Abstract
Transporter associated with antigen processing 2 (Tap-2) is responsible for ATP-dependent transport of peptides from the cytosol to the endoplasmic reticulum, where peptides bind to newly synthesized human leukocyte antigen (HLA) class I molecules, which are essential for cellular immune responses. Epstein-Barr virus (EBV) latent membrane protein 1 (LMP-1) has been shown to induce the expression of Tap-2. In this study, the induction of endogenous Tap-2 by LMP-1 is shown to be associated with and requires the expression of interferon regulatory factor 7 (IRF-7). In DG75 Burkitt's lymphoma (BL) cells, in which LMP-1 induces the expression of IRF-7, LMP-1 induced endogenous Tap-2, and ectopic expression of IRF-7 could enhance the induction. In Akata BL cells, in which LMP-1 could not induce IRF-7, LMP-1 could not induce Tap-2. Addition of IRF-7, which complements the defect in Akata cells, could stimulate the expression of Tap-2. Furthermore, LMP-1 and IRF-7A but not other IRF-7 splicing variants could activate endogenous Tap-2. A Tap-2 promoter reporter construct could be activated by the overexpression of IRF-7A. The activation could be specifically enhanced by LMP-1 and was dependent on an intact interferon-stimulated response element (ISRE) present in the Tap-2 promoter. Also, IRF-7 can bind to the Tap-2 promoter under physiological conditions in vivo, as shown by formaldehyde cross-linking, as well as to the Tap-2 ISRE in vitro, as shown by gel mobility shift assays. Furthermore, LMP-1 facilitates the phosphorylation and nuclear translocation of IRF-7. These data point to the role of IRF-7 as a secondary mediator of LMP-1-activated signal transduction for Tap-2 as follows: LMP-1 stimulates the expression of IRF-7 and facilitates its phosphorylation and nuclear translocation, and then the activated IRF-7 mediates the activation of the cellular Tap-2 gene. The induction of Tap-2 by IRF-7 and LMP-1 may have an important implication for the immune response to EBV and its persistence in vivo.
Infection by Epstein-Barr virus (EBV) may contribute to the development of malignant diseases such as Hodgkin's disease, Burkitt's lymphoma (BL), nasopharyngeal carcinoma, and posttransplant lymphoproliferative diseases (reviewed in references 20, 37, 41, and 44). In vitro, EBV efficiently infects and immortalizes primary B lymphocytes (reviewed in references 19 and 44), and latent membrane protein 1 (LMP-1) expression is required for this immortalization process (18).
LMP-1 is an integral membrane protein with six membrane-spanning domains with a long C-terminal domain, which is located in the cytoplasm (19, 23). LMP-1 acts as a constitutively active receptor-like molecule, which does not need the binding of a ligand (14). The six transmembrane domains mediate oligomerization of LMP-1 molecules in the plasma membrane, a prerequisite for LMP-1 function (11, 14). So far, two domains in the C terminus of LMP-1 have been shown to initiate signaling processes: C-terminal activator region 1 (CTAR-1, amino acids 194 to 231) and region 2 (CTAR-2, amino acids 332 to 386) (16, 29).
LMP-1 can induce a variety of cellular genes that enhance cell survival and adhesive and invasive potential (12, 27, 59, 60, 64). Interestingly, LMP-1 can stimulate the expression of the transporter associated with antigen processing 2 (TAP-2) gene (46). Tap-2 is involved in the ATP-dependent transport of peptides from the cytosol to the endoplasmic reticulum, where peptides bind to newly synthesized HLA class I molecules, which are essential for cellular immune responses (for a review, see references 3 and 57).
Tap-2 appears to be involved in some human diseases. Mutation of Tap-2 may cause defective precessing of HLA I, leading to primary immunodeficiency (55). Tap-2 mutation has been associated with familial bronchiectasis and with susceptibility to Sjögren's syndrome (10, 21). Tap-2B may increase the risk for nickel allergy (52). Tap-2 polymorphism may also be involved in inflammatory bowl disease (15). Understanding the regulation of Tap-2 is essential to elucidate its role in the pathogenesis of these as well as EBV-associated diseases.
The mechanism leading to upregulation of Tap-2 by LMP-1 is currently unknown. Previous results suggest that LMP-1 stimulates the expression of interferon-regulatory factor 7 (IRF-7) (66). IRF-7 was first cloned by its binding activity to the EBV BamHI Q promoter (Qp), used in latent EBV infection for transcription of EBNA-1, and has subsequently proven to be a negative regulator of Qp (65–67). IRF-7 belongs to the IRF family, a group of transcription factors with multiple functions (reviewed in reference 33). The hallmark of this family is the conserved N-terminal DNA-binding domain which has the potential to bind to interferon-stimulated response elements (ISREs) and regulate the activity of promoters containing ISREs. Because there is a putative ISRE in the Tap-2 promoter region, whether IRF-7 is involved in regulation of the Tap-2 gene was investigated. In this paper, we report that IRF-7 is a secondary mediator of LMP-1-triggered signal transduction for activation of the Tap-2 gene.
MATERIALS AND METHODS
Cell culture.
DG75 is an EBV-negative BL cell line (4); BL30 and BL41 are EBV-negative BL lines with EBV-infected counterparts generated by in vitro infection with the P3HR1 strain (BL30-P3HR1 and BL41-P3HR1) or the B95-8 strain (BL30-B95-8 and BL41-B95-8) of EBV (5). Akata is an EBV-positive type I BL cell line, and Jijoye is an EBV-positive type III BL cell line (42, 53). Sav-I and Sav-III are paired EBV-positive BL lines that differ only in their latent infection state (35, 46). X50-7 and B958/CBC are EBV-positive lymphoblastoid cell lines. FaDuHyg is an EBV-negative epithelial cell line (43). T2 is a lymphoblastic cell line with a deletion of the Tap-2 genomic sequence (47, 48). All cell lines were maintained in RPMI 1640 plus 10% fetal bovine serum.
Plasmids and antibodies.
A PCR fragment containing the Tap-2 promoter region, starting at its ISRE and ending at the first coding sequence, was cloned into pBS-CAT (13). The PCR fragment corresponded to nucleotides (nt) 115644 to 116217 of a published sequence (GenBank accession no. X87344) (2). The cloned promoter was identical to the published sequence except for missing 2 T's within a stretch of T's (nt 115965 to 115985). Such a sequence has been found in several PCR clones. We do not know whether this variation was due to a PCR error or a polymorphism. However, this sequence variation seems to play no important role in Tap-2 promoter activity, because results from reporter assays were in agreement with the levels of endogenous Tap-2 RNA (see Fig. 3 to 5). Mutations in the ISRE region (nt 115649 to 115662) were made by PCR with a mutated primer, and mutations were confirmed by sequencing analysis (see Fig. 3A). IRF-7 expression plasmids and IRF-7 antibody have been described (67). pcDNA/CD4 is a human CD4 expression plasmid (gift of Jenny Ting). IRF-7DN (amino acids [aa] 1 to 12 and 103 to 503) is a gift from Tom Maniatis (61). Epidermal growth factor promoter (EGFP)-IRF-7 was cloned by inserting the full-length IRF-7A into the BglII site of the pEGFP-c1 vector (Clontech). The β-galactosidase expression plasmid pCMVβ(6177-1) was purchased from Clontech. LMP-1 expression plasmid pcLMP-1 was a gift from Tomakazu Yoshizaki. The mutant LMP-1 plasmids LMP1-231 and LMPΔ231-387 were gifts from Nancy Raab-Traub (28). The interferon consensus sequence binding protein (ICSBP) and IRF-3 expression plasmids were cloned in the expression plasmid pcDNA3. The IRF-1 (C-20) and IRF-2 (C-19) antibodies were purchased from Santa Cruz Biotechnology, Inc. LMP-1 monoclonal antibody CS1-4 was purchased from Dako. Antitubulin antibody was from Sigma. Tap-2 antibody has been described (58). The IRF-7 C-terminus-specific antiserum was generated by injection of glutathione-S-transferase (GST)-IRF-7B fusion protein (aa 218 to 474) into a rabbit. This IRF-7 antiserum was used only for the experiment shown in Fig. 2C, and the full-length IRF-7B antiserum was used in all other applications (67).
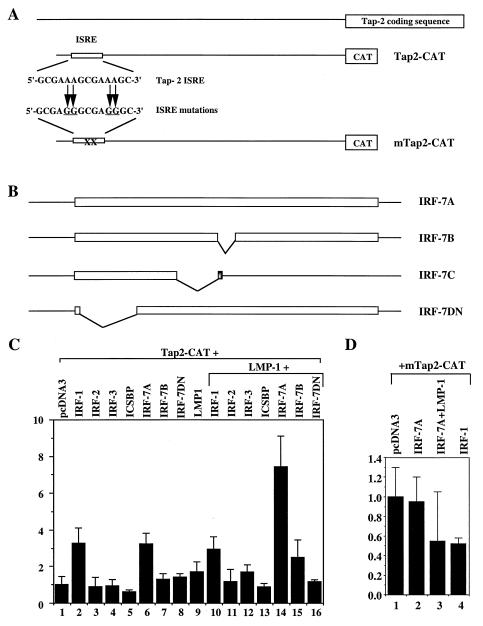
FIG. 3.
Activation of Tap-2 promoter reporter construct by IRF-7. (A) Schematic diagrams of Tap-2 reporter constructs. Top line, Tap-2 genomic region; open rectangle, ISRE. A 573-bp fragment from the Tap-2 promoter region was cloned; the ISRE sequence and mutations are shown. (B) Schematic diagrams of various IRF-7 expression plasmids. IRF-7A, -7B, and -7C are splicing variants of IRF-7. IRF-7DN lacks the DNA-binding domain. (C) Activation of Tap-2 reporter construct by IRF-7 in B cells. Akata cells were transfected with the reporter construct Tap2-CAT together with vector pcDNA-3 (column 1) or expression plasmids for IRF-1 (column 2), IRF-2 (column 3), IRF-3 (column 4), ICSBP (column 5), IRF-7A (column 6), IRF-7B (column 7), IRF-7DN (column 8), or LMP-1 (column 9). Columns 10 to 16, pcLMP-1 was cotransfected with IRF-1, IRF-2, IRF-3, ICSBP, IRF-7A, IRF-7B, and IRF-7DN, respectively. (D) Mutations in ISRE abolish activation by IRF-7. Akata cells were transfected with the reporter construct mTap2-CAT and pcDNA-3 (column 1) or IRF-7A (column 2) or IRF-7A plus pcLMP-1 (column 3) or IRF-1 expression plasmid (column 4). CAT assay results were normalized to β-galactosidase activity. CAT activity is expressed relative to the vector control. Standard deviations are shown.
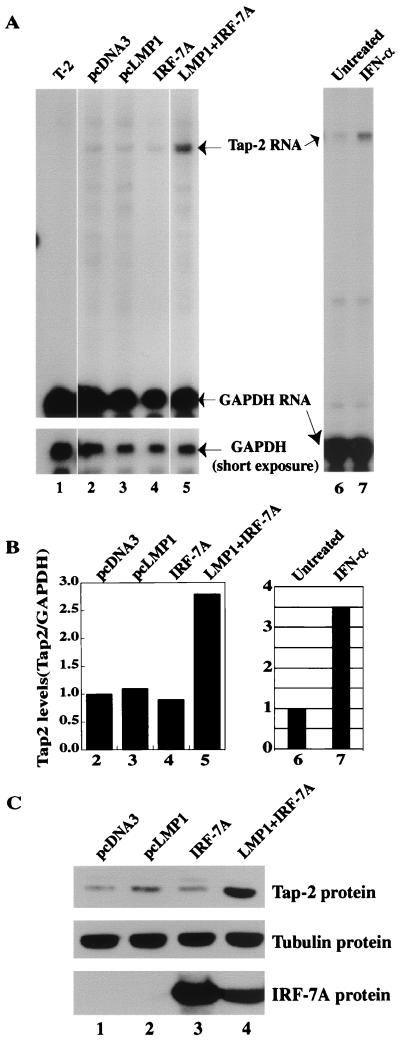
FIG. 5.
IRF-7 and LMP-1 coactivate endogenous Tap-2. (A) RPA was performed with Tap-2 and GAPDH probes. Lane 1, RNA from the T-2 cell line; lanes 2 to 5, RNAs from transfected Akata cells. Transfections: lane 2, pcDNA3; lane 3, pcLMP-1; lane 4, pcDNA-IRF-7A; lane 5, pcLMP-1 plus IRF-7A. Specific protections and undigested probes are shown. Bottom panel, short exposure for GAPDH-protected areas. A representative experiment is shown. (B) Relative Tap-2 levels from panel A. Data were obtained by normalizing Tap-2 RNA levels to GAPDH RNA levels with the use of a PhosphorImager. The column numbers match the lanes in panel A. (C) Western blot analysis of transfected cells with various antibodies. The identity of proteins is indicated.
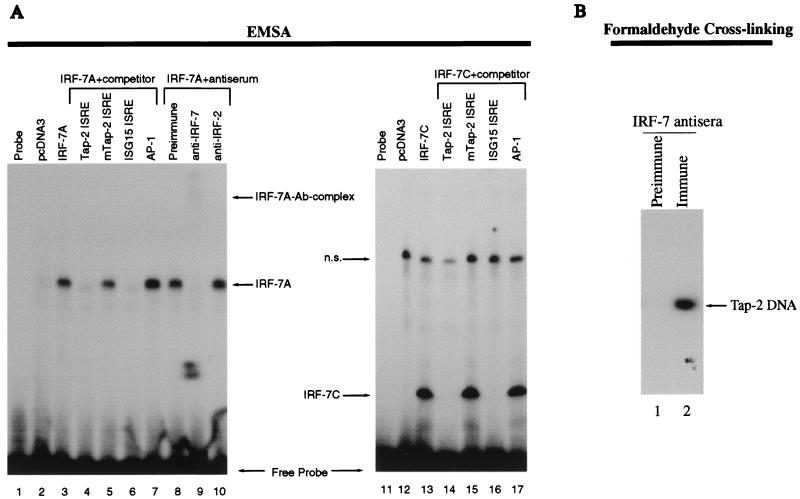
FIG. 2.
IRF-7 binds to Tap-2 promoter. (A) IRF-7 binds to Tap-2 ISRE in vitro. EMSA was performed with the Tap-2 ISRE probe labeled with [α-32P]dCTP. Unlabeled competitors were all added at a 100-fold molar excess over the labeled probe. Lanes 1 and 11, free probe; lane 2, wheat germ lysate containing in vitro-translated protein from plasmid pcDNA3; lanes 3 to 10, wheat germ lysate containing in vitro-translated protein from pcDNA-IRF-7A. The Tap-2 ISRE and the ISRE sequence from the ISG15 gene (ISG15 ISRE) were used as unlabeled competitors in lanes 4 and 6, and the mutated ISRE (mTap2-ISRE) and AP-1 binding site were used for lanes 5 and 7. Preimmune, preimmune serum for IRF-7B protein. Preimmune serum was used for lane 8, and IRF-7 antiserum was used for lane 9. Nonrelevant rabbit polyclonal antibody (Ab) against IRF-2 (Santa Cruz) was used for lane 10. Lane 12, reticulocyte lysates containing in vitro-translated proteins from plasmid pcDNA3; lanes 13 to 17, reticulocyte lysates containing in vitro-translated protein from plasmid pcDNA-IRF-7C.1; Tap-2 ISRE, mTap2-ISRE, ISG15 ISRE, and AP-1 unlabeled competitors were used in lanes 14 to 17, respectively. n.s., nonspecific. (B) IRF-7 binds to Tap-2 in vivo. X50-7 cells were treated with formaldehyde to cross-link proteins bound to DNA. Cross-linked complexes were immunoprecipitated with preimmune (lane 1) and immune (lane 2) antisera to IRF-7. After reversal of the cross-linking, the DNA was then amplified by Tap-2 promoter-specific primers. The PCR products were separated on an agarose gel and analyzed by Southern blot hybridization with the 32P-labeled Tap-2 promoter fragment containing the ISRE as the probe after transfer to a nylon membrane.
Western blot analysis with enhanced chemiluminescence.
Separation of proteins on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed standard methods. After the proteins were transferred to a nitrocellulose or Immobilon membrane, the membrane was blocked with 5% nonfat dry milk in TBST (50 mM Tris [pH 7.5], 200 mM NaCl, 0.05% Tween 20) at room temperature for 10 min. It was then washed briefly with water and incubated with primary antibody in 5% milk in TBST for 1 to 2 h at room temperature or overnight at 4°C. After being washed with TBST three times for 10 min each, the membrane was incubated with the secondary antibody at room temperature for 1 h. It was then washed three times with TBST as before, treated with ECL (Amersham) or SuperSignal (Pierce) detection reagents, and exposed to Kodak XAR-5 film.
Transient transfection, enzyme assays, and isolation of transfected cells.
Cells (107) in 0.5 ml of medium were transfected with the use of a Bio-Rad Gene Pulser (320 V and 925 μF). Two days after transfection, cells were collected for chloramphenicol acetyltransferase (CAT) assay or for isolation of transfected cells. The CAT and β-galactosidase assays were performed essentially as described (22). The CAT assay results were analyzed on a Molecular Dynamics PhosphorImager.
For isolation of transfected cells, cells were collected after transfection, and enrichment for CD4-positive cells was performed with the use of anti-CD4 antibody conjugated to magnetic beads according to the manufacturer's recommendation (Dynal, Inc.). The isolated cells were used for the extraction of total RNA.
RNA extraction and RPA.
Total RNA was isolated from cells with the use of RNease total RNA isolation kit (Qiagen). The RNase protection assay (RPA) was performed with total RNA with the use of either the Lysate RNase protection kit (US Biochemicals, Inc.) (Fig. 1) or the RNase protection kit II (Ambion Inc; all other figures). The hybridization temperature was 37°C for Fig. 1 and 45°C for the rest of the figures. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was supplied by US Biochemicals, Inc. The Tap-2 RPA probe was made by PCR with the primers 5′-GCTCTAGATAATACGACTCACTATAGGGCGACAGACCCAAGCTTGGTACCGAGCTCGGATCCCGCTCTCAGGGAGACAGTCA-3′ and 5′-GCTCTAGACTGGACCTCCCTGCTGCTGGTGGAC-3′. The PCR product contains a T7 promoter, a spacer region, and the Tap-2 first exon sequence (complementary to nt 116227 to 116458 of the sequence published in GenBank [no. X87344]). The PCR product was purified and used directly for synthesis of RNA probe.
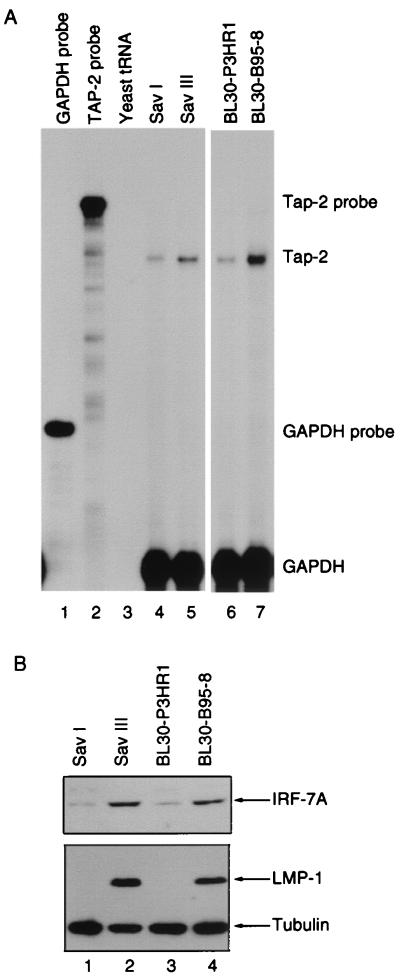
FIG. 1.
Tap-2 RNA is associated with IRF-7 and LMP-1. (A) Tap-2 RNA in various paired cell lines. Tap-2 and GAPDH probes were labeled with [α-32P]UTP and used for RPA. Lanes 1 and 2, undigested Tap-2 and GAPDH probes; lane 3, yeast tRNA; lanes 4 and 5, RNAs from Sav-I and Sav-III cells, respectively; lanes 6 and 7, RNAs from BL30-P3HR1 and BL30-B95-8 cells, respectively. (B) IRF-7 and LMP-1 levels in cell lysates from various cell lines. Equal amounts of protein lysates from cell lines were electrophoresed in SDS–8% PAGE. Western blotting with IRF-7 or LMP-1 and tubulin antibodies was performed.
EMSA.
For the electrophoretic mobility shift assay (EMSA), the Tap-2 ISRE-containing fragment was obtained by annealing two oligonucleotides, 5′-GATCGGAAGCGAAAGCGAAAGCTGCCC-3′ and its complement, with GATC at the 5′ end. The mutated ISRE probe was made exactly the same way with mutated oligonucleotides, as shown in Fig. 3A. The DNA probes were generated by filling in any single-stranded overhang with Klenow enzyme. EMSA was performed essentially as described (65, 67). In vitro-translated reticulate lysates (5 μl) were incubated with 20,000 to 50,000 cpm of labeled probe in a volume of 12.5 μl containing 20 mM HEPES (pH 7.9), 1 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol, 320 μg of poly(dI-dC):poly(dI-dC), and 4% Ficoll-400 for 20 min at room temperature. The samples were separated on a preelectrophoresed 4.8% polyacrylamide gel in 20 mM Tris-borate-EDTA (TBE) buffer. After electrophoresis, gels were dried, followed by autoradiography. When antiserum was needed, 1 μl was added to the reaction mixture. The consensus ISRE oligonucleotide has been described (67). AP-1 binding site competitor was purchased from Promega.
In vitro transcription and translation.
The proteins were made with the TNT coupled transcription and translation kit (Promega) essentially according to the manufacturer's instructions. Wheat germ lysate was used with the plasmid pcDNA-IRF-7A, and rabbit reticulocyte lysate was used with the plasmid pcDNA-IRF-7C.1.
Analysis of DNA-binding activity by in vivo formaldehyde cross-linking.
The cross-linking method is based on a previous publication (36). Cells were fixed with 1/10 volume of 11% formaldehyde solution in 0.1 M NaCl–1 mM EDTA–0.5 mM EGTA–50 mM HEPES (pH 8.0) for 1 h at 4°C. The cross-linking reaction was stopped by adding glycine to 0.125 M. The cells were washed and sonicated, and the cell debris was removed by centrifugation at 15,000 × g for 10 min as described (36). The cell lysates were cleared first by incubating with protein G-agarose beads and preimmune serum. Then immunoprecipitations were performed with preimmune or immune serum specific for the C-terminal region of IRF-7 at 4°C overnight. Immunoprecipitates were collected by protein G-agarose beads and washed three times for 10 min each in 1× phosphate-buffered saline (PBS) solution plus phenylmethylsulfonyl fluoride (PMSF) and another three times for 10 min each in Tris-EDTA (TE) buffer. Finally, the pellets were digested at 65°C overnight in 100 μl of TE plus 0.5% SDS and proteinase K (500 μg/ml). After phenol-chloroform extraction, the DNA was precipitated with glycogen as the carrier. The isolated DNA was used as the template for amplification of the Tap-2-specific region with primers 5′-GAGTTCGGAAGGCCTTGG-3′ (corresponding to nt 115623 to 115640) and 5′-GAAGCAGGAGCGTGGAGT-3′ (complementary to nt 115856 to 115873) (2). The PCR products were separated in a 1.5% agarose gel, transferred to a nylon membrane, and hybridized to labeled Tap-2 promoter probe (nt 115644 to 116217), which was synthesized with random primer and Klenow enzyme by the use of standard methods (49).
Phosphorylation analysis and immunoprecipitation.
293T cells were cotransfected with expression plasmids for IRF-7A and LMP-1 with the use of Effectene (Qiagen). Cells were then labeled with [32P]orthophosphate for 4 h, washed once with 1× TBS, and lysed in a buffer containing 0.5% NP-40, 50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 100 mM NaF, 1 mM sodium orthovanadate, and 1 mM PMSF. Cell lysates were precleared with preimmune serum and protein G-agarose beads under gentle agitation at 4°C for 30 min. The anti-IRF-7 antiserum and protein G-agarose were then added, and lysates were incubated overnight at 4°C. The beads were washed three times with 1 ml of immunoprecipitation washing buffer, resuspended in 50 μl of SDS-PAGE sample loading buffer, and boiled for 5 min. Samples were resolved by electrophoresis on an SDS-polyacrylamide gel and transferred onto an Immobilon membrane. The membrane was dried and autoradiographed. After autoradiography, the membrane was rehydrated with 100% methanol for 30 s and used for Western blotting to visualize the total amount of IRF-7.
Immunofluorescence.
293T cells grown on chamber slides (Lab-TeK) were transfected with plasmids. Twenty-four hours after transfection, the cells were fixed by 60% acetone, stained with DAPI (4′,6′-diamidino-2-phenylindole, 1 μg/ml) in PBS, and mounted with 60% glycerol in PBS. Samples were examined with a Zeiss Axioskope.
RESULTS
Expression of Tap-2 is associated with IRF-7 and LMP-1.
It has been shown recently that EBV LMP-1 protein stimulates the expression of IRF-7 (66). Since LMP-1 can induce a variety of genes, whether IRF-7 is a secondary mediator for some of those induced genes was examined. Tap-2 was especially interesting not only because it is induced by LMP-1, but also because of the putative ISRE in the Tap-2 promoter region (2). With the use of RPA and a specific probe for Tap-2, whether expression of Tap-2 RNA is associated with expression of IRF-7 and LMP-1 was addressed. Sav-I and Sav-III are sister Burkitt's lymphoma lines both derived from a single parental cell line. The paired lines differ only in their types of latency. Sav-III cells, which express LMP-1, have a higher IRF-7 level than Sav-I cells, which do not express LMP-1 (Fig. 1B) (67). Another pair of cell lines, BL30-P3HR1 and BL30-B95-8, were established by infecting the EBV-negative BL30 line with P3HR1 or B95-8 virus, respectively. BL30-P3HR1 expresses very low levels of LMP-1 and IRF-7, whereas BL30-B95-8 cells express high levels of LMP-1 and IRF-7A (Fig. 1B) (66). In both lines, the level of Tap-2 RNA correlates with expression of IRF-7 and LMP-1 (Fig. 1A). Similar results were obtained with another set of paired cell lines, BL41-P3HR1 and BL41-B95-8. Also, Akata cells (low endogenous IRF-7, no LMP-1) have lower Tap-2 RNA levels than Jijoye cells (high IRF-7 and LMP-1) (data not shown). Finally, in EBV-negative BJAB and BL41 cell lines expressing LMP-1, both Tap-2 and IRF-7 protein levels were increased (data not shown) (46, 66). All these data suggest that Tap-2 expression is associated with expression of IRF-7 and LMP-1.
IRF-7 binds to Tap-2 promoter in vitro and in vivo.
A putative ISRE sequence has been identified based on sequence homology in the Tap-2 promoter region (2). Because IRFs have potential to bind to ISRE, whether IRF-7 can bind to the putative Tap-2 ISRE was tested by EMSA. IRF-7 was in vitro translated, and lysate was used for EMSA with labeled Tap-2 ISRE as a probe. As shown in Fig. 2, specific bands appeared when IRF-7A was used for EMSA (lanes 3 to 10). These bands were specific because they disappeared with an excess of unlabeled competitors, such as Tap-2 ISRE and consensus ISRE from the interferon-stimulated gene (ISG) 15 promoter (lanes 4 and 6), but mutated Tap-2 ISRE or nonspecific competitor, such as AP-1 binding site, had no effect (lanes 5 and 7), and DNA-binding activity was not affected when preimmune serum or nonrelevant antibody (anti-IRF-2) was used (lanes 8 and 10); however, specific IRF-7 antibody could block and supershift the IRF-7A–DNA complex (lane 9). Furthermore, IRF-7C, which has only the N-terminal 164 aa (Fig. 3B), can specifically bind to the Tap-2 ISRE sequence (lanes 11 to 17), indicating that the DNA-binding domain of IRF-7 to Tap-2 ISRE is localized in the N-terminal region, as expected (67).
All previous attempts to identify the IRF-7–Tap-2 ISRE complex in cell lysates by EMSA have failed. Interestingly, in these experiments, IRF-2 from the same cell lysates could clearly bind to the Tap-2 ISRE (data not shown), which also confirms the authenticity of the Tap-2 ISRE.
In order to test whether physiological levels of IRF-7 can bind to the Tap-2 promoter in vivo, X50-7 cells, a type III EBV latency cell line with high levels of IRF-7 and LMP-1, were fixed with formaldehyde, and IRF-7-DNA complexes were isolated by immunoprecipitation with IRF-7 antiserum. The DNA recovered from the immunoprecipitates was used as the template for PCR amplification of the Tap-2 promoter region containing the ISRE. The authenticity of the PCR products was verified by Southern blot analysis with the Tap-2 promoter sequence as a probe (see Materials and Methods for details). As shown in Fig. 2B, IRF-7-specific antiserum could specifically precipitate the IRF-7 protein–Tap-2 promoter complex (lane 2). However, preimmune serum did not bring down any Tap-2 DNA (lane 1). Similar results were also obtained with another type III latency cell line, Jijoye (data not shown). From these data, we conclude that IRF-7 is able to bind to the Tap-2 promoter under physiological conditions in vivo.
IRF-7A activates Tap-2 promoter constructs.
The effect of IRF-7 on the promoter activity of the Tap-2 gene was studied using Tap-2 promoter constructs in transient-transfection assays. The Akata cell line was chosen because of its low endogenous expression of IRF-7, and LMP-1 cannot induce IRF-7 in this particular cell line. A Tap-2 promoter construct containing the ISRE sequence, Tap-2-CAT, was cloned according to the published sequence (2). Cotransfection of an IRF-1 or IRF-7-expression plasmid with Tap-2-CAT resulted in activation of the Tap-2 promoter construct (Fig. 3C, columns 2 and 6). However, IRF-2, IRF-3, ICSBP, IRF-7B (which is lacking 29 aa in the middle of the protein), and IRF-7DN (aa 1 to 12 and 103 to 503), which lacks the N-terminal DNA-binding domain of IRF-7 (Fig. 3B) (61, 67), could not activate the Tap-2 promoter construct. Nor could IRF-7C, which lacks the C-terminal domain of IRF-7 by alternative splicing (67), activate the Tap-2 promoter construct (Fig. 3B and data not shown). The activation of the Tap-2 promoter by IRF-7 and IRF-1 was only observed with the intact ISRE sequence; IRF-7 and IRF-1 failed to activate the Tap-2 promoter construct with ISRE mutations (mTap2-CAT) that abolish IRF-7 binding (Fig. 2 and 3D). The Tap-2 ISRE is apparently not essential for the constitutive activity of the Tap-2 promoter, because the ISRE mutations only reduced the constitutive activity slightly (data not shown). These data suggest that both N- and C-terminal domains of IRF-7 are required for transactivation of the Tap-2 promoter and that binding of IRF-7 to the Tap-2 promoter is essential for its activation.
LMP-1 enhances the activation of Tap-2 promoter construct by IRF-7.
Since both IRF-7 and LMP-1 are associated with high levels of Tap-2 expression (Fig. 1), whether LMP-1 can enhance the activation of the Tap-2 promoter by IRF-7 was tested in Akata cells. LMP-1 alone could not activate the Tap-2 construct (Fig. 3C, column 9); however, LMP-1 plus IRF-7 could enhance the activation of the Tap-2 reporter construct (column 14). The enhancement was not due to the increase in IRF-7 expression (see Fig. 4C). LMP-1 plus IRF-7B may also activate TAP-2 (lane 15). LMP-1 plus other IRFs or IRF-7DN did not activate the Tap-2 promoter further (columns 10 to 13 and 16). Once again, the intact ISRE was essential for Tap-2 activation, because ISRE mutations abolished the activation by IRF-7 and LMP-1 (Fig. 3D). These data suggest that LMP-1 specifically enhances the activation of the Tap-2 promoter by IRF-7.
FIG. 4.
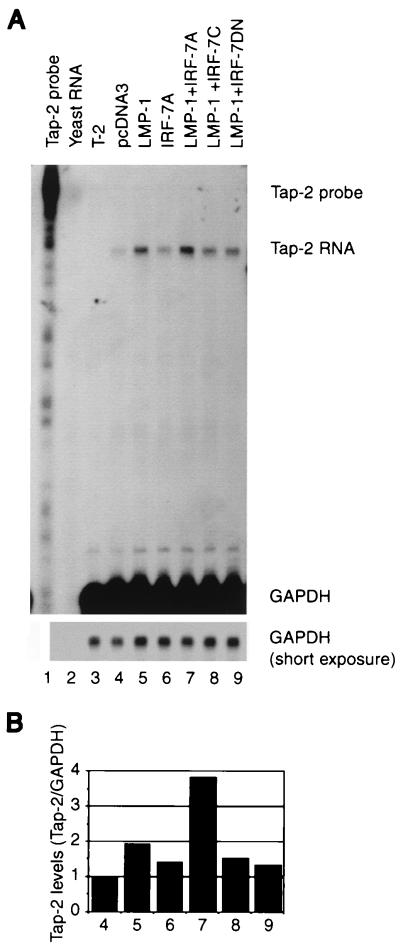
IRF-7 is involved in the activation of endogenous Tap-2 RNA. (A) RPA was performed with Tap-2 and GAPDH probes. Lane 1, undigested Tap-2 probe; lane 2, yeast RNA; lane 3, RNA from T-2 cell line. Lanes 4 to 9, RNAs from transfected and selected DG75 cells; lane 4, pcDNA3; lane 5, pcLMP-1; lane 6, pcDNA-IRF-7A; lanes 7 to 9, pcLMP-1 plus IRF-7A, IRF-7C, and IRF-7DN expression plasmids, respectively. Specific protection of Tap-2 and GAPDH RNAs and undigested probes is indicated. Bottom panel, short exposure for GAPDH-protected areas. (B) Relative Tap-2 levels from panel A. Data were obtained by normalizing Tap-2 RNA levels to GAPDH RNA levels with the use of a PhosphorImager. The column numbers match the lanes in panel A.
IRF-7 is involved in the induction of endogenous Tap-2 RNA by LMP-1.
Whether IRF-7 is involved in the regulation of the endogenous Tap-2 gene was examined. An EBV-negative Burkitt's lymphoma cell line, DG75, was chosen because of transfection efficiency and because LMP-1 can stimulate the expression of IRF-7 in this line (66). Expression plasmids were transfected into the cells along with a CD4 expression plasmid. Two days after transfection, the transfected cells were enriched by magnetic beads conjugated with anti-CD4 antibody (see Materials and Methods for details). Total RNA was isolated, and RPA was employed with specific probes. As shown in Fig. 4, LMP-1 increases Tap-2 RNA about twofold (lane 5). IRF-7A alone has minimal effect on the Tap-2 RNA (lane 6). However, LMP-1 and IRF-7 together increased Tap-2 RNA levels almost fourfold (lane 7). The identity of the protected band as Tap-2 RNA was confirmed by testing RNA from the T-2 cell line, which has a genomic deletion in the Tap-2 gene (lane 3). Tap-2 expression was not increased further by LMP-1 plus IRF-7C, IRF-7DN (Fig. 4, lanes 8 and 9), or IRF-7B (data not shown). IRF-7DN was able to repress the promoter activity of the beta interferon (IFN-β) gene after viral infection (61). IRF-7DN may also repress LMP-1-induced Tap-2 expression (Fig. 4, compare lane 4 to lane 9). These data suggest that IRF-7 is involved in the activation of Tap-2.
IRF-7 and LMP-1 coactivate the endogenous Tap-2 gene.
Whether IRF-7 is involved in the activation of endogenous Tap-2 was further examined in Akata cells, in which LMP-1 cannot induce IRF-7 RNA (66). If LMP-1 activates Tap-2 through IRF-7, then LMP-1 alone would have no effect, but IRF-7 plus LMP-1 would activate the Tap-2 gene in this particular cell line. Expression plasmids were transfected into the cells along with a CD4 expression plasmid. Total RNA was isolated from CD4-positive cells, and RPA was employed with specific probes. As shown in Fig. 5, overexpression of neither LMP-1 nor IRF-7 alone could stimulate the expression of Tap-2 in Akata cells (lanes 3 and 4). However, LMP-1 and IRF-7 together increased the Tap-2 RNA level 2.8-fold (lane 5). As expected, Western blot analysis showed that the level of Tap-2 protein was also increased by expression of LMP-1 and IRF-7 (Fig. 5C). These data suggest that LMP-1 and IRF-7 coactivate endogenous Tap-2.
Because IFN activates Tap-1 and HLA class I genes and because the entire class I system, including Tap-2, is usually upregulated simultaneously (reviewed in references 32, 54, and 63), we tested whether IFN-α can activate Tap-2 in Akata cells. The endogenous Tap-2 RNA was increased 3.5-fold upon IFN treatment (Fig. 5, lanes 6 and 7). If the efficiency of transfection and selection of transfected cells are considered, LMP-1 plus IRF-7 can induce Tap-2 to a level similar to that induced by IFN.
IRF-7A and full-length LMP-1 are required for the activation of Tap-2.
The CAT assay results suggested that IRF-7A but not other IRF-7 splicing variants is an activator of the Tap-2 promoter in Akata cells (Fig. 3). Whether other forms of IRF-7 are capable of activating endogenous Tap-2 was examined by transfection with various forms of IRF-7 along with LMP-1 and monitoring the endogenous Tap-2 levels in Akata cells. As shown in Fig. 6 (lanes 1 to 8), only IRF-7A plus LMP-1 could activate the endogenous Tap-2 gene (lane 3). These data are in agreement with the facts that IRF-7A is a major form of IRF-7 and that LMP-1 primarily stimulates the expression of IRF-7A (66, 67).
FIG. 6.
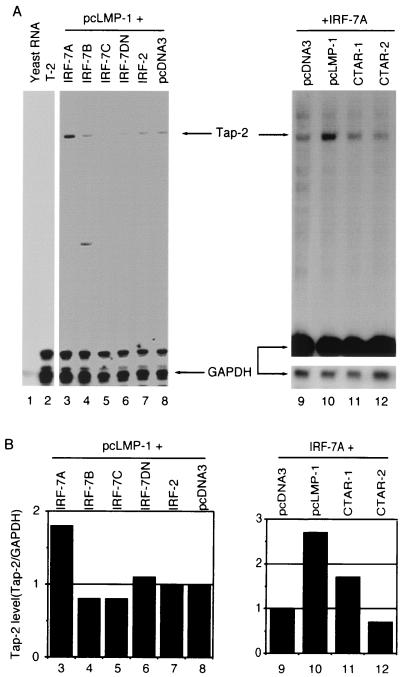
IRF-7A and full-length LMP-1 are required for the efficient activation of endogenous Tap-2. (A) RPA was performed with Tap-2 and GAPDH probes. Lanes 1 and 2, yeast RNA and RNA from the T-2 cell line; lanes 3 to 7, RNAs from transfected Akata cells. Plasmid pcLMP-1 was transfected with pcDNA-IRF-7A (lane 3), IRF-7B (lane 4), IRF-7C (lane 5), IRF-7DN (lane 6), IRF-2 (lane 7), or pcDNA3 (lane 8). Lanes 9 to 12, pcDNA-IRF-7A was transfected with pcDNA3 (lane 9), full-length pcLMP-1 (lane 10), LMP CTAR-1 (lane 11), or CTAR-2 (lane 12). Bottom panel for lanes 9 to 12, short exposure for GAPDH-protected area. Specific protections are shown. A representative experiment is shown. (B) Relative Tap-2 levels from panel A. Data were obtained by normalizing Tap-2 RNA levels to GAPDH RNA levels with the use of a PhosphorImager. The column numbers match the lanes in panel A.
To dissect the domain requirement of LMP-1 for the activation of Tap-2, LMP-1 mutants were tested along with IRF-7A for their ability to increase endogenous Tap-2 RNA in Akata cells. Currently, two major functional domains of LMP-1 have been dissected, namely, CTAR-1 and CTAR-2. These two domains can activate different signaling molecules (16, 29). As shown in Fig. 6 (lanes 9 to 12), full-length LMP-1 is most efficient for the induction of Tap-2 with IRF-7A (lane 2).
LMP-1 enhances phosphorylation and nuclear translocation of IRF-7.
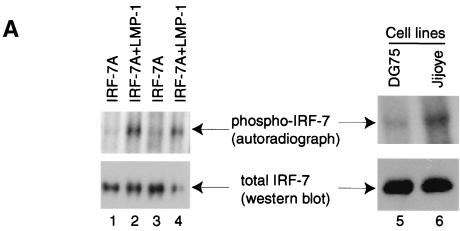
In order to understand the mechanism whereby LMP-1 and IRF-7 coactivate Tap-2, we examined whether LMP-1 regulates IRF-7 in a posttranslational manner. Because LMP-1 is known to activate several important cellular molecules through phosphorylation and IRF-7 may be phosphorylated upon viral infection (24, 61), whether LMP-1 induces the phosphorylation of IRF-7 was examined. Cells transfected with IRF-7 or LMP-1 plus IRF-7 were labeled with [32P]orthophosphate, and cell lysates were used for immunoprecipitation with IRF-7 antiserum. The immunoprecipitates were analyzed by SDS-PAGE and transferred onto a membrane, which was analyzed by autoradiography as well as by Western blot. As shown in Fig. 7A, IRF-7 itself is a phosphoprotein (lanes 1 and 3); however, LMP-1 could enhance the phosphorylation status of IRF-7 (lanes 2 and 4).
FIG. 7.
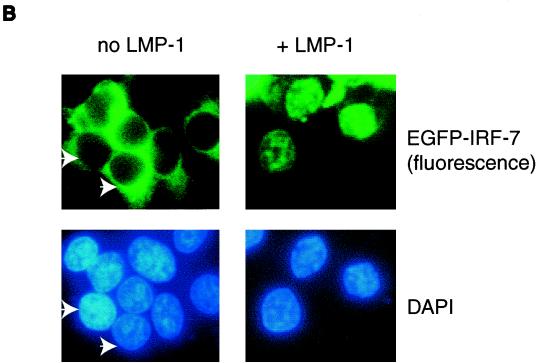
LMP-1 facilitates the phosphorylation and nuclear translocation of IRF-7. (A) LMP-1 facilitates the phosphorylation of IRF-7. 293T cells were transfected with IRF-7A with (lanes 2 and 4) or without (lanes 1 and 3) LMP-1 expression plasmid. At 24 h after transfection, cells were labeled with [32P]orthophosphate, immunoprecipitates were separated in SDS-PAGE and transferred to a membrane, and Western blotting with IRF-7 antibody was done after autoradiography for detection of phospho-IRF-7. Lanes 5 and 6, DG75 and Jijoye cells were labeled with [32P]orthophosphate, respectively. More DG75 cell lysate was needed for immunoprecipitation in order to get equal amounts of total IRF-7. (B) LMP-1 facilitates the nuclear translocation of IRF-7. 293T cells were transfected with EGFP-IRF-7A with or without the LMP-1 expression plasmid. The subcellular localization of IRF-7 was examined 24 h after transfection. Several cells were not transfected with the EGFP-IRF-7A plasmid. Arrows indicate corresponding cells. Magnification, ×400.
Next, whether endogenous LMP-1 can enhance the phosphorylation of IRF-7 was examined. DG75 is an EBV-negative BL cell line with moderate levels of endogenous IRF-7, and Jijoye is an EBV-positive type III latency cell line that expresses LMP-1. These cells were labeled, and phosphorylation of IRF-7 in these cells was examined. Because the endogenous IRF-7 level in DG75 cells was lower than in Jijoye cells, more DG75 cell lysate was used for immunoprecipitation. As shown in Fig. 7A, the IRF-7 in DG75 cells was less phosphorylated than the IRF-7 in Jijoye cells (lanes 5 and 6). Also, highly phosphorylated IRF-7 was readily detectable in other type III cells, such as B958/CBC and X50-7 (data not shown).
Since viral infection may facilitate the nuclear translocation of IRF-7 (1, 61), whether LMP-1 expression affects the subcellular localization of IRF-7 was examined in EBV-negative 293T cells. As shown in Fig. 7B, most of the IRF-7 when expressed alone was localized in the cytoplasm; however, when LMP-1 was present, most of the IRF-7 localized in the nucleus with a punctate appearance. In type III cells, endogenous IRF-7 is also mainly localized in the nucleus (data not shown). Therefore, LMP-1 augments the phosphorylation and nuclear translocation of IRF-7.
DISCUSSION
The signal transduction pathway of LMP-1 has been extensively studied. The involvement of tumor necrosis factor receptor-associated factors and NF-κB in the immediate steps of the signal transduction pathway has been established conclusively (9, 17, 25, 26, 28, 50). While induction of some genes by LMP-1 may result from the direct activation of NF-κB, other genes may need a secondary mediator(s). Here we have shown that the induction of Tap-2 requires IRF-7 as a secondary mediator. First, Tap-2 expression is associated with both IRF-7 and LMP-1 expression (Fig. 1) (46, 66). Second, LMP-1 induces Tap-2 expression in DG75, BJAB, and BL41 cells, in which IRF-7 can be stimulated (Fig. 4) (46) (data not shown). Also, ectopic expression of IRF-7 enhances the induction of Tap-2 by LMP-1 in DG75 cells (Fig. 4). Third, LMP-1 could not induce Tap-2 in Akata cells, in which IRF-7 could not be induced by LMP-1. However, addition of ectopic IRF-7, which artificially restores the defect, could activate the expression of Tap-2 in Akata cells (Fig. 5). Fourth, the Tap-2 promoter construct could be activated by IRF-7 and further enhanced by LMP-1 specifically. Also, the activation of the Tap-2 promoter was dependent on the intact ISRE sequence (Fig. 3). Fifth, IRF-7 could bind specifically to the ISRE in the Tap-2 promoter in vitro and to the Tap-2 promoter under physiological conditions in vivo (Fig. 2). Sixth and finally, LMP-1 facilitates the phosphorylation and nuclear translocation of IRF-7 (Fig. 7).
It is apparent that IRF-7 is the most relevant IRF member for the activation of Tap-2 by LMP-1. First, both IRF-7 and IRF-2 are associated with EBV type III latency, in which levels of Tap-2 are high. IRF-2 could also bind to the Tap-2 ISRE, as determined by EMSA (data not shown). However, neither IRF-2 nor LMP-1 plus IRF-2 could activate Tap-2 expression. Also, LMP-1 cannot induce the expression of IRF-2 (66). Second, LMP-1 could specifically enhance the activation of the Tap-2 promoter by IRF-7 but not by IRF-1, although IRF-1 can activate the Tap-2 promoter reporter (Fig. 3C). Third, other IRFs tested, such as IRF-1, IRF-3, and ICSBP, are not associated with type III latency (34, 66, 67). Fourth and finally, LMP-1 regulates IRF-7 in a posttranslational manner (Fig. 7). Therefore, LMP-1 has an intimate relationship with both the expression and activation of IRF-7.
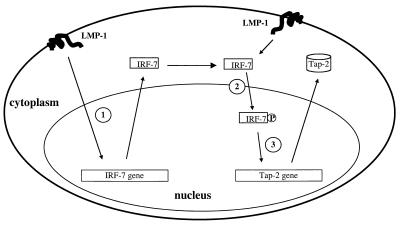
Considering all the existing data, we propose an LMP-1-triggered signal transduction pathway that leads to stimulation of expression of IRF-7. LMP-1 further activates IRF-7 by phosphorylation and nuclear translocation of the protein. Finally, activated IRF-7 mediates the activation of the Tap-2 gene (Fig. 8).
FIG. 8.
Model for induction of the Tap-2 gene by LMP-1. (Step 1) LMP-1 induces the expression of IRF-7. (Step 2) LMP-1 facilitates the phosphorylation and nuclear translocation of IRF-7. (Step 3). The activated IRF-7 mediates the activation of the Tap-2 gene.
The domain analysis of activation of Tap-2 by IRF-7 showed that full-length IRF-7A is required both for activation of the Tap-2 promoter construct and for the increase in endogenous Tap-2 RNA (Fig. 3 and 6). These data suggest that the IRF-7A activation domain, at least in the B-cell lines tested, is located in the C terminus beyond aa 227, because IRF-7B, which lacks aa 227 to 255 in IRF-7A, could not activate Tap-2. This result seems to contradict a previously reported activation domain (corresponding to aa 107 to 224 of IRF-7A) obtained in the L929 mouse fibroblast line (1). The different cell lines that were used for the study may contribute to this discrepancy. In support of such a notion, IRF-7B could efficiently activate the Tap-2 promoter construct in FaDuHyg, an epithelial cell line (data not shown). However, considering that IRF-7 is primarily a lymphoid factor, a conclusion based on B cells may be more relevant to its biological function.
IRF-7 appears to be a secondary mediator for the repression of the EBV latency promoter (Qp) by LMP-1 (66). In this paper, we have shown that IRF-7 is a secondary mediator for Tap-2 activation. The domain requirements of IRF-7 for these two biological effects are quite different. For Qp repression, the N-terminal DNA-binding domain is sufficient (67); however for Tap-2 activation, both the N- and C-terminal domains of IRF-7 are required (Fig. 3 to 6). Whether IRF-7 is involved in the induction of more LMP-1-regulated genes needs to be addressed. Since HLA 1, Tap-1, and Tap-2 are often induced simultaneously for antigen processing (e.g., by treatment with IFN-α, IFN-γ, or lipopolysaccharide), it is tempting to speculate that IRF-7 may also be involved in the regulation of the Tap-1 and HLA I genes, both of which have been shown to be regulated by IRF-1 (39, 62).
What advantage does EBV gain by inducing Tap-2 and other HLA I-related genes? Type III EBV latency, an LMP-1-expressing latency state, is established transiently in primary infection of human B cells in vivo (reviewed in references 19 and 44). Type III cells have enhanced growth, survival, and invasive potential, which allow the EBV-infected cells to proliferate quickly, thereby putting the human host at risk. This rapid proliferative process is checked after the development of EBV-specific primary cytotoxic T lymphocytes (CTL), which eliminate these type III latency cells because of the activation of Tap-2 and other HLA I-related genes by LMP-1, and ensure the safety of the host. In X-linked immunoproliferative disease, in which T-cell activation is defective (51), EBV infection is lethal. Interestingly, EBV in the normal host still survives the CTL attack by establishing a type I-like latency state, in which LMP-1 is not expressed (6, 30, 31, 40, 56). This type I-like latency can escape host immune surveillance, which ensures the survival of the virus. Because the whole process may ensure the survival both of the host and of the virus, a life-long coexistence may thus be maintained between the human host and EBV. In support of such a notion, the LMP-1-positive immunoblastic B-cell lymphomas of the immunosuppressed are highly susceptible to recovered patients' cytolytic T cells or to adoptive CTL therapy (7, 8, 38, 45).
In summary, our data provide direct evidence that IRF-7 is involved in regulation of a crucial immune system gene and is a secondary mediator for the LMP-1 viral protein in modulating the normal functions of the immune and inflammatory responses.
ACKNOWLEDGMENTS
We thank Nancy Raab-Traub, Paula Pitha, Ben Levi, Keiko Ozato, Peter Howley, Tom Maniatis, Ho-sun Park, and Peter van Endert for providing valuable reagents for this work. We also thank Lihong Wu for technical help, Shannon Kenney and Jenny Ting for critical reading of the manuscript, and the UNC sequencing facility.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (AI 42372-01) and from the National Cancer Institute (CA 19014).
REFERENCES
- 1.Au W C, Moore P A, LaFleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 2.Beck S, Kelly A, Radley E, Khurshid F, Alderton R P, Trowsdale J. DNA sequence analysis of 66 kb of the human MHC class II region encoding a cluster of genes for antigen processing. J Mol Biol. 1992;228:433–441. doi: 10.1016/0022-2836(92)90832-5. [DOI] [PubMed] [Google Scholar]
- 3.Belich M P, Trowsdale J. Proteasome and class I antigen processing and presentation. Mol Biol Rep. 1995;21:53–56. doi: 10.1007/BF00990971. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwith Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt-like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 5.Calender A, Billaud M, Aubry J P, Banchereau J, Vuillaume M, Lenoir G M. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc Natl Acad Sci USA. 1987;84:8060–8064. doi: 10.1073/pnas.84.22.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Zou J-Z, DiRenzo L, Winberg G, Hu L-F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr Virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP-1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford D H, Edwards J M. Immunity to Epstein-Barr virus in cyclosporin A-treated renal allograft recipients. Lancet. 1982;i:1469–1470. doi: 10.1016/s0140-6736(82)92479-5. [DOI] [PubMed] [Google Scholar]
- 8.Crawford D H, Sweny P, Edwards J M, Janossy G, Hoffbrand A V. Long-term T-cell-mediated immunity to Epstein-Barr virus in renal-allograft recipients receiving cyclosporin A. Lancet. 1981;i:10–12. doi: 10.1016/s0140-6736(81)90115-x. [DOI] [PubMed] [Google Scholar]
- 9.Devergne O, Hatzivassiliou E, Izumi K, Kaye K, Kleijnen M, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP-1 domain important for B lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donato L, de la Salle H, Hanau D, Tongio M M, Oswald M, Vandevenne A, Geisert J. Association of HLA class I antigen deficiency related to a TAP2 gene mutation with familial bronchiectasis. J Pediatr. 1995;127:895–900. doi: 10.1016/s0022-3476(95)70024-2. [DOI] [PubMed] [Google Scholar]
- 11.Floettmann J E, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerization for NF-κB activation. Oncogene. 1997;15:1851–1858. doi: 10.1038/sj.onc.1201359. [DOI] [PubMed] [Google Scholar]
- 12.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furnari F B, Adams M D, Pagano J S. Regulation of the Epstein-Barr virus DNA polymerase gene. J Virol. 1992;66:2837–2845. doi: 10.1128/jvi.66.5.2837-2845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6130–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heresbach D, Alizadeh M, Bretagne J F, Dabadie A, Colombel J F, Pagenault M, Heresbach L, Berre N, Genetet B, Gosselin M, Semana G. TAP gene transporter polymorphism in inflammatory bowel diseases. Scand J Gastroenterol. 1997;32:1022–1027. doi: 10.3109/00365529709011219. [DOI] [PubMed] [Google Scholar]
- 16.Huen D S, Henderson S A, Croom-Carter S, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 17.Kaye K, Devergne O, Harada J, Izumi K, Yalamanchili R, Kieff E, Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 20.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai S, Kanagawa S, Morinobu A, Takada M, Nakamura K, Sugai S, Maruya E, Saji H. Association of a new allele of the TAP2 gene, TAP2*Bky2 (Val577), with susceptibility to Sjogren's syndrome. Arthritis Rheum. 1997;40:1685–1692. doi: 10.1002/art.1780400919. [DOI] [PubMed] [Google Scholar]
- 22.Laimins L A, Gruss P, Pozzatti R, Khoury G G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984;49:183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebowitz D, Wang D, Kieff E. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller W, Cheshire J, Raab-Traub N. Interaction of tumor necrosis factor receptor-associated factor signaling proteins with the latent membrane protein 1 PXQXT motif is essential for induction of epidermal growth factor receptor expression. Mol Cell Biol. 1998;18:2835–2844. doi: 10.1128/mcb.18.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller W E, Cheshire J L, Baldwin A S, Raab-Traub N. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene. 1997;16:1869–1877. doi: 10.1038/sj.onc.1201696. [DOI] [PubMed] [Google Scholar]
- 27.Miller W E, Earp H S, Raab-Traub N. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller W E, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyashita E M, Yang B, Lam K M C, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 32.Monaco J J, Nandi D. The genetics of proteasomes and antigen processing. Annu Rev Genet. 1995;29:729–754. doi: 10.1146/annurev.ge.29.120195.003501. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 34.Nonkwelo C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 37.Pagano J S. Pathogenesis and treatment of Epstein-Barr virus infection. J Exp Pathol. 1987;3:441–448. [PubMed] [Google Scholar]
- 38.Papadopoulos E B, Ladanyi M, Emanuel D, Mackinnon S, Boulad F, Carabasi M H, Castro-Malaspina H, Childs B H, Gillio A P, Small T N, et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N Engl J Med. 1994;330:1185–1191. doi: 10.1056/NEJM199404283301703. [DOI] [PubMed] [Google Scholar]
- 39.Penninger J M, Sirard C, Mittrucker H W, Chidgey A, Kozieradzki I, Nghiem M, Hakem A, Kimura T, Timms E, Boyd R, Taniguchi T, Matsuyama T, Mak T W. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity. 1997;7:243–254. doi: 10.1016/s1074-7613(00)80527-0. [DOI] [PubMed] [Google Scholar]
- 40.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raab-Traub N, Flynn K, Klein C, Pizza G, De-Vinci C, Occhiuzzi L, Farneti G, Caliceti U, Pirodda E. EBV associated malignancies. J Exp Pathol. 1987;3:449–456. [PubMed] [Google Scholar]
- 42.Ragona G, Ernberg I, Klein G. Induction and biological characterization of the Epstein-Barr virus carried by the Jijoye lymphoma cell line. Virology. 1980;101:553–557. doi: 10.1016/0042-6822(80)90473-0. [DOI] [PubMed] [Google Scholar]
- 43.Reiss M, Munoz-Antonia T, Cowan J M, Wilkins P C, Zhou Z L, Vellucci V F. Resistance of human squamous carcinoma cells to transforming growth factor beta 1 is a recessive trait. Proc Natl Acad Sci USA. 1993;90:6280–6284. doi: 10.1073/pnas.90.13.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 45.Rooney C M, Roskrow M A, Smith C A, Brenner M K, Heslop H E. Immunotherapy for Epstein-Barr virus-associated cancers. J Natl Cancer Inst Monogr. 1998;23:89–93. doi: 10.1093/oxfordjournals.jncimonographs.a024180. [DOI] [PubMed] [Google Scholar]
- 46.Rowe M, Khanna R, Jacob C A, Argaet V, Kelly A, Powis S, Belich M, Croom-Carter D, Lee S, Burrows S R, Trowsdale J, Moss D J, Rickinson A B. Restoration of endogenous antigen processing in Burkitt's lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur J Immunol. 1995;25:1374–1384. doi: 10.1002/eji.1830250536. [DOI] [PubMed] [Google Scholar]
- 47.Salter R D, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant T×B cell hybrid. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salter R D, Howell D N, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP1's association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo M G, Oettgen H, De Vries J E, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 52.Silvennoinen-Kassinen S, Ikaheimo I, Tiilikainen A. TAP1 and TAP2 genes in nickel allergy. Int Arch Allergy Immunol. 1997;114:94–96. doi: 10.1159/000237650. [DOI] [PubMed] [Google Scholar]
- 53.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt's lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukocyte Biol. 1994;56:571–575. doi: 10.1002/jlb.56.5.571. [DOI] [PubMed] [Google Scholar]
- 55.Teisserenc H, Schmitt W, Blake N, Dunbar R, Gadola S, Gross W L, Exley A, Cerundolo V. A case of primary immunodeficiency due to a defect of the major histocompatibility gene complex class I processing and presentation pathway. Immunol Lett. 1997;57:183–187. doi: 10.1016/s0165-2478(97)00072-2. [DOI] [PubMed] [Google Scholar]
- 56.Tierney R, Steven N, Young L, Rickinson A. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7378. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Townsend A, Trowsdale J. The transporters associated with antigen presentation. Semin Cell Biol. 1993;4:53–61. doi: 10.1006/scel.1993.1007. [DOI] [PubMed] [Google Scholar]
- 58.van Endert P M, Tampe R, Meyer T H, Tisch R, Bach J F, McDevitt H O. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity. 1994;1:491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Leibowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 60.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 62.White L C, Wright K L, Felix N J, Ruffner H, Reis L F, Pine R, Ting J P-Y. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity. 1996;5:365–367. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 63.York I A, Goldberg A L, Mo X Y, Rock K L. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol Rev. 1999;172:49–66. doi: 10.1111/j.1600-065x.1999.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 64.Yoshizaki T, Sato H, Furukawa M, Pagano J S. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc Natl Acad Sci USA. 1998;95:3621–3626. doi: 10.1073/pnas.95.7.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Pagano J S. Interferon regulatory factor 2 represses the Epstein-Barr virus BamHI Q latency promoter in type III latency. Mol Cell Biol. 1999;19:3216–3223. doi: 10.1128/mcb.19.4.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L, Pagano J S. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J Virol. 2000;74:5748–5757. doi: 10.1128/jvi.74.3.1061-1068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]