Abstract
Hepadnaviruses replicate by reverse transcription, which takes place in the cytoplasm of the infected hepatocyte. Viral RNAs, including the pregenome, are transcribed from a covalently closed circular (ccc) viral DNA that is found in the nucleus. Inhibitors of the viral reverse transcriptase can block new DNA synthesis but have no direct effect on the up to 50 or more copies of cccDNA that maintain the infected state. Thus, during antiviral therapy, the rates of loss of cccDNA, infected hepatocytes (1 or more molecules of cccDNA), and replicating DNAs may be quite different. In the present study, we asked how these losses compared when woodchucks chronically infected with woodchuck hepatitis virus were treated with L-FMAU [1-(2-fluoro-5-methyl-β-l-arabinofuranosyl) uracil], an inhibitor of viral DNA synthesis. Viremia was suppressed for at least 8 months, after which drug-resistant virus began replicating to high titers. In addition, replicating viral DNAs were virtually absent from the liver after 6 weeks of treatment. In contrast, cccDNA declined more slowly, consistent with a half-life of ∼33 to 50 days. The loss of cccDNA was comparable to that expected from the estimated death rate of hepatocytes in these woodchucks, suggesting that death of infected cells was one of the major routes for elimination of cccDNA. However, the decline in the actual number of infected hepatocytes lagged behind the decline in cccDNA, so that the average cccDNA copy number in infected cells dropped during the early phase of therapy. This observation was consistent with the possibility that some fraction of cccDNA was distributed to daughter cells in those infected hepatocytes that passed through mitosis.
Lamivudine is a potent inhibitor of the hepatitis B virus (HBV) DNA polymerase and can quickly reduce liver injury in HBV carriers (34), apparently by suppressing virus replication. However, the majority of carriers are not cured by lamivudine, and drug-resistant virus emerges in most, often in association with an increase in virus titers towards pretreatment levels (3, 5, 9, 18, 25, 33, 34, 38). Difficulty in completely eliminating HBV stems directly from the mechanism by which this virus reproduces. When a hepadnavirus infects a cell, the incoming viral genome matures into a single covalently closed circular DNA (cccDNA). This cccDNA, located in the nucleus, serves as the template for the transcription of the larger-than-unit-length pregenomic RNA and of the subviral RNA species (8). A virus-encoded reverse transcriptase converts the pregenomic RNA into a partially double-stranded DNA genome in a series of reactions that take place inside virus nucleocapsids (36, 41), which are found in the cytoplasm of the infected cell. The virus nucleocapsids are subsequently enveloped and, after processing of the envelope glycoproteins (7, 30), are released from the cell as mature virions. In a pathway that is negatively regulated by the viral envelope proteins, a fraction of the virus nucleocapsids are transported to the cell nucleus to produce additional copies of cccDNA (37). Estimates of cccDNA copy number range from 5 to 50 or more per hepatocyte (21, 22, 31).
Because the infected state of a hepatocyte is defined by the presence of cccDNA, its stability is important in any consideration of antiviral therapies employing inhibitors of viral DNA synthesis. Cell culture studies with primary hepatocytes, which do not divide, indicated a high degree of cccDNA stability ((32); however, see reference 12). This stability may be a major reason why infections are harder to eliminate by polymerase inhibitors in “healthy” carriers with a slower rate of hepatocyte death and compensatory regeneration than in individuals with active hepatitis. However, it is not known whether cccDNA is lost during mitosis, as proposed, for example, for the Epstein-Barr virus (EBV) plasmid in the absence of EBNA1 (27), or whether it is distributed to daughter cells along with host chromosomes. Loss during mitosis would lead to a rate of cccDNA decline, in the absence of viral DNA synthesis, that would equal approximately twice the rate of infected-cell death. That is, cccDNA would be lost through cell death as well as through division of an infected cell that divided to replace the cell that died. In contrast, retention of cccDNA through the mitotic event would lead to a rate of loss equal to the rate of infected-cell death. However, in the latter case, because of the initially high cccDNA copy number, the fraction of infected hepatocytes would not begin to decline detectably until the average cccDNA copy in the liver dropped to ∼1 to 2 (provided that all cccDNA molecules have the potential to be transcriptionally active). Thus, a long period of treatment, in comparison to the average life time of the infected hepatocyte, would be needed in order to facilitate complete cccDNA loss from an infected liver. Current estimates place the half-life of infected hepatocytes in HBV carriers at between 2 weeks and many months, depending on the severity of hepatitis and, thus, the extent of hepatocyte destruction by the immune system (34).
The present study was carried out in order to determine how cccDNA levels changed in the hepatocyte population during antiviral therapy. For our experiments, we used woodchucks chronically infected with woodchuck hepatitis virus (WHV). As an antiviral agent, we employed L-FMAU [1-(2-fluoro-5-methyl-β-l-arabinofuranosyl) uracil] (11, 34a), which is highly effective against WHV replication in vivo.
We found that L-FMAU produced up to 95 to 99% loss of cccDNA after 30 weeks. The average half-life of the cccDNA over the course of the in vivo experiments that would produce the loss observed after 30 weeks was ∼33 to 50 days, similar to the half-life of hepatocytes estimated from proliferating-cell nuclear antigen (PCNA) staining indices of liver biopsy specimens collected during therapy. Immunoperoxidase and in situ hybridization of liver tissue sections revealed that the fractional loss of infected hepatocytes was ∼5 to 10-fold less than the loss of cccDNA, consistent with the hypothesis that cccDNA is not lost but distributed to daughter hepatocytes during mitosis. Evidence in support of this hypothesis was also recently obtained using primary cultures of woodchuck hepatocytes infected with WHV and induced to proliferate by addition of epidermal growth factor (13).
After about 36 weeks of L-FMAU administration, virus titers in the serum, which had dropped below the limits of detection of our assays (∼106 per ml), began to rise. Prior to this, mutant WHVs were detected with a distribution of nucleoside changes in the active site of the DNA polymerase, some of which were identical to those found after long-term lamivudine therapy (29, 45). The delayed spread of some of these mutants into apparently uninfected hepatocytes suggested that they may have a slower in vivo growth rate in the presence of L-FMAU than wild-type WHV in the absence of drug.
MATERIALS AND METHODS
Woodchucks.
Adult uninfected and WHV-infected woodchucks (Marmota monax), trapped in New York state and in Delaware, respectively, were purchased from Northeastern Wildlife (South Plymouth, N.Y.) and housed in the laboratory animal facility of the Fox Chase Cancer Center (FCCC). Experiments with woodchucks were reviewed and approved by the FCCC Institutional Animal Care and Use Committee. L-FMAU (10 mg per kg of body weight) was administered orally once a day (between 6 and 10 a.m.) in Dyets Woodchuck Control Diet (Dyets, Inc., Bethlehem, Pa.) at a concentration of 10 mg/ml. A total of 11 chronically infected woodchucks were used in this study. Four (343, 344, 345, and 346) had not been previously treated. Another seven, used to evaluate cross resistance between lamivudine and L-FMAU, had received a 14-month dose of lamivudine (44), which had ended 5 months prior to the treatment with either L-FMAU or placebo (Dyets Woodchuck Control Diet without L-FMAU). The sera of this latter group contained WHV genetic variants characteristic of the resistance to lamivudine that had developed in these woodchucks. Serum collection and liver biopsy during the course of these studies were carried out as previously described (22).
L-FMAU treatment of woodchuck primary hepatocytes.
Primary woodchuck hepatocyte cultures were prepared and maintained at 37°C on 60-mm tissue cultures dishes coated with rat tail collagen in a serum-free L15 medium supplemented with insulin, hydrocortisone, and phosphonoacetic acid, as previously described (2, 32). Culture fluids (3 ml) were changed daily. WHV infection of hepatocytes was established by adding 50 μl of serum (ca. 108 virions) from a chronically infected woodchuck at 2 days postseeding. Starting 4 days after WHV infection, L-FMAU was present in culture medium at a 10 μM concentration. Hepatocyte monolayers were harvested at 4, 8, 16, 24, 32, and 40 days after WHV infection and stored at −80°C for subsequent extraction of viral nucleic acids.
Analysis of WHV DNA.
Total DNA and cccDNA isolations from primary hepatocyte cultures and subsequent analyses were performed as described previously (32). Isolation of total DNA and cccDNA from woodchuck liver biopsy specimens was also carried out following a previously described procedure (21). Briefly, approximately 0.05 to 0.1 g of liver tissue was disrupted with a loose-fitting Dounce homogenizer in 1.5 ml of 10 mM Tris-HCl (pH 7.5)–10 mM EDTA. The number of nuclei in each homogenate was determined following staining of aliquots with ethidium bromide and counting in a hemacytometer under fluorescent illumination. Each homogenate was divided into two aliquots, one for extraction of non-protein-bound cccDNA, and the other for total DNA isolation (21, 37). Either 10 μg of total DNA or the cccDNA extracted from 106 liver cells was subjected to electrophoresis through 1.5% agarose gels. (The genome of the woodchuck was assumed to weigh 5 pg per diploid cell.) Known amounts of full-length, cloned viral DNA were electrophoresed as a control. The DNAs were then transferred to a nitrocellulose filter (Schleicher & Schuell, Keene, N.H.) following partial depurination and fragmentation with alkali, similar to the procedure described by Wahl et al. (40), and hybridized with a 32P-labeled WHV DNA probe representing the complete genome. Radioactive signals were quantified using a Fuji phosphoimager (Fuji Corporation, Tokyo, Japan). The amount of viral DNA present in the liver samples was estimated by comparison to the hybridization control and, for the purpose of copy number estimations, is presented as equivalents of full-length, double-stranded WHV DNA.
Serum virus titers.
To measure WHV titers in serum, 50 μl of serum was centrifuged through a 4-ml, 10 to 20% sucrose step gradient containing 150 mM NaCl and 20 mM Tris-HCl (pH 7.5) for 3 h at 50,000 rpm in a Beckman SW60 rotor. The virus pellet was then resuspended in 50 μl of 100 mM NaCl–10 mM Tris-HCl (pH 7.5)–10 mM EDTA–0.1% sodium dodecyl sulfate (SDS)–2 mg of pronase per ml and incubated at 37°C for 1 h. The samples were then subjected to 1.5% agarose gel electrophoresis, transferred to nitrocellulose membranes, hybridized with 32P-labeled WHV probe, and quantified as described above.
Detection of WHV core antigen- and nucleic acid-positive hepatocytes, PCNA-positive hepatocytes, and infiltrates of CD3-positive cells in liver tissue sections.
Biopsy specimens for immunoperoxidase assays and in situ hybridizations were fixed in a 3:1 mixture of ethanol and glacial acetic acid for 20 min at 4°C and then overnight at 4°C in 100% ethanol, followed by dehydration and embedding in paraffin wax. Immunoperoxidase staining for WHV core antigen, PCNA, and CD3-positive leukocytes and in situ hybridization for detection of WHV nucleic acids were carried out as previously described (17, 29).
Genotyping of WHV DNA.
Direct sequencing of PCR products and of cloned PCR products, as well as determination of restriction site polymorphisms, was used for genotyping (45). Virions were collected from 50 μl of woodchuck serum by centrifugation, and viral DNA was released by digestion with a mixture of SDS and pronase, as described above. The pronase-treated mixture was then extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1), and viral DNA was precipitated by the addition of 2 volumes of ethanol. cccDNA for genotyping was extracted from liver tissue as described above, subsequently purified using an alkaline extraction protocol (42), and digested with EcoRI. PCR amplification of a region spanning the active site of the viral DNA polymerase was carried out as described (45). The primers were 5′-AGATTGGTGGTGCACTTCTCTCAGG-3′ (WHV nucleotides 385 to 408) and 5′-CCACGGAATTGTCAGTGCCCAACC-3′ (nucleotides 1474 to 1451), using the numbering system of Kodama et al. (23). The PCR products were purified using a QIAquick kit (Qiagen Inc., Hilden, Germany) according to the manufacturer's instructions, and both strands were sequenced. Selected products were also cloned prior to sequence analysis.
Transfection of HepG2 cells.
For transfection, we used plasmids containing WHV DNA in which transcription of the viral pregenome is directed by a cytomegalovirus immediate-early promoter. The wild-type construct and variants of the wild type containing type I, II, or III mutations of the polymerase active site were reported previously (45). Twenty hours before transfection, HepG2 cells were seeded at 3 × 106 cells per 6-cm tissue culture dish in F-12 minimal essential medium-containing 10% fetal calf serum. L-FMAU at the indicated concentrations was added at this time and maintained at the same concentration in the medium thereafter. Transfection was carried out 1 to 2 days postseeding using a CaPO4 coprecipitation protocol (39). At 4 days posttransfection, the cells were harvested, and WHV core DNA was extracted (45). One quarter of each sample was subjected to Southern blot analysis, as described above.
SDH assay.
Sorbital dehydrogenase (SDH) in serum was determined by Anilytics, Inc., Gaithersburg, Md. Concentrations are expressed in international units per liter.
Histopathology.
Histopathology was done on formalin-fixed liver sections stained with hematoxylin and eosin. Liver injury was graded on a subjective scale. Inflammation was a major determinant, with the other factors, hepatocyte necrosis, vacuolization, biliary hyperplasia, Kupffer cell activation, and variation in hepatocyte nuclear size, influencing the degree of injury. Scoring was as follows: 0, no evidence of liver injury; ±, scant numbers of inflammatory cells in portal tracts with minimal inflammation in the liver parenchyma; 1, mild accumulations of lymphocytes in portal areas and focal accumulations in the parenchyma, with individual hepatocyte necrosis, Kupffer cell aggregates, and variation in hepatocyte nuclear size also present; 2, moderate inflammation of portal tracts with sites of extension into the terminal distributing vasculature; and 3, moderate to extensive inflammatory infiltrate extending from the portal tract into adjacent parenchyma or portal inflammation accompanied by moderate to extensive parenchymal inflammation.
Computational model of redistribution and loss of cccDNA during cell division when viral DNA synthesis is inhibited.
For the purpose of computation, the liver was divided into compartments, each containing cells with a particular number of cccDNA copies. The model was initialized to contain a particular distribution of copies; for example, a truncated Poisson distribution with a mean of 30 copies and truncation limits of 20 and 40 copies can be specified. In the truncated Poisson, 100% of the cccDNA is initially present at a copy number of 20 to 40. Liver size was held constant by adjusting the cell replication rate to compensate for the cell killing rate. Each replicating cell binomially and symmetrically distributes its cccDNA to its two daughters. The sequence of events is: cells are killed, cells divide to repopulate the liver, and cccDNA is redistributed among replicated daughters. In the model, a specified fraction of cccDNA may also be lost during this cell division. In that case, this fraction is removed prior to redistribution of the remaining cccDNA copies. At completion, the program supplies the resulting distribution of cccDNA copies. Other program outputs included graphs of the fraction of cells infected over time, fraction of cccDNA left over time, total cell deaths over time, and the daily cell death rate. (Scripts for the MacIntosh are available upon request from S. Litwin at S_Litwin@fccc.edu.)
RESULTS
L-FMAU treatment did not facilitate loss of cccDNA from WHV-infected primary hepatocyte cultures.
L-FMAU acts on the hepadnavirus reverse transcriptase to inhibit viral DNA synthesis (1). By analogy to EBV, this may occur through noncompetitive binding to the viral polymerase rather than through incorporation into the growing DNA chains (24). The objective of the present study was to characterize the correlation between the decline in infected hepatocytes and cccDNA loss during L-FMAU therapy of chronically infected woodchucks. A pilot study was first carried out to determine if L-FMAU had any unexpected effects on cccDNA stability in primary hepatocyte cultures, which are composed of nondividing hepatocytes. In a previous study with the antiviral agent lamivudine, no loss of cccDNA was observed over that explainable by cell loss from the cultures (32).
Woodchuck hepatocytes were plated and infected with WHV-infected animal serum as described in Materials and Methods. L-FMAU was included in the culture medium starting at 4 days postinfection, at a concentration (10 μM) determined to give maximal inhibition of WHV DNA synthesis. Hepatocyte monolayers were harvested at 4, 8, 16, 24, 32, and 40 days postinfection. Total DNA levels, including replicating DNA and cccDNA, declined as the duration of treatment increased (Fig. 1). This loss appeared to be due largely to the dissociation of adsorbed virus particles from the monolayers. The actual effect on viral DNA synthesis was determined by quantifying the accumulation of single-stranded DNA (SS-DNA), an intermediate in the reverse transcription pathway (28, 36). This analysis indicated that, by day 32, SS-DNA synthesis and accumulation were inhibited about 200-fold or more in the treated compared to the untreated cultures (Fig. 1A).
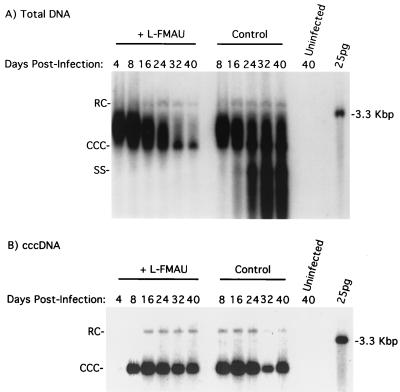
FIG. 1.
L-FMAU does not induce loss of cccDNA from primary cultures of WHV-infected woodchuck hepatocytes. Hepatocyte cultures were infected and treated with L-FMAU, beginning 4 days postinfection, as described in Materials and Methods. Total DNA (A) and cccDNA (B) were extracted from the cells at the indicated times and subjected to Southern blot analysis. Each lane contained one-quarter of the DNA extracted from a 6-cm-diameter tissue culture dish. RC, relaxed circular 3.3-kbp DNA.
In the same experiment, we also examined the effect of L-FMAU on cccDNA levels. At 4 days postinfection, a small amount of cccDNA could be detected, with more detected at 8 days (Fig. 1B). No significant loss of cccDNA from the treated cultures was seen between days 8 and 40 postinfection. A slight decline in cccDNA starting at 24 days postinfection for both the L-FMAU-treated and untreated monolayers was correlated with the gradual loss of cells from the cultures. In summary, a 200-fold suppression in viral DNA replication (Fig. 1A) by L-FMAU did not facilitate a loss of cccDNA from the infected hepatocytes. Assuming that no new cccDNA formation occurred between days 8 and 40, the data would be consistent with a cccDNA half-life of at least 32 days.
Oral administration of L-FMAU inhibited WHV replication and induced a progressive loss of cccDNA in chronically infected woodchucks.
Chu et al. (10) reported that treatment of chronic WHV carriers with L-FMAU at a daily dose of 10 mg per kg of body weight produced a >1,000-fold reduction in serum virus titers within 2 weeks. This dose was used to study the effects of L-FMAU therapy on cccDNA levels in the liver. In our study, four woodchucks chronically infected with WHV (343, 344, 345, and 346) were each treated with L-FMAU by daily oral administration. (Woodchuck 344 died accidentally after 4 weeks of treatment.)
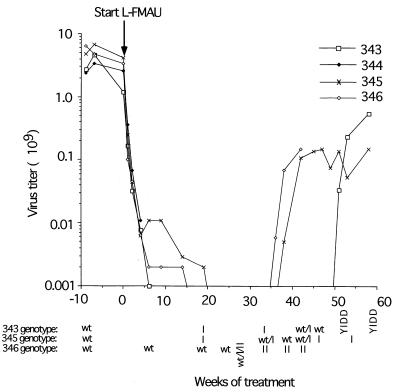
The effects of L-FMAU on WHV replication were confirmed by assaying the serum WHV titers. The WHV titers of the treated woodchucks dropped about 1,000-fold following 4 weeks of L-FMAU treatment (Fig. 2), in agreement with the published results (10). Between ∼20 and 30 weeks, virus titers were near or below the limit of detection of our assay (<106 per ml). Virus titers then escalated towards pretreatment levels in two of three animals between 30 and 40 weeks of treatment and in the third at about 50 weeks. This increase in virus titers followed the appearance of L-FMAU-resistant variants of WHV, as discussed below.
FIG. 2.
Suppression and rebound of viremia during long-term treatment with L-FMAU. Virus titers during treatment with L-FMAU were quantified by Southern blot assays for virus particles in the serum, as described in Materials and Methods. A titer of 106 DNA equivalents per ml represents the lower limit of detection of our assays. Serum samples for genotype analysis were collected 9 weeks before and 6, 19, 24, 27, 33, 38, 42, 45, 51, 53, and 58 weeks after initiation of therapy. The changes in the sequences of the B and C domains of the viral DNA polymerase which define the mutant designations were determined by direct sequencing of PCR products and are illustrated in Fig. 5. wt, wild type.
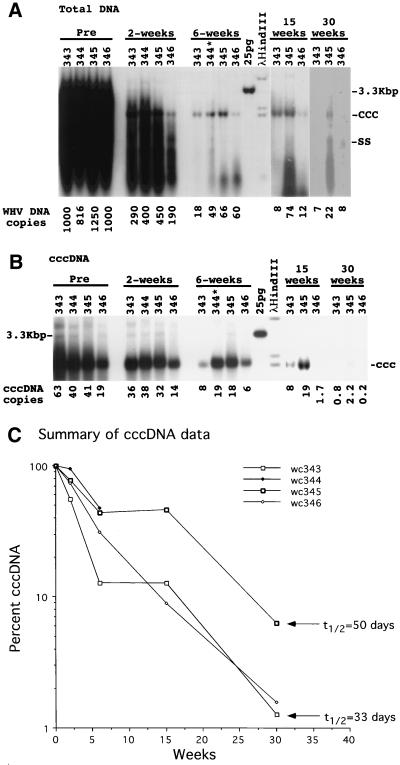
To determine effects on viral DNA accumulation in the liver, total DNA and cccDNA were extracted from the liver during the course of therapy. By 6 weeks, total viral DNA levels had dropped ∼20-fold or more (Fig. 3A) and >100-fold after 30 weeks of treatment (Fig. 3A). Moreover, possibly truncated forms of replicative DNA were now present, migrating near the bottom of the gel. In contrast, the intrahepatic cccDNA levels declined at a much lower rate than replicative DNA (Fig. 3B). While the levels of the DNA replication intermediates were between 1.8 and 6% of pretreatment levels by 6 weeks of therapy (Fig. 3A), cccDNA levels were still between 13 and 44% of pretreatment values (Fig. 3B).
FIG. 3.
Loss of replicating and cccDNA forms of viral DNA during antiviral therapy. Viral DNA was extracted from liver biopsy specimens for analysis of total and cccDNA forms of viral DNA by Southern blot analysis on 1.5% agarose gels, as described in Materials and Methods. Each lane contained either 10 μg of total DNA (A) or a cccDNA-enriched fraction recovered from 106 liver cells (based on nuclear counts in the liver lysates) (B). The average copy numbers of viral DNA per hepatocyte, shown at the bottom of panels A and B, were calculated as equivalents of full-length, double-stranded viral DNA and assume that the liver comprises 70% hepatocytes. The genome of the woodchuck was assumed to weigh 5 pg per diploid cell. The percent loss of cccDNA during treatment is summarized in panel C. The arrows indicate the losses of cccDNA after 30 weeks that would occur via a first-order decay with the indicated half-lives. Woodchuck 344 (wc344) died after 4 weeks of therapy.
By 30 weeks of treatment, cccDNA levels dropped to between 1.2 and 5.4% of pretreatment levels (Fig. 3B). This reduction in cccDNA was consistent with a half-life of 33 to 50 days (Fig. 3C). The PCNA staining data in Table 1 suggest that some of the cccDNA loss must be attributed to the death of infected hepatocytes. In addition, the observed loss of cccDNA is in agreement with the prediction, from cell culture experiments (Fig. 1) (32), that the cccDNA half-life within cells is at least 32 days. We calculate that the observed in vivo loss of cccDNA, assuming no new synthesis, could be explained by an infected-cell death rate of 1.3 to 2.1% per day. The PCNA staining data summarized in Table 1 are in reasonable agreement with this amount of hepatocyte replacement, assuming that PCNA is elevated in the nucleus for about one-third of a 24-h cell cycle. It should, however, be kept in mind that PCNA staining indices do not give a direct measure of rates of cell proliferation, and the duration of the cell cycle is not precisely known. Moreover, processes in addition to cell death may contribute to the total cccDNA loss.
TABLE 1.
Infection and histopathology in L-FMAU-treated woodchucks
| Woodchuck no. | Time | % Virus-positive hepatocytes
|
% Initial cccDNA | Ratio of virus-positive hepatocytes to cccDNA | % PCNA-positive hepatocytesa | % CD3-positive cellsa | Liver injury scoreb | |
|---|---|---|---|---|---|---|---|---|
| In situ hybridization | Anticore staining | |||||||
| 343 | Pretreatment | 100 | >95 | 100 | 1.0 | 0.9 | 15.8 | 1 |
| 2 wk | 100 | >90 | 57 | 1.7 | 0.5 | 19.2 | 1 | |
| 6 wk | 37 | >90 | 13 | 5.3 | 0.6 | 17.2 | ± | |
| 15 wk | 21 | 44 | 13 | 2.5 | 0.1 | 8.9 | 1 | |
| 30 wk | 8 | 11 | 1.3 | 7.3 | 1.3 | 9.4 | 1 | |
| Autopsy | ND | >95 | ND | ND | 1.2 | 8.1 | 1 | |
| 344 | Pretreatment | ND | >95 | 100 | 1.0 | 0.4 | 10.5 | ± |
| 2 wk | ND | >90 | 95 | 1.0 | 0.6 | 10.6 | 1 | |
| 4 wk | ND | >90 | 48 | 2.1 | 1.2 | 6.0 | 3 | |
| 345 | Pretreatment | 100 | >95 | 100 | 1.0 | 0.7 | 27.8 | 2 |
| 2 wk | 100 | >95 | 78 | 1.3 | 0.1 | 34.5 | 1 | |
| 6 wk | 94 | >95 | 44 | 2.2 | 0.3 | 30.3 | 1 | |
| 15 wk | ND | 90 | 46 | 2.0 | 1.3 | 26.3 | ± | |
| 30 wk | 28 | 20 | 5.4 | 4.4 | 0.4 | 19.8 | ± | |
| Autopsy | ND | 95 | ND | ND | 0.8 | 11.0 | 1 | |
| 346 | Pretreatment | 100 | >95 | 100 | 1.0 | 1.0 | 14.0 | 1 |
| 2 wk | 100 | >95 | 74 | 1.4 | ND | 115.3 | ± | |
| 6 wk | 85 | >95 | 43 | 2.2 | 0.1 | 23.3 | 1 | |
| 15 wk | 25 | 29 | 9 | 3.0 | 1.3 | 16.5 | ND | |
| 30 wk | 16 | 11 | 1.2 | 11.2 | <0.01 | 6.2 | 1 | |
| Autopsy | ND | >95 | ND | ND | 0.02 | 6.1 | ND | |
CD3 infiltrates into the hepatic lobule and the percentage of PCNA-positive hepatocyte nuclei were determined as previously described (12, 23). The CD3 percent is the ratio of intralobular CD3-positive cells to hepatocytes, times 100. CD3-positive cells in portal tracts were not included. ND, not determined.
Liver injury scores are explained in Materials and Methods.
cccDNA distribution among infected hepatocytes after antiviral therapy.
Infected hepatocytes contained, on average, ∼20 to 60 copies of cccDNA prior to L-FMAU administration (Fig. 3B). Once viral DNA synthesis is inhibited and the mature replication intermediates are depleted from the cytoplasm, no new cccDNA synthesis should occur. Thus, as cccDNA is lost through cell death, the cccDNA copy number in infected cells should decline through dilution as infected cells divide, provided that the cccDNA can survive through mitosis. The predicted effect on cccDNA distribution, assuming a Poisson distribution of between 20 and 60 copies per hepatocyte and a cccDNA half-life in the liver of 33 to 50 days, can be calculated by computational methods (Materials and Methods). After 30 weeks, 11 to 46% of the hepatocytes should still be infected, even though only 1.2 to 5.4% of the cccDNA remained. Most hepatocytes that remained infected would have fewer than 10 copies of cccDNA.
In contrast, if cccDNA was entirely lost during mitosis of infected hepatocytes, only 1.2 to 5.4% would, with the same total decline in cccDNA, remain infected. Thus, for the same cccDNA loss, there would be a six- to eightfold difference in the number of hepatocytes that remained infected, depending on whether or not cccDNA survived through cell division. In addition, the latter scenario requires only one-third to one-fourth as much cumulative cell death to achieve the same loss of cccDNA.
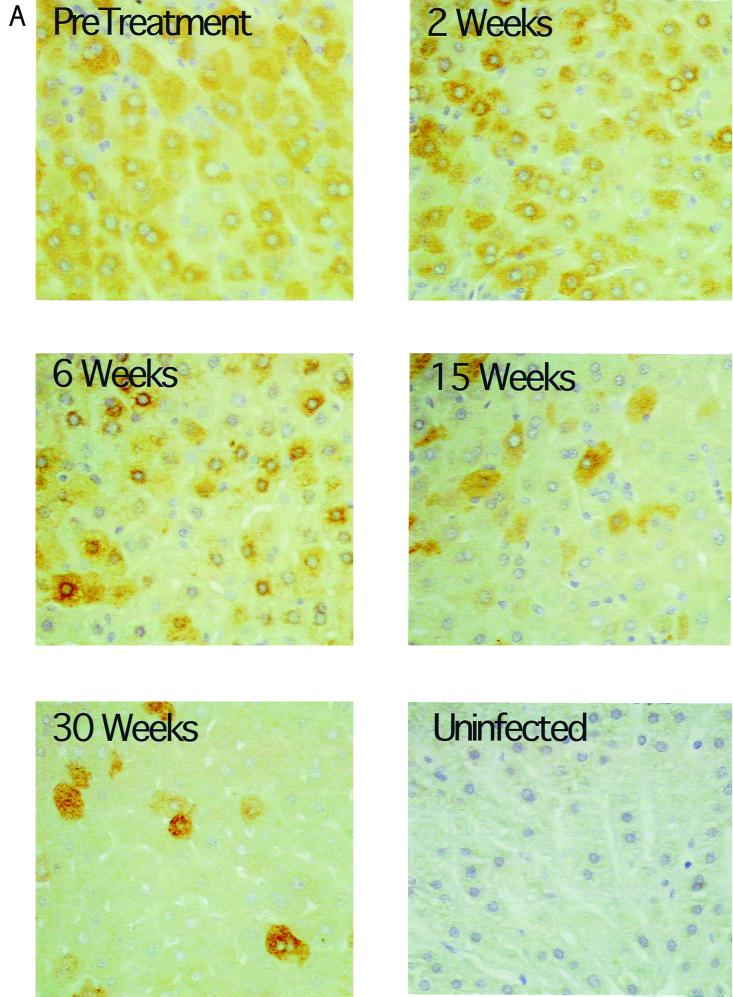
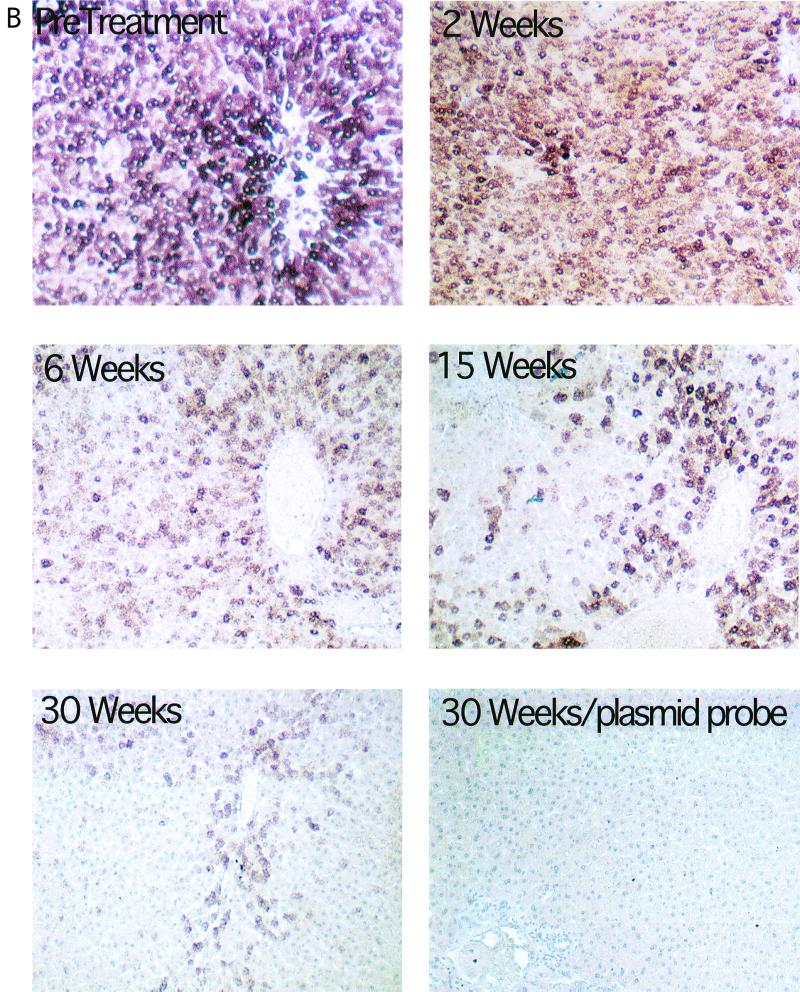
To determine whether average cccDNA levels in “infected” hepatocytes actually did decline during L-FMAU therapy and whether the survival of infected cells was also greater than the survival of cccDNA, we quantified the fraction of infected hepatocytes in tissue sections. This was done using immunoperoxidase detection of viral core antigen and in situ hybridization of viral nucleic acids (Fig. 4). The ratios of the averages of virus antigen and nucleic acid-positive hepatocytes to cccDNA were then calculated and are summarized in Table 1. We observed that the fractional loss of cccDNA greatly exceeded the fractional loss of infected hepatocytes (Table 1), especially taking into consideration the likelihood that our assays (Fig. 4) underestimated the fraction of infected cells in the treated livers. After 30 weeks, the ratio of obviously virus-positive hepatocytes to cccDNA was about 5- to 12-fold higher than before treatment. Therefore, the results are consistent with the hypothesis that some cccDNA survives through mitosis, being distributed to daughter cells.
FIG. 4.
Progressive loss of hepatocytes with detectable levels of WHV core antigen and nucleic acids during antiviral therapy. All liver biopsy specimens were assayed for the fraction of hepatocytes with detectable levels of WHV core antigen and nucleic acids. The results of these analyses for serial liver biopsies from woodchuck 346 are illustrated. Core antigen was detected by an immunoperoxidase assay (A), and viral nucleic acids were detected by in situ hybridization (B). The percentage of positive hepatocytes detected with each assay is summarized in Table 1. Magnification: (A) ×200; (B) ×100. Uninfected liver tissue is shown as a negative control in panel A. In panel B, tissue hybridized with a plasmid-specific probe (no WHV insert) served as the negative control.
L-FMAU-resistant WHV emerged after prolonged treatment.
Treatment of chronically infected woodchucks with lamivudine, a nucleoside analog, leads to a temporary suppression of virus replication. However, within a year, drug-resistant strains of WHV emerge, and virus production increases towards pretreatment levels (29, 45). A similar rebound in viremia was found after 8 to 9 months of treatment with L-FMAU, as summarized in Fig. 2. To determine if the increase in virus titers was associated with the emergence of virus with a mutation(s) in the active sites of the polymerase, direct sequencing of PCR products spanning this region was carried out.
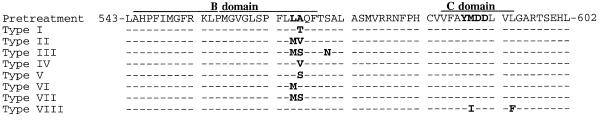
WHV with pol gene mutations were detected 1.5 to 3 months before virus titers began to rise above the limit of detection of the Southern blot assay (∼106 per ml) (Fig. 2). For woodchuck 345, a type I mutation (see Fig. 5 for descriptions of mutation types) was prevalent in serum at least 15 weeks before titers rose detectably (Fig. 2); in fact, at a time when virus titers were still declining. Mutant viruses became detectable in the serum of woodchuck 346 between 6 and 10 weeks before the rise in virus titers. Both type I and II mutations were detected (Fig. 2 and 5). The same mutations have been observed previously in woodchucks treated with lamivudine (45). Again, the mutations were present before the liver biopsy at 30 weeks, at which time many hepatocytes appeared to be no longer infected (Table 1, Fig. 4). In woodchuck 343, a third type of mutation was seen (type VIII; Fig. 5). In this animal, L-FMAU resistance was associated with the initial detection of the type I mutation and the later emergence of a YMDD to YIDD mutation in the C domain of the polymerase (Table 2). This mutation alone confers lamivudine resistance to WHV but has not previously been observed in lamivudine-treated woodchucks (29, 45). This variant had at least two additional polymerase mutations, L467F (numbered as described in reference 45) and L594F.
FIG. 5.
WHV active-site mutations.
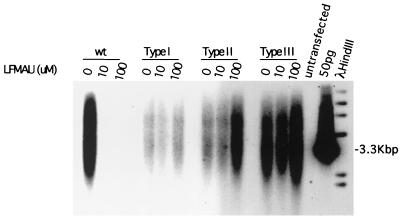
To determine if the type I or II mutations produced resistance to L-FMAU, these mutations were introduced into wild-type WHV DNA (45). The cloned viral DNAs were then transfected into HepG2 cells in the presence or absence of L-FMAU. Both mutants replicated in the presence of 10 and 100 μM L-FMAU, while replication of the wild type was inhibited (Fig. 6). We also tested the type III WHV mutation (Fig. 5), which is associated with resistance to lamivudine and generally emerges after the type I and II mutations (45). This mutation also conferred resistance to L-FMAU.
FIG. 6.
Evidence that mutations that confer lamivudine-resistant DNA synthesis on a laboratory strain of WHV also confer resistance to L-FMAU. Resistance of viral DNA synthesis to L-FMAU was assayed in transfected HepG2 cells, as described in Materials and Methods. The predicted amino acid sequences of the DNA polymerase B and C domains of the type I, II, and III mutations are illustrated in Fig. 5. Evidence that these mutations confer lamivudine resistance has already been published (45).
In view of the observation that mutant WHV was detected in the serum several weeks before the liver biopsy at 30 weeks in all three woodchucks, it was surprising that so many hepatocytes appeared to be virus free (Fig. 4). To determine if the cccDNA detected in the liver at 30 weeks of treatment was residual wild type, presumably in the process of elimination, or mutant, genotyping of the region encoding the Pol active site was carried out using the 30-week samples from the three woodchucks. Only the wild-type sequence was apparent by direct sequencing of PCR products. However, when 96 clones of the PCR products from woodchuck 343 were tested for the mutation, using a restriction site polymorphism, nine were found to have the type I mutation. Thus, at 30 weeks this mutant was reasonably abundant in the liver, with an average copy number of about 0.8 per core antigen-positive hepatocyte (cf. Table 1 and Fig. 3B), but apparently unable to spread efficiently to presumably virus-free hepatocytes (Table 1). A failure to spread quickly within the liver can also be inferred for the mutants detected in woodchucks 345 and 346. This may reflect a slow replication rate for all of these mutants, a possibility that also can be inferred from the data in Fig. 6 and from previously published work (45) (see Discussion).
Evidence for cross-resistance of L-FMAU in woodchucks previously treated with lamivudine.
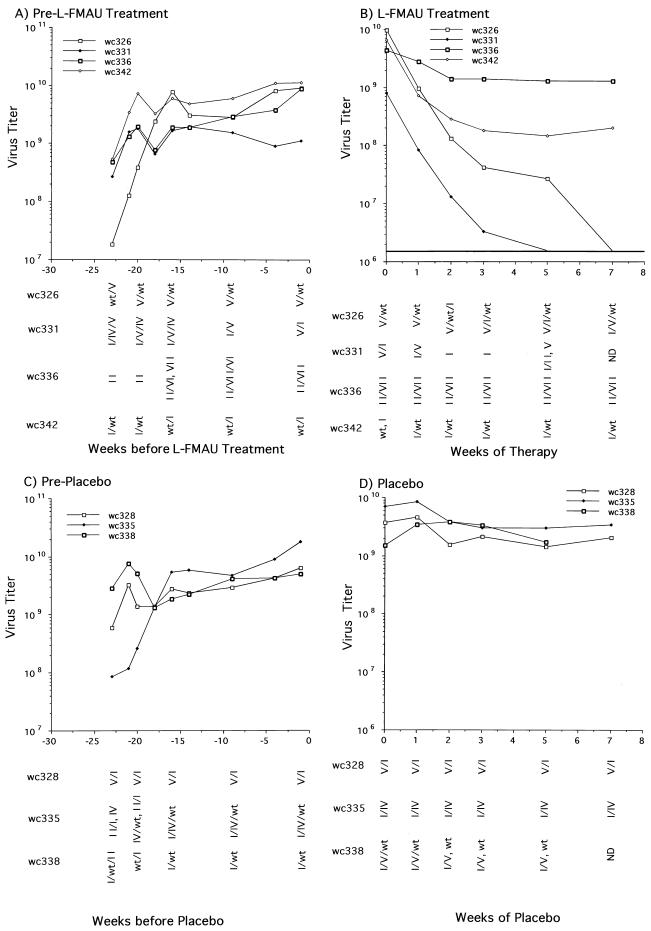
Although the sequence analysis was limited to a small region of the WHV genome, the above results suggested that L-FMAU and lamivudine would show cross-resistance in vivo. To further test this idea, an in vivo study was carried out employing seven woodchucks, six of which had developed resistance to lamivudine during a 14-month trial (44). With woodchuck 326, only a small rise in virus titers was noted toward the end of lamivudine treatment. Six months after the end of lamivudine therapy, four of these woodchucks were administered L-FMAU and three received placebo. The WHV genotypes of each woodchuck in the 5 months between lamivudine and L-FMAU therapy are described in Fig. 5 and 7. All but woodchuck 326 had at least one prevalent virus population with mutations in the Pol active site that had previously been associated with lamivudine resistance in woodchucks (45). The consequences of the type IV to VII mutations on virus DNA replication are still under investigation.
FIG. 7.
Evidence that in vivo infections that are resistant to lamivudine may also be resistant to L-FMAU. WHV-infected woodchucks in which virus titers had rebounded during a previous treatment with lamivudine received a 7-week treatment with L-FMAU (A and B); control animals received the Dyets formula as a placebo (C and D). Virus titers and genotypes in the interval between the end of the lamivudine therapy and the beginning of L-FMAU or placebo administration are shown in panels A and C. In panels A and C, the first serum sample was collected on the day that lamivudine treatment was discontinued. Sequence determinations from direct sequencing of PCR products were validated by cloning and sequencing of PCR products from the zero time points for woodchucks (wc) 326, 331, 336, 342, 335, and 338, as well as the 2-week time point from woodchuck 335. The variants are listed according to their relative abundance in the PCR products, the first being the most abundant. The predicted amino acid sequences of the B and C domains of the DNA polymerase of the various mutants are summarized in Fig. 5. wt, wild type.
Woodchucks 326, 331, 336, and 342 were treated with L-FMAU (10 mg/kg) for 2 months, and 328, 335, and 338 were treated with the placebo. The serum WHV titers of these woodchucks were monitored during the course of treatment and are summarized in Fig. 7. Titers of virus in the woodchucks treated with placebo stayed relatively unchanged (Fig. 7C and D). During L-FMAU treatment, the serum titers in woodchuck 336, harboring a prevalent type II mutant, also remained relatively constant (Fig. 7A and B). Virus titers decreased about 25-fold in woodchuck 342, with a mixture of wild-type and type I mutant virus. In contrast, the decline in virus titers in the two woodchucks harboring a mixture of wild-type and type V mutant virus (no. 326) or type V and type I mutants (no. 331) decreased as rapidly as in woodchucks harboring the wild type as the predominant species (Fig. 2). Residual virus detected in woodchuck 326 after 5 weeks showed a shift towards a wild-type/type I mutant combination, while that in woodchuck 331 showed a shift to a type II variant. Why the type I variant in combination with the wild-type virus appeared to produce greater resistance to L-FMAU (in 342) than in combination with the type V variant (in 331) is unclear. While the type I variant is unable to make the S envelope protein due to a stop codon introduced in the overlapping S gene by the mutation, it is not apparent why this would make any difference in such a short-term experiment. The results might be explainable if the polymerase functioned as a homodimeric protein, with resistance occurring through complementation of wild-type and type I mutant subunits.
L-FMAU treatment was not associated with hepatotoxicity.
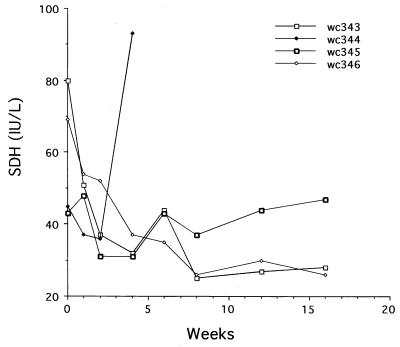
The liver toxicity of L-FMAU treatment was evaluated for each woodchuck by monitoring the serum concentrations of SDH and by analyzing liver tissue biopsy sections for signs of histopathology, including an enhanced PCNA staining index. SDH is an abundant hepatocellular enzyme, and its elevated level in serum is an indication of liver injury (20). As shown (Fig. 8), SDH levels were not increased by L-FMAU administration for up to 19 weeks. Moreover, no signs of enhanced liver injury were observed, either histologically or by enhanced leukocyte infiltration (Table 1). PCNA staining of liver sections also failed to show any pattern of enhanced cell proliferation that might have indicated liver toxicity (Table 1) (e.g., as has been observed during therapy with 2′-carbodeoxyguanosine [14]). These results are thus consistent with the lack of liver injury observed during administration of L-FMAU to duck HBV-infected ducks (1).
FIG. 8.
L-FMAU therapy did not induce a rise in serum SDH levels. Serum samples collected during the first 16 weeks of therapy were assayed for SDH, expressed in international units (IU) per liter (20). The level of SDH in chronically infected woodchucks is generally below 80 IU. No significant elevation was observed except in one woodchuck (wc344) at the time of death, apparently from sepsis unrelated to drug administration.
DISCUSSION
Antiviral therapy of chronic HBV infections presents a novel problem. At the beginning of treatment, every hepatocyte is apparently infected by the virus (4, 6, 16, 17, 19, 21, 22, 35). Moreover, the rate of turnover of this cell population, even with active liver disease, is low (34) (t1/2 > 1 week and, in “healthy” carriers, probably greater than 1 month). Thus, merely inhibiting virus replication would not readily eliminate the virus unless cccDNA, the template for viral RNA synthesis, had a short half-life within infected cells. However, this issue is still controversial. Some studies suggest that the DNA may have a high turnover rate (12, 15). On the other hand, data from the present and other studies suggest that this DNA is highly stable in vivo (14, 26, 43). In particular, these data suggest that if cccDNA has a finite life time, its half-life in the chronically infected liver is similar to that of infected hepatocytes.
One mechanism that would accelerate virus clearance is loss of cccDNA during cell division. In the present study, we sought indirect evidence for loss of cccDNA during mitosis by assaying for declines in the average cccDNA copy number in infected cells during therapy with the nucleoside analog L-FMAU. If this DNA is lost during mitosis and if it does not have any intrinsic instability in nondividing cells, then once virus DNA replication is blocked, the cccDNA copy number in infected cells should, ideally, remain fixed as the liver proliferates. That is, cells would either have lost cccDNA through the process of cell division or retained the original amount because they had not yet divided. The data suggest, however, that the cccDNA was distributed to daughter cells during proliferation of infected hepatocytes, producing the observed decline in the average copy number among cells that remained infected after prolonged therapy. Moreover, loss by mitosis requires that the infected-cell number decline virtually from the beginning of therapy, a possibility inconsistent with the experimental findings. The decline in copy number, by itself, could be explained by a model in which cccDNA is lost during mitosis and is also lost by decay in cells that have survived without division throughout the course of therapy. For instance, the observed results at 30 weeks of therapy could be modeled by an infected-cell death rate of 0.75% per day and a cccDNA half-life of 70 days. However, this model predicts that only 60% of the cells would remain infected after 6 weeks of therapy, a possibility at odds with the overall data.
An alternative possibility is that the low copy number was the result of new infections of cells that had lost existing cccDNA in the presence of L-FMAU. If so, the prevalent cccDNA in the liver might then have a drug resistance genotype. However, after 30 weeks of therapy, at which time the average cccDNA copy number among infected cells had declined at least 5- to 10-fold, the wild-type virus sequence was still prevalent in the cccDNA population. Our data thus favor but do not prove the hypothesis that cccDNA survives through mitosis and is distributed to each daughter cell, resulting in a decline in cccDNA copy number per cell. Data from a recent study (13) of WHV cccDNA survival in primary hepatocyte cultures that were induced to undergo limited proliferation by addition of epidermal growth factor were also consistent with this possibility.
Examination of the data in Fig. 2 and in a previous study (45) revealed an unexpected result. The type I mutation was sometimes detectable as a prevalent species in serum virus at early times in therapy, when virus titers were still declining. Since the same mutation may be associated with the later rebound of virus titers (Fig. 2) (45) and since the type I mutation confers L-FMAU resistance on a laboratory strain of WHV (Fig. 6), the reason for the continued decline at early times is not obvious. Several possibilities, not necessarily mutually exclusive, need to be considered. First, additional mutations outside the sequenced region of the polymerase may contribute to mutant fitness. This was not evident in a previous study, in which the complete pol gene of selected type I mutants was sequenced (45). However, the possibility has not been ruled out. Second, the type I mutant may be a common quasispecies that is generated as a result of errors during reverse transcription of a pregenomic RNA that was transcribed from wild-type cccDNA. In that case, it would be expected that virus titers would continue to decline until a significant fraction of this mutant virus could be converted to cccDNA. Third, the type I mutant may have a low replication rate, which, together with the need for coinfection with a virus producing the viral envelope proteins, may delay its spread to uninfected hepatocytes.
ACKNOWLEDGMENTS
We are grateful to Christoph Seeger, John Taylor, and Jesse Summers for helpful suggestions during this work and for a critical reading of the manuscript, to A. Cywinski and the DNA Sequencing Facility of the FCCC for sequence determinations, and to Wendy Foster (University of Adelaide) for technical assistance. Oligonucleotides were synthesized in the institutional DNA Synthesis Facility under the direction of T. Yeung.
This work was supported by USPHS grants AI-18641, 3P01-CA-4073711S1, and CA-06927 from the National Institutes of Health, by an appropriation from the Commonwealth of Pennsylvania, and by a project grant from the National Health and Medical Research Council of Australia (A.R.J.).
REFERENCES
- 1.Aguesse-Germon S, Liu S H, Chevallier M, Pichoud C, Jamard C, Borel C, Chu C K, Trepo C, Cheng Y C, Zoulim F. Inhibitory effect of 2′-fluoro-5-methyl-beta-l-arabinofuranosyl-uracil on duck hepatitis B virus replication. Antimicrob Agents Chemother. 1998;42:369–376. doi: 10.1128/aac.42.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrich C E, Coates L, Wu T T, Newbold J, Tennant B C, Summers J, Seeger C, Mason W S. In vitro infection of woodchuck hepatocytes with woodchuck hepatitis virus and ground squirrel hepatitis virus. Virology. 1989;172:247–252. doi: 10.1016/0042-6822(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 3.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 4.Barker L F, Chisari F V, McGrath P P, Dalgard D W, Kirschstein R L, Almeida J D, Edgington T S, Sharp D G, Peterson M R. Transmission of type B viral hepatitis to chimpanzees. J Infect Dis. 1973;127:648–652. doi: 10.1093/infdis/127.6.648. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomeusz A, Groenen L C, Locarnini S A. Clinical experience with famciclovir against hepatitis B virus and development of resistance. Intervirology. 1997;40:337–342. doi: 10.1159/000150566. [DOI] [PubMed] [Google Scholar]
- 6.Berquist K R, Peterson J M, Murphy B L, Ebert J W, Maynard J E, Purcell R H. Hepatitis B antigens in serum and liver of chimpanzees acutely infected with hepatitis B virus. Infect Immun. 1975;12:602–605. doi: 10.1128/iai.12.3.602-605.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block T M, Lu X, Mehta A, Park J, Blumberg B S, Dwek R. Role of glycan processing in hepatitis B virus envelope protein trafficking. Adv Exp Med Biol. 1998;435:207–216. doi: 10.1007/978-1-4615-5383-0_20. [DOI] [PubMed] [Google Scholar]
- 8.Buscher M, Reiser W, Will H, Schaller H. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell. 1985;40:717–724. doi: 10.1016/0092-8674(85)90220-x. [DOI] [PubMed] [Google Scholar]
- 9.Chayama K, Suzuki Y, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 10.Chu C K, Du J, Tennant B, Jacob J, Graham L A, Peek S, Korba B, Gerin J L, Witcher J W, Boudinot F D. Pharmacokinetic and pharmacodynamic studies of 1-(2-fluoro-5-methyl-β-l-arabinofuranosyl) uracil (L-FMAU) in the woodchuck model of hepatitis B virus (HBV) infection. Antivir Res. 1999;34:A52. [Google Scholar]
- 11.Chu C K, Ma T, Shanmuganathan K, Wang C, Xiang Y, Pai S B, Yao G Q, Sommadossi J P, Cheng Y C. Use of 2′-fluoro-5-methyl-β-l-arabinofuranosyluracil as a novel antiviral agent for hepatitis B virus and Epstein-Barr virus. Antimicrob Agents Chemother. 1995;39:979–981. doi: 10.1128/aac.39.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Civitico G M, Locarnini S A. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology. 1994;203:81–89. doi: 10.1006/viro.1994.1457. [DOI] [PubMed] [Google Scholar]
- 13.Dandri M, Burda M R, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139–146. doi: 10.1053/jhep.2000.8701. [DOI] [PubMed] [Google Scholar]
- 14.Fourel I, Cullen J M, Saputelli J, Aldrich C E, Schaffer P, Averett D R, Pugh J, Mason W S. Evidence that hepatocyte turnover is required for rapid clearance of duck hepatitis B virus during antiviral therapy of chronically infected ducks. J Virol. 1994;68:8321–8330. doi: 10.1128/jvi.68.12.8321-8330.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genovesi E V, Lamb L, Medina I, Taylor D, Seifer M, Innaimo S, Colonno R J, Standring D N, Clark J M. Efficacy of the carbocyclic 2′-carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob Agents Chemother. 1998;42:3209–3217. doi: 10.1128/aac.42.12.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 17.Guo J-T, Zhou H, Liu C, Aldrich C, Saputelli J, Whitaker T, Barrasa M L, Mason W S, Seeger C. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J Virol. 2000;74:1495–1505. doi: 10.1128/jvi.74.3.1495-1505.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honkoop P, Niesters H G, de Man R A, Osterhaus A D, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B: incidence and patterns. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 19.Hoofnagle J H, Michalak T, Nowoslawski A, Gerety R J, Barker L F. Immunofluorescence microscopy in experimentally induced, type B hepatitis in the chimpanzee. Gastroenterology. 1978;74:182–187. [PubMed] [Google Scholar]
- 20.Hornbuckle W E, Graham E S, Roth L, Baldwin B H, Wickenden C, Tennant B C. Laboratory assessment of hepatic injury in the woodchuck (Marmota monax) Lab Anim Sci. 1985;35:376–381. [PubMed] [Google Scholar]
- 21.Jilbert A R, Wu T T, England J M, Hall P M, Carp N Z, O'Connell A P, Mason W S. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J Virol. 1992;66:1377–1388. doi: 10.1128/jvi.66.3.1377-1388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajino K, Jilbert A R, Saputelli J, Aldrich C E, Cullen J, Mason W S. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J Virol. 1994;68:5792–5803. doi: 10.1128/jvi.68.9.5792-5803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodama K, Ogasawara N, Yoshikawa H, Murakami S. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J Virol. 1985;56:978–986. doi: 10.1128/jvi.56.3.978-986.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kukhanova M, Lin Z Y, Yas'co M, Cheng Y C. Unique inhibitory effect of 1-(2′-deoxy-2′-fluoro-beta-l-arabinofuranosyl)-5-methyluracil 5′-triphosphate on Epstein-Barr virus and human DNA polymerases. Biochem Pharmacol. 1998;55:1181–1187. doi: 10.1016/s0006-2952(97)00598-4. [DOI] [PubMed] [Google Scholar]
- 25.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 26.Luscombe C, Pedersen J, Uren E, Locarnini S. Long-term ganciclovir chemotherapy for congenital duck hepatitis B virus infection in vivo: Effect on intrahepatic-viral DNA, RNA, and protein expression. Hepatology. 1996;24:766–773. doi: 10.1053/jhep.1996.v24.pm0008855174. [DOI] [PubMed] [Google Scholar]
- 27.Mackey D, Sugden B. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol Cell Biol. 1999;19:3349–3359. doi: 10.1128/mcb.19.5.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason W S, Aldrich C, Summers J, Taylor J M. Asymmetric replication of duck hepatitis B virus DNA in liver cells: free minus-strand DNA. Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason W S, Cullen J, Moraleda G, Saputelli J, Aldrich C E, Miller D S, Tennant B, Frick L, Averett D, Condreay L D, Jilbert A R. Lamivudine therapy of WHV-infected woodchucks. Virology. 1998;245:18–32. doi: 10.1006/viro.1998.9150. [DOI] [PubMed] [Google Scholar]
- 30.Mehta A, Block T M, Dwek R A. The role of N-linked glycosylation in the secretion of hepatitis B virus. Adv Exp Med Biol. 1998;435:195–205. doi: 10.1007/978-1-4615-5383-0_19. [DOI] [PubMed] [Google Scholar]
- 31.Miller R H, Robinson W S. Hepatitis B virus DNA forms in nuclear and cytoplasmic fractions of infected human liver. Virology. 1984;137:390–399. doi: 10.1016/0042-6822(84)90231-9. [DOI] [PubMed] [Google Scholar]
- 32.Moraleda G, Saputelli J, Aldrich C E, Averett D, Condreay L, Mason W S. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. 1997;71:9392–9399. doi: 10.1128/jvi.71.12.9392-9399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 34.Nowak M A, Bonhoeffer S, Hill A M, Boehme R, Thomas H C, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Peek, S. F., P. J. Cote, J. R. Jacob, I. A. Toshkov, W. E. Hornbuckle, B. H. Baldwin, F. V. Wells, C. K. Chu, J. L. Gerin, B. C. Tennant, and B. E. Korba. Antiviral activity of clevudine [l-FMAU, 1-(2-fluoro-5-methyl-β-l-arabinofuranosyl) uracil] against WHV replication and gene expression in chronically infected woodchucks (M. monax). Hepatology, in press. [DOI] [PubMed]
- 35.Ponzetto A, Cote P J, Ford E C, Purcell R H, Gerin J L. Core antigen and antibody in woodchucks after infection with woodchuck hepatitis virus. J Virol. 1984;52:70–76. doi: 10.1128/jvi.52.1.70-76.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 37.Summers J, Smith P M, Horwich A L. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutation in HBV DNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 39.van der Eb A J, Graham F L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65:826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- 40.Wahl G M, Stern M, Stark G R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci USA. 1979;76:3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Y, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y-Y, Summers J. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J Virol. 2000;74:5257–5265. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T, Guo J-T, Nunes F A, Molnar-Kimber K L, Wilson J M, Aldrich C E, Saputelli J, Litwin S, Condreay L, Seeger C, Mason W S. Combination therapy with lamivudine and adenovirus causes transient suppression of chronic woodchuck hepatitis virus infections. J Virol. 2000;74:11754–11763. doi: 10.1128/jvi.74.24.11754-11763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou T, Saputelli J, Aldrich C E, Deslauriers M, Condreay L D, Mason W S. Emergence of drug-resistant populations of woodchuck hepatitis virus in woodchucks treated with the antiviral nucleoside lamivudine. Antimicrob Agents Chemother. 1999;43:1947–1954. doi: 10.1128/aac.43.8.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]