Abstract
The vpx gene products of human immunodeficiency virus type 2 (HIV-2) and of the closely related simian immunodeficiency viruses from sooty mangabeys (SIVsm) and macaques (SIVmac) comprise a 112-amino-acid virion-associated protein that is critical for efficient virus replication in nondividing cells such as macrophages. When expressed in the absence of other viral proteins, Vpx localizes to the nuclear membrane as well as to the nucleus; however, in the context of virus replication Vpx is packaged into virions via interaction with the p6 domain of the Gag precursor polyprotein (p55gag). To identify the domains essential for virion incorporation and nuclear localization, site-directed mutations were introduced into the vpx gene of SIVsmPBj1.9 and functionally analyzed. Our results show that (i) mutation of two highly conserved L74 and I75 residues impaired both virion incorporation and nuclear localization of Vpx; (ii) substitution of conserved H82, G86, C87, P103, and P106 residues impaired Vpx nuclear localization but not virion incorporation; (iii) mutations of conserved Y66, Y69, and Y71 residues impaired virion incorporation but not the translocation of Vpx to the nucleus; and (iv) a mutation at E30 (predicted to disrupt an N-terminal α-helix) had no effect on either virion incorporation or nuclear localization of Vpx. Importantly, mutations in Vpx which impaired nuclear localization also reduced virus replication in macaque macrophages, suggesting an important role of the carboxyl terminus of Vpx in nuclear translocation of the viral preintegration complex. Analyzing this domain in greater detail, we identified a 26-amino-acid (aa 60 to 85) fragment that was sufficient to mediate the transport of a heterologous protein (green fluorescent protein [GFP]) to the nucleus. Taken together, these results indicate that virion incorporation and nuclear localization are encoded by two partially overlapping domains in the C-terminus of Vpx (aa 60 to 112). The identification of a novel 26-amino-acid nuclear targeting domain provides a new tool to investigate the nuclear import of the HIV-2/SIV preintegration complex.
One of the features that distinguishes lentiviruses from oncoretroviruses is their genetic complexity. Human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively), and the various simian immunodeficiency viruses (SIVs) which naturally infect more than 20 nonhuman primate species (26) encode several accessory and/or regulatory genes in addition to the structural gag, pol, and env genes that are present in all retroviruses (7, 69). An important step in the early stages of the retrovirus life cycle is the nuclear import of the viral preintegration complex (PIC), a prerequisite for integration of viral DNA into the host genome (4, 11, 17). While nuclear import and integration of oncoretroviral DNA requires breakdown of the nuclear membrane during mitosis, lentiviruses are able to infect nondividing host cells by exploiting cellular nuclear import pathways (37). In the case of HIV-1, the p17 Gag matrix (4, 19), the integrase (17), and Vpr (28, 59, 72) have been implicated as mediators of PIC nuclear translocation, although there is controversy concerning the role of the matrix in this process (14). The matrix and integrase contain classical nuclear localization signals (NLSs) and bind to importin α and importin β for transport to and across the nuclear envelope (4, 18, 59). By contrast, Vpr is believed to contribute to nuclear targeting of the viral PIC by exploiting nonclassical pathways (31). Two discrete Vpr nuclear localization domains have been reported that seem to interact with both proximal and distal components of the nuclear import pathway (31). Vpr has also been shown to bind directly to nucleoporin proteins and to colocalize with importin β in the nuclear membrane, suggesting that it is involved in the docking of the viral PIC to the nuclear pore complex (NPC) (13).
HIV-1 Vpr localizes to the nucleus (9, 31, 40) and the nuclear membrane (13, 41, 72) when expressed in the absence of other viral proteins. Mutational analyses have indicated that two α-helical domains, one each in the N and C termini, and a third arginine-rich domain at the C terminus are all critical for this function (9, 41, 42, 66, 75, 77, 78). Moreover, the α-helical domains are also essential for virion incorporation of Vpr (42, 75). Vpr causes arrest of eukaryotic cells at the G2 stage of the cell cycle (9, 40, 41, 61, 62). This property of Vpr has been mapped to amino acid positions 71 to 82 (9, 40, 41) and may serve to enhance viral gene expression since the HIV-1 long terminal repeat is more active during the G2 phase of the cell cycle (22).
Viruses in the HIV-2/SIVsm/SIVmac lineage contain a vpr gene as well as an evolutionarily related vpx gene. A recent report from our group demonstrated that SIVsm Vpr and Vpx proteins have distinct and noncomplementary functions (12). Vpr induces cell cycle arrest at the G2 stage (12, 65), whereas Vpx is involved in the nuclear import of the viral PIC (12, 54). Vpx is an 18-kDa, 112-amino-acid protein which is highly conserved among divergent isolates of HIV-2 and SIVsm (29, 33, 76). Vpx mutant SIVsm is significantly reduced in its ability to replicate in macaque macrophages (12). Vpx is also essential for efficient in vivo dissemination and spread of SIVsm following mucosal and intravenous infection of macaques (30). Vpx is packaged into virus particles (29, 33, 74, 76) and present in PICs (12). Within viral particles, Vpx seems to be localized within the viral core (35). Virion incorporation is mediated by the p6 domain of the Gag precursor polyprotein, p55gag (1, 53). Vpx has also been shown to bind to single-stranded nucleic acids, although the biological relevance of this finding is presently unclear (29).
Despite its requirement for HIV-2/SIVsm replication in nondividing cells in vitro and in vivo, systematic structure-function analyses of the Vpx protein have not been reported. Two previous studies showed that deletion of amino acid residues 78 to 80, 82 to 87, and 73 to 89 abolished the incorporation of Vpx into virus particles (53, 55). However, it remains unclear whether this was due to altered stability and/or structural conformation of the mutant Vpx proteins. Also, the mechanism by which Vpx mediates the nuclear import of HIV-2/SIVsm PICs remains unknown. Immunofluorescence studies have shown that Vpx localizes to the plasma membrane in cells infected with HIV-2/SIVsm (34) but that, when it is expressed in the absence of other viral proteins, it localizes to the nuclear membrane as well as inside the nucleus (72). More recently, Pancio and coworkers reported that deletion of the proline-rich C terminus of the HIV-2/ROD Vpx protein (carboxyl-terminal 11 amino acids) abrogated its nuclear localization function and attenuated HIV-2 replication in macrophages (54).
Determining how Vpx enters the nucleus is important to ultimately understand its function within in the context of the viral PIC. Proteins that exceed the 40-kDa diffusion limit of the NPC must be actively imported via specific pathways following the recognition of cis-acting targeting sequences, termed NLSs (23, 24, 50). A number of distinct pathways of protein nuclear import have been described. The most extensively characterized is the classical pathway which utilizes monopartite (short stretch of basic residues)- or bipartite (two stretches of basic residues connected by a short linker)-type NLSs (10). These NLSs first interact in the cytoplasm with the import receptors, importin α and importin β, and then dock at saturable sites on the cytoplasmic face of the nuclear pore via importin β (6, 10, 24, 25, 43, 44, 48, 50, 51). The importins and their cargo then translocate through the pore for delivery to the nuclear interior. However, a number of NLSs which do not conform to these consensus sequences have also been described (31, 36, 47, 50). For example, transportin (an importin β-like receptor) recognizes a 38-amino-acid glycine-rich sequence termed M9 and transports a subset of heterogeneous nuclear ribonucleoproteins (hnRNPs) (2, 15, 58, 60). Importin β also directly recognizes arginine-rich NLSs and transports the HIV regulatory proteins Tat and Rev to the nucleus without an importin α intermediate (52, 70). Vpx does not contain sequence elements homologous to any of the previously characterized NLS domains, and it is thus possible that it contains a novel NLS. Alternatively, Vpx may gain access to the nucleus by interacting with another NLS-containing protein.
In this study, we report a systematic mutational analysis of the SIVsm(PBj1.9) vpx gene and present functional data for wild-type and mutant Vpx proteins as well as Vpx-encoding proviral constructs. We confirm that Vpx localizes to the nuclear membrane and the nucleus in mammalian cells when expressed alone or in the context of a green fluorescent protein (GFP) fusion protein. We also describe point mutants of Vpx that segregate virion incorporation and nuclear localization functions. Finally, we describe a highly conserved protein domain (amino acids [aa] 60 to 85) within the carboxyl terminus of Vpx that is sufficient to mediate the transport of a heterologous protein (GFP) to the nucleus.
MATERIALS AND METHODS
Construction of SIVsm(PBj1.9) Vpx mutant proviruses and expression vectors.
Mutational analyses were performed using the infectious molecular clone SIVsm(PBj1.9) (8). The quick-change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to introduce mutations into the PBj1.9 vpx gene (subcloned as an internal SpeI-ClaI DNA fragment). The primers used for the generation of different PBj1.9 vpx mutants were as follows: for VpxE30P, 3′-GACAGAACAGTACCAGAAATAAACAGG-5′ (forward) and 3′-CCTGTTTATTTCTGGTACTGTTCTGTC-5′ (reverse); for VpxY66,69,71A, 3′-GGGATG TCAGTCAGCGCCACTAAAGCCAGAGCCTTGTGCTTGATACAG-5′ (forward) and 3′-CTGTATCAAGCACAAGGCTCTGGCTTTAGTGGCGCTGACTGACATCCC-5′ (reverse); for VpxLI74,75S, 3′-CACTAAATACAGATACTTGTGCTCCTCACAGAAAGCTATGTTTATGC-5′ (forward) and 3′-GC ATAAACATAGCTTTCTGTGAGGAGCACAAGTATCTGTATTTAGTG-5′ (reverse); for VpxH82S, 3′-CAGAAAGCTATGTTTATGTCTTGCAAGAAAGGGTGTAGG-5′ (forward) and 3′-CCTACACCCTTTCTTGCAAGACATAAACATAGCTTTCTG-5′ (reverse); for VpxGC86,87S, 3′-GTTTATGCATTGCAAGAAATCGTCAAGGTGCTTAGGAGGAGAGC-5′ (forward) and 3′- GCTCTCCTCCTAAGCACCTTGACGATTTCTTGCAATGCATAAAC-5′ (reverse); and for Vpx P103,106S, 3′-GCATGGGGCAGGGGGATGGAGACCAGGGTCTCCTCCTTCTCCCCCTCCAGGACTAGC-5′ (forward) and 3′-G CTAGTCCTGGAGGGGGAGAAGGAGGAGACCCTGGTCTCCATCCCC CTGCCCCATGC-5′ (reverse). Mutagenized vpx genes were reinserted into the PBj1.9 proviral vector using a series of subcloning steps. None of the introduced nucleotide substitutions resulted in amino acid changes in overlapping Vif open reading frames. Wild-type and different mutant Vpx expression constructs were generated by inserting PCR-amplified vpx gene fragments (Vpx-BamH [forward; 3′-AATCTCGGATCCGCCGCCACCATGTCAGATCCCAGGGAGAGAAT C-5′] and Vpx-Xho [reverse; 3′-TAGAATCTCGAGTTATGCTAGTCCTGGAGGGGGAGG-5′]) into the mammalian expression vector pCDNA3 (Invitrogen, Carlsbad, Calif.). All introduced mutations were confirmed by DNA sequence analysis.
Construction of Vpx-GFP fusion proteins.
To generate Vpx-GFP fusion constructs, the entire Vpx coding region and three Vpx fragments (Vpx1-63, Vpx64-112, and Vpx60-85) were fused to the carboxyl terminus of GFP and then cloned into the mammalian expression vector pCDNA3. GFP-Vpx fusion constructs were generated by overlap PCR using the following primers: for GFP 3′-CATCTAAAGCTTACCGCCGCCACCATGGTGAGCAAGGGCGAGGAG-5′ (forward) and 3′-CATCTACTCGAGTTACTTGTACAGCTCGTCCAT-5′ (reverse); for GFP–Vpx 3′-ATGGACGAGCTGTACAAGTCCGGACTCAGAATGTCAGATCCCAGGGAGAGAAT-5′ (forward) and 3′-GAGTCCGGACTTGTACAGCTCGTCCATGCC-5′ (reverse); for GFP–Vpx64-112, 3′-ATGGACG AGCTGTACAAGTCCGGACTCAGAGTCAGCTACACTAAATACAGA-5′ (forward); and for GFP–Vpx60-85 3′-TAGAATCTCGAGTTATTTCTTGCAA TGCATAAACATAGCTTTCTGTATCAAGCACAAGTATCTGTATTTAGT GTAGCTGACTGACATCCCCATGGAGCCGCCCTTGTACAGCTCGTCC ATGCCGA-5′ (reverse). All fusion constructs were confirmed by DNA sequence analysis.
Cell culture.
CEMx174 cells were maintained in RPMI 1640 supplemented with 2 mM l-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum (FBS). 293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% FBS. Macaque peripheral blood mononuclear cells (PBMCs) were obtained from heparin-treated whole blood using lymphocyte separation medium (Organon Teknika, Durham, N.C.), stimulated with phytohemagglutinin (4 μg/ml) for 2 to 3 days, and maintained in medium supplemented with interleukin 2 (10 U/ml). Macrophages were purified from unstimulated macaque PBMCs by adherence to plastic as described previously (12). Briefly, 3 × 106 macaque PBMCs were placed in 12-well tissue culture plates in macrophage medium containing 10% autologous macaque serum and conditioned medium to supply growth factors (12). Nonadherent cells were removed after 30 to 60 min of incubation at 37°C, followed by extensive washing with phosphate-buffered saline (PBS). Cells were allowed to differentiate in macrophage medium for 10 to 12 days prior to virus infection.
Transfection and infection.
For the generation of virus stocks, 293T cells were transfected with wild-type and vpx mutant SIVsm(PBj1.9) proviral DNAs (10 μg) using an MBS mammalian transfection kit (Stratagene, La Jolla, Calif.). Forty-eight hours after transfection, cell culture supernatants were collected, clarified by low-speed centrifugation (1,000 × g, 10 min), and analyzed for core antigen (p27gag) content using an SIV core antigen assay (Coulter, Miami, Fla.). CEMx174 and macaque PBMCs were then infected with supernatants containing 10 ng of p27gag, incubated overnight at 37°C in 5% CO2; this incubation was followed by extensive washing to remove residual virus. Infected cells were resuspended in 10 ml of complete T-cell medium (supplemented with 30 U of interleukin-2 per ml). Culture supernatants were split 1:2 every 3 days with fresh medium and aliquots of culture supernatants were frozen at −70°C for p27gag determinations at the conclusion of the experiments. Terminally differentiated macaque macrophages were infected with virion preparations containing 10 ng of p27gag in 12-well plates overnight at 37°C in 5% CO2 and then washed extensively to remove residual virus. Infected cells were adjusted to 2 ml with macrophage medium and incubated at 37°C. Culture supernatant was collected at 3-day intervals and frozen at −70°C for p27gag determinations. For virus rescue experiments, medium was removed at day 21 postinfection and macrophages were cocultivated with 1 × 106 CEMx174 cells in the appropriate medium for 24 h. Nonadherent cells were transferred to T25 tissue culture flasks and incubated for an additional 21 days. Culture supernatants were collected at 3-day intervals for p27gag determinations.
Western blot analysis.
Culture supernatants from transfected cells were clarified of cellular debris by low-speed centrifugation (1,000 × g, 10 min). Virions were then concentrated by ultracentrifugation (125,000 × g, 2 h) through a 20% sucrose cushion, and viral pellets were solubilized in loading buffer (62.5 mM Tris-HCl [pH 6.8], 0.2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol). Viral samples were denatured by boiling and separated on a 15% polyacrylamide gel containing SDS. Following electrophoresis, proteins were transferred to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia, Piscataway, N.J.) by electroblotting, incubated overnight at 4°C in blocking buffer (5% nonfat dry milk in PBS) and then for 2 h at room temperature with the appropriate antibodies, diluted in blocking buffer. Anti-Vpx and anti-Gag monoclonal antibodies were used at dilutions of 1:500 and 1:1,000, respectively (the properties of these antibodies are described in reference 74)). Protein-bound antibodies were probed with horseradish peroxidase-conjugated specific secondary antibodies (at a 1:1,000 dilution), washed extensively, and developed using the enhanced chemiluminescence detection system (Amersham Pharmacia).
Fluorescence microscopy.
HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% FBS, seeded onto Falcon Chamber culture slides (Becton Dickinson, San Diego, Calif.), and transfected with Vpx expression plasmids using Superfect (Qiagen, Santa Clarita, Calif.) according to manufacturer's instructions. Twenty-four to 48 h after transfection, the cells were washed with PBS, fixed with 4% paraformaldehyde–PBS at room temperature for 10 min, and probed with the monoclonal anti-Vpx (74) antibody (1:200) for 90 min at 37°C. Fluorescein isothiocyanate (FITC)-conjugated affinity-purified goat anti-mouse immunoglobulin G (Sigma, St. Louis, Mo.) was used as a secondary antibody to visualize the subcellular localization of Vpx. Texas red-phalloidin was used to stain cytoplasmic actin, and 4,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei (Sigma). To determine the subcellular localization of the GFP-Vpx fusion proteins, cells were fixed with 4% paraformaldehyde for 10 min after transfection and then stained with a rabbit polyclonal antibody specific for the nucleoporin protein p62 (Becton-Dickinson Transduction Laboratories, San Diego, Calif.). Bound antibodies were detected with a Texas red-conjugated goat anti-rabbit secondary antibody, and DAPI was used for nuclear staining. Samples were viewed with an upright Nikon E800 microscope and photographed with a photometrics charge-coupled device camera using Metamorph software.
RESULTS
Construction of mutant vpx genes.
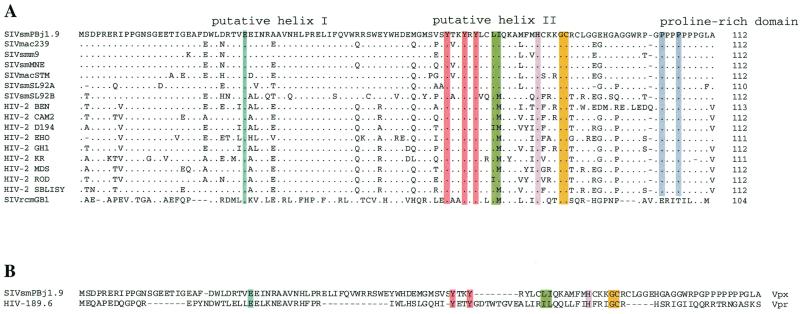
For functional analyses of Vpx, we introduced point mutations into the vpx gene of the infectious molecular clone SIVsm(PBj1.9) as a representative of the HIV-2/SIVsm group of viruses (8). This clone has a complete set of uninterrupted accessory genes, replicates well in macaque macrophages, and represents a primary (PBMC-derived) SIVsm isolate. Protein domains conserved among the vpx gene products of highly divergent HIV-2 and SIV isolates were targeted for mutagenesis. Such domains were identified by aligning the deduced Vpx amino acid sequences of available HIV-1 and SIVsm/SIVmac isolates with the Vpx protein sequence of SIVrcm (Fig. 1A), a highly divergent SIV identified only recently to naturally infect red-capped mangabeys (21, 26; F. Gao, E. Bailes, Y. Li, J. Decker, Y. Chen, F. Simon, E. Nerrienet, S. Souquiere, P. A. Marx, P. Sharp, and B. H. Hahn, Keystone Symposia: novel biological approaches to HIV-1 infeciton based on new insights into HIV biology, abstr. 311, 2000). We also targeted two putative α-helices located between amino acid residues 18 and 49 and 71 and 82, respectively, and introduced amino acid residues that would be predicted to disrupt these structures (Fig. 2). A glutamic acid residue at position 30 in the putative helical domain I (aa 18 to 49) was substituted with a proline residue (E30P) known to have a low potential for supporting α-helical structures (5, 67). Similarly, three conserved tyrosine residues in the putative helical domain II (aa 71 to 82) were replaced with nonpolar alanines (Y66,69,71A), and a leucine and an isoleucine residue were exchanged with serines (LI74,75S) to introduce structural as well as hydrophobic modifications. A conserved histidine residue at position 82 and glycine and cysteine residues at positions 86 and 87, respectively, were also exchanged with serines (H82S and GC86,87S). All but one of these residues are also known to be conserved in the Vpr protein of HIV-1 (Fig. 1B) and have been shown to be important for this protein's virion incorporation and nuclear localization functions (41, 56, 77). Finally, proline-rich motifs have been shown to mediate protein-protein interactions in a wide range of cellular proteins with unrelated functions and have been implicated in viral budding and assembly processes (20, 27). Although the Vpx protein sequence of SIVrcm lacks a proline-rich C terminus (confirmed by analyzing two independent SIVrcm isolates [data not shown]), we replaced two proline residues with serine residues (P103,106S) at the carboxyl terminus of the PBj1.9 Vpx (Fig. 2).
FIG. 1.
(A) Alignment of deduced Vpx protein sequences from divergent HIV-2 and SIV isolates. Sequences are compared to the SIVsm(PBj1.9) Vpx protein sequence. Dots indicate amino acid sequence identity. Dashes represent gaps inserted to improve the alignment. The HIV-2/SIV Vpx sequences shown were obtained from the Los Alamos HIV sequence database (http://hiv-web.lanl.gov/), except for the deduced Vpx protein sequence of SIVrcm(GB1), which is unpublished. (B) Alignment of Vpx [SIVsm(PBj1.9)] and Vpr [HIV-1(89.6)] protein sequences. Groups of amino acid residues chosen for mutagenesis are highlighted in color.
FIG. 2.
Construction of SIVsm(PBj1.9) vpx mutant proviruses. The genetic organization of the SIVsm(PBj1.9) genome is shown at the top, with Vpx substitution mutations indicated at the bottom. None of the amino acid substitutions altered the coding sequences of the overlapping Vif open reading frames.
Effects of mutations on expression and packaging of Vpx.
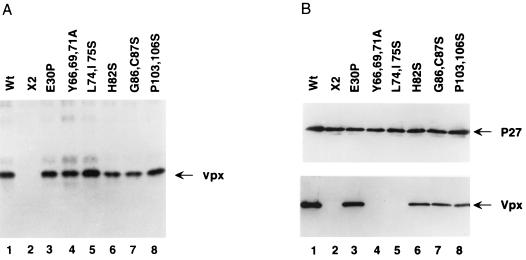
To study the effect of the site-directed Vpx mutations on the expression of this protein in mammalian cells, we employed a vaccinia virus T7-RNA polymerase system (16). vTF7-3-infected HeLa cells were transfected with wild-type or mutant Vpx plasmids, and expression was driven by the bacteriophage T7 promoter. Transfected cells were labeled with 35S, lysed, immunoprecipitated with an anti-Vpx antiserum, and analyzed by SDS-polyacrylamide gel electrophoresis. As expected, cells transfected with plasmids containing wild-type Vpx expressed an 18-kDa protein (Fig. 3A, lane 1). This was also the case for cells transfected with all of the mutant Vpx plasmids (Fig. 3A, lanes 3 to 8). X2, a mutant in which the initiating and internal methionine codons were replaced with threonine and leucine residues and which contained a stop codon at amino acid position 80 (12), was used as a negative control (Fig. 3A, lane 2).
FIG. 3.
Expression and virion incorporation of mutant Vpx proteins. (A) vTF7-3 infected HeLa cells were transfected with Vpx expression plasmids. Transfected cells were labeled with 35S and the cell-associated Vpx proteins were immunoprecipitated with a Vpx-specific monoclonal antiserum. Lane 1, wild-type (Wt); lane 2, X2 (a control construct lacking a functional vpx open reading frame) (12); lanes 3 to 8, mutant constructs, as indicated above lanes. (B) Western blot analysis of SIVsm(PBj1.9) viral particles containing mutant Vpx proteins. 293T cells were transfected with vpx mutant PBj1.9 proviral clones. Virus particles were concentrated from culture supernatants by ultracentrifugation through a 20% sucrose cushion, solubilized in gel loading buffer, and analyzed for protein content by Western blot analysis using Gag (upper panel)- and Vpx (lower panel)-specific antibodies. Lane 1, wild type (Wt); lane 2, negative control (see legend for panel A); lanes 3 to 8, mutant constructs, as indicated above lanes.
We next tested the various mutant Vpx proteins for their ability to package into virions. Mutant vpx genes were introduced back into the SIVsm(PBj1.9) provirus and then transfected into 293T cells. Cell culture supernatants were collected 48 h after transfection and centrifuged over a 20% sucrose cushion, and the viral pellets (normalized by p27gag content) were examined by Western blot analysis (Fig. 3B). Probing with an anti-Gag monoclonal antibody (74) showed that the expression, processing, and assembly of Gag was not affected by the various vpx mutations (Fig. 3B, upper portion). However, probing with a Vpx-specific antiserum revealed the absence of Vpx from two mutants (Y66,69,71A and LI74,75S) (Fig. 3B, lanes 4 and 5). Interestingly, the E30P mutation did not abrogate Vpx packaging, suggesting that the N-terminal α-helix is dispensable for virion incorporation. This was also true for mutations in the carboxyl-terminal region (aa 82 to 112). However, the highly conserved tyrosine residues at positions 66, 69, and 71, as well as the leucine and isoleucine residues at positions 74 and 75, were clearly identified to be critical for Vpx assembly into virus particles.
Carboxyl terminus of Vpx is required for productive macrophage infection.
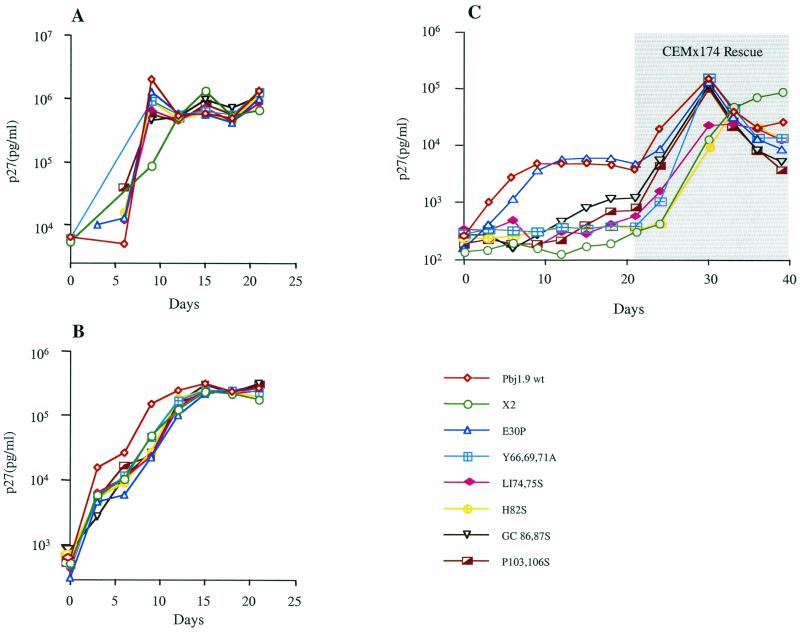
Vpx is essential for efficient HIV-2/SIVsm replication in nondividing cells (12, 54). To map the domains of Vpx involved in this process, we examined the ability of wild-type and mutant SIVsm(PBj1.9) constructs to elicit a spreading infection in monocyte-derived macaque macrophages. All vpx mutant viruses replicated efficiently and to high titers in CEMx174 cells and macaque PBMCs (Fig. 4A and B). However, this was not the case in terminally differentiated macaque macrophage cultures (Fig. 4C). As expected, PBj1.9 mutants that failed to package Vpx proteins were severely impaired in their ability to replicate in macrophages. However, failure to replicate in macrophages was also observed for mutants that were packaged into virus particles at near wild-type quantities: these included H82S, GC86,87S, and P103,106S. In three independent experiments, PBj1.9 mutants with substitution in the carboxyl-terminal half of Vpx replicated poorly in macrophages, while the E30P mutation designed to disrupt the predicted N-terminal α-helix resulted in a growth pattern virtually identical to that of wild-type virus (Fig. 4C). Interestingly, among the Vpx mutants that did package, H82S replicated most poorly in macrophages. The other two constructs, GC86,87S and P103,106S, exhibited some residual replication capacity compared to the X2 control (Fig. 4C). Consistent with previous findings (12), all Vpx mutants were rescued by CEMx174 cocultivation after 21 days (Fig. 4C), indicating persistent low-level infection in macrophage cultures even in the absence of a functional Vpx.
FIG. 4.
Replication kinetics of wild-type and vpx mutant PBj1.9 proviruses. CEMx174 cells (A), macaque PBMCs (B), and terminally differentiated macaque macrophages (C) were infected with the indicated SIVsm(PBj1.9) virus constructs equilibrated by p27gag content (10 ng of p27gag per 106 cells). The isolation and infection of primary macaque PBMCs and macrophages are described in Materials and Methods. Virus replication was assessed by quantifying the amounts of p27gag antigen in culture supernatants at 3-day intervals postinfection. Twenty-one days after infection, adherent macrophages were cocultured for 24 h with 1 × 106 CEMx174 cells. Nonadherent cells were removed and analyzed at 3-day intervals for p27gag antigen production. wt, wild type; X2, negative control.
Subcellular localization of Vpx.
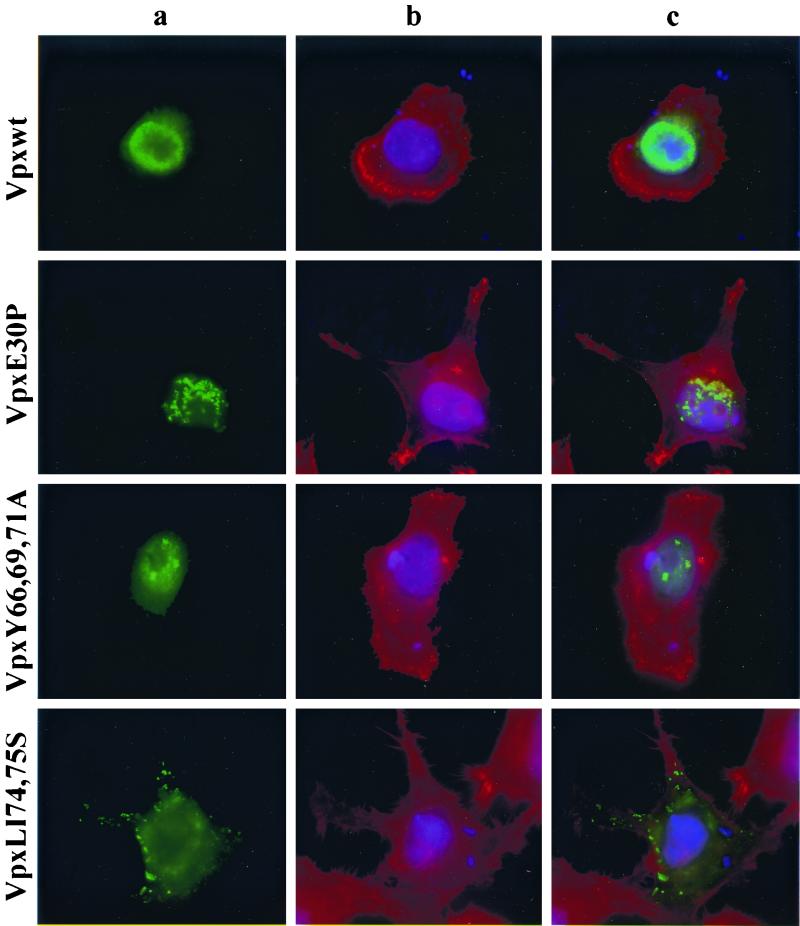
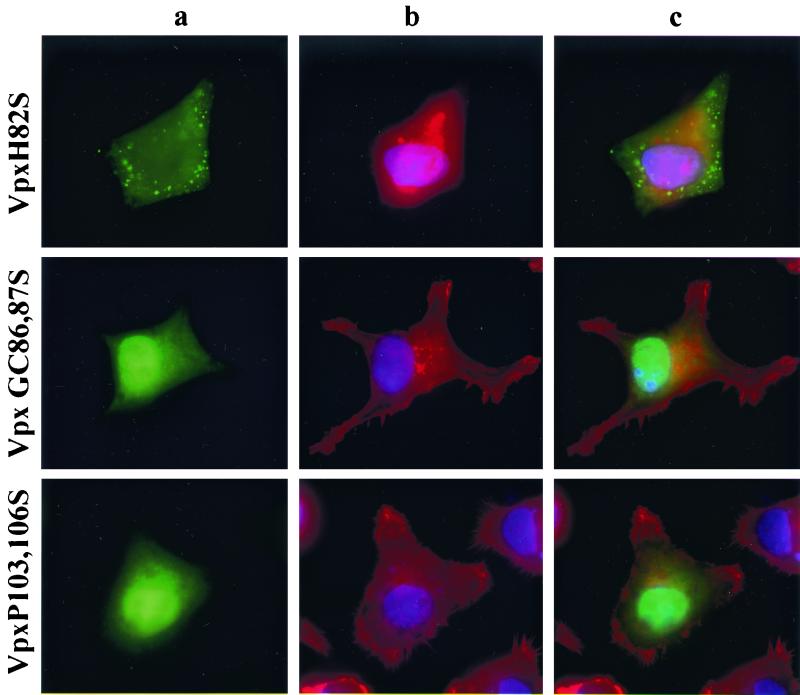
To characterize further the loss-of-function mutants, we determined the subcellular localization of all Vpx proteins using an indirect immunofluorescence assay. HeLa cells were transfected with the wild-type Vpx expression plasmid, fixed with paraformaldehyde, and then stained with an anti-Vpx antiserum (74). As reported previously (72), wild-type Vpx localized primarily to the inside of the nuclear membrane in transfected cells (Fig. 5, top row. The specificity of this staining was demonstrated by the absence of a signal in mock-transfected cells (data not shown). Vpx E30P and Y66,69,71A mutants exhibited a subcellular localization very similar to that of wild-type Vpx although their nuclear staining pattern was punctate (Fig. 5, rows 2 and 3). By contrast, LI74,75S and H82S mutants resulted in a complete loss of nuclear staining, with mutant Vpx proteins detected exclusively in the cytoplasm (Fig. 5, rows 4 and 5). Still another pattern was observed for GC86,87S and P103,106S, which accumulated both in the nucleus and in the cytoplasm (Fig. 5, rows 6 and 7). Although there was some nuclear staining, the diffuse pattern observed for these two Vpx mutants strongly suggested that nuclear import had been perturbed. Of note, there was considerable variation between individual cells in the intensity of fluorescence staining but no obvious correlation between the level of intensity and the patterns of subcellular localization. Taken together, these results along with the replication data in macrophages strongly suggested that a domain in the carboxyl terminus of Vpx, in particular the region spanning amino acids 74 to 82, plays a role in the import of this protein to the nucleus.
FIG. 5.
Subcellular localization of Vpx. vTF7-3 infected HeLa cells were transfected with wild-type and mutant Vpx expression plasmids. Twenty-four hours following transfection, the expressed Vpx proteins were detected by indirect immunofluorescence with an anti-Vpx monoclonal antibody (74) followed by an anti-mouse FITC-conjugated secondary antibody. (a) Localization of Vpx; (b) staining of cytoplasmic (red) and nuclear (blue) compartments with Texas red-phalloidin and DAPI, respectively; (c) superimposition of images shown in panels a and b for each row.
Signal-mediated nuclear import of Vpx.
Some proteins with molecular masses of less than 40 kDa can enter the nucleus by passive diffusion rather than by a signal-mediated process. To distinguish between these two possibilities, we evaluated the import activities of Vpx in the context of a chimeric protein designed to exceed the diffusion limit of the nuclear pore (23, 24, 50). Wild-type Vpx protein (18 kDa) was expressed as a fusion protein with the 28-kDa GFP and analyzed for nuclear localization in transfected HeLa cells (Fig. 6A). We selected GFP as a fusion partner because the fusion protein can be directly visualized in living cells without antibody staining. Further, GFP is known to localize to the nucleus when attached to a functional NLS (64). As shown in Fig. 6B (second row), most of the GFP-Vpx fusion protein accumulated on the inside of the nuclear membrane and the nuclear interior (as demonstrated by costaining of the nuclear membrane with an antibody specific for the nucleoporin protein p62), although some of the fusion protein was also visible on the cytoplasmic face of the nuclear membrane. By contrast, GFP alone was distributed throughout the cytoplasm and the nucleus (Fig. 6B, top row). Importantly, Western blot analysis of whole-cell lysates confirmed that the fusion protein had the appropriate predicted molecular mass (46 kDa) (Fig. 6C, lane 2). Thus, as reported previously (54, 72), Vpx appears to possess a specific signal that mediates its nuclear uptake.
FIG. 6.
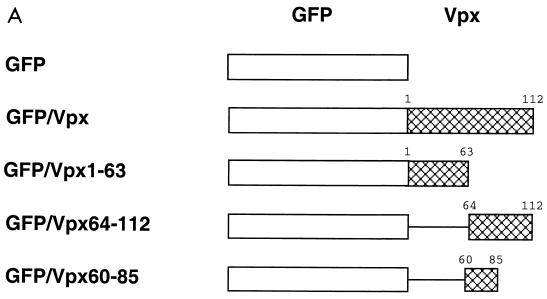
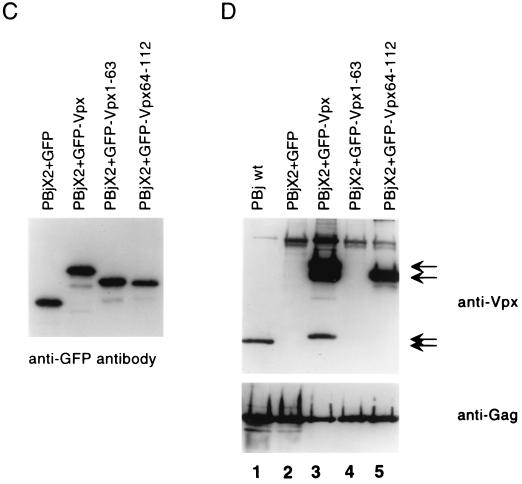
Construction, subcellular localization, expression and packaging of GFP-Vpx fusion proteins. (A) Schematic representation of GFP-Vpx fusion proteins. (B) Subcellular localization of GFP-Vpx fusion proteins in HeLa cells (see Results for details). Subpanels: a, the nuclear membrane was visualized by indirect immunofluorescence using a nucleoporin p62 specific polyclonal antibody, followed by a Texas red-conjugated goat anti-rabbit secondary antibody (red), and DAPI was used for nuclear staining (blue); b, the GFP signal was used to localize the GFP-Vpx fusion proteins; c, superimposition of images shown in subpanels a and b for each row. (C) Expression of GFP-Vpx fusion proteins. 293T cells were transfected with various GFP-Vpx expression plasmids. Lysates of transfected cells were prepared 48 h after transfection and analyzed for fusion protein content by Western blot analysis using an anti-GFP antibody. (D) Packaging of GFP-Vpx fusion proteins. Expression plasmids were cotransfected with the PBj1.9 Vpx− proviral clone X2, and virus particles were concentrated from culture supernatants by ultracentrifugation through a 20% sucrose cushion, solubilized in gel loading buffer, and analyzed for protein content by Western blot analysis using Vpx (top)- and Gag (bottom)-specific monoclonal antibodies. Arrows indicate proteins with Vpx reactivity. Lane 1, wild-type Vpx protein (18 kDa); lane 2, negative (GFP) control; lane 3, full-length GFP-Vpx fusion protein (46 kDa) as well as two smaller proteins (38 kDa and 20 kDa, respectively) likely representing protease cleavage products; lane 4, lack of GFP-Vpx1-63 packaging; lane 5, GFP-Vpx64-112 fusion protein (38 kDa).
Vpx amino acids 60 to 85 are sufficient to mediate transport of a heterologous protein (GFP) to the nucleus.
Using additional GFP fusion constructs, the role of the carboxyl-terminal half of Vpx in nuclear localization was further examined. First, 49 amino acids from the carboxyl terminus were deleted and the remaining 63 amino acids of Vpx were fused to GFP (Fig. 6A). This GFP–Vpx1-63 construct was then transfected into HeLa cells. As shown in Fig. 6B (third row), this fusion protein yielded a diffuse staining pattern very similar to that observed for GFP alone. By contrast, fusion of GFP to the C-terminal half of Vpx (GFP–Vpx64-112) resulted in strong nuclear staining with no reactivity detected in the cytoplasm (Fig. 6B, fourth row). As shown by costaining with the nucleoporin p62 antibody, most of the GFP–Vpx64-112 fusion protein was concentrated on the inside of the nuclear membrane, similar to full-length Vpx. However, some staining was also seen within the nucleus, suggesting that the Vpx64-112 fragment facilitated the transport of the GFP protein to and through the nuclear membrane. The same observations were made when GFP was linked to an even shorter fragment that spanned the evolutionarily most highly conserved protein domain (aa 60 to 85) of Vpx. Again, GFP–Vpx60-85 localized both to the inside of the nuclear membrane and to the nuclear interior (Fig. 6B, bottom row). These results indicate that Vpx amino acids 60 to 85 are sufficient to mediate the nuclear import of a heterologous protein.
Finally, to determine the minimal virion packaging domain of Vpx, we cotransfected the GFP-Vpx, GFP–Vpx1-63, GFP–Vpx64-112, and GFP–Vpx60-85 expression plasmids with a PBj1.9 Vpx− proviral clone and examined the resulting virions by Western blot analysis. The PBj1.9 wild-type construct was also transfected alone as a control. Anti-Vpx antiserum detected the 18-kDa (native) Vpx protein in virions derived from the SIVsm(PBj1.9) wild-type construct. Strongly Vpx-reactive proteins, 46 and 38 kDa in size, were also detected in virions derived by coexpression of PBj1.9 Vpx− with GFP–Vpx and GFP–Vpx64-112 constructs, respectively (Fig. 6D, lanes 3 and 5). However, fusion proteins GFP–Vpx1-63 (Fig. 6D, lane 4) and GFP–Vpx60-85 (data not shown) were not incorporated into virus particles. These results suggest that the N-terminal half of Vpx is not involved in virion incorporation. Moreover, the Vpx60-85 fragment was not sufficient to package a heterologous protein into virus particles, although it contained all residues shown to be critical for virion incorporation by site-directed mutagenesis (Fig. 3).
DISCUSSION
Vpx has two distinct localization properties that direct it either to the nuclear membrane and the interior of the nucleus (72) or, in association with Gag, to budding virus particles at the plasma membrane (1, 29, 33, 53, 74, 76). These two opposite Vpx localizations must be mediated through different protein-protein interactions. In order to identify the amino acid residues and/or motifs involved in these processes, we introduced a number of point mutations into Vpx domains that we found to be highly conserved among divergent isolates of HIV-2, SIVsm/SIVmac, and SIVrcm, as well as between the Vpx proteins of these viruses and the Vpr protein of HIV-1 (Fig. 1). Our data indicate that the carboxyl-terminal half of Vpx encompasses determinants for both virion incorporation and nuclear localization (Fig. 7), as supported by the following: (i) mutation of two highly conserved residues, L74 and I75, impaired both virion incorporation and nuclear localization of Vpx; (ii) substitution of conserved H82, G86, C87, P103, and P106 residues impaired Vpx nuclear localization as well as replication of mutant virus in macaque macrophages; (iii) mutations of conserved Y66, Y69, and Y71 residues impaired packaging of Vpx into virus particles; (iv) a mutation at E30 (predicted to disrupt an N-terminal α-helix) had no effect on either virion incorporation or nuclear localization of Vpx; and (v) a Vpx fragment of 26 amino acids (aa 60 to 85) was found to be sufficient to mediate the transport of a heterologous protein (GFP) to the nucleus.
FIG. 7.
Mutational analysis of the C-terminal half of Vpx. The asterisks indicate amino acid residues that are absolutely essential for the functions indicated; the brackets denote residues that (when mutated) cause some functional impairment. The horizontal bar highlights the fragment of Vpx that is sufficient to mediate the transport of a heterologous protein (GFP) to the nucleus.
Vpx mutants Y66,69,71A and LI74,75S resulted in a total abrogation of Vpx packaging, suggesting that the integrity of the putative helical domain II (aa 71 to 82) is important for Gag interaction. Interestingly, there are two α-helical domains in HIV-1 Vpr, both of which are important for virion incorporation (38, 39, 41, 63, 73, 75). However, this seems not to be the case for Vpx, as the N-terminal half of the protein is clearly dispensable for packaging. The E30P mutant which was designed to cause disruption of helix I packaged at wild-type levels, and the N-terminal half of Vpx failed to mediate virion incorporation of a GFP fusion protein. By contrast, the GFP–Vpx64-112 fusion protein did package, indicating that the C-terminal 49 amino acids are sufficient to target a heterologous protein to the virus particle. Thus, the packaging domain must reside in the C-terminal half of Vpx, although its precise boundaries remain to be determined.
A recent study reported that deletion of amino acids 78 to 80 and 82 to 87 abolished the incorporation of Vpx into virus particles (55). Analyzing substitution mutations between amino acid position 82 and 87, we observed wild-type packaging (Fig. 3B). Hence, the reported impairment of virion incorporation of the latter mutant may have been due to altered protein stability and/or an impaired protein conformation. Deletion mutant Vpx78-80 likely disrupted the putative helical domain II which we found to be critical for packaging. Interestingly, the Vpx60-85 fragment, which spans this region, was not sufficient to mediate the packaging of a heterologous protein. Thus, the minimal packaging domain of Vpx must extend beyond the putative helix II, although it probably does not require the entire C terminus, since deletion of the proline-rich domain does not abrogate Vpx packaging (54). Given their absolute requirement for Vpx packaging, Y66,69,71 as well as L74 and I75 residues in helical domain II may be directly involved in Vpx-Gag interactions. Tyrosine residues are frequently involved in phosphorylation and protein-protein interactions in signal transduction pathways (3, 32, 57). It will be interesting to determine whether phosphorylation of the conserved tyrosine motif (YXXYXY) in Vpx is involved in the association of Vpx to Gag.
We also assessed the replication potential of vpx mutant SIVsm in dividing and nondividing cells. Viruses that contained mutations at positions Y66,69,71, LI74,75, H82, GC86,87, and P103,106 either failed to replicate or grew only very poorly in macaque macrophages (Fig. 4). Vpx Y66,69,71A and LI74,75S mutants are readily explained since these fail to package into virus particles. However, the other mutant proteins were present in virions at near-wild-type levels and thus likely affected the nuclear import properties of the viral PICs, either directly or indirectly. For example, SIVsm virions encoding the Vpx H82S mutation replicated in macaque macrophages as poorly as virions that contained no Vpx at all (Fig. 4). While these results do not formally prove that the H82S mutation rendered the corresponding SIVsm PIC nuclear import defective, the complete loss of nuclear targeting of the H82S mutant protein is consistent with this explanation (Fig. 5, row 5). Of course, other PIC components such as the viral integrase are likely to also play a role in PIC nuclear transport. Thus, future studies will need to examine to what extent nuclear import properties of Vpx govern nuclear import properties of the viral PIC. The Vpx mutant constructs described here should be useful in guiding such studies.
We also identified a 26-amino-acid domain (aa 60 to 85) in the C terminus of Vpx which appears to encode a novel nuclear targeting signal (Fig. 7). This stretch of amino acids is conserved among all Vpx proteins identified to date, including the highly divergent SIVrcm Vpx protein (Fig. 1). Recently, Pancio and coworkers reported that deletion of the C-terminal proline-rich domain (aa 102 to 112) of Vpx resulted in a block of nuclear localization of HIV-2 DNA, thus implicating this particular domain in the nuclear transport of the HIV-2 PIC (54). Our 26-amino-acid nuclear targeting domain lies upstream of this proline-rich region, raising the possibility that Vpx, like HIV-1 Vpr, contains two independent nuclear targeting domains. Alternatively, deletion of the proline-rich domain may have impaired Vpx function indirectly by changing protein structure or conformation. To distinguish between these two possibilities, it will be important to test whether Vpx86-112, like Vpx60-85, can mediate the nuclear transport of a heterologous fusion partner. Nevertheless, we also observed that point mutations in the proline rich domain (Vpx P103,106S) reduced viral replication in macrophages (Fig. 4) and impaired nuclear localization of the corresponding protein (Fig. 5, bottom row). This finding is consistent with an involvement of the proline-rich Vpx C terminus in PIC nuclear import. However, the observed impairment was much less severe than that caused by mutations in the upstream nuclear localization domain, suggesting that the proline-rich domain does not constitute the major nuclear import signal. The lack of a proline-rich domain in the SIVrcm Vpx protein also supports this notion.
The 26-amino-acid domain of Vpx that mediated nuclear targeting of GFP is different not only from classical NLSs (lysine rich) (10) but also from recently identified M9 and hnRNP K protein nuclear shuttling (45, 46, 49) sequences as well as arginine-rich NLS domains (52, 68, 70, 71). Accumulation in the nuclear membrane is not an attribute ordinarily associated with NLSs, although the import factor importin β localizes to the nuclear membrane (23). It is thus possible that Vpx plays a primary role in the docking process of the viral PIC to the nuclear membrane rather than in more distal nuclear import events. Further studies of Vpx and other PIC components will be important to define the mechanism(s) by which HIV-2/SIV preintegration complexes move from one side of the nuclear pore to the other. Since there is presently little consensus concerning the mechanism of nuclear import of lentiviral PICs, the evolutionarily highly conserved 26-amino-acid Vpx nuclear targeting signal identified here is likely to provide a new tool to study the mechanisms that govern early steps in primate lentiviral replication.
ACKNOWLEDGMENTS
We thank Wendy Abbott and Jennifer Wilson for artwork and secretarial assistance.
This work was supported by grants from the National Institutes of Health (POI AI41215 and ROI AI 34748 to B.H.H.) and the Howard Hughes Medical Institute.
REFERENCES
- 1.Accola M A, Bukovsky A A, Jones M S, Göttlinger H G. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnahan P A, Yonemoto W, Greene W C. Cutting edge: SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for down-regulation of CD4. J Immunol. 1999;163:2977–2981. [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–48. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 6.Corbett A H, Silver P A. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 8.Dewhurst S, Embretson J E, Anderson D C, Mullins J I, Fultz P N. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature. 1990;345:636–640. doi: 10.1038/345636a0. [DOI] [PubMed] [Google Scholar]
- 9.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher T M, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier R A, Meyer B E, Simon J H, Fischer U, Albright A V, Gonzalez-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouchier R A, Meyer B E, Simon J H, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 20.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 21.Georges-Courbot M C, Lu C Y, Makuwa M, Telfer P, Onanga R, Dubreuil G, Chen Z, Smith S M, Georges A, Gao F, Hahn B H, Marx P A. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J Virol. 1998;72:600–608. doi: 10.1128/jvi.72.1.600-608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 23.Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 24.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn B H, Shaw G M, De Cock K M, Sharp P M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 27.Harty R N, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson L E, Sowder R C, Copeland T D, Benveniste R E, Oroszlan S. Isolation and characterization of a novel protein (X-ORF product) from SIV and HIV-2. Science. 1988;241:199–201. doi: 10.1126/science.3388031. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch V M, Sharkey M E, Brown C R, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins W R, Hahn B H, Lifson J D, Stevenson M. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang H, Freund C, Duke-Cohan J S, Musacchio A, Wagner G, Rudd C E. SH3 domain recognition of a proline-independent tyrosine-based RkxxYxxY motif in immune cell adaptor SKAP55. EMBO J. 2000;19:2889–2899. doi: 10.1093/emboj/19.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kappes J C, Morrow C D, Lee S-W, Jameson B A, Kent S B H, Hood L E, Shaw G M, Hahn B H. Identification of a novel retroviral gene unique to human immunodeficiency virus type 2 and simian immunodeficiency virus SIVMAC. J Virol. 1989;62:3501–3505. doi: 10.1128/jvi.62.9.3501-3505.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kappes J C, Parkin J S, Conway J A, Kim J, Brouillette C G, Shaw G M, Hahn B H. Intracellular transport and virion incorporation of vpx requires interaction with other virus type-specific components. Virology. 1993;193:222–233. doi: 10.1006/viro.1993.1118. [DOI] [PubMed] [Google Scholar]
- 35.Kewalramani V N, Emerman M. Vpx association with mature core structure of HIV-2. Virology. 1996;218:159–168. doi: 10.1006/viro.1996.0176. [DOI] [PubMed] [Google Scholar]
- 36.LaCasse E C, LaFebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic-acid binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahalingam S, Khan S A, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Identification of residues in the N-terminal acidic domain of HIV-1 Vpr essential for virion incorporation. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 43.Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 44.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michael W M, Siomi H, Choi M, Pinol-Roma S, Nakielny S, Liu Q, Dreyfuss G. Signal sequences that target nuclear import and nuclear export of pre- mRNA-binding proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- 47.Michaud N, Goldfarb D. Microinjected U snRNAs are imported to oocyte nuclei via the nuclear pore complex by three distinguishable targeting pathways. J Cell Biol. 1992;116:851–861. doi: 10.1083/jcb.116.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 50.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 51.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 52.Palmeri D, Malim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pancio H A, Ratner L. Human immunodeficiency virus type 2 Vpx-Gag interaction. J Virol. 1998;72:5271–5275. doi: 10.1128/jvi.72.6.5271-5275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pancio H A, Vander Heyden N, Ratner L. The C-terminal proline-rich tail of human immunodeficiency virus type 2 vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J Virol. 2000;74:6162–6167. doi: 10.1128/jvi.74.13.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park I W, Sodroski J. Amino acid sequence requirements for the incorporation of the Vpx protein of simian immunodeficiency virus into virion particles. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:506–510. [PubMed] [Google Scholar]
- 56.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piguet V, Chen Y L, Mangasarian A, Foti M, Carpentier J L, Trono D. Mechanism of Nef-induced CD4 endocytosis: Nef connects CD4 with the mu chain of adaptor complexes. EMBO J. 1998;17:2472–2481. doi: 10.1093/emboj/17.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 59.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuler W, Wecker K, de Rocquigny H, Baudat Y, Sire J, Roques B P. NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions. J Mol Biol. 1999;285:2105–2117. doi: 10.1006/jmbi.1998.2381. [DOI] [PubMed] [Google Scholar]
- 64.Stauber R H, Pavlakis G N. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology. 1998;252:126–136. doi: 10.1006/viro.1998.9400. [DOI] [PubMed] [Google Scholar]
- 65.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subbramanian R A, Yao X J, Dilhuydy H, Rougeau N, Bergeron D, Robitaille Y, Cohen E A. Human immunodeficiency virus type 1 Vpr localization: nuclear transport of a viral protein modulated by a putative amphipathic helical structure and its relevance to biological activity. J Mol Biol. 1998;278:13–30. doi: 10.1006/jmbi.1998.1685. [DOI] [PubMed] [Google Scholar]
- 67.Tacke E, Schmitz J, Prufer D, Rohde W. Mutational analysis of the nucleic acid-binding 17 kDa phosphoprotein of potato leafroll luteovirus identifies an amphipathic alpha-helix as the domain for protein/protein interactions. Virology. 1993;197:274–282. doi: 10.1006/viro.1993.1588. [DOI] [PubMed] [Google Scholar]
- 68.Tiganis T, Flint A J, Adam S A, Tonks N K. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 69.Trono D. When accessories turn out to be essential. Nat Med. 1998;4:1368–1369. doi: 10.1038/3953. [DOI] [PubMed] [Google Scholar]
- 70.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wecker K, Roques B P. NMR structure of the (1-51) N-terminal domain of the HIV-1 regulatory protein Vpr. Eur J Biochem. 1999;266:359–369. doi: 10.1046/j.1432-1327.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- 74.Wu X, Conway J A, Kim J, Kappes J C. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol. 1994;68:6161–6169. doi: 10.1128/jvi.68.10.6161-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao X-J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu X F, Ito S, Essex M, Lee T H. A naturally immunogenic virion-associated protein specific for HIV-2 and SIV. Nature. 1988;335:262–265. doi: 10.1038/335262a0. [DOI] [PubMed] [Google Scholar]
- 77.Zhao L J, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Specific interaction with a cellular protein. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 78.Zhou Y, Lu Y, Ratner L. Arginine residues in the C-terminus of HIV-1 Vpr are important for nuclear localization and cell cycle arrest. Virology. 1998;242:414–424. doi: 10.1006/viro.1998.9028. [DOI] [PubMed] [Google Scholar]