Abstract
Cystic fibrosis (CF) is a genetic disorder characterized by chronic microbial colonization and inflammation of the respiratory tract (RT), leading to pulmonary exacerbation (PEx) and lung damage. Although the lung bacterial microbiota has been extensively studied, the mycobiome remains understudied. However, its importance as a contributor to CF pathophysiology has been highlighted. The objective of this review is to provide an overview of the current state of knowledge regarding the mycobiome, as described through NGS-based studies, in patients with CF (pwCF).
Several studies have demonstrated that the mycobiome in CF lungs is a dynamic entity, exhibiting a lower diversity and abundance than the bacterial microbiome. Nevertheless, the progression of lung damage is associated with a decrease in fungal and bacterial diversity. The core mycobiome of the RT in pwCFs is mainly composed of yeasts (Candida spp., Malassezia spp.) and molds with lower abundance. Some fungi (Aspergillus, Scedosporium/Pseudallescheria) have been demonstrated to play a role in PEx, while the involvement of others (Candida, Pneumocystis) remains uncertain. The “climax attack” ecological model has been proposed to explain the complexity and interplay of microbial populations in the RT, leading to PEx and lung damage. NGS-based studies also enable the detection of intra- and interkingdom correlations between fungi and bacteria. Further studies are required to ascertain the biological and pathophysiological relevance of these correlations. Finally, with the recent advent of CFTR modulators, our understanding of the pulmonary microbiome and mycobiome in pwCFs is about to change.
Keywords: Cystic fibrosis mycobiome, Next-generation sequencing, Fungi
Introduction
Cystic fibrosis (CF) is one of the most prevalent genetic disorders associated with chronic airway colonization and infections, as well as reduced life expectancy, particularly in Europe, North America and Australia [1, 2]. The reduced mucociliary clearance and inflammation of the airways facilitate the chronic microbial colonization of the respiratory tract (RT) by microorganisms that have adapted to this specific microenvironment [3]. Bacteria, and to a lesser extent, fungi, contribute to recurrent pulmonary exacerbations (PExs), which require anti-infective treatments and result in a progressive decline in respiratory function [1, 4, 5]. For many years, the study of the mycobiota has been based on culture-based studies with a particular focus on the fungi involved in PEx. Aspergillus and Pseudallescheria/Scedosporium have been demonstrated to be the most frequently encountered molds in PEx, typically manifesting as the disease progresses [2, 6]. Rare fungi, including Exophiala dermatitidis, Lomentospora prolificans and Rasamsonia argillacae, have also been linked to PEx in patients with CF (pwCFs) [7]. Candida yeasts and related genera are frequently isolated from oral and respiratory samples of pwCFs, and they are considered to be colonizers [2, 6–8]. Nevertheless, some authors have observed an increased prevalence of PEx and a decline in lung function in patients with persistent Candida colonization. The role of these yeasts remains unclear to date [7, 9–11]. In recent years, culture-independent methods based on next-generation sequencing (NGS), such as metagenomics, have significantly advanced our understanding of the microbiome. It has enabled the identification of species that cannot be cultured or that are present in low abundance, thereby facilitating a more accurate description of fungal (but also bacterial) diversity and the potential correlations between microorganisms within or between kingdoms [12–15]. The principal techniques used are targeted-amplicon NGS (TA-NGS) and shotgun metagenomics (SMg). Both methods have inherent limitations in regard to studying the mycobiome, and it is important to be aware of these limitations when analyzing published studies. The airway mycobiome in pwCFs represents a minor fraction, in terms of quantity and diversity, of the total airway microbiome. Consequently, it is not uncommon for these techniques to underdetect the presence of fungi.
The objective of this review is to describe the current state of knowledge regarding fungal colonization and infection in CF by linking fungal prevalence data obtained from culture-based studies with those obtained from metagenomic analyses. By integrating these two methodologies, we can enhance our understanding of the role of fungi in CF pathogenesis.
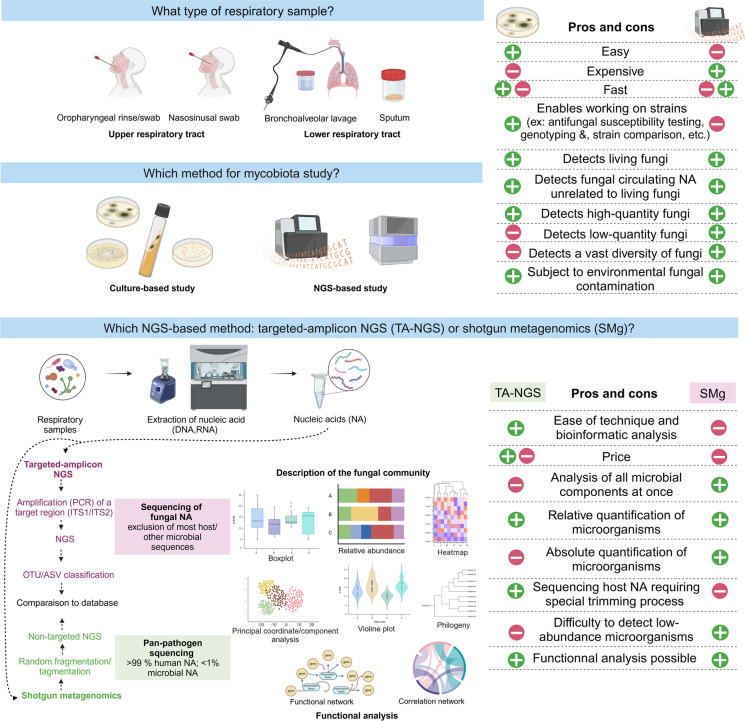
Tools for Metagenomic Studies of the Respiratory Mycobiome
Reference methods used to study the mycobiome in CF or other diseases include targeted-amplicon NGS (TA-NGS) and more recently, shotgun metagenomics (SMg, also called global metagenomics) [16] (Table 1, Fig. 1). These techniques can overcome the biases associated with fungal culture, including fungi that do not grow on standard media, fungal loads that are too low to be detected in culture, and so forth. Consequently, they yield more comprehensive insights into fungal diversity [12, 13, 17, 18]. Nevertheless, certain challenges in investigating the RT mycobiome through NGS-based approaches have been identified. Firstly, the mycobiome represents a small proportion of the total human microbiome. Estimates suggest that it constitutes less than 1% of the microbial metagenome, which itself represents less than 1% of the total host–microbe metagenome [19]. Furthermore, the extraction of fungal nucleic acids is challenging due to the protection of fungal cells by a thick wall composed of complex polysaccharides (chitin, ß-glucan polymers, etc.) and sometimes melanin. It has been demonstrated that significant biases in the diversity of the mycobiome can be observed depending on the extraction technique used [2–4]. In order to obtain a good extraction yield, different lysis methods must be combined including mechanical lysis, which is an essential step and is carried out using beads of different sizes, chemical lysis and enzymatic lysis [21]. When analyzing the mycobiome in pwCFs, it is also important to consider the type of respiratory specimen used (sputum or bronchoalveolar lavage) and its potential dilution, which could affect the detection of the mycobiome. To date, the most commonly used method in studies of the CF RT mycobiome is TA-NGS [16, 23]. TA-NGS usually targets the internal transcribed spacer (ITS) region, which consists of two subregions (ITS1 and ITS2), each 400 bp long (Table 1). The profile of fungal communities may vary depending on the subregion or the primers chosen [24, 25]. The relative abundance of taxa may also be distorted due to the variable number of copies of the ITS region in fungi [26]. SMg, as a non-targeted approach that allows random sequencing of all types of nucleic acids from hosts and microorganisms, can be used to study all components of a microbiome simultaneously including fungi. It also has the advantage of quantifying the abundance of microorganisms in absolute (rather than relative) terms. However, due to the low quantity of fungal sequences compared to those of the host and other microorganisms (the rare biosphere), the depth of sequencing represents a significant challenge in accurately detecting the mycobiome using SMg [19, 27, 28]. Furthermore, fungal genomes exhibit similarities with those of the host, and some fungal sequences are eliminated during bioinformatic analysis. This may result in an underestimation of the mycobiome. In their study, Pienkowska et al. found that only 0.1% of sequences obtained from RT samples of pwCF were assigned to fungi using SMg [28]. To enhance the sensitivity of SMg for fungal detection, enzymatic depletion (DNase/RNase) of human nucleic acids prior to sequencing can be performed, as can an increase of the depth of sequencing and the combination of DNA and RNA detection [27]. To date, SMg has been employed primarily for clinical metagenomics (panpathogen research from a given sample [27, 29]) or to study the bacterial or viral microbiome. A limited number of studies have been published on the RT mycobiome in pwCFs using SMg (n = 9 original articles listed in PubMed NCBI on March 24, 2024). Regardless of the NGS-based approach employed, the accurate taxonomic annotation of fungal sequences remains a challenge [30]. Fungal taxonomy is evolving, and many fungi have multiple names, which may change rapidly. Fungal databases are also far from complete (whether they are sequence or complete genome databases). Furthermore, the use of TA-NGS or SMg necessitates the use of environmental controls (DNA/RNA-free water) in order to detect fungal contamination [16, 27].
Table 1.
Definitions and abbreviations related to NGS-based methods, such as metagenomic analysis, for studying the mycobiome
| ASV | Amplicon Sequence Variant: individual DNA sequences produced by targeted NGS after elimination of poor-quality sequences and use of a sequencing error correction algorithm (e.g., DADA2). All ASV are then assigned to a taxonomic identification. ASVs are an alternative to OTUs |
| Alpha-diversity | An estimate of the diversity of a given sample taking into account richness and evenness. Simpson and Shannon indexes are common alpha-diversity indexes |
| Beta-diversity | An estimate of the difference in community composition between different samples (e.g., two anatomical sites, two individuals, etc.). It can be evaluated through principal component or coordinate analysis (PCA/PCoA) |
| Core mycobiome | Taxa, genera or species present in the majority (generally > 50%) of individuals in the same group with a relative abundance > 0.1% |
| Fungal dysbiosis | Imbalance in the composition of the mycobiota compared with the steady state (eubiosis) detected in healthy subjects |
| Evenness | Evaluation of the distribution of species/taxa/OTUs/ASVs present in a sample, taking into account the relative abundance of the different groups |
| Shannon or Simpson diversity indexes | Indexes measuring the alpha-diversity of a community's composition, taking into account both richness and evenness |
| OTU | Operational Taxonomic Unit: cluster of sequences obtained from targeted-amplicon NGS using a pipeline (e.g., VSEARCH) that clusters sequences with a high percentage of similarity (> 97%). The OTUs are then assigned to a taxonomic identification (or taxon) |
| Richness | Evaluation of the number of species, taxa, OTUs or ASVs present in a sample, without taking into account the relative abundance between the different groups |
| Shotgun metagenomics (SMg) | Global metagenomics: study of all components of microbiome (bacterial, viral, fungal and other eukaryotes) at once using a nontargeted approach |
Fig. 1.
Methods used to study the airway mycobiome in CF patients. Figure adapted from Thornton et al. J Pediatric Infect Dis Soc. 2022 [15] and created with BioRender.com. Abbreviations: ASV amplicon sequence variant; NGS next-generation sequencing; OTU operational taxonomic unit; TA-NGS targeted-amplicon NGS; SMg shotgun metagenomics
It is also important to note that, while metagenomic methods are valuable for studying the diversity of the respiratory mycobiome in pwCF in depth, they are not a replacement for culture-based methods. Indeed, culture-based studies have the advantage of readily identifying the predominant fungal agents in the respiratory tract, distinguishing living fungi from circulating DNA, and facilitating more comprehensive investigations of fungal strains (determination of susceptibility to antifungal agents, search for resistance genes or virulence factors, genotyping for strain comparison, etc.). The culture-based and culture-independent method are therefore ideally suited to complementary use.
Basics of Mycobiota in the Respiratory Tract of PwCFs
A Specific Microenvironment that Favors Colonization by Adapted Microorganisms
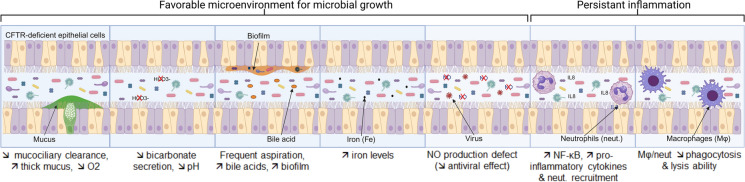
In the airways of pwCFs, dysfunction of the CF transmembrane conductance regulator (CFTR) channel leads to reduced fluidification of mucus on epithelial cells (Fig. 2) [3, 31–33]. A thicker and more viscous mucus leads to an environment enriched with nutrients (such as glycoproteins), a reduction in airway oxygenation (hypoxia), and a defect in mucociliary clearance, which favors the growth of some microorganisms (such as Pseudomonas aeruginosa or anaerobes) [34, 35]. Other specific features of the CF airway microenvironment can provide a breeding ground for chronic colonization with adapted microorganisms. These include a reduction in pH due to a lack of bicarbonates, an increase in bile acid levels due to frequent aspirations that favors biofilm formation, an elevation in iron levels that favors microbial growth, a deficiency in NO production by CFTR-epithelial cells, and so forth (Fig. 2). In addition, pwCFs may exhibit persistent inflammation due to constitutive activation of the NF-kB pathway in CFTR-epithelial cells, which is associated with an excessive innate immune response against chronic colonizing microorganisms. Consequently, the increased production of proinflammatory cytokines and neutrophil recruitment, elevated oxidative stress, impaired macrophage phagocytosis, and increased lysis ability impede the effective microbial clearance, yet exacerbate tissue damage (Fig. 2) [3, 31–33].
Fig. 2.
Airway-specific microenvironment in patients with CF. The different specificities of the CF airway microenvironment favor chronic colonization with adapted microorganisms, allowing possible intra- and interkingdom interactions (through biofilm formation and molecular interactions) and leading to repeated pulmonary exacerbations (PEx). Its association with persistent inflammation leads to lung damage and respiratory failure. Abbreviations: Mφ: macrophage; Neut: neutrophil.
Adapted from Ubags and Marsland, Eur Respir J. 2017 [31]. The figure was created with BioRender.com
Knowledge From Culture-Based Studies Regarding Fungi in the Airways of PwCFs
A substantial body of data regarding the prevalence of colonization or infection by fungi in pwCFs has been collected through culture studies. These data have been collected on a large scale, involving national and international cohorts, such as the European CF Society (ECFS, https://www.ecfs.eu/ecfspr), the French national cohort (“Vaincre la Mucoviscidose” project [2]), the United States CF Foundation Patient Registry [36], and so forth. Candida albicans and Aspergillus fumigatus have been identified as the most prevalent fungi detected in the RT of pwCFs, with prevalence varying with age [2, 36, 37]. In France, in 2022, C. albicans was the most prevalent fungus in the RT of pwCF, with a prevalence of 10–15% in children under the age of 5 and approximately 30% in adolescents and adults (> 15 years) [2]. A. fumigatus was the second most common fungus, with a prevalence of less than 5% in children under 5 years of age, which increased to approximately 15% after 10 years of age [2]. The prevalence of these fungi in the CF population as a whole also varies according to country and center, geographical or environmental criteria [9, 37]. In Europe, during the 2012–2016 period, the prevalence of A. fumigatus was 4% in the Czech Republic, 13% in Poland, 29% in Italy and 42% in Austria [38]. More recently (2019), a Dutch study reported an A. fumigatus prevalence of 56% [39]. The overall prevalence of C. albicans varies from 30% to > 70% in European countries [37, 38]. Other rarer fungi with variable prevalence are found in the RT of pwCFs. The prevalence of the black yeast Exophiala dermatitidis varies from < 1% in the United Kingdom, Italy or the Czech Republic to 15–20% in Sweden [37, 38, 40]. The species from the Scedosporium/Pseudallescheria complex represent the second most common mold in the RT of pwCF (prevalence of 2–17%). Species of the Rasamsonia argillacea complex or Lomentospora prolificans are less prevalent (< 5% for both) [37, 38, 40, 41]. Some authors have also studied the variations in fungal prevalence in the RT of pwCFs according to seasonal or climatic changes [42, 43], as has been demonstrated for CF bacteria such as P. aeruginosa [44]. Furthermore, the prevalence of fungi has been observed to vary depending on the period under study, with a tendency towards a reduction in fungal prevalence over time. For instance, the mean prevalence of A. fumigatus in France was 25% in 2015, 22% in 2021 and 12% in 2022 [2, 45]. These changes may be partially attributed to the novel use of CFTR modulators in the treatment of CF [46, 47].
Role of Yeasts and Filamentous Fungi in CF Lung Mycobiome in the Metagenomic Era
As demonstrated by several authors, the NGS-based approach allows for a more comprehensive description of the mycobiota diversity in pwCFs and enables the detection of fungal colonization at lower RT with greater sensitivity [13, 17]. In a study by Botterel et al., it was observed that the median number of fungal genera detected using TA-NGS in the sputa of pwCFs was 8, while it was 2 according to culture [13]. Delhaes et al. demonstrated that 60% of fungal taxa detected by NGS were undetected in culture [12]. Cuthberston et al. demonstrated that the NGS-based approach allowed for the detection of four major CF fungi (Candida, Aspergillus, Scedosporium and Exophiala) with a prevalence 3–24 times higher than that detected by culture. It is, however, important to note that the positivity cutoff for fungal sequences in this study was particularly low since a sample with only one sequence was considered positive [17].
Diversity, Fungal Burden and Evolution of the Lung Mycobiome in PwCFs Through Age, PEx and Disease Progression
The fungal load and diversity of the RT mycobiome in pwCFs are significantly lower than those of the bacterial mycobiome [13, 14]. However, the total fungal load in pwCFs is higher than that of healthy controls, regardless of age [48]. Additionally, the fungal burden also appears to increase with age, while its diversity decreases with disease progression, as has been previously observed for the bacterial microbiota [12, 13, 15, 48–50]. This reduction in diversity can be observed from the first months after diagnosis (i.e., from birth for some pwCFs) and is correlated with the decline of the respiratory function, which is evaluated through spirometry parameters, forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), and/or the CF-specific questionnaire (CFQ-R [51]) [12, 22, 48–50]. NGS-based studies have also demonstrated the occurrence of a fungal dysbiosis in the lungs of pwCFs compared to controls [13, 17]. Even when compared with patients who have bronchiectasis due to other causes, the diversity of the mycobiome appears to be different, with reduced fungal alpha-diversity in pwCFs [17]. Some authors have also studied the composition and diversity of the mycobiome in pwCFs during the PEx [15, 52]. Hong et al. observed that the global composition of the fungal community differed between pwCF in the stable state and PEx (beta-diversity, Table 1) [52]. Soret et al. analyzed the median alpha-diversity scores of fungal communities during the PEx or in a stable state and demonstrated a decreasing trend in diversity in patients during the PEx [15].
Core Mycobiome in the Respiratory Tract of PwCFs
The definition of the core microbiome may vary depending on the author [53, 54]. However, the fundamental principle is to identify a set of constant and/or stable microbial components within a population, in contrast to the microorganisms of transient flora, which vary according to environmental, individual or exogenous conditions [53]. The core mycobiome is defined as a group of fungal taxa that are shared by most individuals (generally > 50%) and have a relative abundance greater than 0.1%. The initial studies on the core mycobiome were relatively recent and focused primarily on the digestive core mycobiome. Only a few studies have been published regarding the respiratory core mycobiome, whether in healthy subjects or pathological conditions (such as asthma, bronchiectasis, CF, lung cancer, etc.). Moreover, given the variability of the mycobiome across ecological niches, it is of great important to identify the specific site under investigation.
NGS-based studies have shown that the core mycobiome of the lower RT in pwCF is predominantly composed of yeasts belonging to the Saccharomycotina subphylum, including Candida, Debaryomyces, Pichia, and Saccharomyces. These yeasts are frequently associated with Malassezia spp. [15, 17, 48, 52, 55]. Among the Candida species, C. albicans is the most prevalent, followed by Candida dubliniensis and Candida parapsilosis, as also described in culture studies [52, 56]. This core mycobiome is comparable to that observed in the lower RT [57, 58] and the gastrointestinal tract of healthy subjects [24, 59]. Some molds are also detected in the majority of pwCFs (in particular, Aspergillus, Cladosporium, Penicillium) but at lower relative abundances than yeasts [14, 17, 58, 60]. This result differs from what is known from culture, where A. fumigatus has the second highest fungal prevalence after C. albicans [2, 37, 38, 40]. NGS-based studies have revealed a specific picture of the core mycobiota of the RT of pwCF, with an even greater prevalence and diversity of Saccharomycotina yeasts and a much lower abundance of Aspergillus. This may appear counterintuitive, given the established role of Aspergillus in PEx and allergic bronchopulmonary aspergillosis (ABPA) in pwCFs [2, 37, 38, 61]. Other fungi frequently described in pwCFs by culture, such as Scedosporium or Exophiala, are also detected using NGS-based approaches [17]. However, the prevalence of these fungi varies greatly depending on the study, as is also the case in culture-based studies [40, 41, 62], and they may or may not be considered part of the core mycobiome [15, 17, 60]. For instance, Cuthberston et al. reported a prevalence of 48% and 54% for Scedosporium and Exophiala, respectively (samples were considered positive when ≥ 1 fungal sequence was detected), Soret et al. reported a prevalence of 42% and 6%, respectively, and Kramer et al. reported a prevalence of 4% and 4%, respectively [14, 15, 17]. In the study by O’Connor et al., no sequences of Exophiala or Scedosporium were detected using NGS [48]. These discrepancies can be attributed not only to contextual dissimilarities between studies (geographical location of the study, year of the study) but also to variations in the design and the NGS-based protocol employed.
Overall, the core mycobiome described in pwCF is consistent with the hypothesis that the mycobiota of the lower RT is probably of mixed origin. A major part is derived from salivary microaspirations, as has been demonstrated for the bacterial microbiota [63], consistent with yeast predominance resembling the digestive mycobiota. A minor part is derived from inhaled airborne fungal flora (transient flora), which remains in limited quantities in the lung. However, as the mucociliary clearance system is impaired in the RT of pwCFs, the overall abundance of inhaled fungi is higher than that in healthy individuals [17, 48].
Role of Candida and Saccharomycotina Yeasts in the Mycobiota of PwCFs
Although yeasts from the subphylum Saccharomycotina, including Candida spp., are highly prevalent in the upper and lower RTs of pwCFs according to culture and NGS studies, their precise role in pathophysiology remains unclear [7]. C. albicans followed by C. dubliniensis are the most frequently detected Candida species in the RT of pwCFs by culture, and they are mainly considered as colonizers [38, 64]. Nevertheless, some authors have suggested possible association between Candida spp. colonization and a decline in respiratory function and general health [10, 11, 36, 65]. For instance, Chotirmall et al. demonstrated that patients colonized with C. albicans exhibited lower BMIs and decreased FEV1 and FVC [65]. This finding was corroborated in a large cohort of pwCFs by Gileles-Hillel et al., who also observed a greater rate of annual decline in FEV1 in pwCFs chronically colonized with C. albicans [11]. This study also demonstrated that C. albicans colonization was associated with chronic colonization by other CF pathogens, including P. aeruginosa and A. fumigatus. Furthermore, the authors identified potential contributing factors to C. albicans colonization, including pancreatic insufficiency and diabetes [11]. In an NGS-based study, Hong et al. observed an increased rate of C. dubliniensis in patients with a PEx. In addition, a higher ratio of C. dubliniensis to C. albicans was observed in patients with PEx, which was also associated with a lower respiratory score (evaluated through the CFQ-R [51]). The role of Candida spp. in the decline of lung functions remains unclear. One possible explanation could be that the yeasts directly affect the lung parenchyma (e.g., by invading the tissue after switching to hyphae) or exacerbate the local inflammation due to their pro-inflammatory effect, as previously demonstrated for C. albicans in the digestive tract [19, 66]. Candida spp. may also have potential deleterious synergistic association or interaction with other pathogens involved in tissue damage, such as P. aeruginosa. This could occur through the production of mixed biofilm, induction of antimicrobial tolerance, and other mechanisms. Alternatively, Candida spp. might simply have a predilection for already damaged tissues [67–70].
Role of Aspergillus fumigatus and Other Filamentous Fungi in the Mycobiota of PwCFs
Among fungi, A. fumigatus has the greatest impact on pwCF and lung function decline and is implicated in several diseases, particularly in allergic bronchopulmonary aspergillosis (ABPA), Aspergillus sensitization or bronchitis, or even invasive aspergillosis in immunocompromised pwCF (e.g., solid organ transplantation) [2, 7, 61, 71]. The prevalence of ABPA in pwCFs (based on culture-based studies and national CF registries) varies considerably depending on age and country (3–25%), with a pooled prevalence estimated at 8.9% in children and 10.1% in adults (as determined by Maturu et al. from the data of 45 CF studies) [61, 71]. Oher mold species, including Scedosporium/Pseudallescheria spp., Lomentospora prolificans and Rasamsonia spp., have been identified as potential causes of allergic bronchopulmonary disease and may also pose a threat in immunocompromised pwCF [41, 72–76]. In contrast, the clinical role of other fungi associated with CF, such as Exophiala sp. remains to be clearly determined [38, 40, 77] In a Swedish culture-based study, a high prevalence of Exophiala sp. was reported, and an association was found with a more advanced form of the disease [62]. But these results were not confirmed, and a recent Dutch study concluded that Exophiala sp. is probably only a minor pathogen in CF [40].
A Specific Focus on Pneumocystis jirovecii in the RT of PwCFs
NGS-based studies theoretically allow the detection of fungi that are uncultivable, such as Pneumocystis jirovecii. However, the majority of studies employing NGS-based methods do not report the presence of P. jirovecii in RT samples of pwCFs [12–15, 48, 52, 78]. In TA-NGS-based studies, this may be due to the fact that P. jirovecii has only one copy of the rDNA operon (including the ITS region) in contrast to other fungi which have several repeats of the ITS region [26]. In SMg studies, the overall detection of fungal sequences is very low which makes it difficult to provide a precise description of the mycobiome [28, 79]. Only a few authors have studied P. jirovecii in pwCFs, with the majority of studies focusing specifically on this microorganism [60, 80–86]. In a recent multicenter Spanish study on the lung mycobiome in pwCF, the authors added a specific qPCR for P. jirovecii detection (targeting the mt-LSU rRNA region) and reported a 31% prevalence of colonization [60]. Overall, the prevalence of P. jirovecii in pwCF varies considerably between studies [60, 80–87]. For instance, a prevalence of approximately 8% (7.4 and 8.1%) was observed in Germany and in the United Kingdom, while it was of 38.2% in Brazil [83–85]. In France, the prevalence of P. jirovecii exhibited regional variations, with rates below 4% in Brittany and 12% in other regions [80–82]. In Spain, the prevalence reported increased from 21.6% in 2005 to 31% in 2023 [60, 86]. These discrepancies may be explained by the prior exposure of pwCF to cotrimoxazole, as well as differences in the antibiotic protocols used at distinct CF centers [82, 83]. This phenomenon can also result from differences in the age of patients as well as exposure risk factors associated with climate, geographic or environmental context [81]. The role of P. jirovecii in the pathophysiology of CF-related lung decline remains unclear. Green et al. observed an increase in the prevalence of P. jirovecii in pwCF samples during PEx (9.2%) compared with patients in the stable state (2%) [83]. In contrast, Hernández-Hernández et al. reported that pwCF who were colonized with P. jirovecii had less severe lung diseases [80]. In all cases, colonization with P. jirovecii represents a risk of pneumocystosis for pwCFs undergoing lung transplantation [87].
Mycobiome Variations Between Pwcfs According to Geographical Factors or CF Centers
The distribution of CF fungi (other than P. jirovecii) varies according to geographic area or CF center. A geographical gradient of fungal species among European countries has been described by Delhaes et al. in a culture-based study. Candida spp., A. fumigatus, Cladosporium and Penicillium were more prevalent at higher latitudes, while Scedosporium and Lomentospora were more prevalent in southern centers [9]. In a Spanish multicenter study based on multiple Sanger sequencing, Martínez-Rodríguez et al. also observed significant differences in the distribution of fungi between centers (Seville, Madrid, Mallorca), particularly for taxa of lower abundance including Scedosporium spp. and Exophiala spp. [60]. In France, Angebault et al. reported different median relative abundances of A. fumigatus in pwCFs attending two CF centers (unpublished data). As previously discussed for P. jirovecii, these discrepancies in fungal distribution may be attributed to varying exposure levels associated with climate, geographic or environmental factors [41]. Van Rhijn et al. observed that the indoor and outdoor level of Aspergillus spores in a CF center was increased during summer, when temperatures were high and wind speeds low [42, 88]. However, other factors may also contribute to variations in the fungal distribution between centers, including the age of patients (adult, pediatric or mixed population), the type of mutation (homozygous/heterozygous), the severity of the disease, and the differences in antimicrobial protocols from one center to another. To better understand these differences, further international studies including large cohorts of pwCFs from different centers are necessary.
The Lung Mycobiome of PwCFs With Fungal Pulmonary Diseases
To date, only a limited number of studies have used NGS-based methods to describe the dynamics of the mycobiome in pwCFs with pulmonary fungal diseases, including ABPA, Aspergillus sensitization, and fungal bronchitis. Cuthbertson et al. conducted a study in which they compared the mycobiomes of groups of pwCFs with ABPA, fungal bronchitis or no active fungal disease [17]. The authors observed that A. fumigatus was the most predominant taxon only in pwCFs with fungal bronchitis. Furthermore, an increased relative abundance of Scedosporium sp. and Clavispora sp. was observed in pwCFs with ABPA, whereas Exophiala and Candida showed an increased relative abundance in pwCFs with fungal bronchitis [17].
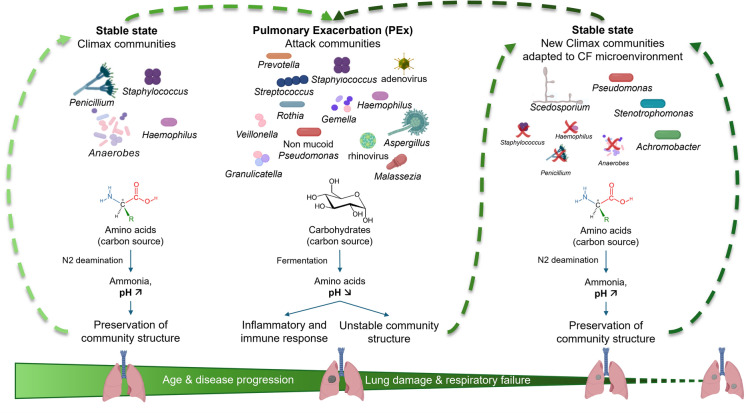
Understanding the “Climax-Attack” Model in CF Airways
The “Climax-Attack” model (CAM) was initially described by Conrad et al. [89]. It proposes an ecological vision that considers the complexity of the lung CF bacterial microbiota and the polymicrobial nature of chronic CF airway infection. It was then refined by Quinn et al., who focused mainly on the functional and metabolic aspects of bacterial communities, and Soret et al., who explored the role of fungi in the model [15, 90]. The CAM proposes the existence of two major functional microbial communities. Microbial interactions between these communities during PEx could explain the changes in the lung microbiota and the progressive reduction in microbial diversity over time (Fig. 3). The climax communities comprise persistent, slow-growing microbial and fungal "keystone" populations that are well adapted to the CF airway microenvironment. The bacterial climax species have metabolic pathways based on amino acid transformation (as a major carbon source) through deamination, which leads to the production of ammonia and an increase in pH, which help maintain community stability [90]. These communities can also produce mixed biofilms, which protect them from the host immune response and antimicrobial treatments. The original or pioneer climax communities are composed of Staphylococcus aureus, Haemophilus sp., respiratory anaerobic bacteria, and possibly also nonpathogenic fungi, such as Penicillium sp. Over time, with the emergence of the attack communities and the recurrence of PEx, the climax communities will evolve, and new species that are better adapted to the specific CF microenvironment will replace the pioneer species. The new climax species include P. aeruginosa (mucoid strains), Stenotrophomonas maltophilia, Achromobacter sp., and possibly fungi such as Scedosporium sp. These species are generally resistant to conventional antimicrobial therapy. On the other hand, attack communities are transient microbial populations that induce strong innate immune responses, leading to acute PEx and lung damage. These communities utilize carbohydrates as an important source of carbon through fermentation pathways, which results in the production of organic acids and a decreased pH. Their emergence favors the instability of the community structure and facilitates the establishment of new climax species. A variety of microorganisms, including viruses (e.g., rhinovirus, influenza, adenovirus, etc.), non-mucoid P. aeruginosa, Staphylococcus sp., Streptococcus sp. and anaerobic bacteria may be part of the attack communities and are often environmentally acquired. Some fungi, such as A. fumigatus, may also be part of attack communities. The species belonging to the attack communities may possess specific virulence factors and elicit intense inflammation. The complete list of species belonging to the climax or attack communities, along with their interrelationships, remains to be established. Some species may be part of both types of communities, such as P. aeruginosa or some anaerobic bacteria, and some species may have changed groups. Some fungal species, such as Candida sp., E. dermatitidis and P. jirovecii, are not yet included in the CAM. In summary, the CAM appears to be an accurate model for describing the interrelationships between two types of polymicrobial communities, which could explain the occurrence of PEx in pwCFs, the progression of the disease, and the change in microbiota diversity over time.
Fig. 3.
The “Climax-Attack” model for CF airways. The “Climax-Attack model” is an ecological model adapted to explain the progressive loss of microbial diversity and the replacement of pioneer communities by adapted communities to CF airways. The figure was adapted from the articles of Quinn et al. npj Biofilms Microbiomes. 2016; Soret et al. Sci Rep. 2020; Conrad et al. Am J Respir Cell Mol Biol 2013 [15, 89, 90] and created, in part, with BioRender.com
Intra- and Interkingdom Correlation Networks in PwCFs
NGS-based studies of the mycobiome (and microbiome) allow the detection of possible positive or negative intra- or interkingdom correlations (co-occurrence or co-exclusion between fungi and/or bacteria). This is achieved by constructing a correlation matrix from the entire set of microbial data. The correlation matrix can then be represented graphically as a correlation network [91]. Some authors have constructed correlation networks for the bacterial microbiome of pwCFs, but few have done so for the mycobiome or to study possible bacterial-fungal correlations [15, 48, 90]. In their study of the fungal and bacterial microbiomes of 33 pwCFs, Soret et al. observed several interkingdom correlations (at the genus level) between fungi and bacteria. In particular, they observed three clusters centered on Aspergillus, Candida and Scedosporium, which showed positive or negative correlations with different bacteria [15]. Among all the statistical correlations identified, the authors selected one between Aspergillus and Streptococcus for in vitro testing to determine the biological relevance and potential for a real interaction. Their findings demonstrated that Aspergillus growth was enhanced by two species of Streptococcus. Angebault et al. also constructed correlation networks based on the fungal and bacterial microbiomes of 56 pwCFs, identifying other correlations between fungi and bacteria (unpublished data). To date, an increasing number of interactions between microorganisms (intra- or interkingdom) have been explored in the CF context. These interactions can vary widely in nature and include direct physical interactions, mixed biofilm production, quorum-sensing-related interactions, bacterial release of volatile organic compounds, competition for nutrients and siderophores, changes in susceptibility to antimicrobial agents, and so forth [69, 92]. For example, several interactions between A. fumigatus and P. aeruginosa or S. maltophilia or between C. albicans and P. aeruginosa have been elucidated [67, 68, 92–97]. Intra- and interkingdom correlation analysis based on NGS microbiome studies may highlight possible unknown interactions between microorganisms, but these interactions must be tested in experimental models to verify their biological relevance.
The New Era of CFTR Modulators and Their Effects on the Mycobiome
Symptomatic therapies have long been employed to treat inflammation, lung dysfunction and chronic airway infections in pwCFs. The advent of CFTR modulators has led to remarkable improvements in lung function and health status in approximately 90% of the CF population, depending on the mutation(s) carried [98–100]. Among other effects, the use of CFTR modulators has led to significant changes in the respiratory bacterial microbiome of pwCFs including increased bacterial richness, alpha diversity and reduced rates of specific pathogens, such as P. aeruginosa [101–104]. Regarding fungi, different authors have demonstrated that CFTR modulators have the potential to reduce the rate of colonization by Aspergillus [32, 47, 103–105]. Heltshe et al. reported a significant reduction in the number of pwCFs positive for Aspergillus in their sputa during the year following the introduction of CFTR potentiator, ivacaftor [104]. Bessonova et al. utilized the CF national registries from the USA and the UK to report a decrease in the prevalence of Aspergillus colonization between patients treated with and without ivacaftor [105]. Frost et al. observed a reduction in the prevalence of Aspergillus colonization over three years in patients treated with ivacaftor [103]. Chesnay et al. also observed a reduction in Aspergillus-positive cultures in pwCFs under combined CFTR modulators (elexacaftor/tezacaftor/ivacaftor) [47]. The authors also demonstrated a reduction in the amount of anti-Aspergillus precipitating IgG and total IgE [47]. This finding suggests a potential effect of CFTR modulators on APBA and Aspergillus sensitization [47]. In an experimental study, Currie et al. showed that CFTR modulators do not have a direct fungicidal effect on Aspergillus. Nevertheless, they have been demonstrated to reduce the production of reactive oxygen species by CF phagocytes in the presence of Aspergillus [106]. The available data on changes in the overall mycobiome (taxonomic composition, diversity, and dynamics) due to CFTR modulators are currently limited [32]. Following the bacterial and fungal microbiota of 41 pwCFs for six months after the introduction of CFTR modulators (lumacaftor/ivacaftor), Enaud et al. observed no changes in the fungal (or bacterial) load, α- or β-fungal (bacterial) diversity or the abundance of specific taxa [107]. In contrast, Hong et al. observed an increased fungal α-diversity among pwCFs treated with CFTR modulators as compared to those without [52]. It is hypothesized that the observed changes in the respiratory mycobiome are linked to improved lung function and reduced chronic inflammation resulting from the restoration of CFTR channel functions. Further studies on the long-term effects of CFTR modulators on the lung mycobiome are necessary.
Conclusion
To date, the NGS-based approaches used for studying the mycobiome of the RT in pwCFs are mostly TA-NGS. For the time being, SMg methods have limitations in describing the lung mycobiome due to the difficulty in precisely identifying microbial communities of low abundance (low fungal load). Many variations have been described in mycobiome studies. This can be explained not only by geographical data but also by variations in the design and the NGS-based protocol chosen. The pathophysiological role of Candida spp. and Saccharomycotina yeasts, as well as P. jirovecii in the context of CF, remains to be determined. As for the bacterial microbiome, the diversity of the RT mycobiome decreases over time in pwCFs, as recurrent PEx occur and lung damage progresses. NGS-based studies permit the identification of correlations between microorganisms within and between kingdoms, which may lead to the discovery of novel interactions between microorganisms in the CF context. The climax-attack model has been proposed to describe the complexity of the lung CF bacterial and fungal microbiota and explain their dynamics of change over time. With the use of CFTR modulators, significant changes in the mycobiome (and microbiome) of pwCFs are anticipated in the coming years. Consequently, it will be essential to continue investigating the mycobiome of RT in pwCFs.
Key Research Questions About the RT Mycobiome in CF
Technical improvement of metagenomics protocols (design, universal targets, databases, bioinformatics pipelines, standardization, etc.) for mycobiome analysis
Contribution of TA-NGS and SMg to determining prevalence of yeast and filamentous fungi compared to culture-based methods
Use of RT mycobiome (detected through TA-NGS or SMg) for diagnostic or prognostic purposes in CF
Understanding the role of all prevalent fungi in CF (including Candida, Malassezia, Exophiala, P. jirovecii, etc.)
Geographic environment, age, exacerbation or CFTR modulators in the evolution of RT mycobiome
Bacterial and fungal associations in the RT of pwCF: mixed biofilms and therapeutic implications
Authors Contribution
CA wrote the manuscript. FB supervised and corrected the manuscript.
Use of generative AI for manuscript preparation
All content was written by CA and FB. English grammar and turns of phrase were revised with the help of DeepL and Curie.
Funding
This review was written as an addendum to a study entitled "Ecologie et dynamique des communautés bactériennes et fongiques des voies aériennes supérieures des patients atteints de mucoviscidose”, which received a grant from the “Vaincre La Mucoviscidose” Foundation (Grant Number: 2013/2013–047/03, https://www.vaincrelamuco.org/). The funders were not involved in the review of the literature or the preparation of the manuscript.
Declarations
Competing interest
The authors have no conflicts of interest to declare.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elborn JS. Cystic fibrosis. Lancet. 2016;388:2519–31. [DOI] [PubMed] [Google Scholar]
- 2.Vaincre la Mucoviscidose. Registre français de la mucoviscidose - Bilan des données 2022. https://www.vaincrelamuco.org/registredelamuco. 2022.
- 3.Rossi GA, Morelli P, Galietta LJ, Colin AA. Airway microenvironment alterations and pathogen growth in cystic fibrosis. Pediatr Pulmonol. 2019;54:497–506. [DOI] [PubMed] [Google Scholar]
- 4.Flume PA, VanDevanter DR. Cystic fibrosis: definition, severity and impact of pulmonary exacerbations. In: Burgel P-R, Contoli M, López-Campos JL, editors. Acute Exacerbations of Pulmonary Diseases. European Respiratory Society; 2017. p. 25–37. 10.1183/2312508X.10015716. [Google Scholar]
- 5.Thornton CS, Parkins MD. Microbial epidemiology of the cystic fibrosis airways: past, present, and future. Semin Respir Crit Care Med. 2023;44:269–86. [DOI] [PubMed] [Google Scholar]
- 6.Tracy MC, Moss RB. The myriad challenges of respiratory fungal infection in cystic fibrosis. Pediatr Pulmonol. 2018;53:S75-85. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz C, Hartl D, Eickmeier O, Hector A, Benden C, Durieu I, et al. Progress in definition, prevention and treatment of fungal infections in cystic fibrosis. Mycopathologia. 2018;183:21–32. [DOI] [PubMed] [Google Scholar]
- 8.Francis F, Enaud R, Soret P, Lussac-Sorton F, Avalos-Fernandez M. MucoFong investigation group, et al. new insights in microbial species predicting lung function decline in CF: lessons from the MucoFong project. J Clin Med. 2021;10:3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delhaes L, Touati K, Faure-Cognet O, Cornet M, Botterel F, Dannaoui E, et al. Prevalence, geographic risk factor, and development of a standardized protocol for fungal isolation in cystic fibrosis: results from the international prospective study “MFIP.” J Cyst Fibros. 2019;18:212–20. [DOI] [PubMed] [Google Scholar]
- 10.Chotirmall SH, O’Donoghue E, Bennett K, Gunaratnam C, O’Neill SJ, McElvaney NG. Sputum Candida albicans presages FEV1 decline and hospital-treated exacerbations in cystic fibrosis. Chest. 2010;138:1186–95. [DOI] [PubMed] [Google Scholar]
- 11.Gileles-Hillel A, Shoseyov D, Polacheck I, Korem M, Kerem E, Cohen-Cymberknoh M. Association of chronic Candida albicans respiratory infection with a more severe lung disease in patients with cystic fibrosis. Pediatr Pulmonol. 2015;50:1082–9. [DOI] [PubMed] [Google Scholar]
- 12.Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community-implications for therapeutic management. PLoS ONE. 2012;7:e36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botterel F, Angebault C, Cabaret O, Stressmann FA, Costa J-M, Wallet F, et al. Fungal and bacterial diversity of airway microbiota in adults with cystic fibrosis: concordance between conventional methods and ultra-deep sequencing, and their practical use in the clinical laboratory. Mycopathologia. 2018;183:171–83. [DOI] [PubMed] [Google Scholar]
- 14.Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham W-R, Höfle MG. Cohort study of airway mycobiome in adult cystic fibrosis patients: differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol. 2015;53:2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soret P, Vandenborght L-E, Francis F, Coron N, Enaud R, Avalos M, et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci Rep. 2020;10:3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton CS, Acosta N, Surette MG, Parkins MD. Exploring the cystic fibrosis lung microbiome: making the most of a sticky situation. J Pediatric Infect Dis Soc. 2022;11:S13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuthbertson L, Felton I, James P, Cox MJ, Bilton D, Schelenz S, et al. The fungal airway microbiome in cystic fibrosis and non-cystic fibrosis bronchiectasis. J Cyst Fibros. 2021;20:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott G, Walsh A, Crispie F, Frost S, Greally P, Cotter PD, et al. Insights into the adolescent cystic fibrosis airway microbiome using shotgun metagenomics. Int J Mol Sci. 2024;25:3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iliev ID, Cadwell K. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology. 2021;160:1050–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angebault C, Payen M, Woerther P-L, Rodriguez C, Botterel F. Combined bacterial and fungal targeted amplicon sequencing of respiratory samples: does the DNA extraction method matter? PLoS ONE. 2020;15: e0232215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huseyin CE, Rubio RC, O’Sullivan O, Cotter PD, Scanlan PD. The Fungal Frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front Microbiol. 2017;8:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angebault C, Ghozlane A, Volant S, Botterel F, d’Enfert C, Bougnoux M-E. Combined bacterial and fungal intestinal microbiota analyses: impact of storage conditions and DNA extraction protocols. PLoS ONE. 2018;13: e0201174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangneux J-P, Guegan H, Vandenborght L-E, Buffet-Bataillon S, Enaud R, Delhaes L. A European ECMM-ESCMID survey on goals and practices for mycobiota characterisation using next-generation sequencing. Mycoses. 2019;62:1096–9. [DOI] [PubMed] [Google Scholar]
- 24.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dellière S, Dannaoui E, Fieux M, Bonfils P, Gricourt G, Demontant V, et al. Analysis of microbiota and mycobiota in fungal ball rhinosinusitis: specific interaction between Aspergillus fumigatus and Haemophilus influenza? Journal of Fungi. 2021;7:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahimana A, Francioli P, Blanc DS, Bille J, Wakefield AE, Hauser PM. Determination of the copy number of the nuclear rDNA and beta-tubulin genes of Pneumocystis carinii f. sp. hominis using PCR multicompetitors. J Eukaryot Microbiol. 2000;47:368–72. [DOI] [PubMed] [Google Scholar]
- 27.d’Humières C, Salmona M, Dellière S, Leo S, Rodriguez C, Angebault C, et al. The potential role of clinical metagenomics in infectious diseases: therapeutic perspectives. Drugs. 2021. 10.1007/s40265-021-01572-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pienkowska K, Pust M-M, Gessner M, Gaedcke S, Thavarasa A, Rosenboom I, et al. The cystic fibrosis upper and lower airway metagenome. Microbiol Spectr. 2023;11: e0363322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez C, Jary A, Hua C, Woerther P-L, Bosc R, Desroches M, et al. Pathogen identification by shotgun metagenomics of patients with necrotizing soft-tissue infections. Br J Dermatol. 2020;183:105–13. [DOI] [PubMed] [Google Scholar]
- 30.Lind AL, Pollard KS. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome. 2021;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubags NDJ, Marsland BJ. Mechanistic insight into the function of the microbiome in lung diseases. Eur Respir J. 2017;50:1602467. [DOI] [PubMed] [Google Scholar]
- 32.Bercusson A, Jarvis G, Shah A. CF fungal disease in the age of CFTR modulators. Mycopathologia. 2021;186:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. 2012;18:509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granchelli AM, Adler FR, Keogh RH, Kartsonaki C, Cox DR, Liou TG. Microbial interactions in the cystic fibrosis airway. J Clin Microbiol. 2018;56:e00354-e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renner S, Nachbaur E, Jaksch P, Dehlink E. Update on respiratory fungal infections in cystic fibrosis lung disease and after lung transplantation. Journal of Fungi. 2020;6:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz C, Bouchara J-P, Buzina W, Chrenkova V, Dmeńska H, de la Pedrosa EGG, et al. Organization of patient management and fungal epidemiology in cystic fibrosis. Mycopathologia. 2018;183:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel TGP, Tehupeiory-Kooreman M, Melchers WJG, Reijers MH, Merkus P, Verweij PE. Evaluation of a new culture protocol for enhancing fungal detection rates in respiratory samples of cystic fibrosis patients. J Fungi (Basel). 2020;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Jong CCM, Slabbers L, Engel TGP, Yntema JB, van Westreenen M, Croughs PD, et al. Clinical relevance of Scedosporium spp. and Exophiala dermatitidis in patients with cystic fibrosis: a nationwide study. Med Mycol. 2020;58:859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravenel K, Guegan H, Gastebois A, Bouchara J-P, Gangneux J-P, Giraud S. Fungal colonization of the airways of patients with cystic fibrosis: the role of the environmental reservoirs. Mycopathologia. 2024;189:19. [DOI] [PubMed] [Google Scholar]
- 42.van Rhijn N, Coleman J, Collier L, Moore C, Richardson MD, Bright-Thomas RJ, et al. Meteorological factors influence the presence of fungi in the air; A 14-month surveillance study at an adult cystic fibrosis center. Front Cell Infect Microbiol. 2021;11: 759944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziesing S, Suerbaum S, Sedlacek L. Fungal epidemiology and diversity in cystic fibrosis patients over a 5-year period in a national reference center. Med Mycol. 2016;54:781–6. [DOI] [PubMed] [Google Scholar]
- 44.Psoter KJ, De Roos AJ, Wakefield J, Mayer JD, Rosenfeld M. Seasonality of acquisition of respiratory bacterial pathogens in young children with cystic fibrosis. BMC Infect Dis. 2017;17:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaincre la Mucoviscidose. Registre français de la mucoviscidose - Bilan des données 2016. http://www.vaincrelamuco.org/sites/default/files/registre_francais_de_la_mucoviscidose_-_bilan_2015_v4.pdf. 2016.
- 46.Regard L, Martin C, Da Silva J, Burgel P-R. CFTR modulators: current status and evolving knowledge. Semin Respir Crit Care Med. 2023;44:186–95. [DOI] [PubMed] [Google Scholar]
- 47.Chesnay A, Bailly É, Cosson L, Flament T, Desoubeaux G. Advent of elexacaftor/tezacaftor/ivacaftor for cystic fibrosis treatment: what consequences on Aspergillus-related diseases? Preliminary insights J Cyst Fibros. 2022;21:1084–5. [DOI] [PubMed] [Google Scholar]
- 48.O’Connor JB, Mottlowitz M, Kruk ME, Mickelson A, Wagner BD, Harris JK, et al. Network analysis to identify multi-omic correlations in the lower airways of children with cystic fibrosis. Front Cell Infect Microbiol. 2022;12: 805170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. 2017;72:1104–12. [DOI] [PubMed] [Google Scholar]
- 50.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015. 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quittner AL, Sawicki GS, McMullen A, Rasouliyan L, Pasta DJ, Yegin A, et al. Psychometric evaluation of the cystic fibrosis questionnaire-revised in a national sample. Qual Life Res. 2012;21:1267–78. [DOI] [PubMed] [Google Scholar]
- 52.Hong G, Daniel SG, Lee J-J, Bittinger K, Glaser L, Mattei LM, et al. Distinct community structures of the fungal microbiome and respiratory health in adults with cystic fibrosis. J Cyst Fibros. 2023;22:636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neu AT, Allen EE, Roy K. Defining and quantifying the core microbiome: challenges and prospects. Proc Natl Acad Sci. 2021;118: e2104429118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risely A. Applying the core microbiome to understand host–microbe systems. J Anim Ecol. 2020;89:1549–58. [DOI] [PubMed] [Google Scholar]
- 55.Enaud R, Vandenborght L-E, Coron N, Bazin T, Prevel R, Schaeverbeke T, et al. The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms. 2018. 10.3390/microorganisms6010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lepesqueur LSS, Tanaka MH, de Lima G MG, Chiba SM, Mota AJ, Santos SF, et al. Oral prevalence and antifungal susceptibility of Candida species in cystic fibrosis patients. Arch Oral Biol. 2020;116:104772. [DOI] [PubMed] [Google Scholar]
- 57.Ali NABM, Ivan FX, Aogáin MM, Narayana JK, Lee SY, Lim CL, et al. The healthy airway mycobiome in individuals of Asian descent. Chest. 2021;159:544–8. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen LDN, Viscogliosi E, Delhaes L. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol. 2015;6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delavy M, Sertour N, Patin E, Le Chatelier E, Cole N, Dubois F, et al. Unveiling Candida albicans intestinal carriage in healthy volunteers: the role of micro- and mycobiota, diet, host genetics and immune response. Gut Microbes. 2023;15:2287618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martínez-Rodríguez S, Friaza V, Girón-Moreno RM, Gallego EQ, Salcedo-Posadas A, Figuerola-Mulet J, et al. Fungal microbiota dynamics and its geographic, age and gender variability in patients with cystic fibrosis. Clin Microbiol Infect. 2023;29:539.e1-539.e7. [DOI] [PubMed] [Google Scholar]
- 61.Armstead J, Morris J, Denning DW. Multi-country estimate of different manifestations of aspergillosis in cystic fibrosis. PLoS ONE. 2014;9: e98502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kondori N, Gilljam M, Lindblad A, Jonsson B, Moore ERB, Wenneras C. High rate of Exophiala dermatitidis recovery in the airways of patients with cystic fibrosis is associated with pancreatic insufficiency. J Clin Microbiol. 2011;49:1004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. MBio. 2017;8:e02287-e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baxter CG, Moore CB, Jones AM, Webb AK, Denning DW. IgE-mediated immune responses and airway detection of Aspergillus and Candida in adult cystic fibrosis. Chest. 2013;143:1351–7. [DOI] [PubMed] [Google Scholar]
- 65.Chotirmall SH. Candida albicans in cystic fibrosis: “opening statements presented, let the trial begin.” Pediatr Pulmonol. 2016;51:445–6. [DOI] [PubMed] [Google Scholar]
- 66.d’Enfert C, Kaune A-K, Alaban L-R, Chakraborty S, Cole N, Delavy M, et al. The impact of the fungus-host-microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol Rev. 2021;45:060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alam F, Blackburn SA, Davis J, Massar K, Correia J, Tsai H-J, et al. Pseudomonas aeruginosa increases the susceptibility of Candida albicans to amphotericin B in dual-species biofilms. J Antimicrob Chemother. 2023;78:2228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alam F, Catlow D, Di Maio A, Blair JMA, Hall RA. Candida albicans enhances meropenem tolerance of Pseudomonas aeruginosa in a dual-species biofilm. J Antimicrob Chemother. 2020;75:925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santos-Fernandez E, Martin-Souto L, Antoran A, Areitio M, Aparicio-Fernandez L, Bouchara J-P, et al. Microbiota and fungal-bacterial interactions in the cystic fibrosis lung. FEMS Microbiol Rev. 2023;47:029. [DOI] [PubMed] [Google Scholar]
- 70.Bigot J, Ruffin M, Guitard J, Vellaissamy S, Thorez S, Corvol H, et al. Effect of flagellin pre-exposure on the inflammatory and antifungal response of bronchial epithelial cells to fungal pathogens. J Fungi (Basel). 2022;8:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maturu VN, Agarwal R. Prevalence of Aspergillus sensitization and allergic bronchopulmonary aspergillosis in cystic fibrosis: systematic review and meta-analysis. Clin Exp Allergy. 2015;45:1765–78. [DOI] [PubMed] [Google Scholar]
- 72.Rammaert B, Puyade M, Cornely OA, Seidel D, Grossi P, Husain S, et al. Perspectives on Scedosporium species and Lomentospora prolificans in lung transplantation: results of an international practice survey from ESCMID fungal infection study group and study group for infections in compromised hosts, and European confederation of medical mycology. Transpl Infect Dis. 2019;21: e13141. [DOI] [PubMed] [Google Scholar]
- 73.Abdolrasouli A, Bercusson AC, Rhodes JL, Hagen F, Buil JB, Tang AYY, et al. Airway persistence by the emerging multi-azole-resistant Rasamsonia argillacea complex in cystic fibrosis. Mycoses. 2018;61:665–73. [DOI] [PubMed] [Google Scholar]
- 74.Zouhair R, Rougeron A, Razafimandimby B, Kobi A, Bouchara J-P, Giraud S. Distribution of the different species of the Pseudallescheria boydii/Scedosporium apiospermum complex in French patients with cystic fibrosis. Med Mycol. 2013;51:603–13. [DOI] [PubMed] [Google Scholar]
- 75.Giraud S, Pihet M, Razafimandimby B, Carrère J, Degand N, Mely L, et al. Geosmithia argillacea: an emerging pathogen in patients with cystic fibrosis. J Clin Microbiol. 2010;48:2381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spahr JE, Love RB, Francois M, Radford K, Meyer KC. Lung transplantation for cystic fibrosis: current concepts and one center’s experience. J Cyst Fibros. 2007;6:334–50. [DOI] [PubMed] [Google Scholar]
- 77.Mills R, Rautemaa-Richardson R, Wilkinson S, Patel L, Maitra A, Horsley A. Impact of airway Exophiala spp. on children with cystic fibrosis. J Cyst Fibros. 2021;20:702–7. [DOI] [PubMed] [Google Scholar]
- 78.Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome. 2014;2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dmitrijeva M, Kahlert CR, Feigelman R, Kleiner RL, Nolte O, Albrich WC, et al. Strain-resolved dynamics of the lung microbiome in patients with cystic fibrosis. MBio. 2021;12:e02863-e2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernández-Hernández F, Fréalle E, Caneiro P, Salleron J, Durand-Joly I, Accoceberry I, et al. Prospective multicenter study of Pneumocystis jirovecii colonization among cystic fibrosis patients in France. J Clin Microbiol. 2012;50:4107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nevez G, Robert-Gangneux F, Pougnet L, Virmaux M, Belleguic C, Deneuville E, et al. Pneumocystis jirovecii and cystic fibrosis in brittany. France Mycopathologia. 2018;183:81–7. [DOI] [PubMed] [Google Scholar]
- 82.Gal SL, Héry-Arnaud G, Ramel S, Virmaux M, Damiani C, Totet A, et al. Pneumocystis jirovecii and cystic fibrosis in France. Scand J Infect Dis. 2010;42:225–7. [DOI] [PubMed] [Google Scholar]
- 83.Green HD, Bright-Thomas RJ, Mutton KJ, Guiver M, Jones AM. Increased prevalence of Pneumocystis jirovecii colonisation in acute pulmonary exacerbations of cystic fibrosis. J Infect. 2016;73:1–7. [DOI] [PubMed] [Google Scholar]
- 84.Sing A, Geiger AM, Hogardt M, Heesemann J. Pneumocystis carinii carriage among cystic fibrosis patients, as detected by nested PCR. J Clin Microbiol. 2001;39:2717–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pederiva MA, Wissmann G, Friaza V, Morilla R, de La Horra C, Montes-Cano MA, et al. High prevalence of Pneumocystis jirovecii colonization in Brazilian cystic fibrosis patients. Med Mycol. 2012;50:556–60. [DOI] [PubMed] [Google Scholar]
- 86.Respaldiza N, Montes-Cano MA, Dapena FJ, de la Horra C, Mateos I, Medrano FJ, et al. Prevalence of colonisation and genotypic characterisation of Pneumocystis jirovecii among cystic fibrosis patients in Spain. Clin Microbiol Infect. 2005;11:1012–5. [DOI] [PubMed] [Google Scholar]
- 87.Bonnet P, Le Gal S, Calderon E, Delhaes L, Quinio D, Robert-Gangneux F, et al. Pneumocystis jirovecii in patients with cystic fibrosis: a review. Front Cell Infect Microbiol. 2020;10: 571253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loeffert ST, Melloul E, Dananché C, Hénaff L, Bénet T, Cassier P, et al. Monitoring of clinical strains and environmental fungal aerocontamination to prevent invasive aspergillosis infections in hospital during large deconstruction work: a protocol study. BMJ Open. 2017. 10.1136/bmjopen-2017-018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conrad D, Haynes M, Salamon P, Rainey PB, Youle M, Rohwer F. Cystic fibrosis therapy: a community ecology perspective. Am J Respir Cell Mol Biol. 2013;48:150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quinn RA, Whiteson K, Lim YW, Zhao J, Conrad D, LiPuma JJ, et al. Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes. 2016;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peschel S, Müller CL, von Mutius E, Boulesteix A-L, Depner M. NetCoMi: network construction and comparison for microbiome data in R. Brief Bioinform. 2020;1706:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Debourgogne A, Monpierre L, Sy KA, Valsecchi I, Decousser J-W, Botterel F. Interactions between bacteria and Aspergillus fumigatus in airways: from the mycobiome to molecular interactions. J Fungi (Basel). 2023;9:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Briard B, Mislin GLA, Latgé J-P, Beauvais A. Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J Fungi (Basel). 2019. 10.3390/jof5020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Melloul E, Roisin L, Durieux M-F, Woerther P-L, Jenot D, Risco V, et al. Interactions of Aspergillus fumigatus and Stenotrophomonas maltophilia in an in vitro mixed biofilm model: does the strain matter? Front Microbiol. 2018. 10.3389/fmicb.2018.02850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roisin L, Melloul E, Woerther P-L, Royer G, Decousser J-W, Guillot J, et al. Modulated response of Aspergillus fumigatus and Stenotrophomonas maltophilia to antimicrobial agents in polymicrobial biofilm. Front Cell Infect Microbiol. 2020;10: 574028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Brien S, Fothergill JL. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol Lett. 2017. 10.1093/femsle/fnx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ostapska H, Le Mauff F, Gravelat FN, Snarr BD, Bamford NC, Van Loon JC, et al. Co-operative Biofilm Interactions between Aspergillus fumigatus and Pseudomonas aeruginosa through secreted galactosaminogalactan exopolysaccharide. J Fungi (Basel). 2022;8:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Graeber SY, Mall MA. The future of cystic fibrosis treatment: from disease mechanisms to novel therapeutic approaches. Lancet. 2023;402:1185–98. [DOI] [PubMed] [Google Scholar]
- 99.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. Lumacaftor–Ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373:220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Konstan MW, McKone EF, Moss RB, Marigowda G, Tian S, Waltz D, et al. Assessment of safety and efficacy of long-term treatment with combination lumacaftor and ivacaftor therapy in patients with cystic fibrosis homozygous for the F508del-CFTR mutation (PROGRESS): a phase 3, extension study. Lancet Respir Med. 2017;5:107–18. [DOI] [PubMed] [Google Scholar]
- 101.Harris JK, Wagner BD, Zemanick ET, Robertson CE, Stevens MJ, Heltshe SL, et al. Changes in airway microbiome and inflammation with ivacaftor treatment in patients with cystic fibrosis and the G551D mutation. Ann Am Thorac Soc. 2020;17:212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Graeber SY, Boutin S, Wielpütz MO, Joachim C, Frey DL, Wege S, et al. Effects of lumacaftor–ivacaftor on lung clearance index, magnetic resonance imaging, and airway microbiome in Phe508del homozygous patients with cystic fibrosis. Ann Am Thorac Soc. 2021;18:971–80. [DOI] [PubMed] [Google Scholar]
- 103.Frost FJ, Nazareth DS, Charman SC, Winstanley C, Walshaw MJ. Ivacaftor is associated with reduced lung infection by key cystic fibrosis pathogens. A cohort study using national registry data. Ann Am Thorac Soc. 2019;16:1375–82. [DOI] [PubMed] [Google Scholar]
- 104.Heltshe SL, Mayer-Hamblett N, Burns JL, Khan U, Baines A, Ramsey BW, et al. Pseudomonas aeruginosa in cystic fibrosis patients with G551D-CFTR treated with ivacaftor. Clin Infect Dis. 2015;60:703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bessonova L, Volkova N, Higgins M, Bengtsson L, Tian S, Simard C, et al. Data from the US and UK cystic fibrosis registries support disease modification by CFTR modulation with ivacaftor. Thorax. 2018;73:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Currie AJ, Main ET, Wilson HM, Armstrong-James D, Warris A. CFTR modulators dampen aspergillus-induced reactive oxygen species production by cystic fibrosis phagocytes. Front Cell Infect Microbiol. 2020;10:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Enaud R, Lussac-Sorton F, Charpentier E, Velo-Suárez L, Guiraud J, Bui S, et al. Effects of lumacaftor–ivacaftor on airway microbiota-mycobiota and inflammation in patients with cystic fibrosis appear to be linked to Pseudomonas aeruginosa chronic colonization. Microbiol Spectr. 2023;11: e0225122. [DOI] [PMC free article] [PubMed] [Google Scholar]