Abstract
Tyrosine kinase inhibitors (TKIs) have revolutionized the treatment landscape for chronic myeloid leukemia (CML). However, TKI resistance poses a significant challenge, leading to treatment failure and disease progression. Resistance mechanisms include both BCR::ABL1-dependent and BCR::ABL1-independent pathways. The mechanisms underlying BCR::ABL1 independence remain incompletely understood, with CML cells potentially activating alternative signaling pathways, including the AKT/mTOR and JAK2/STAT5 pathways, to compensate for the loss of BCR::ABL1 kinase activity. This study explored tumoral VISTA (encoded by VSIR) as a contributing factor to TKI resistance in CML patients and identified elevated tumoral VISTA levels as a marker of resistance and poor survival. Through in vitro and in vivo analyses, we demonstrated that VSIR knockdown and the application of NSC-622608, a novel VISTA inhibitor, significantly impeded CML cell proliferation and induced apoptosis by attenuating the AKT/mTOR and JAK2/STAT5 pathways, which are crucial for CML cell survival independent of BCR::ABL1 kinase activity. Moreover, VSIR overexpression promoted TKI resistance in CML cells. Importantly, the synergistic effect of NSC-622608 with TKIs offers a potent therapeutic avenue against both imatinib-sensitive and imatinib-resistant CML cells, including those harboring the challenging T315I mutation. Our findings highlight the role of tumoral VISTA in mediating TKI resistance in CML, suggesting that inhibition of VISTA, particularly in combination with TKIs, is an innovative approach to enhancing treatment outcomes in CML patients, irrespective of BCR::ABL1 mutation status. This study not only identified a new pathway contributing to TKI resistance but also revealed the possibility of targeting tumoral VISTA as a means of overcoming this significant clinical challenge.
Keywords: Tumoral VISTA, TKI resistance, NSC-622608, Chronic myeloid leukemia
INTRODUCTION
Chronic myeloid leukemia (CML) is characterized by the presence of the Philadelphia chromosome (Ph+), which harbors the oncogenic BCR::ABL1 fusion gene derived from the t(9;22) (q34;q11) chromosomal translocation (Jabbour and Kantarjian, 2022; Scalzulli et al., 2022). Tyrosine kinase inhibitors (TKIs), which act as single agents, have achieved remarkable success in treating CML, with an 80% 10-year survival rate achieved by binding to the kinase BCR::ABL1 and inhibiting downstream phosphorylation (Jabbour and Kantarjian, 2022). However, treatment failure can be observed in 20-25% of CML patients, occurring either dependent or independent of the kinase BCR-ABL1. Mutations in fusion genes classified as BCR::ABL1-dependent mechanisms impair TKI binding to target kinases and lead to TKI drug resistance. Notably, the “gatekeeper” mutation T315I is more prevalent than other mutations and has been associated with a poor response to newly developed TKIs (Liu et al., 2021). Despite the efficacy of first- and second-generation TKIs such as imatinib (IM) and dasatinib (Das), over 20% of CML patients exhibit resistance or intolerance (De Novellis et al., 2021). The third-generation TKI olverembatinib (OL) demonstrates efficacy against T315I but has notable toxicity (Jiang et al., 2022).
Moreover, BCR::ABL1-independent mechanisms have been shown to occur in the later stages of CML progression (Stuckey et al., 2022). In CML cells, the loss of BCR::ABL1 kinase activity can be compensated for by activation of alternative signaling pathways to overcome the inhibition of BCR::ABL1. The loss of BCR::ABL1 kinase activity can be compensated for by activation of alternative signaling pathways in CML cells, including the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway and phosphoinositide 3-kinase (PI3K)/AKT pathway, thus overcoming the inhibition of BCR::ABL1 (Loscocco et al., 2019). Consequently, CML cells can proliferate and survive despite effective BCR::ABL1 inhibition, and the JAK/STAT and PI3K/AKT pathways are some of the pathways that contribute to TKI resistance (Alves et al., 2021). Recently, immune checkpoint receptor dysregulation has been reported at CML diagnosis, but its role in maintaining remission after TKI cessation is poorly understood (Irani et al., 2023). The inhibitory immune checkpoint VISTA (V-domain Ig suppressor of T-cell activation), which is encoded by VSIR, is expressed in tumor cells and diverse immune cells (Zheng et al., 2022). Previous studies have demonstrated that VISTA is overexpressed in many cancers, including ovarian and endometrial cancers, particularly those with low immunotherapy responses or acquired resistance (Mulati et al., 2019; Mortezaee et al., 2022). Interestingly, in addition to being a receptor, VISTA is also a ligand (Muller et al., 2020). Understanding the molecular mechanisms underlying the therapeutic effect of anti-VISTA agents is crucial, given that human clinical trials are currently in progress. To date, the role of VISTA as a ligand and its effect on myeloid cells are poorly understood (Schaafsma et al., 2022), while its efficacy in treating myeloid malignancies and its relative contributions to proliferative signaling and function remain unclear (Hosseinkhani et al., 2021).
This study aimed to elucidate the functional role of tumoral VISTA in CML. In the present study, we observed that the expression of tumoral VISTA in CML cells was correlated with TKI resistance and poor survival. Our results revealed that knockdown of VISTA not only impaired CML proliferation but also induced apoptosis. Transcriptomic analyses revealed that VISTA regulated the JAK2/STAT5 and PI3K/AKT/mTOR signaling pathways, which are involved in CML cell survival. Furthermore, we explored whether NSC-622608 (a small molecule ligand inhibitor of VISTA (Gabr and Gambhir, 2020)) induces the apoptosis of CML cells with or without the T315I mutation both in vivo and ex vivo. Taken together, our findings demonstrate that the inhibition of tumoral VISTA may overcome TKI resistance in CML patients and may be an effective treatment in combination with TKIs.
MATERIALS AND METHODS
Cell culture
CML cell lines, namely, KBM5, KBM5-T315I (harboring the T315I mutation in the BCR::ABL1 fusion gene), K562 (cryopreserved at Zhujiang Hospital (Guangzhou, China) and K562-R (K562 resistant to imatinib, purchased from CTCC, CTCC-0107-NY), were cultivated in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C and 5% carbon dioxide. Moreover, K562-R and KBM5-T315I cells were cultured in the aforementioned medium supplemented with 1.0 μM imatinib (IM), and IM was subsequently removed before the experimental procedures, followed by a wash-out period lasting 2–3 days. Prior to experimentation, the cell lines were subjected to short tandem repeat (STR) profiling to confirm their mycoplasma-free status. Only cells with fewer than 15 passages were chosen for experimental purposes.
VSIR expression profiles in CML
We analyzed the expression profiles of VSIR in CML patients using several publicly available datasets, namely, GSE130404, GSE44589, GSE33224, GSE51082, GSE77191, and GSE24739. These datasets comprise a total of 247 bone marrow samples from CML patients and 167 samples from individuals with other hematologic malignancies, all collected at the time of diagnosis. The data were generated using Affymetrix HG-U133A arrays, and CEL files were processed using custom CDF annotation packages.
Furthermore, we explored the correlation between VSIR expression and outcomes of TKI treatment by analyzing 388 CML samples from the GSE44589, GSE24493, GSE203442, GSE33224, and GSE130404 datasets. All microarray data were sourced from the GEO database (http://www.ncbi.nih.gov/geo) in MINiML file format. Box plots were constructed using the boxplot function, and PCA plots were generated using the ggord package in R software (version: 4.3.2, https://www.r-project.org/).
Differential expression analysis of mRNAs was conducted using the limma package in R. The adjusted p value was applied to correct for false-positive results in GEO datasets, with the threshold for differential expression set at “Adjusted p<0.05 and Log (Fold Change) >1 or Log (Fold Change)<−1.”
To decipher the potential functions of the identified targets, functional enrichment analysis was performed. The Gene Ontology (GO) tool was utilized for annotating genes based on molecular function (MF), biological pathways (BP), and cellular components (CC). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis provided insights into gene functions and high-level genome functional information. For a comprehensive understanding of mRNA carcinogenesis, the clusterProfiler package (version: 3.18.0) in R was used to analyze GO functions and enrich KEGG pathways. Heatmaps were generated using the pheatmap package in R.
Establishment and characterization of cell lines with stable VSIR knockdown or overexpression
To suppress the expression of VSIR, we conducted knockdown experiments by transfecting CML cells with lentiviral vectors (GV493-hU6-MCS-CBh-gcGFP-IRES-puromycin) expressing small hairpin RNA (shRNA). The sequences of the annealed oligonucleotide fragments encoding short hairpin RNA were as follows:
sh-VSIR-1: ccCTGACTCTCCAAACTTTGA;
sh-VSIR-2: ccGTATTCCCTGTATGTCTGT;
and sh-VSIR-3: cgGATGGACAGCAACATTCAA
Increased expression of VSIR in CML cells was achieved using a lentiviral vector (Ubi-MCS-CBh-gcGFP-IRES-puromycin) expressing VSIR that was subsequently introduced into the cells. Empty lentiviral vectors were used as controls. The transfected cells were treated with 2 μg/mL puromycin on the third day posttransfection, and stable VSIR knockdown and overexpression were confirmed through quantitative real-time polymerase chain reaction (qRT‒PCR) and Western blot assays.
qRT‒PCR
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions and then reverse transcribed to cDNA using a HiScript® III RT SuperMix kit (Vazyme, Nanjing, China). qRT‒PCR was performed using SYBR® Green Premix (Accurate Biology, Changsha, China), which targets the VSIR, JAK2 and STAT5 genes, with GAPDH included as an internal amplification control. The primer sequences for the genes used were as follows:
VSIR-F: TCATCCTGCTCCTGGTCTACAA;
VSIR-R: CTTGAATGTTGCTGTCCATCCG;
JAK2-F: TGCCGGTATGACCCTCTACA
JAK2-R: ACCAGCACTGTAGCACACTC
STAT5-F: CACAGATCAAGCAAGTGGTC
STAT5-R: CTGTCCATTGGTCGGCGTAA
GAPDH-F: GCACCGTCAAGGCTGAGAAC
GAPDH-R: TGGTGAAGACGCCAGTGGA.
Cell viability assay
The proliferation of CML cells across various groups was assessed using a Cell Counting Kit-8 (CCK-8) assay kit (Beyotime, Shanghai, China) following the manufacturer’s instructions. In brief, CML cells were exposed to Das, OL, NSC622608, Das+NSC622608, or OL+NSC622608 for 24, 48, or 72 h. Subsequently, each well was treated with 10 μL of CCK-8 reagent, and the optical density (OD) was measured at 450 nm after 4 h of incubation at 37°C.
Immunofluorescence assay
The cells were initially spread on slides, fixed for 10 min with paraformaldehyde, and permeabilized for 15 min with Triton X-100. Subsequently, the cells were blocked with 2% BSA for 1 h at 4°C, incubated overnight with the primary antibody at 4°C, and then incubated for 1 h with a FITC-labeled secondary antibody at 37°C (Invitrogen, Carlsbad, CA, USA). Finally, the cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 15 min at 37°C, and the slides were sealed with glycerin and subjected to fluorescence microscopy.
Western blot analysis
Proteins were extracted from cells using RIPA buffer, quantified, and subjected to Western blotting as previously described (Chen et al., 2018). Initially, protein samples were obtained through cell lysis, followed by separation through SDS‒PAGE. The separated proteins were then transferred to a membrane. To prevent nonspecific binding, the membrane was blocked. Subsequently, the membrane was incubated with a antibodies (VISTA, GAPDH, PI3K, mTOR, p-mTOR, AKT, p-AKT, 4EBP1, p70s6k, STAT5, p-STAT5, Bcl2, Bad, Bax, Caspase3, Cleaved caspase3 and Cleaved PARP), followed by washing to remove unbound antibodies. Next, an HRP-conjugated secondary antibody from a different source was added, followed by another round of washing to remove unbound secondary antibodies. Binding antibodies were visualized using a specific substrate, and Western blot images were obtained using an imaging system.
RNA sequencing (RNA-seq)
Total RNA was extracted from KBM5-sh-con/KBM5-sh-VSIR-3 and KBM5-T315I/KBM5-T315I+NSC622608 (20 μM, 72 h) cells using an RNAiso Plus Kit according to the manufacturer’s instructions and sequenced on an Illumina HiSeq 3000 platform (Illumina, San Diego, CA, USA). We identified DEGs using the edge R package based on RefSeq ID and analyzed pathway enrichment using the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Flow cytometry
Cell apoptosis was evaluated using double staining with Annexin V and PI (Keygen, Nanjing, China). The apoptosis rate was determined by flow cytometry (Beckman, Pasadena, CA, USA, Cytoflex), and cells showing Annexin V or Annexin V-plus-PI expression were considered apoptotic.
Animal experiments
The animal experiments adhered to the Laboratory Animal Care Guidelines of Southern Medical University (Guangzhou, China). Seven-week-old athymic BALB/c nu/nu female mice (18-20 g) were obtained from the animal experimental center of Zhujiang Hospital and used in the study. Briefly, 2×106 KBM5-T315I cells in 100 μL of PBS were subcutaneously injected into the mice. The mice were then treated with KBM5-T315I with NSC-622608 (10 mg/kg) or PBS as a control via intraperitoneal (i.p.) injection every two days. The tumor volumes were limited to 3000 mm3. Upon completion of the experiment, the animals were sacrificed, the tumors were removed, and the tumor volumes were calculated as follows: V=0.5×L×W2, where L and W represent length and width, respectively. At the conclusion of the experimental phase, the tumor tissues were meticulously excised, and the subsequent steps involved specific procedures for histopathological examination, including hematoxylin and eosin (HE) staining and Ki67 immunostaining. Briefly, paraffin-embedded tissue sections were meticulously sliced and subjected to the standard process of deparaffinization and hydration. A subset of these sections was subjected to HE staining, facilitating comprehensive observation of the overall tissue architecture and morphology. Simultaneously, another set of sections underwent immunostaining with Ki67 antibody, enabling the identification and labeling of actively proliferating cells within the cell cycle, thereby yielding valuable insights into cellular proliferation dynamics. After staining, the tissue sections were subjected to subsequent steps involving dehydration, clarification, and mounting. The final stained sections were meticulously examined under a microscope, allowing for a detailed analysis of tissue structure through HE staining and the assessment of cellular proliferation activity through Ki67 staining. This methodological approach in immunohistochemical analysis provided critical histological information, contributing substantively to the broader scope of research exploration. The research was approved by the ethics committee of the Zhujiang hospital (NO. LAEC-2022-128).
Statistical analysis
All experiments were performed in triplicate, and the results are expressed as the means and standard deviations. Statistical analyses were conducted using GraphPad Prism software version 5.0, with one-way analysis of variance (ANOVA) followed by Tukey’s HSD test used to compare means among groups. Statistical significance was determined at *p<0.05, **p<0.01, and ***p<0.001.
RESULTS
Tumoral VISTA in CML patients and cells involved in TKI resistance and poor survival
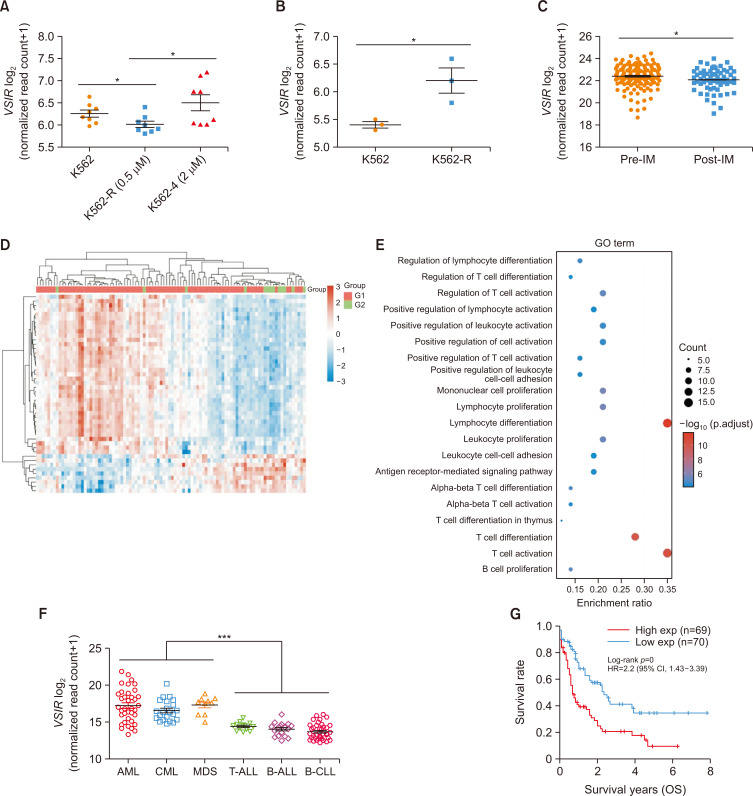
To explore the relationship between VISTA expression and the therapeutic effect of TKIs, we analyzed the GSE203442 dataset and found that imatinib-resistant K562 (K562-R) cells exhibited greater VSIR expression than did IM-sensitive K562 cells (p<0.05, Fig. 1A). Similarly, K562 cells that were resistant to TKIs (K562R) had greater expression of VSIR than TKI-sensitive K562 cells in GSE208314 (p<0.05, Fig. 1B). We subsequently analyzed the transcriptional landscapes from the GSE44589 dataset and found lower VSIR expression in imatinib (IM)-treated patients (n=63) than in nontreated CML patients (n=135) (p<0.05, Fig. 1C). Among the 183 CML patients who received IM treatment in the GSE130404, GSE44589, and GSE33224 datasets, 124 patients exhibited a drug response (IM-R), while 59 patients showed no response (IM-NR). Using the limma package in R software to study the differentially expressed mRNAs, a GO enrichment analysis was performed to analyze the functional distribution of the differentially expressed mRNAs between IM-R and IM-NR. We found that the DEGs were mainly enriched in terms of T-cell activation (Fig. 1D, 1E). Transcriptional profiles of VSIR in hematologic malignant samples across the GSE51082 dataset revealed higher levels in myeloid cancers (AML, CML, and MDS) than in lymphoid malignant cancers (T-ALL, Pre-B-ALL, and B-ALL) (Fig. 1F, p<0.001). Finally, considering the long survival period of patients in the chronic phase of CML and the lack of statistical data on survival rates, we analyzed the effect of VSIR expression on survival time in acute myeloid leukemia, which has a greater association with CML than other leukemias. High VSIR expression was associated with a shorter survival time in patients (log-rank p<0.001) (Fig. 1G).
Fig. 1.
Tumoral VISTA in CML patients and cells is involved in TKI resistance and poor survival. (A) Scatter plot showing the relative expression of VSIR in K562 cells resistant to 2 μM IM (n=8) compared with K562 cells resistant to 0.5 μM IM (n=8) and K562 cells (n=8) based on the GSE203442 dataset (p<0.05). (B) The relative expression of VSIR in K562 cells resistant to TKIs (K562-R cells, n=3) compared with that in K562 cells (n=3) was analyzed in GSE208314 (p<0.05). (C) VSIR expression in the GSE44589 dataset was lower in IM-treated patients (n=63) than in untreated CML patients (n=135) (p<0.05). (D) There were 183 CML patients who received IM treatment in the GSE130404, GSE44589 and GSE33224 datasets, among which 124 patients achieved a drug response (IM-R) and 59 patients showed no response (IM-NR). A heatmap of the differential gene expression between IM-R and IM-NR and the top 50 upregulated genes and the top 50 downregulated genes are shown (red indicates upregulated genes; blue indicates downregulated genes). (E) The abscissa indicates the gene ratio, and the enriched pathways are presented on the ordinate. GO analysis of potential targets of DGE between IM-R and IM-NR based on the Cluster Profiler package in R software (version: 4.3.2). Colors represent the significance of differential enrichment, and the size of the circles represents the number of genes; the larger the circle is, the greater the number of genes. According to the enrichment results, p<0.05 or FDR <0.05 was considered to indicate a meaningful pathway (an enrichment score of −log10 (P) greater than 1.3). (F) The GSE51082 dataset was used to verify VSIR mRNA levels in hematologic malignancies, and the scatter plot shows that the levels of VSIR expression were greater in AML (n=37), CML (n=22) and MDS (n=10) than in T-ALL (n=13), Pre-B-ALL (n=17) and B-CLL (n=41) (p<0.001). (G) Survival time of AML patients with different expression levels of VSIR (log-rank p<0.001). Asterisks (*) indicate significance, *** indicates p<0.001, and * indicates p<0.05.
Tumoral VISTA expression profiles in CML patients and cells
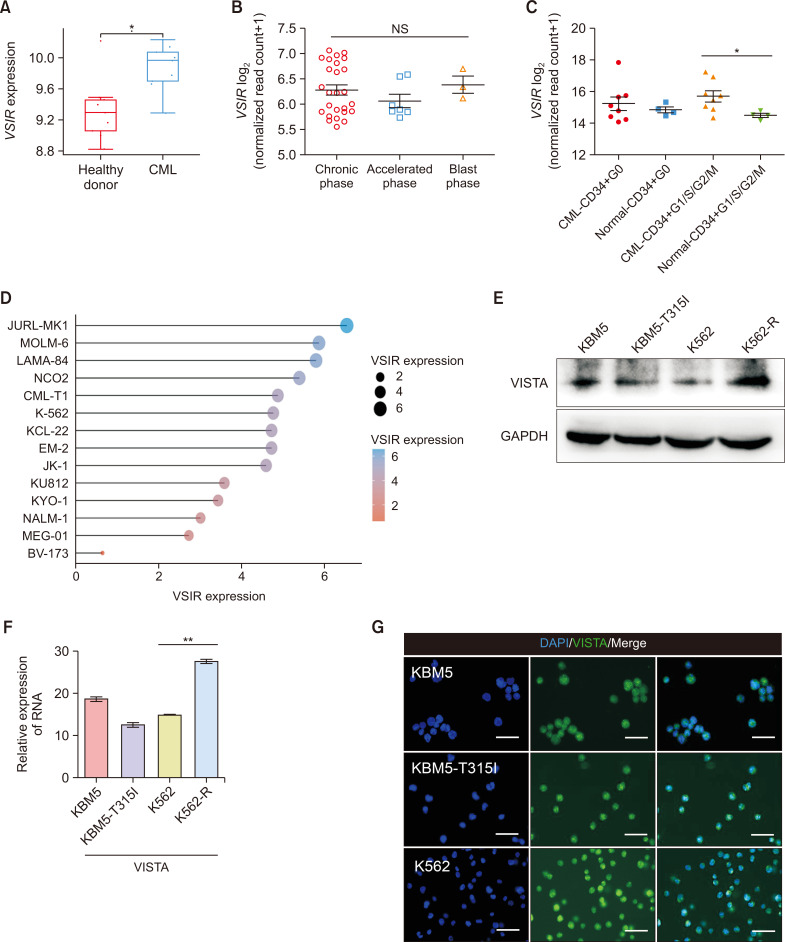
Analysis of the transcriptional profiles of VSIR through the GSE33075 dataset showed that the expression of VSIR in CML patients (n=9) was greater than that in healthy donors (n=9) (Fig. 2A, p<0.05). Subsequently, we analyzed 40 CML samples from GSE77191 and detected VSIR expression during the chronic, accelerated, and blast phases of CML (Fig. 2B). Screening of the GSE24739 dataset revealed that the VSIR expression of CD34+ G1/S/G2/M cells was greater in CML patients than in normal individuals (Fig. 2C, p<0.05). Next, the CML cell line mRNA expression matrix was obtained from the CCLE dataset (https://portals.broadinstitute.org/ccle). The analysis was performed using the R v4.0.3 software package ggplot2 (v3.3.3) (Fig. 2D). We employed Western blot and qRT‒PCR analyses to explore the levels of VISTA expression in KBM5, KBM5-T315I, K562 and K562-R cells and found that CML cells expressed VISTA at both the protein and RNA levels (Fig. 2E, 2F). We also performed immunofluorescence analysis to determine the localization of VISTA in CML cells and found that it was expressed in both the cytoplasm and nucleus (Fig. 2G).
Fig. 2.
The expression of VISTA in CML patients and cells. (A) Scatter plot showing the levels of VSIR expression in CML patients (n=9) and healthy donors (n=9) in the GSE33075 dataset (p<0.05); (B) Patterns of VSIR expression in 40 CML samples from the GSE77191 dataset across three phases (chronic, accelerated, and blast phases). (C) The VSIR of CD34+ G1/S/G2/M (n=4) CML patients was greater than that of normal CD34+ G1/S/G2/M (n=8) CML patients based on the GSE24739 dataset (p<0.05). (D) The cell line mRNA expression matrix of CML cells was obtained from the CCLE dataset (https://portals.broadinstitute.org/ccle). The analysis was performed with the R v4.0.3 software package ggplot2 (v3.3.3). (E, F) Protein and mRNA levels of VISTA in CML cells (KBM5, KBM5-T315I, K562 and K562-R cells) determined via Western blotting and qRT‒PCR. (G) Immunofluorescence results showing the localization of VISTA (green) in both the cytoplasm and nucleus of CML cells (blue: DAPI, bar: 50 μm). Asterisks (*) indicate significance, ** indicates p<0.01, * indicates p<0.05, and NS indicates no significance.
Effect of second- and third-generation TKIs on CML cell lines
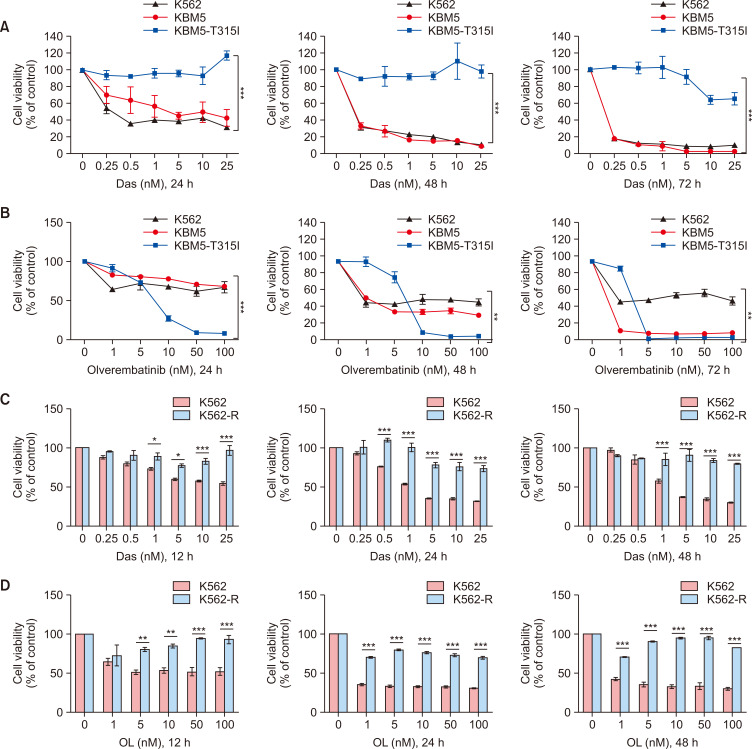
To assess the cytotoxic effects of second- and third-generation TKIs, we initially treated KBM5, KBM5-T315I, K562 and K562-R cells with dasatinib (Das, a second-generation TKI) and olverembatinib (OL, a third-generation TKI) in a dose-dependent manner. Subsequently, a CCK-8 assay was performed to analyze cell viability. Das significantly suppressed the survival of K562 and KBM5 cells but had no substantial impact on KBM5-T315I cells at 24 h, 48 h, or 72 h posttreatment (Fig. 3A, p<0.001). Notably, OL significantly reduced the viability of all CML cell lines, particularly KBM5-T315I (Fig. 3B, p<0.01). As expected, K562-R cells incubated with Das (0-25 nM) or OL (0-100 nM) for 12 h, 24 h or 48 h were more insensitive to Das (Fig. 3C, p<0.001) or OL (Fig. 3D, p<0.001) than were K562 cells.
Fig. 3.
The effect of second- and third-generation TKIs on CML cell lines. (A) CCK-8 assays showed that Das significantly suppressed the viability of K562 and KBM5 cells at 24 h, 48 h and 72 h but had no effect on KBM5-T315I cells (p<0.001). (B) CCK-8 assays showed that OL suppressed the viability of K562, KBM5 and KBM5-T315I cells at 24 h, 48 h and 72 h (p<0.01). (C, D) K562 and K562-R cells were treated with Das and OL, and a CCK-8 assay was used to detect the OD at 450 nm. Asterisks (*) indicate significance, *** indicates p<0.001, ** indicates p<0.01, * indicates p<0.05, and NS indicates no significance.
Tumoral VISTA promotes CML cell proliferation and TKI resistance
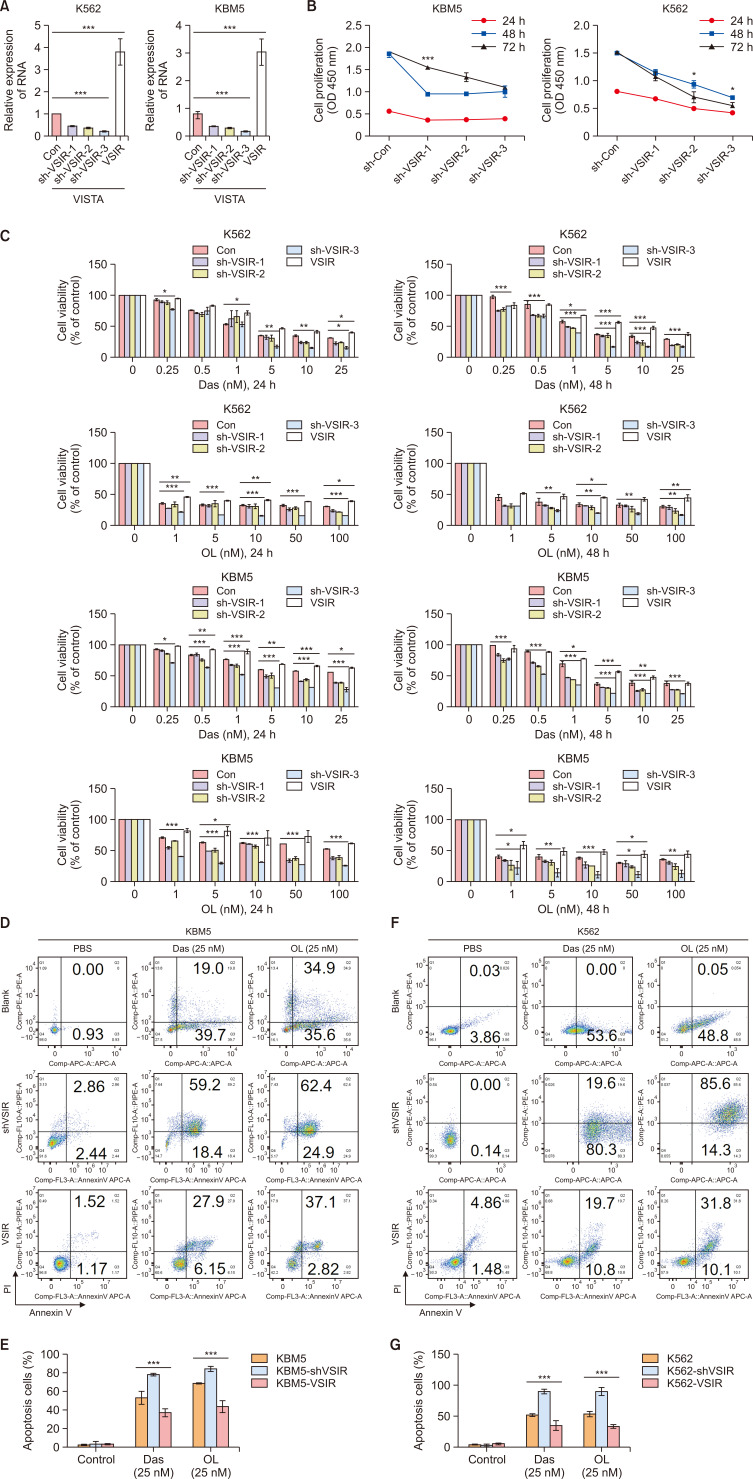
We elucidated the mechanism underlying the action of VSIR in CML cells by knocking down (sh-VSIR1, sh-VSIR2 and sh-VSIR3) and overexpressing (VSIR) VSIR in KBM5 and K562 cells through lentiviral transfection. The relative mRNA expression of VSIR in K562 and KBM5 cells decreased in sh-VSIR-transfected cells and increased in VSIR-transfected cells compared to sh-con-transfected cells (p<0.001) (Fig. 4A). Moreover, the CCK-8 assay results showed that VSIR knockdown inhibited the proliferation of K562 and KBM5 cells (Fig. 4B, p<0.001, p<0.05). To observe the correlation between VISTA and TKI resistance, K562-sh-VSIR1, sh-VSIR2, sh-VSIR3, and VSIR cells and KBM5-sh-VSIR1, sh-VSIR2, sh-VSIR3, and VSIR cells were incubated with Das (0-25 nM) or OL (0-100 nM) for 24 h or 48 h. Interestingly, knocking down VISTA (sh-VSIR1, sh-VSIR2 and sh-VSIR3) improved the killing effects of Das (0.25, 0.5, 1, 5, 10, 25 nM, 24 h and 48 h, p<0.001) and OL (1, 5, 10, 50, 100 nM, 24 h and 48 h, p<0.05); in contrast, CML cells that overexpressed VISTA (K562-VSIR and KBM5-VSIR) were more resistant to Das (0.25, 0.5, 1, 5, 10, 25 nM, 24 h and 48 h, p<0.001) and OL (1, 5, 10, 50, 100 nM, 24 h and 48 h, p<0.05) than were K562 and KBM5 cells (Fig. 4C). To analyze the effects of Das and OL on the apoptosis of K562-sh-VSIR, VSIR, KBM5-sh-VSIR and VSIR cells, we conducted flow cytometry to detect the percentage of apoptotic cells. Flow cytometry analysis revealed that Das and OL induced more apoptosis in K562-sh-VSIR and KBM5-sh-VSIR cells than in K562 and KBM5 cells, while overexpression of VISTA decreased the percentage of apoptotic cells induced by Das (25 nM, p<0.001) and OL (25 nM, p<0.001) (Fig. 4D-4G).
Fig. 4.
Tumoral VISTA promotes CML cell proliferation and TKI resistance. (A) The relative mRNA expression of VSIR in sh-VSIR-transfected K562 and KBM5 cells and sh-con cells (p<0.001). (B) CCK-8 results showing that VSIR knockdown inhibited the proliferation of KBM5 (p<0.001) and K562 cells (p<0.05). (C) CCK-8 and flow cytometry showed that knockdown of VISTA (sh-VSIR1, sh-VSIR2 and sh-VSIR3) improved the killing effects of Das (0.25, 0.5, 1, 5, 10, 25 nM, 24 h and 48 h, p<0.001) and OL (1, 5, 10, 50, 100 nM, 24 h and 48 h, p<0.05); in contrast, CML cells overexpressing VISTA (K562-VSIR and KBM5-VSIR) were more resistant to Das (0.5, 1, 5, 10, 25 nM, 24 h and 48 h, p<0.001) and OL (1, 5, 10, 50, 100 nM, 24 h and 48 h, p<0.05). (D-G) Flow cytometry and rate of apoptosis cells of K562 and KBM5 cells (Control, sh-VSIR and VSIR) treated with Das (25 nM) and OL (25 nM). Asterisks (*) indicate significance, *** indicates p<0.001, ** indicates p<0.01, * indicates p<0.05, and NS indicates no significance.
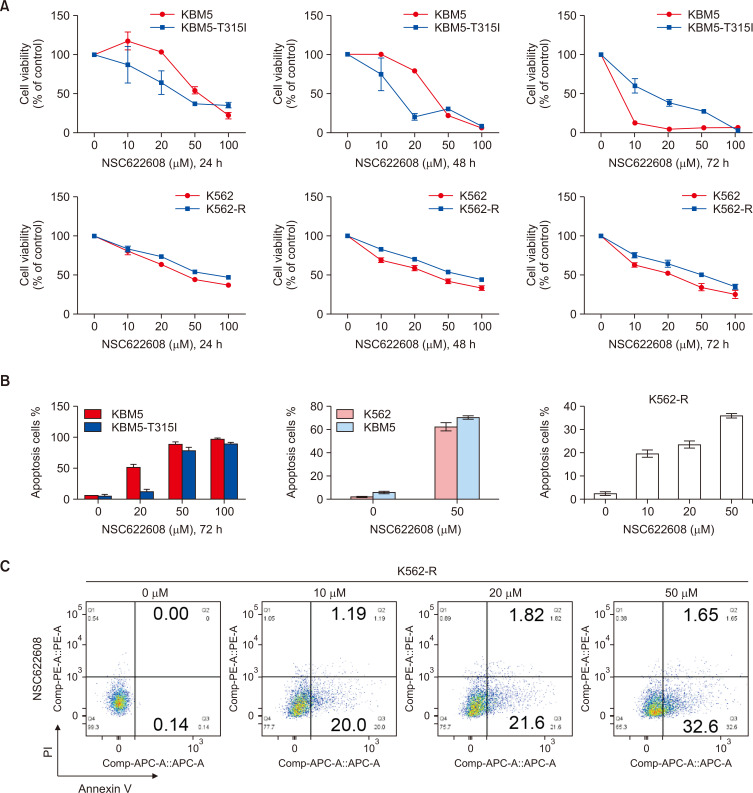
NSC-622608 inhibits the proliferation of CML cells and induces apoptosis
NSC-622608 is a recently discovered small molecule ligand inhibitor of VISTA, and its impact on hematologic malignancies remains unknown. To further study the effect of NSC-622608 on CML cells, we treated KBM5, KBM5-T315I, K562 and K562-R cells with NSC-622608 at 0, 5, 10, 20, 50, and 100 μM for 24 h, 48 h and 72 h. In this study, the CCK-8 results showed a marked inhibition of cell proliferation in a dose-dependent manner (Fig. 5A). To analyze the effects of NSC622608 on the apoptosis of CML cells, we conducted flow cytometry analysis to detect the percentage of apoptotic cells. The flow cytometry results revealed that NSC-622608 (50 μM) induced significant apoptosis in KBM5 (78%), K562 (62%), and KBM5-T315I (60%) cells (Fig. 5B). More importantly, NSC-622608 induced the apoptosis of K562-R cells in a dose-dependent manner, and the percentage of apoptotic cells was 38% after treatment with NSC-622608 (50 μM) for 24 h (Fig. 5C).
Fig. 5.
NSC-622608 inhibits the proliferation of CML cells and induces apoptosis. (A) CCK-8 assay results showing the inhibitory effect of NSC-622608 (0, 10, 20, 50 and 100 μM) on the proliferation of KBM5 and KBM5-T315I cells. (B, C) Flow cytometry results showing that NSC-622608 could induce significant apoptosis in CML cells, especially K562-R cells.
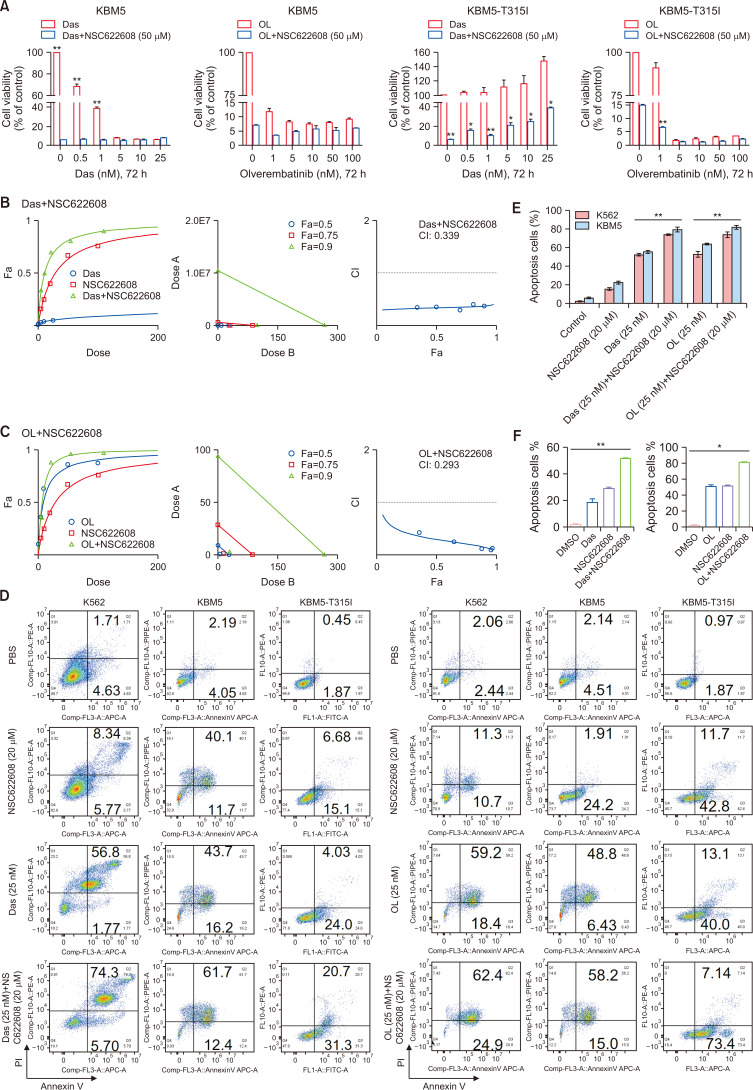
NSC-622608 synergizes with TKIs to promote the apoptosis of CML cells
To analyze the effect of the combination treatment, we treated KBM5 and KBM5-T315I cells with Das alone (0, 0.5, 1, 5, 10, or 25 nM), OL alone (0, 1, 5, 10, 50, or 100 nM) or in combination with NSC-622608 (0, 5, 10, 20, 50, or 100 μM) at different concentrations for 72 h (Fig. 6A). The results demonstrate that the combination of Das or OL with NSC-622608 (50 μM) synergistically inhibited the growth of KBM5 and KBM5-T315I cells (Fig. 6A). We conducted a drug synergy experiment to explore the synergistic effect of TKIs and NSC-622608. As shown in Fig. 6B and 6C, Das and NSC-622608 synergistically inhibited the growth of KBM5-T315I cells (combination index, CI: 0.339). Moreover, the combination of NSC-622608 with OL inhibited the proliferation of KBM5-T315I cells, proving more effective than a single drug (CI: 0.293, Fig. 6C). To analyze the effects of the combination of TKIs and NSC-622608 on apoptosis, we conducted flow cytometry analysis to detect the percentage of apoptotic cells in the different groups. The flow cytometry results revealed more apoptotic cells in the combination group than in the Das, OL or NSC-622608 groups (Fig. 6D-6F, p<0.05 and p<0.01).
Fig. 6.
NSC-622608 synergizes with TKIs to promote apoptosis in CML cells. (A) CCK-8 results showing the effect of combination treatment with TKIs (das: 0, 0.5, 1, 5, 10, and 25 nM; OL: 0, 1, 5, 10, 50, and 100 nM) and NSC-622608 (50 μM) on KBM5 and KBM5-T315I cells (p<0.01, p<0.05). (B, C) Drug synergy experiment on TKIs and NSC-622608, and the data were analyzed by CompuSyn: additive effect (CI=1), synergistic effect (CI<1), and antagonistic effect (CI>1). Das and NSC622608 (CI=0.339), Das and NSC622608 (CI=0.293), (D-F) Flow cytometry results showing the greatest proportion of apoptotic cells in the combination, TKI and NSC622608 groups compared with the single drug group (p<0.01, p<0.05; bar: 50 μm). Asterisks (*) indicate significance, ** indicates p<0.01, * indicates p<0.05, and NS indicates no significance.
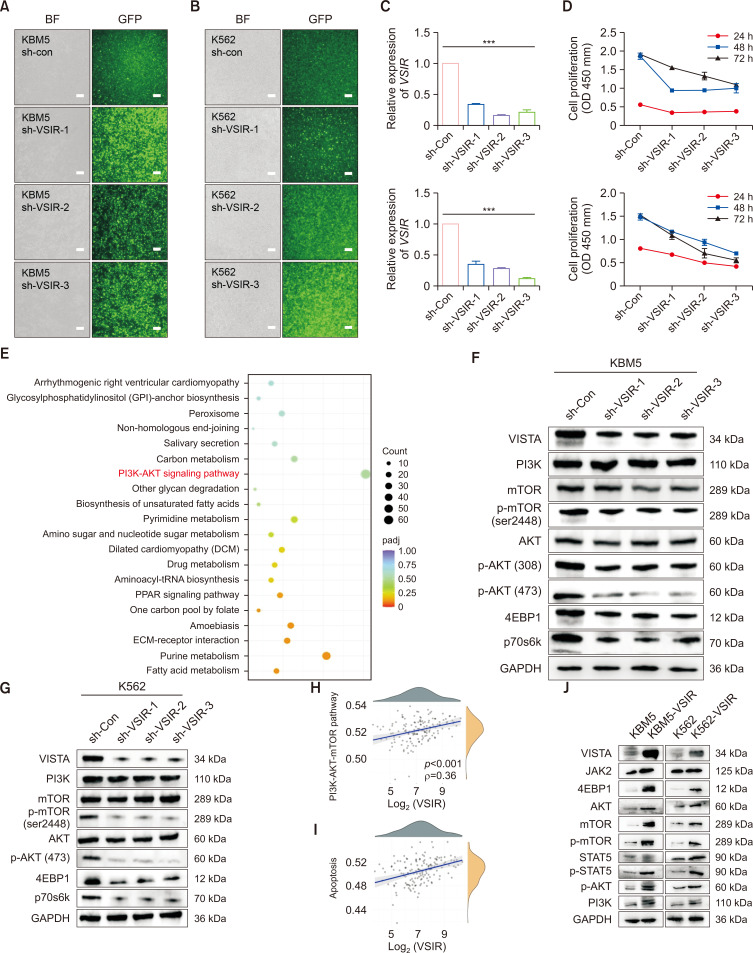
VISTA promotes the proliferation of CML cells through regulation of the PI3K/AKT/mTOR pathway ex vivo
We elucidated the mechanism underlying the action of VSIR in CML cells by knocking down its expression in KBM5 and K562 cells through lentiviral transfection (Fig. 7A, 7B). The relative mRNA level of VSIR decreased in the sh-VSIR-transfected cells compared to the sh-con cells (p<0.001) (Fig. 7C). Moreover, the CCK-8 assay results showed that VSIR knockdown inhibited the proliferation of K562 and KBM5 cells (Fig. 7D, p<0.001, p<0.05). Next, we utilized RNA-seq (KBM5-sh-con vs. KBM5-sh-VSIR-3) to quantify the expression of downstream genes and pathways. KEGG pathway analysis revealed that the DEGs were mainly enriched in pathways regulating PI3K-AKT signaling, purine metabolism, carbon metabolism, and pyrimidine metabolism, among others (Fig. 7E). VSIR knockdown inhibited the proliferation of KBM5 and K562 cells by inhibiting the activation of the PI3K/AKT/mTOR pathway (Fig. 7F, 7G). VISTA expression is regulated through a multifaceted signal transmission system that involves transcriptional, translational, and posttranslational processes, necessitating the use of both qRT‒PCR and Western blot analyses for accurate assessment. Understanding these complex regulatory mechanisms is crucial for elucidating the role of VISTA in immune modulation and its therapeutic implications. To further validate the correlations between the VSIR gene and the PI3K/AKT/mTOR pathway, we explored RNA-sequencing expression profiles and corresponding clinical information downloaded from the TCGA dataset (https://portal.gdc.com). Spearman correlation analysis revealed significant correlations between the VSIR gene and the PI3K/AKT/mTOR (p<0.001, ρ=0.36) and apoptosis (p<0.001, ρ=0.38) pathway scores (Fig. 7H, 7I). The positive correlation between VSIR expression and apoptosis pathway activation, shown in Fig. 7H and 7I, in conjunction with its role in reducing cell proliferation, underscores the dual functionality of VSIR in cell regulation. This discrepancy can be attributed to the context-dependent role of VSIR in CML cells, where its reduction may simultaneously suppress proliferative signals while activating apoptotic pathways under specific conditions. As expected, Western blot analysis revealed that the overexpression of VISTA increased the activation of the PI3K/AKT/mTOR pathway (Fig. 7J).
Fig. 7.
VSIR promotes the proliferation of CML cells through regulation of the PI3K/AKT/mTOR pathway ex vivo. (A, B) KBM5 and K562 cells were transfected with FITC-conjugated sh-VSIR vectors. Significant green fluorescence was observed in sh-VSIR-transfected KBM5 cells (bar: 100 μm). (C) The relative mRNA expression of VSIR in sh-VSIR-transfected cells and sh-con cells (p<0.001). (D) CCK-8 results showing that VSIR knockdown inhibited the proliferation of KBM5 (p<0.001) and K562 cells (p<0.05). (E) KEGG pathway analysis showing enrichment of differentially expressed genes (DEGs) between KBM5-sh-con and KBM5-sh-VSIR-3. The enriched KEGG signaling pathways were selected to demonstrate the primary biological actions of the DEGs. For the enrichment results, p<0.05 or FDR<0.05 was considered to indicate a meaningful pathway (an enrichment score of −log10 (P) greater than 1.3). (F, G) Western blots showing that knockdown of VSIR in KBM5 and K562 cells significantly downregulated the PI3K/AKT/mTOR signaling pathway (n=3). (H, I) The correlations between the VSIR gene and the PI3K/AKT/mTOR (p<0.001, ρ=0.36) and apoptosis (p<0.001, ρ=0.38) pathway scores were analyzed with Spearman’s correlation coefficient. (J) Western blots showing that VSIR overexpression in KBM5 and K562 cells significantly increased the activity of the PI3K/AKT/mTOR pathway. Asterisks (*) indicate significance, *** indicates p<0.001.
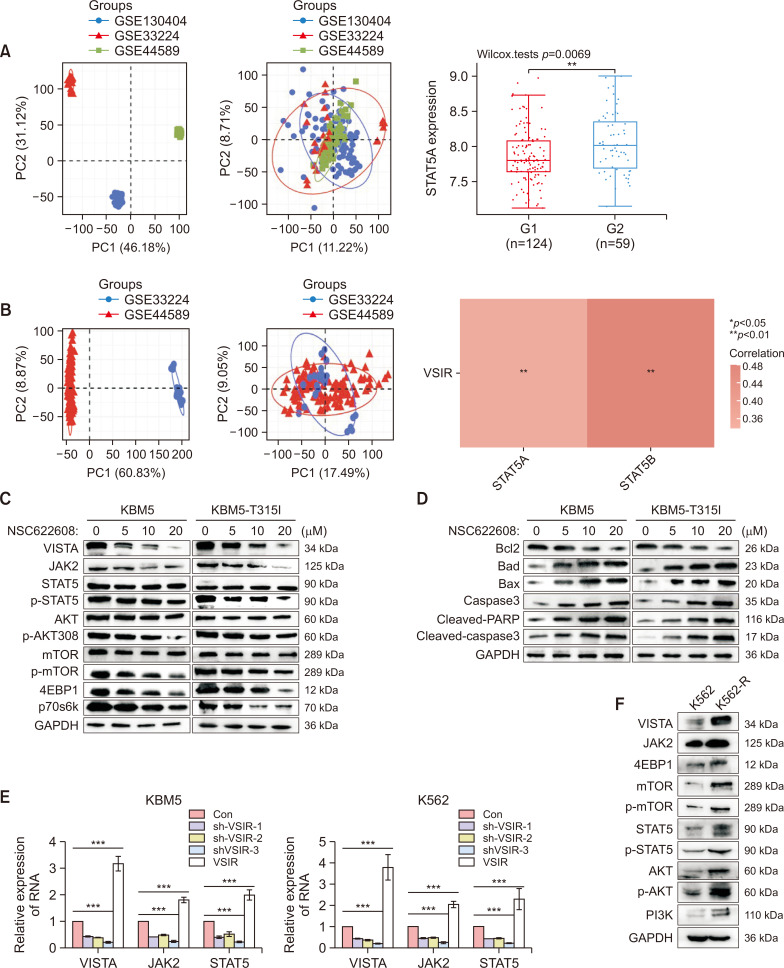
NSC-622608 overcomes TKI resistance via downregulation of the AKT/mTOR and JAK2/STAT5 pathways
We assessed the correlation between the expression of VSIR and that of STAT5 in 183 CML patients from the GSE130404, GSE44589 and GSE33224 datasets. Initially, the PCA results before batch removal for multiple datasets indicated complete separation without intersection. After batch removal, the PCA results demonstrated an intersection of the three datasets, enabling subsequent analysis (Fig. 8A, 8B). There were 124 patients who achieved a drug response (IM-Rs), and 59 patients had no response (IM-NRs). Among the 183 patients, 124 exhibited a drug response (IM-R), while 59 showed no response (IM-NR). The relative expression of STAT5 in IM-NR patients was greater than that in IM-R patients (p<0.001) (Fig. 8A). The correlations between the expression of VSIR and that of STAT5A and STAT5B were analyzed using Spearman’s correlation analysis. Spearman’s correlation analysis revealed a positive correlation with both STAT5A (p<0.01) and STAT5B (p<0.01) (Fig. 8B). Next, we comprehensively analyzed the mechanism underlying the anticancer activity of NSC622608 in CML cells by assessing changes in the expression of related signaling proteins after treatment with NSC622608 (0, 5, 10, or 20 μM) for 48 h through Western blot analysis. Compared with those in the control group, the levels of JAK2, STAT5, p-STAT5, AKT, p-AKT, p-mTOR, 4EBP1, and p70s6k significantly decreased after NSC622608 treatment (Fig. 8C). We also observed an increase in the mitochondria-induced apoptosis pathway with increasing NSC622608 concentration. Furthermore, Bcl-2 levels were significantly decreased, whereas Bax, Bad, cleaved PARP, and cleaved caspase-3 levels were significantly increased after NSC622608 treatment (Fig. 8D). To further analyze these changes, we conducted qPCR and found a decrease in JAK2 and STAT5 mRNA expression along with VSIR knockdown (Fig. 8E). We also explored the differences in the AKT/mTOR and JAK2/STAT5 pathways between K562 and K562-R cells, and the AKT/mTOR and JAK2/STAT5 pathways were more strongly activated in K562-R cells than in K562 cells (Fig. 8F).
Fig. 8.
NSC-622608 overcomes TKI resistance via downregulation of the AKT/mTOR and JAK2/STAT5 pathways. (A) PCA results before batch removal for multiple datasets. Different colors represent different datasets. As shown in the schematic diagram, the three CML datasets (GSE130404, GSE44589, and GSE33224) (n=183) were separated without any intersection. PCA results after batch removal, as shown in the schematic diagram, showing the intersection of the three datasets, which can be used in subsequent analysis. There were 124 patients who achieved a drug response (IM-Rs), and 59 patients had no response (IM-NRs). The relative expression of STAT5 in the IM-NR group was greater than that in the IM-R group (p<0.001). (B) The correlations between the expression of VSIR and that of STAT5A and STAT5B were analyzed with Spearman’s correlation. The density curve on the right represents the trend in the distribution of STAT5A and STAT5B, and the upper density curve represents the trend in the distribution of VSIR expression. The abscissa and ordinate represent genes, and different colors represent different correlation coefficients (red represents a negative correlation); the darker the color is, the stronger the correlation. (C) Western blots showing decreased expression levels of the JAK2, STAT5, p-STAT5, AKT, p-AKT, p-mTOR, 4EBP1 and p70s6k proteins after treatment with NSC-622608 (0, 5, 10, 20 μM) in KBM5 and KBM5-T315I cells for 48 h. (D) At 48 h, the mitochondria-induced apoptosis pathway was increased in a concentration-dependent manner after treatment with NSC-622608 (0, 5, 10, or 20 μM) in KBM5 and KBM5-T315I cells. (E) qPCR revealed a decrease in JAK2 and STAT5 mRNA expression following VSIR knockdown. (F) The AKT/mTOR and JAK2/STAT5 pathways were more strongly activated in K562-R cells than in K562 cells according to Western blot analysis. Asterisks (*) indicate significance, *** indicates p<0.001, ** indicates p<0.01, * indicates p<0.05, and NS indicates no significance.
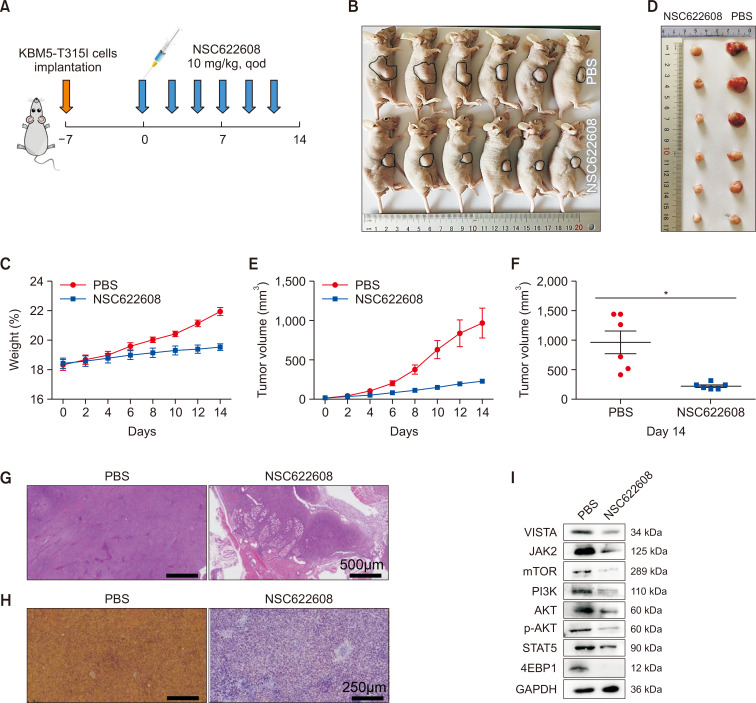
NSC-622608 inhibits the growth of CML tumors in vivo
To evaluate the impact of NSC-622608 on the expansion of CML cells, we established a mouse xenograft model by subcutaneously injecting KBM5-T315I cells into athymic BALB/c nu/nu female mice (Fig. 9A). Tumors were confirmed, and tumor growth was measured. Subsequent analysis revealed that mice treated with NSC-622608 exhibited significantly smaller tumor sizes and growth rates than those in the PBS group (Fig. 9B-9F). Furthermore, additional staining of tumor tissues with HE and immunohistochemistry for Ki67 were performed. The results consistently revealed a pronounced decrease in the tumor growth rate in the mice treated with NSC-622608 (Fig. 9G, 9H). We next compared the expression of proteins involved in the AKT/mTOR and JAK2/STAT5 pathways between the PBS and NSC-622608 groups, and Western blot analysis of tumor tissue samples revealed that the AKT/mTOR and JAK2/STAT5 pathways were downregulated in the NSC-622608 group (Fig. 9I).
Fig. 9.
Effect of NSC-622608 on the progression of KBM5-T315I tumors in vivo. (A) Protocol scheme of BALB/c nu/nu female mice engrafted with KBM5-T315I cells. (B, C, E) Tumor volumes and body weights were measured every other day for 14 days. (D, F) Tumor volumes after extraction from mice at the end of the experiment. (G) H&E staining of tumor tissues from each group. (H) Immunohistochemical staining for Ki67. (I) Western blot analysis of tumors revealed downregulation of the AKT/mTOR and JAK2/STAT5 pathways in the NSC-622608 group. Asterisks (*) indicate significance, * indicates p<0.05, and NS indicates no significance.
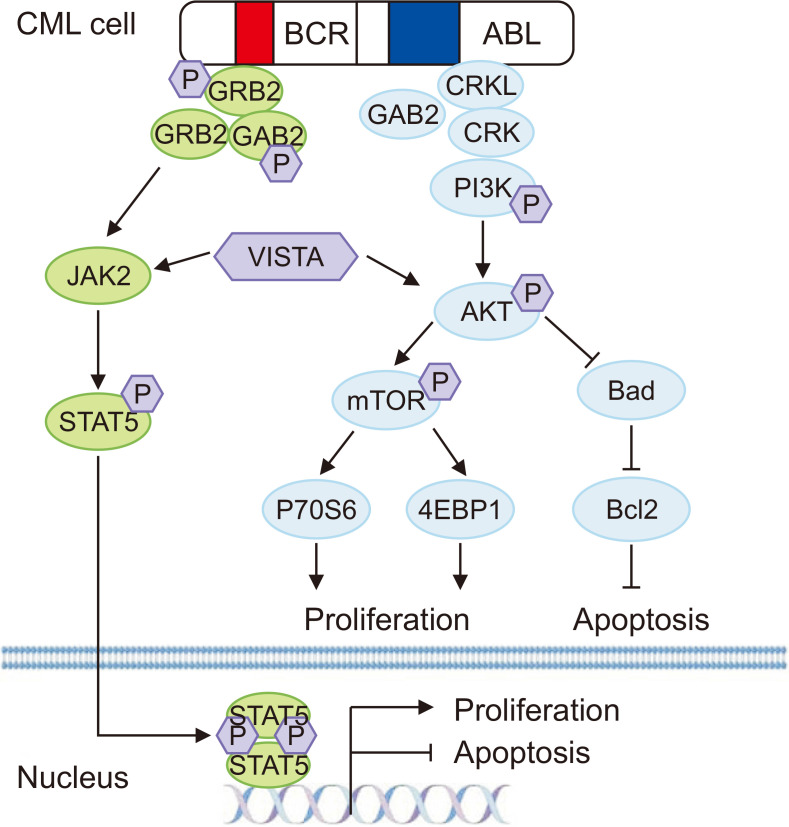
In summary, VISTA facilitated the proliferation and suppressed the apoptosis of CML cells by activating the AKT/mTOR and JAK2/STAT5 pathways and increasing the expression of the antiapoptotic protein Bcl2. NSC-622608, a novel VISTA inhibitor, attenuated the phosphorylation of the AKT/mTOR and JAK2/STAT5 pathways by inhibiting VISTA activation. This inhibition reduced the proliferation of CML cells and induced their apoptosis (Fig. 10).
Fig. 10.
Mechanism of tumoral VISTA facilitation of the proliferation of CML cells and suppression of apoptosis. Tumoral VISTA activated the AKT/mTOR and JAK2/STAT5 pathways and increased the expression of the antiapoptotic protein Bcl2. NSC-622608, a novel VISTA inhibitor, attenuated the phosphorylation of the AKT/mTOR and JAK2/STAT5 pathways by inhibiting VISTA activation. This inhibition resulted in reduced proliferation of CML cells and induced apoptosis in CML cells.
DISCUSSION
The persistent challenge of TKI resistance in the treatment of CML remains a significant hurdle, despite advances in targeted therapies. The exploration of tumoral VISTA as a contributory factor in this resistance paradigm presents a novel therapeutic target, potentially broadening the horizon for overcoming resistance mechanisms. This study not only revealed the pivotal role of VISTA in mediating resistance through the AKT/mTOR and JAK2/STAT5 pathways but also revealed that NSC622608, a VISTA inhibitor, is a promising adjunct to existing TKI therapies (Pagliuca et al., 2022; Schaafsma et al., 2023). By demonstrating the feasibility of sensitizing TKI-resistant CML cells through VISTA inhibition, our findings could herald a new era in CML management, particularly for patients with mutations such as T315I, which is notorious for conferring resistance to conventional TKIs (Alves et al., 2021).
The mechanisms underlying TKI resistance in CML are multifaceted, encompassing both BCR-ABL1-dependent and BCR-ABL1-independent pathways. Among the latter, alternative signaling pathways, such as the AKT/mTOR and JAK2/STAT5 pathways, have been implicated in sustaining cell proliferation and survival, independent of BCR-ABL1 kinase activity (Chorzalska et al., 2018). Our research highlights a novel aspect of this resistance mechanism, pinpointing the overexpression of tumoral VISTA as a significant contributor to the activation of these alternative pathways. The finding that VISTA expression is correlated with poor survival outcomes in CML patients further emphasizes its potential as a therapeutic target.
Given the central role of immune checkpoints in cancer pathogenesis, the involvement of VISTA, an immune checkpoint molecule, in CML resistance to TKIs adds an intriguing layer to our understanding of the disease’s complexity. This finding suggests that the immune microenvironment, through mechanisms such as immune checkpoint regulation, plays a crucial role in mediating treatment resistance. This insight opens new avenues for research focusing on the interplay between immune modulation and oncogenic signaling in CML (Patel et al., 2017). VISTA, an inhibitory immune checkpoint expressed in tumoral and immune cells (Zheng et al., 2022), is reportedly highly expressed in a range of malignancies, especially those that are less responsive to immunotherapy (Mulati et al., 2019; Mortezaee et al., 2022). Studies have shown that VISTA is expressed in some types of human tumors, including endometrial, kidney, ovarian, lung, and colorectal cancers (Villarroel-Espindola et al., 2018; Xie et al., 2018; Hong et al., 2019; Mulati et al., 2019). In ovarian cancer cells, VISTA expression is correlated with overall survival (OS) and progression-free survival (Zong et al., 2021), and higher VISTA expression is associated with lymph node metastasis (Liao et al., 2018). Esophageal squamous cell carcinoma exhibits high VISTA expression in 85% of tumor tissues, which is positively correlated with overall patient survival (Chen et al., 2021). In triple-negative breast cancer patients, VISTA is expressed in 18.5% of tumor cells in untreated patients (Mortezaee et al., 2022). In hematologic malignancies, VISTA binds to galectin-9 secreted by AML cells, suppressing granzyme B release, caspase activity, and the expression of apoptotic proteins such as caspase-9 in AML cells (Yasinska et al., 2020). Despite its expression in AML blasts and MDSCs, whether VISTA possesses immunosuppressive properties or serves as a marker of myeloid origin remains unclear (Wang et al., 2018). The role of VISTA as a ligand on myeloid cells has been largely ignored (Schaafsma et al., 2022). Our study revealed higher VISTA expression at both the protein and RNA levels in myeloid (AML, CML, and MDS) than in lymphoid (T-ALL, Pre-B-ALL, and B-ALL) malignancies. Immunofluorescence revealed that VISTA was localized in the cytoplasm and nucleus of CML cells, and its expression was correlated with the therapeutic effect of TKIs. Exposure of KBM5 and KBM5-T315I cells to Das (25 nM) and OL (25 nM) resulted in the downregulation of VISTA, although the latter effect was not significant. These results suggest that VISTA not only functions as an immune checkpoint but is also involved in TKI treatment. In contrast to other members of the B7 superfamily, VISTA reportedly functions both as a ligand and as a receptor. Mechanistically, VISTA has 30 intramolecular hydrogen bonds between its loop and the F-strand, along with a disulfide bond (Cys51/Cys113) forming a unique elongated loop (Im et al., 2022).
The discovery of the ability of NSC-622608 to inhibit tumoral VISTA and its subsequent impact on TKI-resistant CML cells represents a significant breakthrough. This small molecule inhibitor not only curtails CML cell proliferation but also enhances the efficacy of TKIs, offering a dual-faceted approach to overcoming resistance. Following the observation of the inhibitory effects of NSC-622608 on tumoral VISTA and its ability to enhance TKI efficacy, our investigations into the effects of VISTA knockdown combined with TKI therapy revealed additional effects of VISTA on CML resistance. These findings highlight the synergistic potential of the integration of VISTA targeting with TKI treatments, emphasizing a broader strategy for overcoming TKI resistance. The synergy observed suggests that VISTA modulation might not only disrupt CML survival pathways but also enhance TKI sensitivity, marking a significant step forward in addressing the complexities of TKI resistance. Alves (Alves et al., 2021) highlighted the complexity of TKI resistance mechanisms, suggesting the importance of novel targets such as VISTA. Loscocco (Loscocco et al., 2019) provided context for BCR-ABL-independent resistance pathways, emphasizing the relevance of additional therapeutic targets. These findings underscore the relevance of VISTA as a therapeutic target and align with the increasing recognition of the roles of immune checkpoints in cancer resistance mechanisms. Schaafsma (Schaafsma et al., 2023) discussed the targeting of immune checkpoints such as VISTA to overcome cancer resistance, supporting the combined use of VISTA inhibitors with TKIs. The differential anticancer efficacy observed between NSC622608 treatment and direct VISTA targeting, as well as the comparison with dasatinib, underscores the multifaceted nature of CML treatment resistance (Alves et al., 2021). The superior performance of NSC-622608 suggests that it may exert broader effects beyond VISTA inhibition, potentially impacting other survival pathways. This could explain the enhanced efficacy over direct VISTA processing. Moreover, the difference in efficacy between the NSC622608 and dasatinib treatment groups highlights the unique mechanisms of action of these compounds, with NSC622608 affecting immune checkpoint regulation and possibly offering a synergistic effect when combined with direct BCR-ABL inhibition by dasatinib. Furthermore, the observed decrease in cell proliferation upon VISTA knockdown, as demonstrated in Fig. 6D, underscores the potential of targeting VISTA to inhibit the growth of CML cells. This approach, particularly in combination with TKIs, represents a promising strategy for enhancing therapeutic efficacy against resistant CML cells (Patel et al., 2017). This synergy between NSC622608 and TKIs against resistant CML cells supports the concept that immune modulation, alongside direct oncogenic targeting, could offer a comprehensive approach to mitigate resistance. The synergistic effect observed when NSC-622608 was combined with TKIs underscores the potential of targeting VISTA as a strategy to increase the sensitivity of CML cells to TKIs. Furthermore, the ability of NSC-622608 to induce apoptosis in CML cells, including those with the T315I mutation, highlights its potential as a versatile therapeutic agent capable of addressing one of the most challenging aspects of CML treatment (Krishnan et al., 2023).
The promising results of NSC-622608 in preclinical models warrant further exploration, particularly through clinical trials designed to assess its safety and efficacy in humans (Meenakshi Sundaram et al., 2019). These studies are crucial for determining the optimal dosing, potential side effects, and real-world effectiveness of combining NSC-622608 with TKIs in CML patients, especially those with resistant disease.
Moreover, a deeper understanding of the molecular interactions between VISTA and the AKT/mTOR and JAK2/STAT5 pathways could unveil additional targets within these signaling cascades (Chorzalska et al., 2018). This knowledge might lead to the development of combination therapies that more effectively disrupt the complex network of pathways contributing to TKI resistance (Lussana et al., 2018).
Given the role of the immune microenvironment in CML progression and treatment resistance, future research should also consider the impact of VISTA inhibition on immune cells within the tumor milieu (Marce et al., 2015). Investigating how NSC-622608 and similar agents influence the interaction between CML cells and the immune system could provide insights into the broader immunomodulatory effects of targeting VISTA in cancer (Eide and O’Hare, 2015).
Our study provides compelling evidence of the role of tumoral VISTA in mediating TKI resistance in CML and introduces a novel therapeutic approach to overcome this challenge. By targeting VISTA with the small molecule inhibitor NSC-622608, the proposed strategy not only addresses a key mechanism of resistance but also enhances the efficacy of existing TKI therapies (Deng et al., 2019). This dual approach represents a significant advancement in the quest to improve outcomes for CML patients, particularly those with limited treatment options. As we continue to unravel the complex interplay between oncogenic signaling and the immune microenvironment, the potential for novel therapeutic strategies, such as the one presented here, will undoubtedly expand, offering new hope to patients battling CML and other cancers (Talati and Pinilla-Ibarz, 2018).
ACKNOWLEDGMENTS
This research was supported by the the National Natural Science Foundation of China (No. 82270233), Frontier Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (No. 2018GZR110105014), The Science and Technology Program of Guangzhou (No. 202201011041) and the National Natural Science Foundation of China (No. U2001224).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Data will be made available on request.
AUTHOR CONTRIBUTIONS
Kexin Ai, Mu Chen and Zhao Liang performed research and analyzed the data; Xiangyang Ding and Yang Gao evaluated and collected the clinic data; Honghao Zhang and Suwan Wu contributed to the writing of the paper; Yanjie He and Yuhua Li designed the research.
REFERENCES
- Alves R., Goncalves A. C., Rutella S., Almeida A. M., De Las Rivas J., Trougakos I. P., Sarmento Ribeiro A. B. Resistance to tyrosine kinase inhibitors in chronic myeloid leukemia-from molecular mechanisms to clinical relevance. Cancers (Basel) 2021;13:4820. doi: 10.3390/cancers13194820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Feng R., He B., Wang J., Xian N., Huang G., Zhang Q. PD-1H expression associated with CD68 macrophage marker confers an immune-activated microenvironment and favorable overall survival in human esophageal squamous cell carcinoma. Front. Mol. Biosci. 2021;8:777370. doi: 10.3389/fmolb.2021.777370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xie X., Wu A., Wang L., Hu Y., Zhang H., Li Y. A synthetic cell-penetrating peptide derived from nuclear localization signal of EPS8 exerts anticancer activity against acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2018;37:12. doi: 10.1186/s13046-018-0682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorzalska A., Ahsan N., Rao R. S. P., Roder K., Yu X., Morgan J., Tepper A., Hines S., Zhang P., Treaba D. O., Zhao T. C., Olszewski A. J., Reagan J. L., Liang O., Gruppuso P. A., Dubielecka P. M. Overexpression of Tpl2 is linked to imatinib resistance and activation of MEK-ERK and NF-kappaB pathways in a model of chronic myeloid leukemia. Mol. Oncol. 2018;12:630–647. doi: 10.1002/1878-0261.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Novellis D., Cacace F., Caprioli V., Wierda W. G., Mahadeo K. M., Tambaro F. P. The TKI era in chronic leukemias. Pharmaceutics. 2021;13:2201. doi: 10.3390/pharmaceutics13122201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Li J., Sarde A., Lines J. L., Lee Y. C., Qian D. C., Pechenick D. A., Manivanh R., Le Mercier I., Lowrey C. H., Varn F. S., Cheng C., Leib D. A., Noelle R. J., Mabaera R. Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Immunol. Res. 2019;7:1079–1090. doi: 10.1158/2326-6066.CIR-18-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide C. A., O'Hare T. Chronic myeloid leukemia: advances in understanding disease biology and mechanisms of resistance to tyrosine kinase inhibitors. Curr. Hematol. Malig. Rep. 2015;10:158–166. doi: 10.1007/s11899-015-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabr M. T., Gambhir S. S. Discovery and optimization of small-molecule ligands for V-domain Ig suppressor of T-cell activation (VISTA) J. Am. Chem. Soc. 2020;142:16194–16198. doi: 10.1021/jacs.0c07276. [DOI] [PubMed] [Google Scholar]
- Hong S., Yuan Q., Xia H., Zhu G., Feng Y., Wang Q., Zhang Z., He W., Lu J., Dong C., Ni L. Analysis of VISTA expression and function in renal cell carcinoma highlights VISTA as a potential target for immunotherapy. Protein Cell. 2019;10:840–845. doi: 10.1007/s13238-019-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinkhani N., Derakhshani A., Shadbad M. A., Argentiero A., Racanelli V., Kazemi T., Mokhtarzadeh A., Brunetti O., Silvestris N., Baradaran B. The role of V-domain Ig suppressor of T cell activation (VISTA) in cancer therapy: lessons learned and the road ahead. Front. Immunol. 2021;12:676181. doi: 10.3389/fimmu.2021.676181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E., Sim D. Y., Lee H. J., Park J. E., Park W. Y., Ko S., Kim B., Shim B. S., Kim S. H. Immune functions as a ligand or a receptor, cancer prognosis potential, clinical implication of VISTA in cancer immunotherapy. Semin. Cancer Biol. 2022;86:1066–1075. doi: 10.1016/j.semcancer.2021.08.008. [DOI] [PubMed] [Google Scholar]
- Irani Y. D., Kok C. H., Clarson J., Shanmuganathan N., Branford S., Yeung D. T., Ross D. M., Hughes T. P., Yong A. S. Association of TIM-3 checkpoint receptor expression on T cells with treatment-free remission in chronic myeloid leukemia. Blood Adv. 2023;7:2364–2374. doi: 10.1182/bloodadvances.2022008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E., Kantarjian H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2022;97:1236–1256. doi: 10.1002/ajh.26642. [DOI] [PubMed] [Google Scholar]
- Jiang Q., Li Z., Qin Y., Li W., Xu N., Liu B., Zhang Y., Meng L., Zhu H., Du X., Chen S., Liang Y., Hu Y., Liu X., Song Y., Men L., Chen Z., Niu Q., Wang H., Lu M., Yang D., Zhai Y., Huang X. Olverembatinib (HQP1351), a well-tolerated and effective tyrosine kinase inhibitor for patients with T315I-mutated chronic myeloid leukemia: results of an open-label, multicenter phase 1/2 trial. J. Hematol. Oncol. 2022;15:113. doi: 10.1186/s13045-022-01334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Schmidt F., Nawaz Z., Venkatesh P. N., Lee K. L., Ren X., Chan Z. E., Yu M., Makheja M., Rayan N. A., Lim M. G. L., Cheung A. M. S., Bari S., Chng W. J., Than H., Ouyang J., Rackham O., Tan T. Z., Hwang W. Y. K., Chuah C., Prabhakar S., Ong S. T. A single-cell atlas identifies pretreatment features of primary imatinib resistance in chronic myeloid leukemia. Blood. 2023;141:2738–2755. doi: 10.1182/blood.2022017295. [DOI] [PubMed] [Google Scholar]
- Liao H., Zhu H., Liu S., Wang H. Expression of V-domain immunoglobulin suppressor of T cell activation is associated with the advanced stage and presence of lymph node metastasis in ovarian cancer. Oncol. Lett. 2018;16:3465–3472. doi: 10.3892/ol.2018.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang Y., Huang H., Lei X., Tang G., Cao X., Peng J. Recent advances in Bcr-Abl tyrosine kinase inhibitors for overriding T315I mutation. Chem. Biol. Drug Des. 2021;97:649–664. doi: 10.1111/cbdd.13801. [DOI] [PubMed] [Google Scholar]
- Loscocco F., Visani G., Galimberti S., Curti A., Isidori A. BCR-ABL independent mechanisms of resistance in chronic myeloid leukemia. Front. Oncol. 2019;9:939. doi: 10.3389/fonc.2019.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussana F., Intermesoli T., Stefanoni P., Rambaldi A. Mechanisms of resistance to targeted therapies in chronic myeloid leukemia. Handb. Exp. Pharmacol. 2018;249:231–250. doi: 10.1007/164_2017_81. [DOI] [PubMed] [Google Scholar]
- Marce S., Cortes M., Zamora L., Cabezon M., Grau J., Milla F., Feliu E. A thirty-five nucleotides BCR-ABL1 insertion mutation of controversial significance confers resistance to imatinib in a patient with chronic myeloid leukemia (CML) Exp. Mol. Pathol. 2015;99:16–18. doi: 10.1016/j.yexmp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Meenakshi Sundaram D. N., Jiang X., Brandwein J. M., Valencia-Serna J., Remant K. C., Uludag H. Current outlook on drug resistance in chronic myeloid leukemia (CML) and potential therapeutic options. Drug Discov. Today. 2019;24:1355–1369. doi: 10.1016/j.drudis.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Mortezaee K., Majidpoor J., Najafi S. VISTA immune regulatory effects in bypassing cancer immunotherapy: updated. Life Sci. 2022;310:121083. doi: 10.1016/j.lfs.2022.121083. [DOI] [PubMed] [Google Scholar]
- Mulati K., Hamanishi J., Matsumura N., Chamoto K., Mise N., Abiko K., Baba T., Yamaguchi K., Horikawa N., Murakami R., Taki M., Budiman K., Zeng X., Hosoe Y., Azuma M., Konishi I., Mandai M. VISTA expressed in tumour cells regulates T cell function. Br. J. Cancer. 2019;120:115–127. doi: 10.1038/s41416-018-0313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Victoria Lai W., Adusumilli P. S., Desmeules P., Frosina D., Jungbluth A., Ni A., Eguchi T., Travis W. D., Ladanyi M., Zauderer M. G., Sauter J. L. V-domain Ig-containing suppressor of T-cell activation (VISTA), a potentially targetable immune checkpoint molecule, is highly expressed in epithelioid malignant pleural mesothelioma. Mod. Pathol. 2020;33:303–311. doi: 10.1038/s41379-019-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca S., Gurnari C., Zhang K., Kewan T., Bahaj W., Mori M., Nautiyal I., Rubio M. T., Ferraro F., Maciejewski J. P., Wang L., Visconte V. Comprehensive transcriptomic analysis of VISTA in acute myeloid leukemia: insights into its prognostic value. Int. J. Mol. Sci. 2022;23:14885. doi: 10.3390/ijms232314885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. B., O'Hare T., Deininger M. W. Mechanisms of resistance to ABL kinase inhibition in chronic myeloid leukemia and the development of next generation ABL kinase inhibitors. Hematol. Oncol. Clin. North Am. 2017;31:589–612. doi: 10.1016/j.hoc.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalzulli E., Carmosino I., Bisegna M. L., Martelli M., Breccia M. CML resistant to 2nd-generation TKIs: mechanisms, next steps, and new directions. Curr. Hematol. Malig. Rep. 2022;17:198–205. doi: 10.1007/s11899-022-00683-3. [DOI] [PubMed] [Google Scholar]
- Schaafsma E., Croteau W., ElTanbouly M., Nowak E. C., Smits N. C., Deng J., Sarde A., Webber C. A., Rabadi D., Cheng C., Noelle R., Lines J. L. VISTA targeting of T-cell quiescence and myeloid suppression overcomes adaptive resistance. Cancer Immunol. Res. 2023;11:38–55. doi: 10.1158/2326-6066.CIR-22-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaafsma E., Croteau W., ElTanbouly M. A., Nowak E. C., Smits N. C., Deng J., Sarde A., Webber C. A., Rabadi D., Cheng C., Noelle R., Lines J. L. VISTA targeting of T-cell quiescence and myeloid suppression overcomes adaptive resistance. Cancer Immunol. Res. 2022;11:38–55. doi: 10.1158/2326-6066.CIR-22-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey R., Lopez Rodriguez J. F., Gomez-Casares M. T. Discontinuation of tyrosine kinase inhibitors in patients with chronic myeloid leukemia: a review of the biological factors associated with treatment-free remission. Curr. Oncol. Rep. 2022;24:415–426. doi: 10.1007/s11912-022-01228-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talati C., Pinilla-Ibarz J. Resistance in chronic myeloid leukemia: definitions and novel therapeutic agents. Curr. Opin. Hematol. 2018;25:154–161. doi: 10.1097/MOH.0000000000000403. [DOI] [PubMed] [Google Scholar]
- Villarroel-Espindola F., Yu X., Datar I., Mani N., Sanmamed M., Velcheti V., Syrigos K., Toki M., Zhao H., Chen L., Herbst R. S., Schalper K. A. Spatially resolved and quantitative analysis of VISTA/PD-1H as a novel immunotherapy target in human non-small cell lung cancer. Clin. Cancer Res. 2018;24:1562–1573. doi: 10.1158/1078-0432.CCR-17-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jia B., Claxton D. F., Ehmann W. C., Rybka W. B., Mineishi S., Naik S., Khawaja M. R., Sivik J., Han J., Hohl R. J., Zheng H. VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML. Oncoimmunology. 2018;7:e1469594. doi: 10.1080/2162402X.2018.1469594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Huang J., Qiao Q., Zang W., Hong S., Tan H., Dong C., Yang Z., Ni L. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol. Immunother. 2018;67:1685–1694. doi: 10.1007/s00262-018-2227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasinska I. M., Meyer N. H., Schlichtner S., Hussain R., Siligardi G., Casely-Hayford M., Fiedler W., Wellbrock J., Desmet C., Calzolai L., Varani L., Berger S. M., Raap U., Gibbs B. F., Fasler-Kan E., Sumbayev V. V. Ligand-receptor interactions of galectin-9 and VISTA suppress human T lymphocyte cytotoxic activity. Front. Immunol. 2020;11:580557. doi: 10.3389/fimmu.2020.580557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Zhang K., Zhang X., Xiao Y., Wang T., Jiang S. Development of inhibitors targeting the V-domain Ig suppressor of T cell activation signal pathway. J. Med. Chem. 2022;65:11900–11912. doi: 10.1021/acs.jmedchem.2c00803. [DOI] [PubMed] [Google Scholar]
- Zong L., Yu S., Mo S., Zhou Y., Xiang Y., Lu Z., Chen J. High VISTA expression correlates with a favorable prognosis in patients with colorectal cancer. J. Immunother. 2021;44:22–28. doi: 10.1097/CJI.0000000000000343. [DOI] [PubMed] [Google Scholar]