Abstract
The use of monoclonal antibodies for the control of drug resistant nosocomial bacteria may alleviate a reliance on broad spectrum antimicrobials for treatment of infection. We identify monoclonal antibodies that may prevent infection caused by carbapenem resistant Acinetobacter baumannii. We use human immune repertoire mice (Kymouse platform mice) as a surrogate for human B cell interrogation to establish an unbiased strategy to probe the antibody-accessible target landscape of clinically relevant A. baumannii. After immunisation of the Kymouse platform mice with A. baumannii derived outer membrane vesicles (OMV) we identify 297 antibodies and analyse 26 of these for functional potential. These antibodies target lipooligosaccharide (OCL1), the Oxa-23 protein, and the KL49 capsular polysaccharide. We identify a single monoclonal antibody (mAb1416) recognising KL49 capsular polysaccharide to demonstrate prophylactic in vivo protection against a carbapenem resistant A. baumannii lineage associated with neonatal sepsis mortality in Asia. Our end-to-end approach identifies functional monoclonal antibodies with prophylactic potential against major lineages of drug resistant bacteria accounting for phylogenetic diversity and clinical relevance without existing knowledge of a specific target antigen. Such an approach might be scaled for a additional clinically important bacterial pathogens in the post-antimicrobial era.
Subject terms: Bacterial infection, Bacterial host response, Antimicrobial resistance
In this work, the authors describe immunisation of human antibody immune repertoire transgenic mice as a surrogate to define protective human antibody reactivity to clinically diverse, antibiotic resistant Acinetobacter baumannii.
Introduction
The sustained overreliance on antimicrobials over the last ~70 years has created a problem; many commonly used antibacterial agents are no longer effective against pathogenic bacteria. Consequently, antimicrobial resistance (AMR) represents one of the greatest current challenges in infectious diseases. The devastating impact of AMR can be highlighted by outbreaks of multi-drug resistant (MDR) and extensively-drug resistant (XDR) bacteria in high dependency hospital units1,2. These outbreaks, commonly associated with bacteraemia leading to sepsis, are reported in high- and low-income countries alike and are dominated by bacterial organisms belonging to the ESKAPE group of pathogens3. One of the most problematic ESKAPE pathogens is the Gram-negative coccobacilli Acinetobacter baumannii. Infections with A. baumannii typically occur following trauma, surgery, catheterisation, and endotracheal intubation, and are associated with high mortality and morbidity4,5. Recent decades have seen a widespread increase in MDR/XDR phenotypes of A. baumannii1. Isolates are now commonly resistant to β-lactams, aminoglycosides, fluoroquinolones, and are frequently resistant to colistin, the drug of last resort6. A. baumannii is currently the leading cause of AMR-attributable death in Southeast Asia, east Asia, and Oceania3. Of great concern across these, as well as global nosocomial settings are carbapenem-resistant A. baumannii (CRAB) and infected patients have increased mortality7. Given the emerging importance of this bacterial pathogen, the World Health Organisation identified A. baumannii as critical priority for the development of new antimicrobials8.
The recent COVID-19 pandemic has highlighted the role that monoclonal antibodies (mAbs) can play in treating/preventing infectious disease and how new technologies can be deployed to identify functional mAbs9. We considered that mAbs specifically targeting A. baumannii lineages causing highest mortality and morbidity in hospital settings could be developed for pre-exposure prophylaxis, as stand-alone therapies or as adjuncts to current antimicrobials. One challenge with developing antibodies targeting bacterial pathogens is establishing the target antigen(s). This problem may be overcome using antigen-agnostic screening approaches. The sustained use of bacterial vaccines, where antibody represents the most robust protective correlate, suggests interrogation of the human B cell repertoire could result in identification of protective mAbs10. One approach is to isolate antibodies from people recovering from bacterial infection, but there is no indication that these antibodies will be protective, indeed the person being infected in the first place may suggest a failure of antibody mediated protection. An alternative is to use mouse models, but the mouse repertoire does not accurately reproduce the human B cell response. A third route is to use transgenic, human immune repertoire mice, because of well characterised routes of immunisation and accessibility to the target-specific B cell populations11. For A. baumannii, mAbs targeting various surface carbohydrate antigens have been identified as being potentially protective12, but thus far, the full breadth of membrane protein targets that may induce protective antibody remains underexploited13.
A reliable approach for raising novel mAbs is through immunisation, but the antigens used need to reflect the infectious bacteria, and may lead to a bias; consequently, a broad cocktail of antigens should maximise antibody discovery. Like other Gram-negative bacteria, A. baumannii produce outer membrane vesicles (OMVs) in culture via blebbing of the outer membrane. OMVs mediate bacterial protein secretion, cell-to-cell communication and play active roles in pathogenesis, immunomodulation, and horizontal bacterial gene transfer14,15. Although the composition of OMVs is not entirely representative of the cell membrane from which they derive16, OMVs generated from A. baumannii contain polymeric carbohydrate antigens and many bacterial membrane proteins17,18. OMVs are highly immunogenic, are in development as human vaccines19–21, and we have recently demonstrated that immunisation with OMVs can protect mice from A. baumannii infection17. Therefore, OMVs may be exploited to generate antibodies that target the bacterial cell membrane and protect against infection.
Here we utilised the Kymouse platform18, with additional genetic modifications to allow expression of fully human antibodies22, to probe the human antibody repertoire response after immunisation with purified OMVs pooled from different clades of clinically relevant A. baumannii. The aim was to develop a pipeline allowing mAb isolation and characterisation that could be applied more widely to a range of bacterial pathogens.
Results
The immunisation of Kymouse platform mice with OMVs induces robust and reproducible cross-reactive human antibodies
To identify mAbs capable of binding bacterial antigens with potential to mediate protection, we immunised the Kymouse with OMVs isolated from well characterised, phylogenetically relevant, clinical isolates of A. baumannii. These A. baumannii were isolated from patients with ventilator associated pneumonia (VAP) in Ho Chi Minh City, Vietnam emerging after the clinical introduction of carbapenems in 200823, and belonged to the globally dominant carbapenem-resistant A. baumannii lineage clonal complex 2 (CC2)23,24. We selected isolates from Vietnam as Southeast Asia represents a region where AMR-attributable death is dominated by A. baumannii. To reflect the genetic diversity among the CC2 lineage after the emergence of carbapenem resistance, we selected isolates representing four of five sub-lineages from this collection for subsequent experiments (BAL 084, BAL 191, BAL 215 and BAL 276). The diversity of the selected clones is shown relative to that seen for the widely studied and publicly available CC2-typed isolates25 NCTC13302 and NCTC13424, the CC1-typed isolate ATCC-BAA-1710, and the ATCC17978 reference (Fig. 1a).
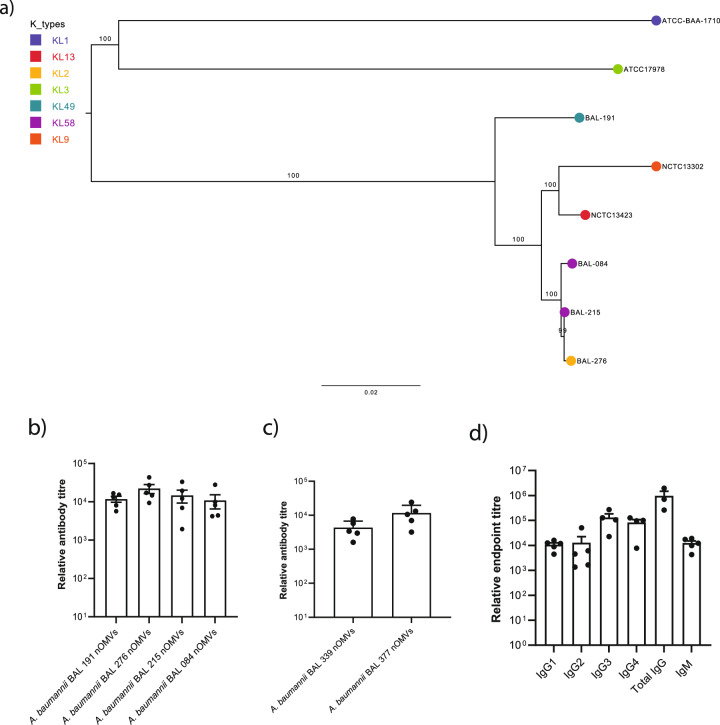
Fig. 1. Polyclonal reactivity of Kymouse platform mouse serum raised against OMVs generated from phylogenetically selected A. baumannii.
a Phylogenetic relationship between BAL 084, BAL 191, BAL 215 and BAL 276 and the publicly available CC2, CC1-typed and reference isolates available from culture collections (ATCC and NCTC). The phylogeny was built using whole genome sequence data from the calling of 57844 single nucleotide polymorphisms from 2879 core genes using RAxML (Randomised Axelerated Maximum Likelihood- v8.12.8) using a general time reversible (GTR) model of nucleotide substitution (ASC-GTRGAMMA) and a “Lewis” method of ascertainment bias correction. One hundred bootstrap pseudo-replicate analysis was applied to measure the robustness of the ML phylogeny. The numbers on the tree branches indicate the bootstrap support values while the scale bar reveals the number of nucleotide substitutions per site. The coloured circles at the node tips represent the different capsule types (KLs). b Relative antibody titre measured by indirect ELISA of sera from Kymouse platform mice (n = 5; n defined as serum analysed from a single mouse) immunised with pooled OMVs generated from native, genetically unmodified Acinetobacter baumannii clinical isolates (nOMV) versus sera from 5 non-immunised Kymouse platform mice. c Relative antibody titre similarly measured by indirect ELISA against separate nOMVs generated from phylogenetically related individual isolates not contained in the pool used for immunisation. d Relative titres of human IgG1-4 and IgM subclasses represented in Kymouse platform mouse sera (n = 5; n defined as serum analysed from a single mouse) versus sera from 5 non-immunised Kymouse platform mice against pooled nOMVs used for immunisation. Data for (b, c, d) are presented as mean values ± SEM.
OMVs were generated and extracted from the four selected isolates; the integrity of the individual OMV preparations were confirmed by negative staining with 1% uranyl actetate followed by transmission electron microscopy (TEM) and exhibited membranous vesicular particles ranging in size from 20 to 100 nm (Supplementary Fig. 1a). The OMVs were comprised of protein antigens with a range of molecular weights as indicated by SDS-PAGE (Supplementary Fig. 1b). To generate human polyclonal antibody responses, Kymouse platform mice (n = 5) were immunised with 1 µg of a pooled OMV cocktail and boosted with the same immunogen 35 days later. Mice were sacrificed 7 days after the boost (day 42) and the serum was analysed for total IgG against the pooled OMVs by ELISA. There was comparable reactivity to all four OMVs contained in the OMV cocktail among individual mice. Antibody endpoint titres, measured as dilution of serum required to show antibody binding signal equivalent to naïve serum from non-immunised Kymouse platform mice, were ~104, indicating robust polyclonal responses directed to OMVs (Fig. 1b). Comparable titres were observed against purified OMVs from A. baumannii isolates not represented in the OMV cocktail used for immunisation (Fig. 1c), signifying that the polyclonal response incorporated cross-reactive antibodies. The prime-boost OMV regimen induced robust IgG1, IgG2, IgG3, and IgG4 subclass responses, as well as IgM (Fig. 1d). Therefore, immunising Kymouse with an OMV cocktail induced a human antibody response with evidence of cross reactivity to several phylogenetically defined A. baumannii isolates.
Labelled OMVs can be used to isolate antigen specific B cells for characterisation
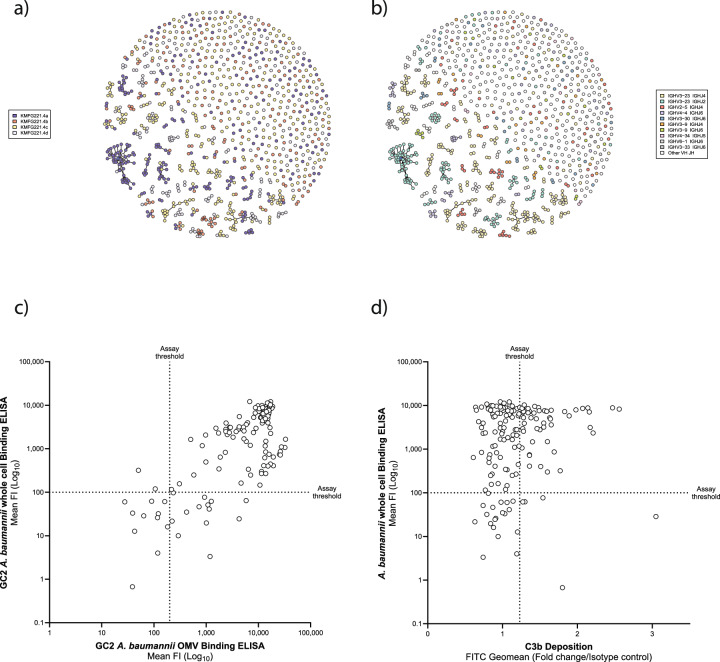
To identify B cells expressing A. baumannii specific B cell antigen receptors (BCRs), we labelled OMV with FM4-64 lipophilic dye as a bait to sort single B cells from OMV-immunised Kymouse. Using the sorting and gating strategy shown in Supplementary Fig. 2, we isolated 2087 single CD19+/B220+ B cells with capacity to bind at least one of the four OMVs used for immunisation. Analysis of the IgH/L sequences from isolated B cells identified 951/2,087 (45.5%) successful heavy and light chain paired reads, indicating an intact B cell receptor (BCR); with 530/2087 (25.4%) identified as IgG. Analysis using IntelliSellect™ software (Kymab’s proprietary software platform to perform sequence clustering) clustered 372/2087 (17.8%) of the sequenced BCRs, based on variable heavy and light (VH/VL) sequence relation. These clusters suggested convergent targeting of antibody germline gene and evolution between the different immunised mice, as evidenced by clusters containing related sequences from more than one mouse (where multiple colours are seen in a cluster, Fig. 2a). BCR sequences were derived from multiple germline IGHV and IGHJ combinations, indicating broad engagement of Ig germline genes (Fig. 2b).
Fig. 2. Antibody sequence repertoire analysis from OMV-immunised Kymouse platform mice.
a A sunspot plot showing antibody sequence clustering, where spot colour indicates the individual mouse immunised with pooled OMVs from which the antibody sequence originated (n = 4). Individual mice (indicated by nominative code identifier in the index) are represented by colour-matched code in the index. b Sunspot plot showing antibody sequence clustering based on an individual germline IGHV and IGHJ combination utilised. Germline IGHV and IGHJ combination utilised are represented by colour-matched code in the index. c Plot of signal from indirect human IgG1 ELISAs against the OMV cocktail (x-axis) versus unfixed pooled bacteria from which the immunising OMVs were derived (y-axis). d Deposition of the C3b protein on OMVs as a surrogate for primary initiation of the complement cascade (x-axis) versus indirect human IgG1 ELISAs against unfixed pooled bacteria from which the immunising OMVs were derived (y-axis).
We down-selected heavy and light chain pairs (IgH/L) to produce a collection of antibodies for further analysis. To increase stringency, we selected paired VH and VL sequences that were unique in amino acid sequence, with evidence of affinity maturation (≥2 amino acid mutations from germline across VH/VL). 291 VH/VL paired sequences were selected for DNA synthesis, construction on a human IgG1 scaffold and transfection into HEK 293 cells for small-scale (100–500 µg/ml) mAb expression. We added a further six mAbs to this selection derived from VH/VL sequences from bone marrow plasma cells from equivalently immunised Kymouse (n = 5) that could not be sorted by cell surface specific IgG, but instead were sorted based on the plasma cell marker CD138. We included these in the screen as we considered they may exhibit target-agnostic functional activity.
Isolated antibodies from humanised mice can bind A baumannii-derived OMVs and trigger complement
179 of the 297 selected mAbs bound to the immunising OMVs at a detectable concentration above background (Fig. 2c). We also assessed the ability of the mAbs to bind to whole, unfixed pooled bacteria from which the immunising OMVs were derived. 136/297 of the mAbs bound intact bacteria and OMVs (Fig. 2c). Having selected antibodies that bound bacterial cells, we next assessed their functionality. C3b protein deposition on the bacterial membrane is a surrogate for initiation of the complement cascade, inferring the ability of the mAb to induce complement mediated bacterial killing26. We used flow cytometry to detect C3b deposition on the surface of FM4-64 stained OMVs. We identified 76 mAbs that demonstrated C3b deposition above the background (Fig. 2d).
Isolated monoclonal antibodies can bind diverse CC2-typed A. baumannii isolates
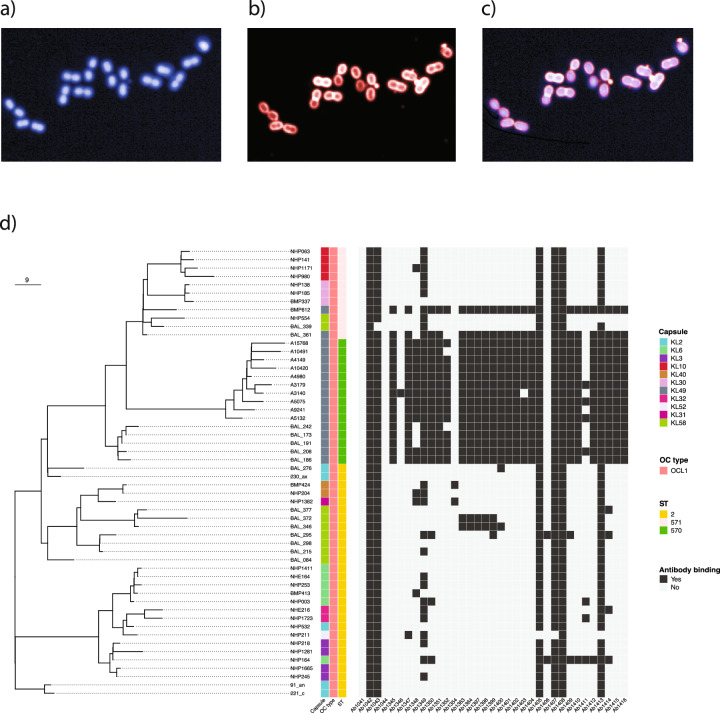
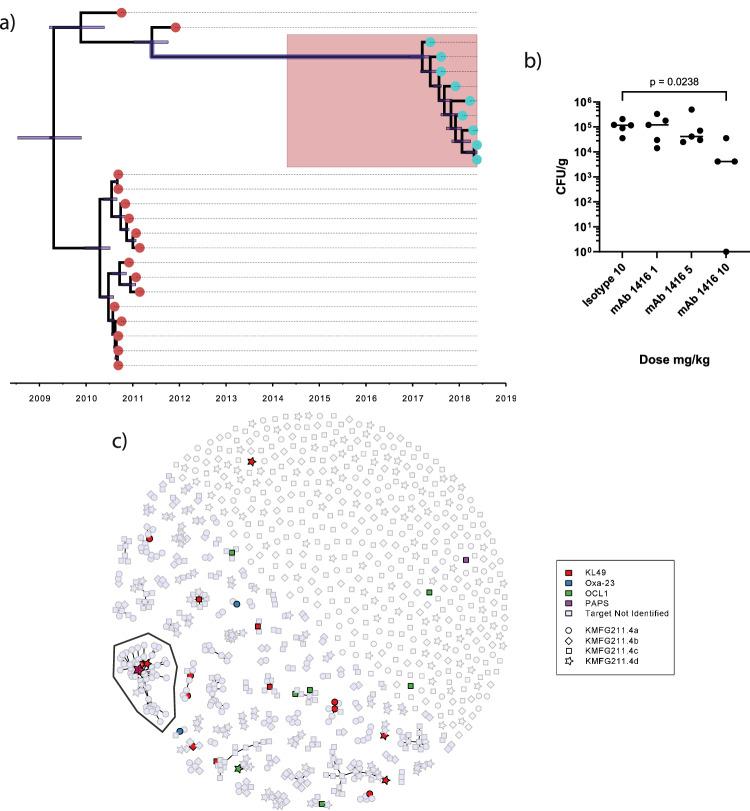
Having observed that the isolated mAbs could effectively bind to bacterial cells, we sought to investigate whether the mAbs could bind to a diverse collection of CC2 A. baumannii. We measured mAb binding across a collection of A. baumannii isolated in ICUs in Vietnam between 2003 and 2018, that reflected the diversity of circulating CC2 isolates23,27,28. The collection encompassed 11 distinct capsule antigen types (KL2, 3, 6, 10, 30, 31, 32, 40, 49, 52 and 58) and a single lipooligosaccharide antigen type (OCL1). Visualising mAb binding at single cell resolution, we defined single bacteria using DAPI and measured surface mAb binding using Alexa Fluor 647 intensity (Fig. 3a–c) using a high-content imaging (HCI) system8. Of the 35 mAbs tested, we identified six that bound almost all CC2 isolates in the collection (≥98%), one mAb that bound the majority of isolates (72%), and two with some degree of cross-specificity (30% and 15% respectively) (Fig. 3d). Seven of these antibodies utilised the IGHV2-5 IGHJ4 germline gene combination, one utilised IGHV3-9 IGHJ6 and one utilised IGHV3-9 IGHJ4. We also identified a group of 18 mAbs that bound to bacteria producing KL49 typed capsule antigen. These 18 mAbs utilised either the IGHV3-23 IGHJ2 (n = 8) or the IGHV3-23 IGHJ4 germline gene combinations (n = 10) (Supplementary Fig 3).
Fig. 3. Analysis of mAb binding by HCI to a curated collection of CC92 A. baumannii strains isolated in Vietnam between 2003 and 2018.
Individual bacteria were stained and segmented using DAPI which stains intracellular bacterial DNA (a), antibody binding was measured by the indirect signal from binding human IgG1 mAb using an Alexa Fluor 647 labelled secondary anti-human IgG1 (b) overlay of both signals (c). d Bacterial surface mAb binding at 1 μg/ml across the entire phylogenetically characterised CC92 collection. Binding was defined as positive where the Alexa Fluor 647 mean intensity per well was >500 relative fluorescent units (RFU).
Isolated mAb targeted both protein and non-protein antigens
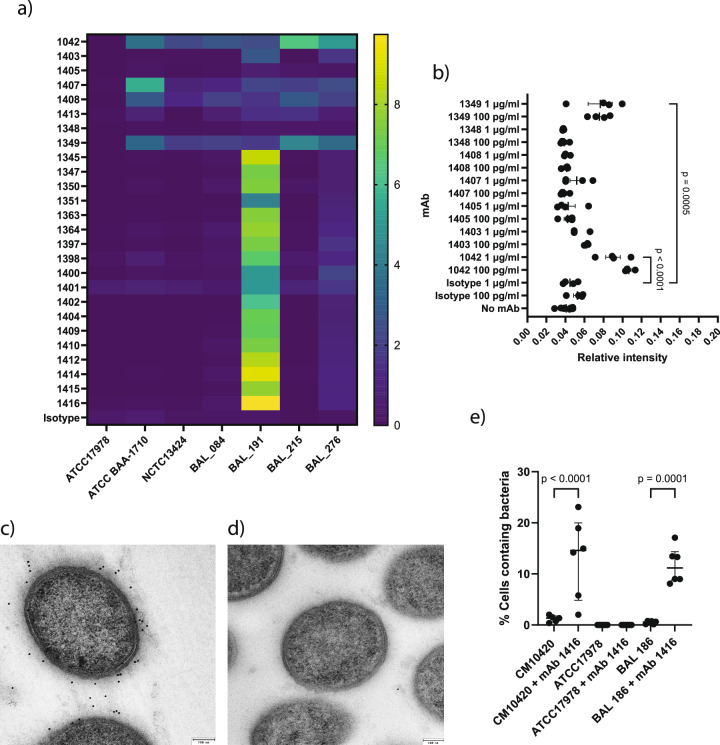
To identify the targets of the broadly-reactive mAbs, we constructed a phage expression library incorporating genomic DNA from A. baumannii BAL 084 and BAL 276 as selected representative strains of the immunising panel and used expression cloning to detect mAb-recognised proteins9,10. Using this library (Supplementary Fig 4a), we identified two mAbs (mAb 1348 and mAb 1349) that bound plaques on bacterial plates made by phage expressing DNA from Oxa-23, a class D β-lactamase conferring resistance to carbapenems29. These mAbs were confirmed to bind a ~ 30 kDa band corresponding to the molecular weight of Oxa-23 on Western blotting of SDS-PAGE resolved bacterial lysates from Oxa-23 encoding A. baumannii (BAL 084, BAL 191, BAL 215, BAL 276 and NCTC13424), but not a bacterial lysate of ATCC17978 lacking a class D β-lactamase or a bacterial lysate of NCTC13302 expressing class D β-lactamase Oxa-2511 (Supplementary Fig 4b).
The remainder of the mAbs from this broadly reactive group failed to bind phage after repeated screens. However, six mAbs bound to ~10 kDa target from bacterial lysates after Western blotting (Supplementary Fig 4c). Because lipooligosaccharide (LOS) from Acinetobacter runs as a single band on SDS-PAGE at ~10 kDa30, we hypothesised that these mAbs were binding LOS. Non-proteinaceous carbohydrate extracts containing both capsule and LOS were generated from multiple Acinetobacter isolates, resolved by SDS-PAGE (Supplementary Fig 5a) and subjected to immunoblotting with individual mAbs. All six mAbs bound a 10 kDa band, corresponding with LOS isolated from OCL1 type strains but showed no binding to an OCL2 variant (ATCC 17978) (Supplementary Fig 5b), indicating that these mAbs bound to LOS of OCL1 type. We additionally analysed non-proteinaceous carbohydrate extracts with the panel of 18 mAbs specifically binding isolates genotyped as producing KL49 capsule. We found that all individual mAbs in this panel bound a high molecular weight band corresponding with organisms elaborating the KL49 capsular polysaccharide; notably, no binding was observed for non-KL49 isolates (Supplementary Fig 5c). Furthermore, these mAbs bound this high molecular band in similarly tested whole cell lysates of genotypic KL49 isolates (n = 6); an isotype control showed no reactivity (Supplementary Fig 6a). Therefore, we concluded that these 18 mAbs specifically targeted the KL49 capsular antigen.
OCL-1, Oxa-23 and capsule targeting antibodies bind live bacteria and trigger complement
As high content imaging was performed on formalin-fixed bacteria, we sought to confirm binding of mAbs targeting OCL-1, Oxa-23 and KL49 on live bacteria by indirect ELISA. We compared binding of this panel of mAbs to the bacterial isolates used to generate OMVs for immunisation, the publicly available CC2 typed isolate NCTC13424, the CC1 isolate ATCC-BAA-1710, and the ATCC17978 reference (Fig. 4a). The OCL-1 binding mAbs (1042, 1403, 1407, 1408 and 1413) typically bound across isolates producing OCL-1 but not ATCC-17978 producing OCL-2. mAb 1349 bound Oxa-23 producing isolates but not ATCC-17978, which does not express class D β-lactamases. All KL49 specific mAbs bound BAL 191 which produced KL49-typed capsule antigen but not ATCC17978 (KL3), ATCC BAA1710 (KL1), NCTC13424 (KL13), or BAL 084, and BAL 215 (both KL58).
Fig. 4. Functional binding of mAbs targeting LOS (OCL-1), Oxa-23 and KL49 typed capsule on live A. baumannii.
a Analysis of mAb binding to live bacterial cells by indirect ELISA. Heat map indicates background corrected signal as relative fluorescent units (RSU) from an indirect ELISA evaluating mAb binding against live bacteria at 1 μg/ml. Signal intensity expressed as the mean of independent assay replicates (n = 3). b Ability of selected mAbs at 1 μg/ml and 100 pg/ml concentrations to trigger C3b deposition when binding live BAL 276 bacteria as measured by indirect ELISA targeting C3b. Relative intensity of fluorescent signal shows mean, and SEM (n = 4; n is a single biological replicate for each bacterial isolate analysed in a single experiment). Statistical analysis was performed using Dunnett’s one-way ANOVA to compare multiple groups with p shown as a numerical value when p ≤ 0.05. c Transmission electron microscopy of ultrathin sections of KL49 producing strain CM10420 and (d) KL-3 producing strain ATCC1798, labelled with mAb 1416 and gold-conjugated anti-human IgG. Labelling via mAb binding to KL49 was observed for CM10420 in (c), but not for ATCC17978. e Uptake of strain CM10420 by THP-1 cells and the alternative KL49 expressing strain BAL 186, in the presence or absence of mAb 1416 as measured by High Content Imaging (n = 6; n is a single biological replicate for each bacterial isolate analysed in a single experiment). In both cases percentage THP-1 cells per well analysed containing bacteria increased in the presence of mAb 1416. Bar represents median with interquartile range. Statistical analysis was performed using an ordinary one-way ANOVA with p shown as a numerical value when p ≤ 0.05.
We additionally addressed whether binding of mAb to live bacteria could functionally trigger activation of the human complement pathway on the bacterial surface. We measured C3b deposition on the bacterial surface after incubation of live BAL 276 in the presence of human serum and 1 μg/ml or 100 pg/ml mAb using an indirect ELISA. We observed elevated C3b deposition when BAL 276 was incubated with either mAb 1042 or mAb 1349 at both 1 μg/ml or 100 pg/ml for both mAbs individually (Fig. 4b). These data indicate that mAb 1042 and mAb 1349, which bind OCL1 and bacterial membrane-associated Oxa-23 respectively, can trigger the human complement system and have the potential to orchestrate functional immunological responses against A. baumannii in vivo.
To further investigate function of a capsule targeting antibody, we assayed binding and uptake of bacteria via opsonophagocytosis with mAb 1416. As visualised by immunogold labelling of ultrathin sections prepared for transmission electron microscopy, mAb 1416 bound the KL49-producing CM10420 (Fig. 4c) but not the KL3 producing ATCC17978 (Fig. 4d). The mAb 1416 mediated the increased uptake of CM10420 into THP-1 cells (Fig. 4e). These results were replicated with the alternative KL49 expressing strain BAL 186 while increased bacterial uptake by THP-1 cells was not observed in any of the same conditions for ATCC17978. Taken together, these data demonstrate that binding of mAb 1416 to KL49-typed capsule results in enhanced opsonophagocytic uptake by phagocytic cells.
Assessment of target engagement for prophylactic protection in vivo
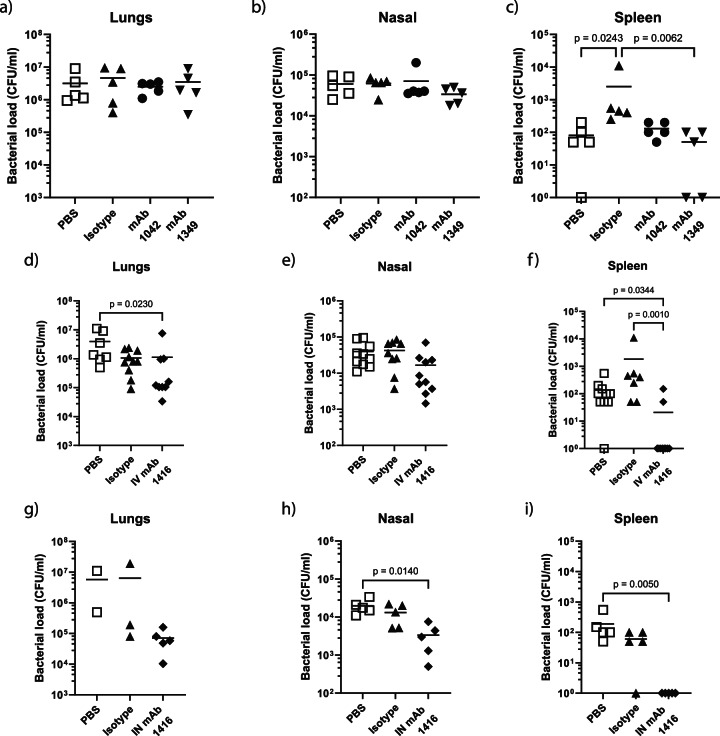
We tested selected mAbs for in vivo efficacy in a pre-exposure prophylaxis intranasal mouse challenge model using BAL 191. This challenge model permitted the evaluation of effects of administering mAbs via intravenous (IV) or intranasal (IN) routes on bacterial burden in the upper and lower respiratory tract as well as systemic spread of bacteria to the spleen in mice 24 h after infection. Compared with untreated mice or mice treated with an isotype control mAb, we did not observe any reduction in BAL 191 bacterial burden in the lungs (Fig. 5a) or nasal wash (Fig. 5b) of mice treated with 10 mg/kg mAb 1042 targeting OCL1 or mAb 1349 targeting Oxa-23 delivered IV. There was a significant reduction in bacteria recovered from the spleen (Fig. 5c). The administration of an IV dose of mAb 1416 of 10 mg/kg reduced bacterial burden in the lung compared to untreated mice (Fig. 5d) but not in the nasal wash (Fig. 5e). There was a significant reduction in the spleen compared to untreated or mAb isotype control mice (Fig. 5f). Intranasal administration of 10 mg/kg mAb 1416 mice did not reduce bacterial burden in the lungs compared to untreated mice or isotype control mice, although we acknowledge that low numbers of mice in the latter two groups limits interpretation of this study (Fig. 5g). But it reduced it in the nasal wash (Fig. 5h) and provided complete protection from bacterial dissemination to the spleen after challenge (Fig. 5i).
Fig. 5. Ability of mAbs to protect in vivo against A. baumannii BAL 191.
Adult BALB/c mice were intravenously dosed with 10 mg/kg of 1042, 1349, isotype control mAb or left untreated on day -1 and day 0, challenged on day 0 with 5 × 107 CFU A. baumannii isolate BAL 191 and culled 24 h later. Bacterial loads at 24 h in lungs (a), nasal wash (b) and spleen (c) were enumerated to evaluate protection. In a subsequent study, mice intravenously or intranasally dosed with 10 mg/kg of 1416 or isotype control were similarly evaluated for protection using the KL49 isolate BAL 191. Bacterial loads at 24 h in lungs and spleen of intravenously dosed (d, e, f) and intranasally dosed mice (g, h, i) were evaluated. Statistical analysis was performed using a Mann–Whitney test with p shown as a numerical value when p ≤ 0.05. The data presented in (a–c) are a single study (n = 5; n defined as an individual mouse); the data presented in (d–f) represent 2 experiments combined (n = 5 per study, n = 10 total); (g–i) represent a single n = 5 study. Bar represents mean value for each data set.
Targeting KL49-typed A. baumannii for pre-exposure prophylaxis by passive immunisation
A. baumannii within our study panel included isolates recovered from neonates diagnosed with probable or culture-confirmed sepsis in Vietnam between January 2017 and June 201827. Most of these isolates (n = 16) were typed as KL49 by WGS. Critically, these neonatal sepsis-associated A. baumannii were found to be ancestrally related to the KL49-producing BAL 191 used to generate the OMV for the primary mouse immunisations (Fig. 6a). To test the potential of mAb 1416 to protect against contemporaneous KL49-typed isolates, we selected an individual isolate associated with a case of neonatal sepsis (CM10420) and tested whether mAb 1416 given prophylactically could protect mice after intranasal challenge with CM10420. Mice were prophylactically treated with 10, 5 or 1 mg/kg of mAb 1416 and challenged 24 h later with A. baumannii CM10420 (Fig. 6b). We observed a dose response, with the A. baumannii CFU/lung decreasing with higher doses of mAb 1416 with a significant reduction in bacterial load in the lungs of mice 24 h post challenge for mice given 10 mg/kg 1416 compared to mice administered 10 mg/kg of an isotype control mAb (Fig. 6b). These data confirm that a mAb raised to OMVs generated from a KL49-typed isolate inducing VAP in adults exhibited prophylactic activity against an KL49-typed isolate causing neonatal sepsis isolated approximately a decade later.
Fig. 6. Ability of mAbs to protect mice against neonatal sepsis causing strains of A. baumannii.
a Temporal phylogenetic characterisation of carbapenem-resistant A. baumannii strains isolates from Vietnamese healthcare settings between 2003 and 2018. Phylogeny was built using whole genome sequence data from the calling of single nucleotide polymorphisms from core genes using RAxML (Randomised Axelerated Maximum Likelihood). b Adult C57BL6 mice were intravenously dosed with 10, 5 or 1 mg/kg of 1416 and 10 mg/kg isotype control mAb on day -1 and day 0, challenged on day 0 with 5 × 107 CFU A. baumannii isolate CM10420. Bacterial loads at 24 h in lungs were enumerated as CFU/gram of lung tissue (CFU/g) to evaluate protection. Statistical analysis was performed using a Mann–Whitney test with p shown as a numerical value when p ≤ 0.05. The data presented are from a single study (n = 5; n defined as an individual mouse). c Sunspot plot showing antibody sequence clustering for different bacterial antigen targets. Colour of symbol in index indicates target bound by individual mAb. Symbol shape in index indicates individual mouse from which corresponding VH/VL sequence pair was derived (indicated by nominative code identifier in the index). The mAb 1416 is indicated in a large cluster (indicated by enclosed region with blue Asterix).
Discussion
Here, we present an end-to-end process which identified mAbs binding across the A. baumannii CC2 diversity, a lineage which is largely representative of globally circulating A. baumannii. We demonstrated that these mAbs have downstream prophylactic potential. Notably, our approach was target agnostic and contributes to the understanding of the human B-cell epitope landscape that is accessible to antibody on the surface of A. baumannii. We down selected various mAbs based on their ability to bind the intact bacterial membrane and trigger C3b deposition. A proportion of the studied mAbs bound to targets accessible on CC2 A. baumannii as well as a phylogenetically related group of isolates expressing the KL49 capsular antigen. A combination of a humanised mouse platform, clinically relevant isolates of bacteria and in vitro followed by in vivo screening enabled us to identify several new human antibodies against CC2 A. baumannii; this approach could be routinely exploited for other bacterial pathogens.
We identified mAbs that could bind Oxa-23, the major A. baumannii carbapenemase enzyme, and OCL-1, both of which are highly conserved across the CC2 A. baumannii lineage. Oxa-23 is therefore a targetable and accessible antigen located in the bacterial membrane. NDM-1, a metallo-β-lactamase, which correspondingly confers resistance against carbapenems, is anchored to the bacterial membrane by lipidation, conferring extended enzymatic activity via packaging in OMVs31,32. We speculate that Oxa-23 demonstrates enhanced enzymatic activity via an analogous mechanism and may be further accessible when exposed to carbapenems, highlighting a hypothetical synergistic interaction between an antimicrobial and a mAb that could be useful from a therapeutic standpoint. The identification of OCL-1 as an exposed surface antigen was unsurprising as CC2 A. baumannii generally synthesise this LOS subtype33. The third major mAb target we identified was the capsular target KL49. KL49 and two additional capsular subtypes, K2 and K58, were highly represented in the isolates used to generate our combined OMV immunogen. Notably, we only identified mAbs binding KL49, which may reflect our VH/VL sequencing approach, increased antigenicity of KL49 in comparison to other capsular subtypes, or a proportionally increased relative abundance of KL49 on OMVs produced by BAL 191. In the mAb binding experiments using live bacteria, mAbs targeting OCL1 and Oxa-23 produced reduced binding signals in comparison to the non-KL49 isolates, suggesting potential masking of other surface antigens. Recent studies have shown that acapsular A. baumannii are highly susceptible to serum or antimicrobial killing34,35. Similarly, specific A. baumannii K-subtypes, such as KL49, may act as a barrier preventing bactericidal activity of mAbs binding cell membrane-proximal targets such as LOS and Oxa-23.
Using an established mouse model for A. baumannii lung infection17 we found that pre-exposure prophylactic protection was not afforded by mAbs directed against the two cross-reactive targets, LOS and Oxa-23. Further studies could investigate different routes of infection to investigate the breadth of protection. Wider interrogation of the human B cell repertoire directed against these two targets using this approach may result in identification of additional mAbs that are capable of protection in vivo. We were able to demonstrate that mAb 1416 directed against KL49 had a dose dependent effect on reducing the number of bacteria in the lung post challenge when used in a pre-exposure prophylactic setting. Furthermore, and pertinent for the development of such antibodies against relevant pandemic antimicrobial resistant lineages, we were able to show pre-exposure prophylactic protection against two distinct clinical KL49 organisms isolated a decade apart in different ICUs in the same city. This suggests the epitope targeted mAb 1416 on KL49 has not varied in this timeframe due to immune pressure. Despite this, A. baumannii causing nosocomial infections produce many distinct capsule subtypes likely necessitating a cocktail of capsule-targeting mAbs to obtain sufficient strain coverage for successful therapeutic use. Several mAbs targeting capsular polysaccharides of A. baumannii have been developed including a bi-specific format and combinations of these show broader binding across A. bamannii isolates and show in vivo protection36,37. Production of KL49-typed capsule has been linked to individual clones of A. baumannii defined by sequence type (ST) with increased virulence in humans. An emerging endemic clone typed as ST457 associated with increased mortality identified in Southern China as well as LAC-4, typed as ST10, an outbreak strain from the USA, both produce KL49 capsule38,39. Therefore, mAbs targeting KL49 may have utility in outbreak scenarios or be added to cocktails of mAbs or bi-specific antibodies aiming for broad strain coverage.
The ultimate utility of the approach we present lies in the ability to generate functional antibodies from the human B cell repertoire across phylogenetically characterised diversity against clinically relevant A. baumannii at a single timepoint. Our approach is reinforced by the identification of a protective human mAb directed against the KL49 capsule, widely present on emerging A. baumannii clones in hospital settings, most notably in China and Southeast Asia, where the burden of disease is the greatest. Future work should expand the screening process beyond preclinical studies into small scale clinical testing to inform down selection for manufacture; combining this work with human challenge studies may accelerate clinical development. Expansion of this approach should combine genomic surveillance and organism characterisation with the identification of mAbs for bacterial pandemic preparedness as we rapidly enter the post-antimicrobial era.
Methods
All research performed complies with ethical regulations, receiving institutional approval for all study protocols. Ethical approval for the study contributing Acinetobacter isolates and metadata was provided by the Oxford Tropical Research Ethics Committee (OxTREC 35–16) and the Ethics Committee of Children’s Hospital 1 (CH1) (73/GCN/ BVND1). Written informed consent from a parent or guardian was a prerequisite for enrolment into the study. The study was registered with the International Standard Randomised Controlled Trial Number ISRCTN69124914. All research involving Kymouse platform mice was carried out under United Kingdom Home Office License 70/8718 with all procedures receiving approval of the Wellcome Trust Sanger Institute Animal Welfare and Ethical Review Body. Mouse studies carried out at Imperial college were performed in accordance with the United Kingdom’s Home Office guidelines under animal study protocol number P4EE85DED. All work was approved by the Animal Welfare and Ethical Review board at Imperial College London and followed NC3Rs guidelines. Mouse studies carried out at the University of Cambridge were performed under project licence P653704A5 with university approval by a local Animal Welfare and Ethical Review Board (AWERB).
Bacterial isolates
A. baumannii ATCC17978 and ATCC-BAA-1710 were purchased from the American Typed Culture Collection, USA. A. baumannii NCTC13302 and NCTC13424 were purchased from the National Collection of Typed Cultures, United Kingdom. Clinical A. baumannii isolates BAL 84, BAL 173, BAL 186, BAL 191, BAL 208, BAL 215, BAL 276, BAL 242, BAL 339 BAL 361 and BAL 377 were cultured from ICU patients in Ho Chi Minh City, Vietnam. CM10420 was isolated from the blood of a neonate with sepsis at Children’s hospital 1 in Ho Chi Minh City, Vietnam. All bacterial isolates were cultured in Lysogeny broth (LB) at 37 °C before use.
Generation and characterisation of outer membrane vesicles
200 ml of LB was inoculated with a single colony of A. baumannii and cultured overnight at 37 °C with shaking at 200 rpm. Supernatants were harvested by centrifugation at 3200 rpm for 30 min at 4 °C. Supernatants containing OMVs were filtered under vacuum through a 0.45 μm membrane. Filtered supernatants were then centrifuged at 100,000 × g in a Beckmann Optima L-100 ultracentrifuge for 2 h at 4 °C. OMV pellets were resuspended in 100 μl sterile Phosphate-buffered saline (PBS) and checked for sterility by plating an aliquot on non-selective LB-agar. OMV preparations were analysed for integrity by scanning electron microscopy and analysed for protein content using the Pierce BCA Protein Assay (Thermo Fisher Scientific, US). OMVs were analysed for lipid content by adding a 10 μl aliquot to a solution of FM4-64 (2.2 μg/ml in PBS; Thermo Fisher Scientific, US) and measuring fluorescence at Ex/Em 520 nm/700 nm with an Envision Plate Reader (Perkin Elmer, US).
Mouse immunisations
The genetic background of Kymouse platform mice is a randomised mixture of 129S7 and C57BL/6J strains. All mice were housed on a 12-h light/dark cycle at 20–24 °C with 55% ± 10% humidity. 6–12-week-old Male and female Kymouse platform mice were immunised via intraperitoneal route with 1 μg of pooled OMVs generated from A. baumannii BAL 084, BAL 191, BAL 215 and BAL 276 (0.25 μg of each OMV) in PBS and boosted with the same dose via intraperitoneal route 5 weeks later.
Indirect ELISA of live bacterial isolates and outer membrane vesicles
For analysis of live bacterial isolates, 100 μl bacteria in PBS (OD600nm of 1.0) was added to wells of a black 96 well sterile microplate (Greiner Bio-One) and incubated at room temperature overnight. Supernatants were removed and wells were blocked with 100 μl PBS containing 1% Bovine serum albumin (BSA). Blocking buffer was removed from wells, 50 μl of PBS containing 1 or 0.1 μg/ml monoclonal antibody was added to each well and plates were incubated at room temperature for 1 h. Plates were centrifuged at 900 × g for 2 min at 4 °C, supernatants were removed, and wells were washed with 100 μl PBS. 100 μl of PBS containing Europium-labelled anti-human IgG (1:500, DELFIA, Perkin Elmer, US) was added to each well and plates were incubated at room temperature for 1 h. Plate wells were washed twice with 100 μl PBS and 50 μl of enhancement solution (DELFIA, Perkin Elmer, US) was added to each well and optically clear plate seals (StarLab, UK) were added to plates. Fluorescence intensity at Ex/Em 320 nm/620 nm was then measured using an Envision Plate Reader (Perkin Elmer, US).
Isolation of specific OMV-binding B cells
Spleen and inguinal lymph nodes from immunised Kymouse platform mice were processed to single cell suspensions. Cells were centrifuged at 400 × g for 10 min at 4 °C and resuspended in FACS buffer (PBS, 1% FBS, 1 mM EDTA, 25 mM HEPES). Cells were stained for B220 (BUV395, BD Biosciences, Cat-563793, RA3-6b2, lot-7177756, 1/200), CD19 (BUV395, BD Horizon, Cat-563557, Clone-1D3, lot-8206694, 1/200), IgM (BV605, Biolegend, Cat-314526, Clone-MHM88, Lot-B268198, 1/100), IgG (BV421, BD Horizon, Cat-562581, Clone-X40, Lot-9079965, 1/40), mouse lambda (PE, Southern Biotech, Cat-1175-09L, Clone-JC5-1, Lot-G1714-VL87C, 1/200) and for CD8a (BV510, Biolegend, Cat-100752, Clone-53-6.7, Lot-B284151 1/500), CD4 (BV510, Biolegend, Cat-100449, Clone-GK1.5, Lot-B248587, 1/500), Ly-6G (BV510, Biolegend, Cat-127633, Clone RB6-8C5, Lot-B307304, 1/500), F4/80 (BV510, Biolegend, Cat-123135, Clone-BM8, Lot-B25669, 1/500) and CD11c (BV510, BD Biosciences, Cat-562949, Clone-HL3, Lot-8011606, 1/500). Cells were also stained with pooled OMVs generated from A. baumannii BAL 084, BAL 191, BAL 215 and BAL 276 stained with FM4-64. Cells were then subjected to FACS using a BD FACS Aria Fusion flow cytometer (Beckton Dickinson, US) with individual B220+ CD19+ IgG+ OMV-FM464+ cells transferred to individual wells of 96-well plates.
Bioinformatic selection of IgH/IgL chain pairs and mAb isolation
The variable heavy (VH) and variable light (VL) sequences of Ig genes were amplified by RT-PCR and Ilumina sequencing libraries were generated for sequencing using an Ilumina MiSeq. IgH/IgL pairs were selected from sequencing data by clustering analysis using Intellisellect™ software and selected VH and VL sequences were synthesised by Twist Biosciences (San Francisco, USA). Synthesised VH/VL gene fragments were cloned into a vector allowing incorporation in a human IgG1 backbone and expressed in HEK293 cells (Expi293™, Gibco, Thermo Fisher Scientific, US. Validation carried out by manufacturer) with culture supernatants collected and IgG1 quantified using an Octet RH16 (Sartorius, Germany). Antibodies that bound pooled OMVs, pooled live bacterial cells and triggered complement activation were re-expressed in CHO-3E7 cells (National Research Council, Canada NRC file 11992. Validation carried out by supplier) and purified using columns containing MabSelect SuRe LX resin (GE Healthcare, US) to allow use for subsequent assays.
C3b deposition assays
A flow cytometry-based assay to measure C3b deposition was developed based on a modified form of a previously published method40. For bead coating, AbC™ negative capture beads (Invitrogen, Thermo Fisher Scientific, US) were added dropwise into 120 μl of pooled FM464-stained OMVs in PBS (100 μg/ml) with the mixture incubated at room temperature for 10 min. HEK293 cell supernatants containing 100 μg/ml IgG1 construct, or human IgG1 isotype control (Sigma Aldrich, US) were added to 96-well plates and 20 μl of FM464-stained OMV beads were added to each well. Plates were incubated at 37 °C for 3 h. 100 μl of guinea pig complement protein (Cedarlane, Canada) diluted in gelatine veronal buffer (Boston Bioproducts, US) were added to wells of a V-bottomed plate. The beads were transferred to the V-bottom plate and incubated at 37 °C for 15 min and washed three times in PBS containing 1% BSA. C3b deposition on beads was then measured by adding 50 μl of FITC-labelled anti-Guinea pig complement C3 (Cat-855385, MP Biomedicals, US) diluted 1:100 in PBS containing 1% BSA, incubating at room temperature for 15 min, washing beads twice and finally resuspending in 150 μl, and measuring bead FITC signal by flow cytometry using an Attune Cytometer (Thermo Fisher Scientific, US).
For ELISA based C3b deposition on live bacterial cells, bacterial cells were coated on 96-well plates as described above. Plates were centrifuged and resuspended in 70 μl RPMI supplemented with 10% heat-inactivated foetal bovine serum (Thermo Fisher Scientific, US). Individual monoclonal antibodies were added to wells at a final concentration of 1 or 0.1 μg/ml and incubated at room temperature for 45 min. 20 μl normal human complement serum (Pel-Freeze) was added to each well and plates were incubated at 37 °C for 15 min with shaking (500 rpm). Bacteria were washed with ice-cold PBS, resuspended in 50 μl PE-conjugated anti-human C3 (Cat-CL7636PE, Cedarlane, Canada), diluted 1:1000 in PBS and incubated in the dark at 4 °C for 30 min. Bacteria were washed with ice-cold PBS and fluorescence intensity was measured at Ex/Em 496 nm/576 nm using a Varioskan LUX microplate reader (Thermo Fisher Scientific, US).
High-content antibody binding assays and Opera Phenix image analysis
A collection of CC2-typed carbapenem-resistant A. baumannii clinical isolates cultured from VAP and neonatal sepsis ICU patients in multiple Vietnamese hospitals23,27,28 was compiled by the Cambridge Institute of Therapeutic Immunology and Infectious Diseases as representative of bacterial diversity. Ethical approval for the studies contributing Acinetobacter isolates and metadata was provided by the Oxford Tropical Research Ethics Committee (OxTREC 35–16) and the Ethics Committee of Children’s Hospital 1 (CH1) (73/GCN/ BVND1). Written informed consent from a patient or parent or guardian of a minor was a prerequisite for enrolment into all studies where bacterial isolates were collected. For high-content screening of antibody binding and imaging analysis41 bacteria were added to CellCarrier Ultra 96 plates (Perkin Elmer. US) and left to adhere at 37 °C for 2 h. The supernatant was removed and adhered bacteria were fixed with 4% paraformaldehyde, then washed with PBS. Antibodies were diluted to 1 μg/ml in PBS + 1% BSA and added to the plate for 1 h at room temperature, after which it was replaced by 2 μg/ml Alexa Fluor 647 Goat Anti-Human IgG (Thermo Fisher Scientific, US) plus 2 μg/ml DAPI (Thermo Fisher Scientific, US) in PBS + 1% BSA for 30 min in the dark. The plate was washed once with PBS, then imaged on an Opera Phenix (Perkin Elmer, US) using the Alexa Flour 647 and DAPI channels and the 63x water immersion objective. Image analysis was carried out using the Perkin Elmer Harmony version 4.8. Individual bacteria were segmented using the DAPI staining, and the Alexa Fluor 647 intensity was measured for each bacterial cell. An Alexa Fluor 647 average intensity per well of >500 relative fluorescent units (RFU) was set as a positive binding threshold based on background and negative controls.
Opsonophagocytic assay and Opera Phenix image analysis
THP-1 cells (ATCC TIB-202™. Validation carried out by supplier) were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat inactivated foetal bovine serum (Gibco, Thermo Fisher Scientific, US). Cells were seeded on PhenoPlate 96-well plates (Perkin Elmer) at 2.5 × 105/ml, incubated at 37 °C for 48 h and stimulated for 24 h 10 ng/mL phorbol-12-myristate-13 acetate (PMA). Bacteria in PBS (OD600nm of 1.0) was diluted 1/200 in RPMI 1640 medium (Gibco) supplemented with 10% heat inactivated foetal bovine serum (Gibco, Thermo Fisher Scientific, US), 0, 2 or 5 μg/ml mAb 1416 added and incubated at 37 °C with shaking at 200 rpm for 1 h. Media in 96-well plates containing PMA-activated THP-1 cells was then replaced with a mixture of 100 μl of bacteria in media. 96-well plates were then incubated at 37 °C with 5% CO2 for 1 h. Cells were then washed with PBS, fixed with PBS containing 4% paraformaldehyde, washed again and permeabilised with PBS containing 0.1% Triton X-100 for 10 min. After washing, cells were blocked with 10% BSA and cell surface capsule of all internalised bacteria was detected with 2 μg/ml mAb 1416 followed by 2 μg/ml Alexa464-conjugated goat anti-human IgG (Thermo Fisher Scientific, US) in 1% BSA in the dark for 1 h. Cells were then washed and stained with 20 μg/ml DAPI (Sigma Aldrich, US) and 2 μg/ml CellMask Orange (Invitrogen, Thermo Fisher Scientific, US). Plates were imaged on an Opera Phenix (Perkin Elmer, US) using a 40x objective from three fields per well with four z-stacks. THP-1 cells were segmented using DAPI and CellMask Orange, and cells containing internalised bacteria were then enumerated using an Opera Phenix using the Alexa Fluor 647 and DAPI channels and the 40x objective from three fields per well with four z-stacks. Image analysis was carried out using the Perkin Elmer Harmony version 4.8.
Transmission electron microscopy
Samples were prepared by loading single bacterial colonies cultured on agar plates into planchettes. Planchettes were filled with 1-hexadecene and rapidly frozen using a BALTEC HPM010 high-pressure freezer. Samples were then freeze-substituted with acetone solution containing 0.1% tannic acid and 0.5% glutaraldehyde for 7 h. Samples were then treated in acetone containing 1% osmium tetroxide for 72 h, rinsed at room temperature and embedded in Lowicryl HM20 resin for ultrathin sectioning. Ultrathin sections were mounted on formvar-coated grids and labelled with mAb 1416 (20 μg/ml) for 1 h and Goat Anti-Human IgG H&L (12 nm Gold) secondary antibody (Abcam, UK) for 30 min. Ultrathin sections were then contrasted with uranyl acetate and lead citrate prior to imaging using a 120 kV FEI Spirit Biotwin TEM, equipped with a Tietz F4.16 CCD digital camera.
Construction of A. baumannii whole genome expression libraries
Bacteriophage Lambda ZAPII libraries (Agilent Technologies, US) containing random 1–2 kbp DNA fragments of BAL 084 and BAL 276 A. baumannii genomes were constructed and screened by expression cloning according to manufacturer’s instructions and as previously described42,43. Nitrocellulose membrane lifts from plated library phage plaques were probed with individual monoclonal antibodies at 1 μg/ml and bound antibody detected using alkaline phosphatase conjugated Anti-Human IgG1 Hinge (SouthernBiotech, US) diluted 1:2500 in PBS-Tween20: PBS supplemented with 0.1% (v/v) Tween 20 (Sigma Aldrich, US). After washing three times with PBS-Tween20, membranes were developed using Western Blue Stabilised Substrate for Alkaline Phosphatase (Promega, UK).
Immunoblotting of bacterial whole cell lysates
100 μl of overnight bacterial cultures normalised to an OD600nm of 1.0 were centrifuged at 900 × g for 1 min and bacterial pellets were resuspended in 20 μl of Pierce™ MagicMark™ XP reducing sample buffer (Thermo Fisher Scientific, US). Samples were boiled at 95 °C for 5 min and cooled on ice. Samples were loaded onto a Mini-PROTEAN TGX gel (Bio-Rad Laboratories, US) alongside Precision Plus Protein Dual Colour Standards (Bio-Rad Laboratories, US). Gels were then resolved at 100 V for 45 min. Protein from resolved gels was blotted onto nitrocellulose membranes using a Trans-Blot Turbo Mini Nitrocellulose Transfer Pack (Bio-Rad Laboratories, US). Nitrocellulose membranes were blocked for 1 h at room temperature in PBS-Tween20 containing 5% (w/v) non-fat dried milk powder. Membranes were incubated in PBS-Tween20, 5% (w/v) non-fat dried milk containing 1 μg/ml monoclonal antibody for 1 h at room temperature, washed with PBS-Tween20 and developed by incubation with 1:2000 goat anti-human IgG conjugated to HRP (Invitrogen, Thermo Fisher Scientific, US) for 1 h at room temperature, followed by incubation in ECL prime western blotting reagent for 5 min in the dark. Blots were visualised and imaged using a ChemiDoc Imaging System (Bio-Rad Laboratories, US).
Generation of bacterial non-proteinaceous carbohydrate extracts
Bacterial pellets from 1 ml of normalised overnight bacterial culture were resuspended in 200 μl Tris-HCl (pH 8) containing 2% (w/v) SDS and boiled for 5 min. 800 μl Tris-HCl (pH 8) containing 300 U/ml Benzonase nuclease (Millipore) was added and incubated at 37 °C for 1 h. Proteinase K (Qiagen) was then added to a final concentration of 500 μg/ml and incubated at 55 °C for 4 h. Samples were run on a Mini-PROTEAN TGX gel alongside Precision Plus Protein Dual Colour Standards. Gels were washed 3 times with distilled water and removal of protein was confirmed by Imperial protein stain (Thermo Fisher Scientific, US). Capsule polysaccharide was visualised by staining gels with Alcian Blue solution (Sigma-Aldrich, US) diluted 1:7 in 3% acetic acid for 1 h and washed in distilled water overnight. To detect lipo-oligosaccharide (LOS), gels were fixed with 50% methanol, 5% acetic acid for 30 min and treated using a Pro-Q Emerald 300 Lipopolysaccharide Gel Stain kit according to manufacturer’s instructions. Stained gels were visualised and imaged using a ChemiDoc Imaging System (Bio-Rad Laboratories, US). Resolved carbohydrate extracts were blotted onto nitrocellulose membranes and probed for monoclonal antibody binding as described above.
Murine challenge studies
Murine challenge studies carried out at Imperial college used 6–12-week-old male and female BALB/c mice. Murine challenge studies carried out at the University of Cambridge used 6–12-week-old male and female C57BL6 mice. All mice were housed on a 12-h light/dark cycle at 20–24 °C with 55% ± 10% humidity. Mice were intravenously injected with either 1, 5 or 10 mg/kg of monoclonal antibodies 1042, 1349 or 1416 on day −1 and day 0 before being challenged intranasally with BAL 191 or CM10420 on day 0. For intranasal administration of monoclonal antibody, mice were given 10 mg/kg 1416 intranasally on day −1 and day 0 prior to intranasal challenge with BAL 191 on day 0.
Overnight cultures of BAL 191 or CM10420 were diluted 1:100 in 20 ml LB and grown at 37 °C with shaking at 200 rpm to an OD600 of 0.7. Bacterial pellets from these cultures were washed in PBS and resuspended in PBS to generate an inoculum of 5 × 108 CFU/ml. Mice were anaesthetised via inhalation of vaporised isoflurane and challenged intranasally with 100 µl of this inoculum. Mice were culled 24 h later via intraperitoneal injection with pentobarbital and nasal lavage fluid, lung and spleen homogenate were then serially diluted 1:10 and plated on to HiCrome Acinetobacter agar plates with MDR supplement. Plates were incubated at 37 °C overnight and individual colonies were enumerated to determine the bacterial load as CFU/ml for each organ.
Statistical analyses were performed using Graph Pad Prism V9.3.1. The Shapiro-Wilk test for normality was performed to determine normal distribution before performing unpaired t tests or Mann–Whitney tests to determine significant differences between the 1042, 1349 or 1416 monoclonal-treated mice and antibody isotype control-treated mice.
Statistics and reproducibility
For all in vitro studies a minimum sample size of n = 4 was employed in all cases. Individual experiments were repeated three times to assess reproducibility and an individual representative experiment was presented. Statistically significant difference between multiple groups was determined using Dunnett’s one-way ANOVA. Comparison of individual groups for statistically significant differences was performed using a Mann–Whitney test. For all in vivo mouse studies, study size was defined in each institutional animal according to power calculations for delivery of a conclusive outcome (n = 5). For mouse studies, the Shapiro-Wilk test for normality was performed to determine normal distribution before performing unpaired t tests or Mann–Whitney tests to determine significant differences between individual groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by grants from the Bill & Melinda Gates Foundation, OPP1159947 (P.K.) and INV-040928 (S.T.R.), the UK Medical Research Council Newton Fund (M.E.T.), the Viet Nam Ministry of Science and Technology (M.E.T.), and Wellcome Senior Research Fellowship 215515/Z/19/Z (S.B.). Additional support was provided by the Academy of Medical Sciences (M.E.T.), the Health Foundation (M.E.T.), and the National Institute for Health Research Cambridge Biomedical Research Centre (M.E.T. and G.D.). The funders did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualisation-S.T.R., S.B., P.K., Data curation-A.K., S.H., D.G., K.H., S.O’.L., J.B.S., P.N., I.A., E.A., S.C., S.F. Formal analysis-S.T.R., S.B., A.K., P.N., J.B.S., J.S.T., S.O’.L., S.H., S.W., E.A., S.C., S.F. Methodology-T.N.T.N., N.D.T., L.R., B.S., N.F.O., D.M.S., C.P., Resources-P.K., G.D., H.C., J.S.T., V.W., C.P., M.E.T., Visualisation-S.W., Q.Z., L.R., Writing- Original draft-S.T.R. and S.B., Writing-Review and editing-JST and JBS with input from all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All VH/VL sequence data generated in this study are provided in the file: Source data_Manuscript Figs under Figs. 2a and b. All VH/VL paired sequences used to synthesize monoclonal antibodies and using the same unique identifiers in the manuscript are available via the published patent filing WO2023094628A1. Source data are provided with this paper.
Competing interests
The authors declare the following competing interests. J.B.S., Q.Z., and S.T.R. are employees of Kymab Ltd, a Sanofi company and may have held or continue to hold stock options or shares in Sanofi. S.O’.L., J.B.S., A.K., S.W., S.T.R., and S.B. are listed as inventors on a published patent WO2023094628A1 covering monoclonal antibodies generated by this work. S.O’.L., A.K., I.A., B.S., H.C., V.W., N.F.O., D.M.S., C.P., S.W., and P.K. were employees of Kymab, a Sanofi Company within the last three years and may have held or continue to hold stock options or shares in Sanofi. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Stephen Baker, Aishwarya Krishna, Sophie Higham.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52357-8.
References
- 1.Ibrahim, S., Al-Saryi, N., Al-Kadmy, I. M. S. & Aziz, S. N. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep.48, 6987–6998 (2021). 10.1007/s11033-021-06690-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dijkshoorn, L., Nemec, A. & Seifert, H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol.5, 939–951 (2007). 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 3.Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet399, 629–655 (2022). 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong, D. et al. Clinical and pathophysiological overview of acinetobacter infections: a century of challenges. Clin. Microbiol. Rev.30, 409–447 (2017). 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg, A. Y., Seifert, H. & Paterson, D. L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev.21, 538–582 (2008). [DOI] [PMC free article] [PubMed]
- 6.Doi, Y., Husain, S., Potoski, B. A., McCurry, K. R. & Paterson, D. L. Extensively Drug-Resistant Acinetobacter baumannii.Emerg. Infect. Dis.15, 980–982 (2009). 10.3201/eid1506.081006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemos, E. V. et al. Carbapenem resistance and mortality in patients with Acinetobacter baumannii infection: systematic review and meta-analysis. Clin. Microbiol. Infect.20, 416–423 (2014). 10.1111/1469-0691.12363 [DOI] [PubMed] [Google Scholar]
- 8.Jesudason, T. WHO publishes updated list of bacterial priority pathogens. Lancet Microbe 100940. 10.1016/j.lanmic.2024.07.003 (2024). [DOI] [PubMed]
- 9.Ambrose, N. et al. Neutralizing monoclonal antibody use and COVID-19 infection outcomes. Jama Netw. Open6, e239694 (2023). 10.1001/jamanetworkopen.2023.9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Micoli, F., Bagnoli, F., Rappuoli, R. & Serruto, D. The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol.19, 287–302 (2021). 10.1038/s41579-020-00506-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson, E. et al. Characterisation of the immune repertoire of a humanised transgenic mouse through immunophenotyping and high-throughput sequencing. Elife12, e81629 (2023). 10.7554/eLife.81629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen, T. B. et al. Monoclonal antibody therapy against acinetobacter baumannii. Infect. Immun.89, e00162–21 (2021). 10.1128/IAI.00162-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker, S., Kellam, P., Krishna, A. & Reece, S. Protecting intubated patients from the threat of antimicrobial resistant infections with monoclonal antibodies. Lancet Microbe1, e191–e192 (2020). 10.1016/S2666-5247(20)30126-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sartorio, M. G., Pardue, E. J., Feldman, M. F. & Haurat, M. F. Bacterial outer membrane vesicles: from discovery to applications. Annu. Rev. Microbiol.75, 1–22 (2021). 10.1146/annurev-micro-052821-031444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwechheimer, C. & Kuehn, M. J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol.13, 605–619 (2015). 10.1038/nrmicro3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villageliu, D. N. & Samuelson, D. R. The role of bacterial membrane vesicles in human health and disease. Front. Microbiol.13, 828704 (2022). 10.3389/fmicb.2022.828704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higham, S. L. et al. Intranasal immunisation with Outer Membrane Vesicles (OMV) protects against airway colonisation and systemic infection with Acinetobacter baumannii 1 1 Current addresses: KF UCL. J. Infecti.10.1016/j.jinf.2023.02.035 (2023). [DOI] [PubMed]
- 18.Lee, E.-C. et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol.32, 356–363 (2014). 10.1038/nbt.2825 [DOI] [PubMed] [Google Scholar]
- 19.Zhu, Antenucci, Zhuang, Villumsen, Fabio, Bojesen, Kasper Rømer & Anders, Miki Bacterial outer membrane vesicles as a versatile tool in vaccine research and the fight against antimicrobial resistance. mBio12, e0170721 (2021). 10.1128/mBio.01707-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balhuizen, M. D., Veldhuizen, E. J. A. & Haagsman, H. P. Outer membrane vesicle induction and isolation for vaccine development. Front. Microbiol.12, 629090 (2021). 10.3389/fmicb.2021.629090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jan, A. T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: a perspective update. Front. Microbiol.8, 1053 (2017). 10.3389/fmicb.2017.01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls, A. C. et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell183, 1367–1382.e17 (2020). 10.1016/j.cell.2020.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz et al. Repeated local emergence of carbapenem-resistant Acinetobacter baumannii in a single hospital ward. Micro. Genom.2, e000050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karah, N., Sundsfjord, A., Towner, K. & Samuelsen, Ø. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Update15, 237–247 (2012). 10.1016/j.drup.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 25.Wand et al. Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J. Med. Microbiol.61, 470–477 (2012). 10.1099/jmm.0.037523-0 [DOI] [PubMed] [Google Scholar]
- 26.Zarantonello, A., Revel, M., Grunenwald, A. & Roumenina, L. T. C3‐dependent effector functions of complement. Immunol. Rev.313, 120–138 (2023). 10.1111/imr.13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toan, N. D. et al. Clinical and laboratory factors associated with neonatal sepsis mortality at a major Vietnamese children’s hospital. Plos Glob. Public Health2, e0000875 (2022). 10.1371/journal.pgph.0000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, L. W. et al. Genomic characterisation of multidrug-resistant Escherichia coli, Klebsiella pneumoniae, and acinetobacter baumannii in two intensive care units in Hanoi, Viet Nam: a prospective observational cohort study. Lancet Microb.3, e857–e866 (2022). 10.1016/S2666-5247(22)00181-1 [DOI] [PubMed] [Google Scholar]
- 29.Antunes, N. T. et al. Class D β-Lactamases: are they all carbapenemases? Antimicrob. Agents Chemother.58, 2119–2125 (2014). 10.1128/AAC.02522-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenyon, Nigro., Johanna, J., Hall, Steven, J. & Ruth, M. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS ONE9, e107833 (2014). 10.1371/journal.pone.0107833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez, M. M. B. et al. On the offensive: the role of outer membrane vesicles in the successful dissemination of new delhi Metallo-β-lactamase (NDM-1). mBio12, e0183621 (2021). 10.1128/mBio.01836-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González, L. J. et al. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol.12, 516–522 (2016). 10.1038/nchembio.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyres, K. L. et al. Identification of acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom.6, e000339 (2020). [DOI] [PMC free article] [PubMed]
- 34.Altamirano, G. et al. Phage-antibiotic combination is a superior treatment against Acinetobacter baumannii in a preclinical study. EBioMedicine80, 104045 (2022). 10.1016/j.ebiom.2022.104045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdelkader, k. et al. The specific capsule depolymerase of phage PMK34 sensitizes acinetobacter baumannii to Serum Killing. Antibiotics11, 677 (2022). 10.3390/antibiotics11050677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen, T. B. et al. Development of a bispecific antibody targeting clinical isolates of acinetobacter baumannii. J. Infect. Dis.227, 1042–1049 (2023). 10.1093/infdis/jiac499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slarve, M. et al. Therapeutic, humanized monoclonal antibody exhibits broad binding and protective efficacy against acinetobacter baumannii. Antimicrob. Agents Chemother.67, e00086–23 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou, H.-Y. et al. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci. Rep.5, 8643 (2015). 10.1038/srep08643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, K. et al. An Emerging Clone (ST457) of acinetobacter baumannii Clonal Complex 92 with enhanced virulence and increasing endemicity in South China. Clin. Infect. Dis.67, S179–S188 (2018). 10.1093/cid/ciy691 [DOI] [PubMed] [Google Scholar]
- 40.Fischinger, S. et al. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods473, 112630 (2019). 10.1016/j.jim.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maes, M. et al. A novel therapeutic antibody screening method using bacterial high-content imaging reveals functional antibody binding phenotypes of Escherichia coli ST131. Sci. Rep.10, 12414 (2020). 10.1038/s41598-020-69300-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reece, S. T. et al. ML0405 and ML2331 are antigens of Mycobacterium leprae with potential for diagnosis of leprosy. Clin. Vaccin. Immunol.13, 333–340 (2006). 10.1128/CVI.13.3.333-340.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lodes, D. et al. A. Expression cloning. Methods Mol. Med.94, 91–106 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All VH/VL sequence data generated in this study are provided in the file: Source data_Manuscript Figs under Figs. 2a and b. All VH/VL paired sequences used to synthesize monoclonal antibodies and using the same unique identifiers in the manuscript are available via the published patent filing WO2023094628A1. Source data are provided with this paper.