiple defects that impair replication (44). However, while increasingly attenuated viruses may be safer, theoretically there appear to be “threshold effects” in that viruses that are more severely impaired in their ability to replicate in vivo have not been able to generate protective immunity (44, 64, 80, 98). Approaches that reduce viral pathogenicity while preserving an immune response would be highly desirable, and in this regard, strategies that alter the presentation of viral proteins to the host immune system could be advantageous. Our findings indicate that the SIV TM cytoplasmic tail, and in particular this Tyr-based sorting motif, can be an additional target for attenuation strategies. Additional studies to determine the mechanism of this effect, its impact on the host immune response, and the ability of Tyr-721 mutants to generate protective immunity will be of considerable interest.
ACKNOWLEDGMENTS
We thank Peter Daly (Chiron Corp.) for performing bDNA assays and Beth Haggarty, Jackie Stallworth, Teresa Wiltrout, and Pamela May for expert technical assistance.
This work was supported by PHS grants RO1 AI33854 (J.A.H.), AI32377 (P.N.F.), AI46246 (M.H.M.), and P30-AI27767 for the flow cytometry core of the UAB Center for AIDS Research, a grant from the United Kingdom Medical Research Council (M.M.), and funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-56000. M.M. and J.A.H. were also supported by a NATO collaboration grant. J.D.D. was supported by NIH Medical Scientist Training Program grant GM07170.
REFERENCES
- 1.Aiken C, Konner J, Landau N R, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Weiskopf E, Greenough T C, Gaddis N C, Auerbach M R, Malim M H, O'Brien S J, Walker B D, Sullivan J L, Desrosiers R C. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almond N, Stott J. Live attenuated SIV—a model of a vaccine for AIDS. Immunol Lett. 1999;66:167–170. doi: 10.1016/s0165-2478(98)00153-9. [DOI] [PubMed] [Google Scholar]
- 4.Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 5.Baba T W, Liska V, Khimani A H, Ray N B, Dailey P J, Penninck D, Bronson R, Greene M F, McClure H M, Martin L N, Ruprecht R M. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 6.Baur A S, Sawai E T, Dazin P, Fantl W J, Cheng-Mayer C, Peterlin B M. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1:373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 7.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adapter complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boge M, Wyss S, Bonifacino J S, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adapter. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 9.Bowers K, Pelchen-Matthews A, Höning S, Vance P J, Creary L, Haggarty B S, Romano J, Ballensiefen W, Hoxie J A, Marsh M. The simian immunodeficiency virus envelope glycoprotein contains multiple signals that regulate its cell surface expression and endocytosis. Traf. 2000;1:661–674. doi: 10.1034/j.1600-0854.2000.010810.x. [DOI] [PubMed] [Google Scholar]
- 10.Brander C, Walker B D. T lymphocyte responses in HIV-1 infection: implications for vaccine development. Curr Opin Immunol. 1999;11:451–459. doi: 10.1016/S0952-7915(99)80076-4. [DOI] [PubMed] [Google Scholar]
- 11.Brown A, Wang X, Sawai E, Cheng-Mayer C. Activation of the PAK-related kinase by human immunodeficiency virus type 1 Nef in primary human peripheral blood lymphocytes and macrophages leads to phosphorylation of a PIX-p95 complex. J Virol. 1999;73:9899–9907. doi: 10.1128/jvi.73.12.9899-9907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrington M, Dean M, Martin M P, O'Brien S J. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8:1939–1945. doi: 10.1093/hmg/8.10.1939. [DOI] [PubMed] [Google Scholar]
- 13.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 14.Chowers M Y, Spina C A, Kwoh T J, Fitch N J, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clements J E, Montelaro R C, Zink M C, Amedee A M, Miller S, Trichel A M, Jagerski B, Hauer D, Martin L N, Bohm R P. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J Virol. 1995;69:2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 17.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 18.Collins K L, Baltimore D. HIV's evasion of the cellular immune response. Immunol Rev. 1999;168:65–74. doi: 10.1111/j.1600-065x.1999.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 19.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature (London) 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 20.Cosson P. Direct interaction between the envelope and matrix. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 21.Craig H M, Pandori M W, Guatelli J C. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cranage M P, Whatmore A M, Sharpe S A, Cook N, Polyanskaya N, Leech S, Smith J D, Rud E W, Dennis M J, Hall G A. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology. 1997;229:143–154. doi: 10.1006/viro.1996.8419. [DOI] [PubMed] [Google Scholar]
- 23.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 24.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 25.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 26.de Ronde A, Klaver B, Keulen W, Smit L, Goudsmit J. Natural HIV-1 NEF accelerates virus replication in primary human lymphocytes. Virology. 1992;188:391–395. doi: 10.1016/0042-6822(92)90772-h. [DOI] [PubMed] [Google Scholar]
- 27.Deschambeault J, Lalonde J P, Cervantes-Acosta G, Lodge R, Cohen E A, LeMay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desrosiers R C. Prospects for live attenuated HIV. Nat Med. 1998;4:982. doi: 10.1038/1949. [DOI] [PubMed] [Google Scholar]
- 29.Desrosiers R C. Strategies used by human immunodeficiency virus that allow persistent viral replication. Nat Med. 1999;5:723–725. doi: 10.1038/10439. [DOI] [PubMed] [Google Scholar]
- 30.Desrosiers R C, Lifson J D, Gibbs J S, Czajak S C, Howe A Y, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fauci A S. The AIDS epidemic—considerations for the 21st century. N Engl J Med. 1999;341:1046–1050. doi: 10.1056/NEJM199909303411406. [DOI] [PubMed] [Google Scholar]
- 34.Flaherty M T, Hauer D A, Mankowski J L, Zink M C, Clements J E. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J Virol. 1997;71:5790–5798. doi: 10.1128/jvi.71.8.5790-5798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature (London) 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 38.Gauduin M C, Glickman R L, Ahmad S, Yilma T, Johnson R P. Characterization of SIV-specific CD4+ T-helper proliferative responses in macaques immunized with live-attenuated SIV. J Med Primatol. 1999;28:233–241. doi: 10.1111/j.1600-0684.1999.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna Z, Kay D G, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 41.Hirsch V M, Dapolito G, Hahn A, Lifson J, Montefiori D, Brown C R, Goeken R. Viral genetic evolution in macaques infected with molecularly cloned simian immunodeficiency virus correlates with the extent of persistent viremia. J Virol. 1998;72:6482–6489. doi: 10.1128/jvi.72.8.6482-6489.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson R P, Desrosiers R C. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr Opin Immunol. 1998;10:436–443. doi: 10.1016/s0952-7915(98)80118-0. [DOI] [PubMed] [Google Scholar]
- 43.Johnson R P, Glickman R L, Yang J Q, Kaur A, Dion J T, Mulligan M J, Desrosiers R C. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J Virol. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson R P, Lifson J D, Czajak S C, Cole K S, Manson K H, Glickman R, Yang J, Montefiori D C, Montelaro R, Wyand M S, Desrosiers R C. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 46.Kestler H W, III, Ringler D J, Kazayasu M, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 47.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 48.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 49.Kodama T, Wooley D P, Naidu Y M, Kestler III H W, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of SIVmac transmembrane molecule increases envelope glycoprotein expression on cells and virions. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Learmont J C, Geczy A F, Mills J, Ashton L J, Raynes-Greenow C H, Garsia R J, Dyer W B, McIntyre L, Oelrichs R B, Rhodes D I, Deacon N J, Sullivan J S. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 52.Letvin N L. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 53.Levy J A. The value of primate models for studying human immunodeficiency virus pathogenesis. J Med Primatol. 1996;25:163–174. doi: 10.1111/j.1600-0684.1996.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 54.Lodge R, Göttlinger H, Gabuzda D, Cohen E A, LeMay G. The intracytoplasmic domain of gp41 mediated polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lodge R, Lalonde J P, LeMay G, Cohen E A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, VanRompay K K, Reay E, Antipa L, Pedersen N C, Marthas M L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luciw P A, Shaw K E, Shacklett B L, Marthas M L. Importance of the intracytoplasmic domain of the simian immunodeficiency virus (SIV) envelope glycoprotein for pathogenesis. Virology. 1998;252:9–16. doi: 10.1006/viro.1998.9467. [DOI] [PubMed] [Google Scholar]
- 58.Luo T, Livingston R A, Garcia J V. Infectivity enhancement by human immunodeficiency virus type 1 Nef is independent of its association with a cellular serine/threonine kinase. J Virol. 1997;71:9524–9530. doi: 10.1128/jvi.71.12.9524-9530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mangasarian A, Piguet V, Wang J K, Chen Y L, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marsh M, McMahon H T. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 62.Marsh M, Pelchen-Matthews A, Hoxie J A. Endocytosis and lentiviral pathogenesis. Trends Biochem Sci. 1997;7:1–4. doi: 10.1016/S0962-8924(97)20038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marthas M L, Ramos R A, Lohman B L, Van Rompay K K, Unger R E, Miller C J, Banapour B, Pedersen N C, Luciw P A. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marthas M L, Sutjipto S, Higgins J, Lohman B, Torten J, Luciw P A, Marx P A, Pedersen N C. Immunization with a live, attenuated simian immunodeficiency virus (SIV) prevents early disease but not infection in rhesus macaques challenged with pathogenic SIV. J Virol. 1990;64:3694. doi: 10.1128/jvi.64.8.3694-3700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 66.Miller C J, McChesney M B, Lu X, Dailey P J, Chutkowski C, Lu D, Brosio P, Roberts B, Lu Y. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic SIVmac239. J Virol. 1997;71:1911–1921. doi: 10.1128/jvi.71.3.1911-1921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montefiori D C, Baba T W, Li A, Bilska M, Ruprecht R M. Neutralizing and infection-enhancing antibody responses do not correlate with the differential pathogenicity of SIVmac239delta3 in adult and infant rhesus monkeys. J Immunol. 1996;157:5528–5535. [PubMed] [Google Scholar]
- 69.Mukherjee S, Ghosh R N, Maxfield F R. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 70.Murakami T, Freed E O. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74:3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nilsson C, Makitalo B, Thorstensson R, Norley S, Binninger-Schinzel D, Cranage M, Rud E, Biberfeld G, Putkonen P. Live attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS. 1998;12:2261–2270. doi: 10.1097/00002030-199817000-00006. [DOI] [PubMed] [Google Scholar]
- 73.O'Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 74.Ohno H, Aguilar R C, Fournier M C, Hennecke S, Cosson P, Bonifacino J S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 75.Piguet V, Schwartz O, Le Gall S, Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 76.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 77.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 78.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of the HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 79.Rud E W, Cranage M, Yon J, Quirk J, Ogilvie L, Cook N, Webster S, Dennis M, Clarke B E. Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J Gen Virol. 1994;75:529–543. doi: 10.1099/0022-1317-75-3-529. [DOI] [PubMed] [Google Scholar]
- 80.Ruprecht R M, Baba T W, Rasmussen R, Hu Y, Sharma P L. Murine and simian retrovirus models: the threshold hypothesis. AIDS. 1996;10(Suppl. A):S33–S40. doi: 10.1097/00002030-199601001-00005. [DOI] [PubMed] [Google Scholar]
- 81.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 82.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M F, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawai E T, Baur A, Struble H, Peterlin B M, Levy J A, Cheng-Mayer C. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmid S L. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 85.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 86.Shacklett B L, Weber C J, Shaw K E, Keddie E M, Gardner M B, Sonigo P, Luciw P A. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J Virol. 2000;74:5836–5844. doi: 10.1128/jvi.74.13.5836-5844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skowronski J, Parks D, Mariani R. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 1993;12:703–713. doi: 10.1002/j.1460-2075.1993.tb05704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith S M, Holland B, Russo C, Dailey P J, Marx P A, Connor R I. Retrospective analysis of viral load and SIV antibody responses in rhesus macaques infected with pathogenic SIV: predictive value for disease progression. AIDS Res Hum Retroviruses. 1999;15:1691–1701. doi: 10.1089/088922299309739. [DOI] [PubMed] [Google Scholar]
- 89.Sodora D L, Sheridan K E, Marx P A, Connor R I. Immunization with a live, attenuated simian immunodeficiency virus vaccine leads to restriction of viral diversity in Rhesus macaques not protected from pathogenic challenge. J Virol. 1999;73:4443–4446. doi: 10.1128/jvi.73.5.4443-4446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stevenson M. Molecular mechanisms for the regulation of HIV replication, persistence and latency. AIDS. 1997;11(Suppl. A):S25–S33. [PubMed] [Google Scholar]
- 92.Stott J, Hu S L, Almond N. Candidate vaccines protect macaques against primate immunodeficiency viruses. AIDS Res Hum Retroviruses. 1998;14(Suppl. 3):S265–S270. [PubMed] [Google Scholar]
- 93.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 94.Tao B, Fultz P N. Pathogenicity and comparative evolution in vivo of the transitional quasispecies SIVsmmPBj8. Virology. 1999;259:166–175. doi: 10.1006/viro.1999.9759. [DOI] [PubMed] [Google Scholar]
- 95.Tencza S B, Mietzner T A, Montelaro R C. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res Hum Retroviruses. 1997;13:263–269. doi: 10.1089/aid.1997.13.263. [DOI] [PubMed] [Google Scholar]
- 96.Tencza S B, Miller M A, Islam K, Mietzner T A, Montelaro R C. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J Virol. 1995;69:5199–5202. doi: 10.1128/jvi.69.8.5199-5202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ten Haaft P, Verstrepen B, Uberla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ui M, Kuwata T, Igarashi T, Miyazaki Y, Tamaru K, Shimada T, Nakamura M, Uesaka H, Yamamoto H, Hayami M. Protective immunity of gene-deleted SHIVs having an HIV-1 Env against challenge infection with a gene-intact SHIV. J Med Primatol. 1999;28:242–248. doi: 10.1111/j.1600-0684.1999.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 99.Veazey R S, DeMaria M, Chalifoux L V, Shvetz D E, Pauley D R, Knight H L, Rosenzweig M, Johnson R P, Desrosiers R C, Lackner A A. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 100.Wang J K, Kiyokawa E, Verdin E, Trono D. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc Natl Acad Sci USA. 2000;97:394–399. doi: 10.1073/pnas.97.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whatmore A M, Cook N, Hall G A, Sharpe S, Rud E W, Cranage M P. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wyand M S, Manson K, Montefiori D C, Lifson J D, Johnson R P, Desrosiers R C. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J Virol. 1999;73:8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu X N, Laffert B, Screaton G R, Kraft M, Wolf D, Kolanus W, Mongkolsapay J, McMichael A J, Baur A S. Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor zeta chain. J Exp Med. 1999;189:1489–1496. doi: 10.1084/jem.189.9.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]