Abstract
Tigecycline is a last-resort drug used to treat serious infections caused by multidrug-resistant bacteria. tet(X4) is a recently discovered plasmid-mediated tigecycline resistance gene that confers high-level resistance to tigecycline and other tetracyclines. Since the first discovery of tet(X4) in 2019, it has spread rapidly worldwide, and as a consequence, tigecycline has become increasingly ineffective in the clinical treatment of multidrug-resistant infections. In this study, we identified and analyzed tet(X4)-positive Escherichia coli isolates from duck farms in Hunan Province, China. In total, 976 samples were collected from nine duck farms. Antimicrobial susceptibility testing and whole-genome sequencing (WGS) were performed to establish the phenotypes and genotypes of tet(X4)-positive isolates. In addition, the genomic characteristics and transferability of tet(X4) were determined based on bioinformatics analysis and conjugation. We accordingly detected an E. coli strain harboring tet(X4) and seven other resistance genes in duck feces. Multi-locus sequence typing analysis revealed that this isolate belonged to a new clone, and subsequent genetic analysis indicated that tet(X4) was carried in a 4608-bp circular intermediate, flanked by ISVsa3-ORF2-abh elements. Moreover, it exhibited transferability to E. coli C600 with a frequency of 10-5. The detection of tet(X4)-harboring E, coli strains on duck farms enhances our understanding of tigecycline resistance dynamics. The transferable nature of the circular intermediate of tet(X4) contributing to the spread of tigecycline resistance genes poses a substantial threat to healthcare. Consequently, vigilant monitoring and proactive measures are necessary to prevent their spread.
Keywords: antimicrobial drug resistance, tigecycline, tet(X4), Escherichia coli, duck
1. Introduction
Antimicrobial resistance (AMR) has been recognized by the World Health Organization (WHO) as a major threat to global public health (O’Neill, 2016; Liu et al., 2024). Tigecycline is a glycylcycline antibiotic with an expanded spectrum of activity that was developed to address the global threat of emerging antibiotic resistance (Yahav et al., 2011). It is considered a last-resort treatment option for severe infections caused by multidrug-resistant bacteria (MDR) (Jenner et al., 2013), including Enterobacteriaceae strains resistant to carbapenem (CRE), Staphylococcus aureus resistant to methicillin (MRSA), and vancomycin-resistant Enterococcus (VRE) (Tasina et al., 2011; Sheu et al., 2019; Chen et al., 2020; Wu et al., 2022; Yaghoubi et al., 2022). However, the emergence of resistance genes from the tet(X) family [specifically tet(X4)] confers high resistance against tigecycline, posing significant challenges for successful treatment (Sun et al., 2019; Xu et al., 2022). Positive tet(X4) strains were first reported in porcine isolates of Escherichia coli in 2019. These strains can confer high-level resistance to all tetracycline antibiotics, including fourth-generation tetracycline drugs (eravacycline and omadacycline) recently approved by the United States Food and Drug Administration (FDA) (He et al., 2019). To date, tet(X4) genes have mainly been identified in ColE2-like, IncA/C2, IncX1, IncQ, IncFII, IncHI1, and other types of plasmids ranging in size from 9 to 330 kb with different replicons, which substantially enhances tet(X4) transferability (Ding et al., 2020; Aminov, 2021; Yu et al., 2021; Li et al., 2023). tet(X4) is present worldwide within a range of bacterial species that have been isolated from diverse ecological niches, including human and animal feces and the environment, among which, Acinetobacter spp., Enterobacteriaceae, and E. coli have been identified as the predominant reservoirs of tet(X4) (Zhang et al., 2022).
Although in recent years, the epidemiology and transmission of tet(X4) have been extensively reported, most of this research has been limited to pig farms and related production lines (Wang et al., 2022; Wu et al., 2022). Thus, conducting large-scale epidemiological and functional studies is essential to gain a better understanding of the prevalence and dissemination of tet(X4) in poultry. To this end, in the present study, we collected 976 samples from duck farms in Hunan Province, China, to investigate the epidemic and transmission characteristics of bacterial strains positive for tet(X4). Herein, we report the isolation of E. coli positive for tet(X4) from duck farms.
2. Methods
2.1. Collection of samples and bacterial isolation
In July 2022, a total of 976 non-duplicate samples were collected from nine duck farms in Hunan province, China, comprising 677 duck feces and 299 environmental-related samples. All samples were collected using sterile swabs and suspended in centrifuge tubes containing 1.5 mL of Luria–Bertani (LB) broth. Having immediately placed the samples in ice boxes, they were transported to the laboratory, wherein they cultured overnight at 37°C in a chromogenic medium suitable for urine cultures (Comagal, Shanghai, China) containing 4 µg/mL tigecycline. Colonies of different colors that subsequently developed were subjected to polymerase chain reaction (PCR) analysis to establish whether the selected isolates contained tet(X) resistance genes. Tet(X)-F/tet(X)-R primers (F: 5′-CCGTTGGACTGACTATGGC-3′, R: 5′-TCAACTTGCGTGTCGGTAA-3′) were used for Sanger sequencing (Sun et al., 2020). Subsequently, bacteria were identified at the species level based on 16S rRNA sequencing and comparison of sequencing results with the sequences of GenBank database reference strains (Gimenez-Miranda et al., 2023).
2.2. Antimicrobial susceptibility testing
The antimicrobial sensitivity of the tet(X4)-positive isolates to nine antibiotics (tigecycline, chloramphenicol, nalidixic acid, florfenicol, trimethoprim-sulfamethoxazole, cefotaxime, colistin, meropenem, and amikacin) was determined using the broth microdilution method according to the Clinical and Laboratory Standards Institute 2020 guidelines (CLSI M100-S30), with the exception of the break-point for tigecycline. The resistance break-point of tigecycline was interpreted as >2 mg/L according to the European Committee on Antimicrobial Susceptibility Testing guidelines (EUCAST, version 14.0) (https://www.eucast.org/clinical_breakpoints) (accessed on December 15th 2022). The quality control strain was E. coli ATCC 25922.
2.3. Whole-genome sequencing and bioinformatics analysis
The genomic DNA of tet(X4)-positive isolates was extracted using a TIANamp Bacteria DNA Kit (Tiangen Biotech, China), and DNA libraries of these isolates were constructed using Illumina HiSeq 2500 (Annoroad Genomics Co.) and nanopore sequence platforms. The draft genome sequences of E. coli positive for tet(X4) were assembled using Unicycler (https://github.com/rrwick/Unicycler) (accessed on January 10th, 2023). The assembled genome sequences were annotated using PATRIC3.6.9 (https://patricbrc.org/) (accessed on January 11th, 2023). Sequence types (ST), AMR genes, and plasmid replicon types were identified using the CGE server (https://cge.cbs.dtu.dk/services/) (accessed on January 11th, 2023). Easyfig v2.2.3 (http://mjsull.github.io/Easyfig) (accessed on January 12th, 2023) was used to generate linear comparison figures and visualize the comparative genetic characteristics.
2.4. Conjugation assay
A conjugation experiment was performed to verify the transferability of tet(X4) in a tet(X4)-positive isolate (Yang et al., 2023) using streptomycin-resistant E. coli C600 as a recipient and the tet(X4)-positive strain as the donor. The donor and recipient bacteria were mixed in LB broth in a 1:3 ratio, and the mixture was transferred to a fresh LB agar plate with 0.22-μm sterile filter paper, followed by incubation at 37°C for 16 h. The transconjugants were detected by culturing on Mueller–Hinton plates containing 1000 mg/L streptomycin and 2 mg/L tigecycline. Finally, the transconjugants were verified by performing PCR using ERIC primers (F: 5′-ATGTAAGCTCCTGGGGATTCAC-3′, R: 5′-AAGTAAGTGACTGGGGTGAGCG-3′) (Yang et al., 2023). The frequency of conjugation transfer was calculated as the ratio of the number of transconjugants to the total number of recipients.
3. Results and discussion
3.1. Bacterial isolates
A strain positive for tet(X4), E. coli e6cp, was isolated from a duck fecal sample, representing a total prevalence of tet(X4) positivity in fecal samples of 0.14% (1/677). Comparatively, a previous study that assessed the prevalence of E. coli positive for tet(X4) on intensive pig and chicken farms in Hunan Province (Yang et al., 2023) detected six positive strains at a positivity rate of 2.3% (6/257). tet(X4) has similarly been also detected on pig and chicken farms in Jiangsu (18.24%, 24/159) (Wang et al., 2023), Shandong (66.7%, 40/60) (He et al., 2019), and Shanghai (12.24%, 6/49) (Wang et al., 2022) provinces. In humans, the prevalence of tet(X4) has gradually increased from a detection rate of 10.1% (11/109) (Ding et al., 2020) to 18.8% (147/782) (Zeng et al., 2022).
The prevalence of tet(X4)-positive strains in this study is thus somewhat lower than that which has been reported in previous studies on chicken and pig farms in China. We suspect that this discrepancy in prevalence rates could be attributed to the adoption of standardized management practices throughout all the assessed duck farms, including hatching and finishing, along with enhancements in captive breeding environments. Furthermore, implementing antimicrobial management and adhering to veterinary guidelines may have contributed to reducing antimicrobial resistance (Coyne et al., 2019).
3.2. Drug resistance phenotype and genotype
The resistance phenotype of the isolate reported in this study was determined by performing antimicrobial susceptibility tests using the broth microdilution method, with the minimum inhibitory concentrations (MICs) determined and compared with the resistance break-point according to the CLSI 2020 guidelines. The tet(X4)-positive E. coli exhibited resistance to a range of antibiotics, namely, chloramphenicol (>512 mg/L), nalidixic (16 mg/L), florfenicol (256 mg/L), trimethoprim-sulfamethoxazole (>16 mg/L), and tigecycline (8 mg/L), but was sensitive to cefotaxime (<0.125 mg/L), colistin (0.125 mg/L), meropenem (0.03 mg/L), and amikacin (2 mg/L) ( Table 1 ).
Table 1.
Characterization of the tet(X4)-positive Escherichia coli.
| Strain | Species | Source | Sequence type | Antimicrobial agents (mg/L) 1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGC | CPL | NAL | FFC | STX | CTX | COL | MEM | AMK | ||||
| e6cp | E. coli | Duck feces | ST10776 | 8 | > 512 | > 16 | 256 | > 16 | < 0.125 | 0.125 | 0.03 | 2 |
1 Abbreviations and antibiotic resistance break-points according to the Clinical and Laboratory Standards Institute (CLSI) 2020 guidelines: TGC: tigecycline (R > 2 mg/L), CPL, chloramphenicol (R > 8 mg/L); NAL, nalidixic acid (R > 4 mg/L); FFC, florfenicol (R > 16 mg/L); SXT, trimethoprim–sulfamethoxazole (R > 4 mg/L); CTX, cefotaxime (R > 2 mg/L); COL, colistin (R > 2 mg/L); MEM, meropenem (R > 8 mg/L;, AMK, amikacin (R >16 mg/L). Bold underlined numbers indicate resistance to the corresponding antimicrobial agents.
On the basis of whole-genome sequencing (WGS) and subsequent standalone BLAST analysis against ResFinder, we identified eight resistance genes coexisting in the isolated strain, namely, tet(X4), floR, tet(A), bla TEM-1B, aph(6)-Id, dfrA14, qnrS1, and sul3. Only aph(6)-Id, blaTEM-1B, qnrS1, and sul3 were identified in IncX1 plasmid. Notably the tet(X4) gene was carried within a separate circular. Most of these genes were consistent with the resistance phenotype, although there were some exceptions, For example, despite the presence of the resistance genes, aph(6)-Id and bla TEM-1B, the strain did not exhibit resistance to amikacin, cefotaxime, or meropenem, and conferred resistance only to older-generation antibiotics. In contrast, a high level of chloromycetin resistance (>512 mg/L) was observed in the absence of any corresponding resistance genes.
3.3. Genetic environments and transferability of tet(X4)
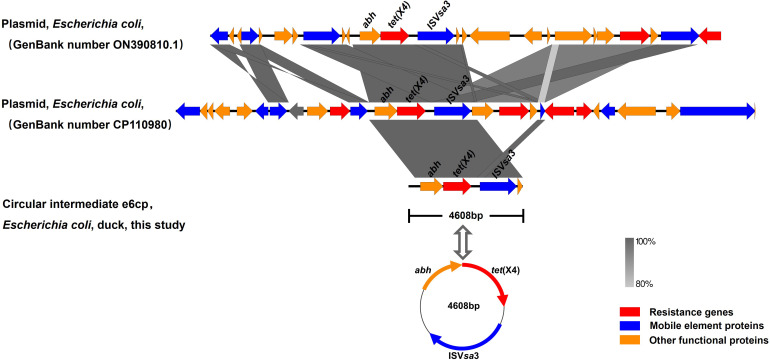
The genome sequence of the tet(X4)-positive strain was determined based on WGS, and bioinformatic analysis revealed that the strain is characterized by a multidrug resistance gene phenotype and belongs to a new type of sequence, ST10776. This discovery thus provides evidence to indicate that the spread of tet(X4) is not attributable to clonal dissemination ( Table 1 ). Splicing results indicated that tet(X4) is located within a circular intermediate (4608 bp). The sequence upstream of tet(X4) contains an alpha/beta hydrase (abh) and an ISVsa3 (mobile element protein), which form the genetic environment of the ISVsa3-ORF2-abh-tet(X4) circular intermediate ( Figures 1 and 2 ). It was highly consistent with p34AB plasmid (170312-bp) from E.coli in 2019 (He et al., 2019), which contains a highly mobile 5586-bp region “ISVsa3-ORF2-abh-tet(X4)-ISVsa3”. Reverse PCR has demonstrated that the region “ISVsa3-ORF2-abh-tet(X4)-ISVsa3” could form a circular intermediate “ISVsa3-ORF2-abh-tet(X4)”, which insert to other ISVsa3 positive plasmid and mediated transfer (He et al., 2019). In this study, the existence of the circular intermediate “ISVsa3-ORF2-abh-tet(X4)” was directly discovered for the first time through Whole-genome sequencing.
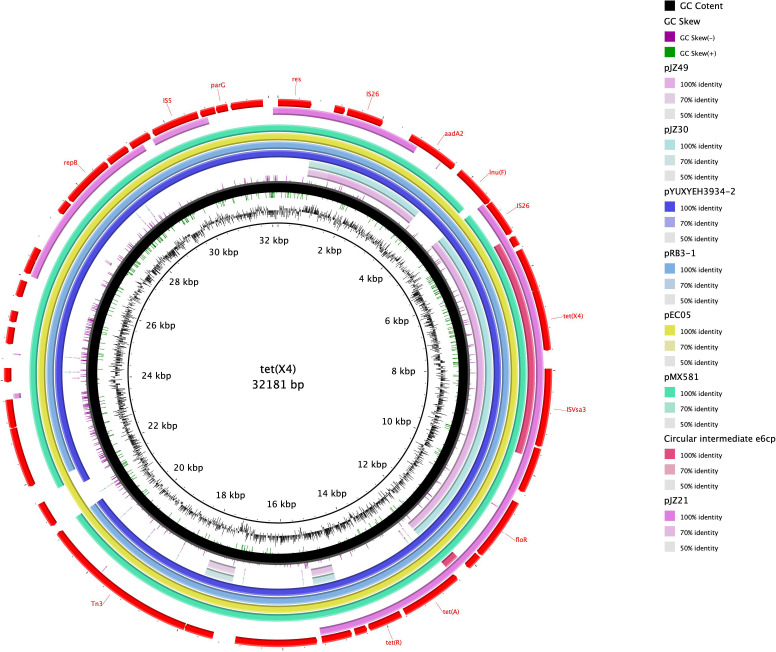
Figure 1.
Circular comparison of the tet(X4)-bearing Circular intermediate e6cp with other similar plasmids obtained from the NCBI database. The outermost ring indicates the reference plasmid with the respective gene positions. The tet(X4)-bearing Circular intermediate e6cp is shown in pink.
Figure 2.
Comparison of the genetic context of tet(X4) with those of closely-related sequences. The position and orientation of genes are indicated by arrows and labeled with gene names. Resistance genes, mobile element proteins, and other functional proteins are indicated by red, blue, and orange arrows, respectively.
Upon comparison of the genetic environment, the backbone of the circular intermediate e6cp identified in the present study was found to exhibit 100% coverage and 99% identity with pJZ49-tet(X4) (GenBank accession number ON390810) in Proteus mirabilis isolated from swine and 94% coverage and 99% identity with pYUXYEH3934-2 (GenBank accession number CP110980) in E. coli isolated from Homo sapiens ( Figure 2 ). Moreover, the circular intermediate e6cp.showed 100% identity and 6.3% coverage with pJZ21 (GenBank accession number ON390807.1), which was isolated from swine ( Figure 1 ). The circular intermediate, ISVsa3-ORF2-abh-tet(X4) identified by WGS in the present study would tend to indicate that tet(X4) will continue to spread and pose a potential risk to public health. Furthermore, similar genetic contexts in diverse sources and strains are conducive to the ongoing spread of this resistance gene.
ISVsa3 is an important mobile element that mediates the horizontal translocation of tet(X4) between plasmids (Aminov, 2021; Li et al., 2021; Wang et al., 2022). To investigate whether ISVsa3 could mediate tet(X4) gene transmissibility, we subjected the positive strain to conjugation experiments with E. coli C600, and accordingly found that the strain successfully transferred tet(X4) to E. coli C600 at a frequency of 10-5. The result of the antimicrobial susceptibility test revealed that the MIC values of these transconjugants increased from 0.5 mg/L to 4 mg/L, thereby conferring resistance to tigecycline. These results imply that the tet(X4) gene of the positive strain is harbored within a conjugative plasmid, the transmission of which is mediated by ISVsa3 elements. At 4608 bp in size, this circular intermediate is relatively and simple in structure, and is characterized by enhanced mobility and transferability, thereby indicating that the horizontal dissemination of tet(X4) via conjugative circular intermediate or other mobilizable genetic elements may have occurred on the duck farm from which the E. coli e6cp strain was isolated.
In summary, to the best of our knowledge, this is the first study in which the tet(X4) tigecycline resistance gene has been detected on a duck farm in Hunan Province. Our investigation of the prevalence and characteristics of tet(X4) indicated that this gene was detected exclusively in duck fecal samples at a positivity rate of 0.14% (1/677), which is lower than that previously reported. Furthermore, tet(X4) was established to be harbored a circular intermediate (4608 bp) within the genetic environment of the ISVsa3-ORF2-abh-tet(X4) elements and could be transferred to other chromosomes and plasmids. Compared with previously reported tet(X4)-carrying plasmids, we found that this circular intermediate was inserted into different bacterial sequence types and conferred high-level resistance to tigecycline. Considering the management model of duck farms, each farm maintains fixed breeders that do not intermingle with others, and the use of medications on these farms adheres strictly to veterinarian-prescribed protocols. We hypothesize that (1) the establishment of informational supervision throughout the industry chain is conducive to the prevention and control of antimicrobial resistance, and (2) horizontal transmission plays an important role in the spread of the tet(X4) gene and is a potential transmission route that facilitates the spread of antimicrobial resistance genes. However, the number of positive tet(X4) samples in this study was insufficiently large to clarify the transmission mechanism. Moreover, the circular intermediate of tet(X4) identified in study is highly mobile and transferable, thereby highlighting the threat posed by the spread of genes with resistance to tigecycline within this duck farm, and potentially other farms. Consequently, it is imperative to strengthen the scientific use of antibiotics in poultry farm and conduct daily monitoring of tigecycline resistance genes.
Acknowledgments
We express our gratitude for the support and assistance from the College of Basic Medical Sciences, Xiangnan University, and the Veterinary Medicine Engineering Center, College of Animal Medicine, Hunan Agricultural University.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Scientific Research Fund of the Hunan Provincial Education Department, China (Grant No. 22C0569); The Natural Science Foundation of Hunan Province (Grant No.2024JJ7516); National College Students Innovation and Entrepreneurship Training Program, China (Grant No. S202310545009); and National College Students Innovation and Entrepreneurship Training Program, Hunan province (Grant No. 3750).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The requirement of ethical approval was waived by Experimental animal ethics review committee of Xiangnan University for the studies involving animals because this study did not carry out experimental research on animals and did not affect the welfare of animals. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
CJ: Writing – original draft, Conceptualization, Formal analysis, Funding acquisition, Validation. JY: Writing – original draft, Conceptualization, Investigation, Software. GX: Writing – review & editing, Data curation, Methodology. NX: Writing – review & editing, Methodology. JH: Writing – review & editing, Methodology. YY: Methodology, Writing – review & editing. ZS: Writing – review & editing, Supervision. YL: Writing – review & editing, Data curation, Visualization.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aminov R. (2021). Acquisition and spread of antimicrobial resistance: A tet(X) case study. Int. J. Mol. Sci. 22, 3905. doi: 10.3390/ijms22083905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distribution and genomic characterization of tigecycline-resistant tet(X4)-positive escherichia coli of swine farm origin. (Accessed 8 August 2023). [DOI] [PMC free article] [PubMed]
- Chen C., Cui C.-Y., Yu J.-J., He Q., Wu X.-T., He Y.-Z., et al. (2020). Genetic diversity and characteristics of high-level tigecycline resistance tet(X) in acinetobacter species. Genome Med. 12, 111. doi: 10.1186/s13073-020-00807-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne L., Arief R., Benigno C., Giang V. N., Huong L. Q., Jeamsripong S., et al. (2019). Characterizing antimicrobial use in the livestock sector in three south east asian countries (Indonesia, Thailand, and Vietnam). Antibiotics (Basel) 8, 33. doi: 10.3390/antibiotics8010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Saw W.-Y., Tan L. W. L., Moong D. K. N., Nagarajan N., Teo Y. Y., et al. (2020). Emergence of tigecycline- and eravacycline-resistant tet(X4)-producing enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75, 3480–3484. doi: 10.1093/jac/dkaa372 [DOI] [PubMed] [Google Scholar]
- Gimenez-Miranda L., Samhouri B. F., Wolf M. J., Anderson D. K., Midthun D. E., Lim K. G., et al. (2023). Diagnostic yield of 16S ribosomal ribonucleic acid gene-based targeted metagenomic sequencing for evaluation of pleural space infection: A prospective study. Mayo Clin. Proc. Innov. Qual Outcomes 7, 373–381. doi: 10.1016/j.mayocpiqo.2023.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T., Wang R., Liu D., Walsh T. R., Zhang R., Lv Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2 [DOI] [PubMed] [Google Scholar]
- Jenner L., Starosta A. L., Terry D. S., Mikolajka A., Filonava L., Yusupov M., et al. (2013). Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 110, 3812–3816. doi: 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Peng K., Yin Y., Sun X., Zhang W., Li R., et al. (2021). Occurrence and molecular characterization of abundant tet(X) variants among diverse bacterial species of chicken origin in Jiangsu, China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.751006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang P., Xiao X., Li R., Wang Z. (2023). Genomic characterization of tigecycline-resistant tet(X4)-positive E. Coli in slaughterhouses. Veterinary Microbiol. 276, 109606. doi: 10.1016/j.vetmic.2022.109606 [DOI] [PubMed] [Google Scholar]
- Liu Y.-Y., Lu L., Yue C., Gao X., Chen J., Gao G., et al. (2024). Emergence of plasmid-mediated high-level tigecycline resistance gene tet(X4) in enterobacterales from retail aquatic products. Food Res. Int. 178, 113952. doi: 10.1016/j.foodres.2024.113952 [DOI] [PubMed] [Google Scholar]
- O’Neill J. (2016). Tackling drug-resistant infections globally: final report and recommendations. The Review on Antimicrobial Resistance. Available at: https://amr-review.org/Publications.html. [Google Scholar]
- Sheu C.-C., Chang Y.-T., Lin S.-Y., Chen Y.-H., Hsueh P.-R. (2019). Infections caused by carbapenem-resistant enterobacteriaceae: an update on therapeutic options. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen C., Cui C.-Y., Zhang Y., Liu X., Cui Z.-H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Cui M., Zhang S., Liu D., Fu B., Li Z., et al. (2020). Genomic epidemiology of animal-derived tigecycline-resistant escherichia coli across China reveals recent endemic plasmid-encoded tet(X4) gene. Commun. Biol. 3, 412. doi: 10.1038/s42003-020-01148-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasina E., Haidich A.-B., Kokkali S., Arvanitidou M. (2011). Efficacy and safety of tigecycline for the treatment of infectious diseases: A meta-analysis. Lancet Infect. Dis. 11, 834–844. doi: 10.1016/S1473-3099(11)70177-3 [DOI] [PubMed] [Google Scholar]
- Wang Q., Lei C., Cheng H., Yang X., Huang Z., Chen X., et al. (2022). Widespread dissemination of plasmid-mediated tigecycline resistance gene tet(X4) in enterobacterales of porcine origin. Microbiol. Spectr. 10, e0161522. doi: 10.1128/spectrum.01615-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lu M.-J., Wang Z.-Y., Jiang Y., Wu H., Pan Z.-M., et al. (2022). Tigecycline-resistant escherichia coli ST761 carrying tet(X4) in a pig farm, China. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.967313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., He R., Qin M., Yang Y., Chen J., Feng Y., et al. (2022). Identification of plasmid-mediated tigecycline-resistant gene tet (X4) in enterobacter cloacae from pigs in China. Microbiol. Spectr. 10, e02064–e02021. doi: 10.1128/spectrum.02064-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhou Y., Niu S., Liu Z., Zou Y., Yang Y., et al. (2022). A novel inhibitor of monooxygenase reversed the activity of tetracyclines against tet(X3)/tet(X4)-positive bacteria. eBioMedicine 78, 103943. doi: 10.1016/j.ebiom.2022.103943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi S., Zekiy A. O., Krutova M., Gholami M., Kouhsari E., Sholeh M., et al. (2022). Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 41, 1003–1022. doi: 10.1007/s10096-020-04121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahav D., Lador A., Paul M., Leibovici L. (2011). Efficacy and safety of tigecycline: A systematic review and meta-analysis. J. Antimicrobial Chemotherapy 66, 1963–1971. doi: 10.1093/jac/dkr242 [DOI] [PubMed] [Google Scholar]
- Yang J., Xiao G., Xiao N., Jiang Z., Jiang C., Li Y., et al. (2023). Characteristics of tet(X4)-producing escherichia coli in chicken and pig farms in hunan province, China. Antibiotics (Basel) 12, 147. doi: 10.3390/antibiotics12010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Cui C.-Y., Kuang X., Chen C., Wang M.-G., Liao X.-P., et al. (2021). Prevalence of tet(X4) in escherichia coli from duck farms in southeast China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.716393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Deng L., Zhou X., Zhang C., Hu Z., Chen Y., et al. (2022). Prevalence and risk factors of tet(X4)-positive enterobacteriaceae in human gut microbiota. J. Glob Antimicrob. Resist. 31, 15–21. doi: 10.1016/j.jgar.2022.07.014 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wen J., Wang Y., Wang M., Jia R., Chen S., et al. (2022). Dissemination and prevalence of plasmid-mediated high-level tigecycline resistance gene tet (X4). Front. Microbiol. 13. doi: 10.3389/fmicb.2022.969769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.