Abstract
Stable packaging cell lines expressing the rep and cap genes for recombinant adeno-associated virus type 2 (rAAV-2) assembly constitute an attractive alternative to transient transfection protocols. We recently characterized a stable HeLa rep-cap cell clone (HeRC32) and demonstrated that upon vector transfection and adenovirus infection, efficient rAAV assembly correlated with a 100-fold amplification of the integrated rep-cap sequence with the inverted terminal repeats (ITRs) deleted. We now report a more detailed analysis of this phenomenon and highlight the key cellular and viral factors involved. Determination of the rep-cap copy number of HeRC32 cells indicated that maximum rep-cap amplification occurred between 24 and 48 h following adenovirus infection. Analysis by pulsed-field gel electrophoresis of adenovirus-infected HeRC32 cells indicated that amplified rep-cap sequences were found in an extrachromosomal form. Amplification of the rep-cap sequence with the ITRs deleted was not dependent on adenovirus replication and still occurred when the highly specific adenovirus polymerase was inactivated. In contrast, amplification was inhibited in the presence of aphidicolin, indicating that cellular polymerases were needed. Our study also documented that among the adenovirus gene products, the DNA-binding protein (DBP) was essential, since rep-cap amplification was severely abrogated when HeRC32 cells were infected at a nonpermissive temperature with an adenovirus mutant encoding a thermosensitive DBP. Furthermore, expression of DBP alone in HeRC32 cells was sufficient to induce a sustained level of rep-cap amplification. Finally, immunofluorescence analysis showed that HeRC32 cells expressing the DBP also simultaneously expressed the Rep proteins, suggesting a possible involvement of the latter in rep-cap amplification. Indeed, the lack of detectable amplification in an adenovirus-infected stable rep-cap HeLa cell clone unable to produce Rep proteins further supported that, among the viral gene products, both the DBP and Rep proteins are necessary to induce the targeted amplification of the integrated rep-cap sequences in the absence of the AAV ITRs.
Adeno-associated virus type 2 (AAV-2) is a human parvovirus that has attracted increasing interest because of its use as a gene transfer vector (23, 32). The viral genome consists of a 4.7-kb single-stranded DNA molecule which is composed of two 145-base inverted terminal repeats (ITRs) flanking two open reading frames (ORFs), rep and cap. The ITRs constitute the viral sequences required in cis for DNA replication and encapsidation. The rep ORF contains two promoters (p5 and p19) and encodes four regulatory Rep proteins (1). The two larger Rep proteins, Rep 78 and Rep 68, are involved in all aspects of the viral life cycle, including regulation of gene expression and DNA replication. They recognize a specific binding site present in the ITRs (the Rep binding site), and they can nick the origin of replication in a strand- and sequence-specific fashion (7, 17, 27, 38). All of the Rep proteins also possess ATPase and helicase activities (31, 40, 41). These activities are essential to the initiation of AAV DNA replication. The two smaller Rep proteins, Rep 52 and Rep 40, are required for single-stranded DNA accumulation and encapsidation (6, 11). The cap gene is regulated by the p40 promoter and encodes three structural proteins, VP1, VP2, and VP3, which constitute the capsid.
To undergo a productive infection, AAV requires the presence of a helper virus, adenovirus or herpesvirus. The helper virus, for instance, adenovirus, plays a role in nearly every step of the AAV life cycle by promoting AAV gene expression and DNA replication. The critical adenovirus factors involved in the helper effect are the products of the E1a, E1b, E4 (orf6), and E2a genes and the VA1 RNA (2). Among these early adenovirus proteins, the one encoded by the E2a gene, the DNA binding protein (DBP), was shown to be directly implicated in AAV DNA replication by stimulating the processivity of DNA polymerization (35), possibly by stabilizing single-strand templates for replication (36).
Recombinant AAV vectors (rAAV) used for gene therapy are derived from the wild-type virus by deleting the rep and cap ORFs and replacing them with the transgene and the transcriptional control elements. The only viral sequences retained in the vector are the ITRs. To assemble rAAV, the rep and cap genes are usually provided in trans by transfecting cells with a plasmid harboring the AAV genome with the ITRs deleted together with the vector plasmid. Adenovirus helper activities can be provided either by adenovirus infection or by transfection of a plasmid encoding the critical adenovirus gene products (15). Several variations of this production scheme have been developed, including the use of herpesviruses to provide helper functions (10).
Recently, several studies reported the use of packaging cell lines expressing the rep and cap genes for rAAV production. The cell lines previously described are all derived from HeLa cells and harbor one to several copies of the AAV genome with the ITRs deleted stably integrated in the chromosomes. rAAV is assembled following transfection of the AAV vector plasmid and adenovirus infection (8, 9, 18). Alternatively, the vector can be provided by an adenovirus with E1 deleted, which is then used to infect the packaging cell line (13, 21).
We previously described a HeLa-derived packaging cell line (HeRC32) which harbors one copy of an AAV genome with the ITRs deleted (3, 28). Upon vector transfection and wild-type adenovirus infection, we have found that efficient rAAV assembly correlated with a 100-fold amplification of the rep-cap genome (3). This observation was supported by a similar finding reported by Liu et al. (21).
The present study was undertaken to further investigate this phenomenon. Determination of the rep-cap copy number of HeRC32 cells indicated that maximum rep-cap amplification occurred between 24 and 48 h following adenovirus infection. A more detailed analysis by pulsed-field gel electrophoresis indicated that amplified rep-cap sequences were found in an extrachromosomal form. Cellular, but not the adenovirus, polymerase activities were required for amplification to proceed. We also documented that the DBP is the essential and sufficient adenovirus gene product, since expression of DBP alone in HeRC32 cells was able to induce rep-cap amplification. Finally, we also confirmed that Rep proteins were involved in the establishment of the phenomenon, since HeRC32 expressing DBP alone also expressed Rep proteins. Furthermore, rep-cap genome amplification was abrogated in a stable HeLa clone harboring a deleted rep-cap genome that was unable to produce Rep proteins.
MATERIALS AND METHODS
Cell lines and viruses.
HeRC32, 293RC21, and TERC21 cell clones were obtained by cotransfecting plasmid pspRC, which harbors the rep-cap genome with the ITRs deleted (bp 190 to 4484 of wild-type AAV), with plasmid PGK-Neo, conferring resistance to G418 on HeLa, 293, and TE671 (a human medulloblastoma cell line) cells, respectively. The ΔRep-HeLa cell clone was obtained using the pRCtag/Δ plasmid, in which 350 bp located at the 5′ end of the rep-cap genome (corresponding to nucleotides [nt] 191 to 540 of the wild-type AAV) was deleted. The isolation and characterization of HeRC32 and 293RC21 cells have been described elsewhere (3). TERC21 and ΔRep-HeLa cells were similarly characterized and shown to have one or less than one integrated rep-cap copy per cell genome. The B50 cell line, kindly provided by J. Wilson (University of Pennsylvania), is a HeLa-derived cell clone harboring a stably integrated, rep-cap genome with the ITRs deleted (13). The adenoviruses used were wild-type adenovirus type 5 (Ad5) (ATCC VR-5) and two thermosensitive strains, one with a mutation in the E2a gene (Ad.ts125) and one with a mutation in the E2b gene (Ad.ts149) (12). Adenoviruses were produced and titrated on 293 cells using standard procedures (14). The absence of revertants in the purified stock of Ad.ts125 and Ad.ts149 was tested at a nonpermissive temperature. The absence of contaminating wild-type AAV in the three parental cell lines (HeLa, 293, and TE671) and the adenovirus stocks was determined by PCR analysis using rep primers.
Plasmids.
To obtain the CMVDBP construct, plasmid pMSG-DBP-EN (19) was digested with KpnI, filled in with T4 polymerase, and subsequently digested with HindIII. The resulting band containing the E2a gene was gel purified and inserted into the blunt-ended pRC/CMV plasmid (Promega) which had been digested with HindIII and XbaI. Plasmid pspRC (3) contained the AAV genome with the ITRs deleted (nt 190 to 4484 of wild-type AAV) and was obtained by excising the rep-cap fragment from plasmid psub201 by XbaI digestion (29) and by inserting it in the XbaI site of plasmid pSP72 (Promega). The pRCtag/Δ plasmid contains a rep-cap sequence with 350 bp deleted (nt 191 to 540 of the wild-type AAV) followed at the 3′ end of the AAV sequences by 404 bp from φX174DNA.
Analysis of total genomic DNA by Southern blotting.
Total DNA was extracted by lysing the cells in a 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–100 mM NaCl–1% sodium dodecyl sulfate (SDS) solution containing 500 μg of proteinase K (Boehringer Mannheim)/ml. After overnight digestion at 50°C, the DNA was extracted twice with phenol-chloroform and precipitated.
For analysis, DNA was digested with the enzyme indicated, run on a 1% agarose gel, and transferred under alkaline conditions (NaOH at 0.4 N) to a Hybond N+ membrane (Amersham). The membrane was hybridized to a fluorescein-labeled probe (Gene Images random prime labeling module; Amersham) and incubated overnight at 65°C. The following day, the membrane was washed in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) (Research Organics)–0.1% SDS, and then in 0.1× SSC–0.1% SDS, for 15 min at 65°C each time. The membrane was then processed according to the manufacturer's protocol (Gene Images CDP-star detection module; Amersham) and exposed to autoradiography film.
Analysis of total genomic DNA by pulsed-field gel electrophoresis.
Cells were harvested by trypsinization, washed with phosphate-buffered saline (PBS) (KCl at 2.5 mM, KH2PO4 at 1.5 mM, NaCl at 137 mM, Na2HPO4 at 8 mM [pH 7.4]) at 37°C, resuspended at 4 × 107 cells/ml, and gently mixed with an equal volume of a 1% solution of low-melting-point agarose (SeaPlaque; FMC Bioproducts) in Mg2+-Ca2+-free PBS precooled at 50°C. The mixture was allowed to solidify in the cold, and agarose-cell plugs were then treated with proteinase K (2 mg/ml) in the presence of 1% SDS. After washing, the plugs were stored at 4°C in 20 mM Tris buffer–5 mM EDTA (pH 8.0). For digestion, the plugs were incubated for 6 h at 37°C with 50 U of enzyme in a total volume of 300 μl per plug. Electrophoresis was carried out using 1% agarose gels (SeaKem ME agarose [FMC Bioproducts] in 0.5× TBE buffer [90 mM Tris, 90 mM borate, 2 mM EDTA [pH 8.0]) at 6 V/cm for 14 to 20 h with a switching time of 50 to 90 s, using recirculating 0.5× TBE. After ethidium bromide (EtBr) staining and UV visualization, the DNA was transferred to a Hybond N+ membrane under alkaline conditions (NaOH at 0.4 N). The membrane was treated and hybridized as described above.
Immunofluorescence analysis.
Immunofluorescence analysis was performed on 5 × 104 cells seeded on glass slides. After being washed for 5 min in PBS, the cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature and then permeabilized with 2% Triton X-100 in PBS for 20 min at room temperature (RT). After a wash in PBS, the cells were incubated with 2% bovine serum albumin (BSA) in PBS for 20 min at RT and then incubated with the appropriate antibody. The primary antibody was diluted in PBS–0.1% Tween and incubated for 1 h with the fixed cells at RT. The monoclonal anti-DBP mouse antibody (kindly provided by A. Levine [26]) was diluted 1/10, and the polyclonal anti-Rep guinea pig antibodies (kindly provided by J. Kleinschmidt [39]) were diluted 1/100. Next, the slides were washed in PBS and then incubated with a fluoresceinated anti-mouse antibody (Amersham) and a rhodamine-conjugated anti-guinea-pig antibody diluted 1/200 and 1/50, respectively, in PBS–0.1% Tween for 1 h at RT in the dark. After a wash in PBS, the cells were embedded in Vectashield mounting medium (Vector Laboratories, Inc.) and analyzed using a confocal Leica DMiRBE microscope.
Fluorescent in situ hybridization (FISH) analysis.
To obtain metaphase spread, exponentially growing cells were treated with colcemid (40 ng/ml) for 1 h at 37°C. After trypsinization and centrifugation, the cell pellets were resuspended in 75 mM KCl for 35 min at 37°C. After addition of a cold methanol-acetic acid (3:1) solution, cells were pelleted, then resuspended in the same fixative solution for 10 min at 4°C, and finally dropped onto slides. Slides were air dried, and the DNA was denatured in 70% formamide–2× SSC (pH 7.0) for 1 min at 75°C. Slides were then dehydrated in an ice-cold ethanol series (70, 85, and 100% for 1 min each) and air dried. Hybridization was performed overnight at 37°C using a fluorescein-labeled probe according to the manufacturer's protocol (Nick Translation Reagent Kit; Vysis Inc.). Slides were then washed sequentially in 2× SSC for 2 min at 75°C and in 2× SSC–0.1% Triton for 2 min at RT. After being air dried in the dark, slides were dehydrated and mounted with an antifade 4′,6′-diamidino-2-phenylindole (DAPI) solution. Hybridization signals were visualized by using a Zeiss Axioplan 2 fluorescence microscope with a oil immersion objective.
RESULTS
AAV rep-cap gene amplification is induced preferentially in adenovirus-infected HeLa-derived cell clones.
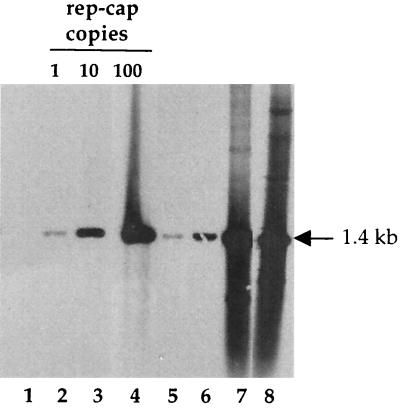
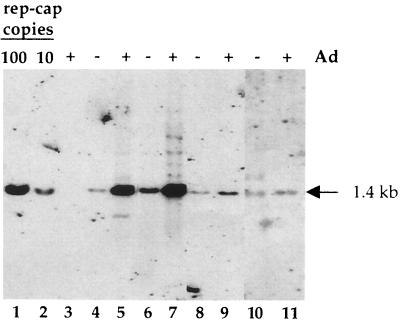
The initial observation underlying this study was made using a HeLa-derived cell clone harboring one integrated copy of rep-cap genome with an ITR deletion (HeRC32 cells) (3). When HeRC32 cells were infected with wild-type adenovirus, the integrated rep-cap copies underwent a dramatic amplification, leading to a 100-fold increase in the rep-cap copy number, as evidenced by Southern blot analysis of total DNA and hybridization with a rep probe (Fig. 1). The determination of the rep-cap copy number at different time points indicated that amplification occurred mainly between 24 and 48 h following adenovirus infection. After the 48-h time point, no significant increase was detected. To exclude the possibility that this phenomenon was due to an intrinsic property of the HeRC32 cell clone, the same analysis was performed with another HeLa-derived rep-cap cell clone (B50), which harbors five integrated rep-cap copies (13). Despite the different origin of the B50 cells, rep-cap sequences were similarly amplified following adenovirus infection (Fig. 2, lanes 6 and 7). Interestingly, the number of rep-cap copies found in the B50 cells after adenovirus infection was similar to that measured in HeRC32 cells, suggesting that the level of amplification was not dependent upon the initial rep-cap copy number (Fig. 2; compare lanes 5 and 7). The same results were obtained using a cap probe (data not shown), indicating that the entire rep-cap genome had undergone amplification. In addition, other cellular or viral endogenous sequences such as those corresponding to the elongation factor 1-α (EF1-α), bilirubin glycuronyl transferase 1 (BGT1), and human papillomavirus (HPV) genes were not found to be amplified upon adenovirus infection (data not shown), suggesting that the amplification phenomenon was restricted to rep-cap-containing sequences.
FIG. 1.
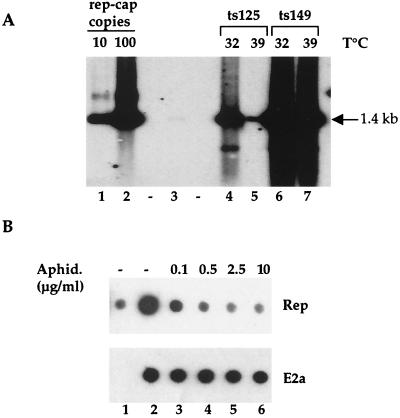
Kinetics of rep-cap amplification upon adenovirus infection. HeRC32 cells were infected with Ad5 at an MOI of 50. Total genomic DNA extracted at 24, 48, and 72 h postinfection was digested with PstI and analyzed on a Southern blot using a rep probe (1.4 kb) obtained by digesting plasmid pspRC with PstI. The position of the expected 1.4-kb rep band is indicated. The standard samples with 1, 10, and 100 rep-cap copies per cell were obtained by adding 36, 360, and 3,600 pg, respectively, of plasmid pspRC to 10 μg of total genomic DNA from noninfected HeLa cells. Lane 1, DNA from adenovirus-infected HeLa cells; lanes 2, 3, and 4, standard rep-cap genome copies; lane 5, DNA from noninfected HeRC32 cells; lanes 6, 7, and 8, DNA extracted from HeRC32 cells 24, 48, and 72 h post-adenovirus infection, respectively.
FIG. 2.
Analysis of rep-cap amplification in different stable rep-cap cell clones. The stable rep-cap cell clones analyzed are HeRC32, B50 (derived from HeLa cells [13]), 293RC21 (derived from 293 cells), and TERC21 (derived from TE671 cells). rep-cap amplification was analyzed as described in the legend to Fig. 1 following adenovirus infection of the cells at an MOI of 50 (for HeLa-derived cells), 10 (for 293-derived cells), or 25 (for TE671-derived cells). Lanes 1 and 2, standard rep-cap genome copies; lane 3, DNA from adenovirus-infected HeLa cells; lanes 4, 6, 8, and 10, DNA from noninfected HeRC32, B50, 293RC21, and TERC21 cells, respectively; lanes 5, 7, 9, and 11, DNA from adenovirus-infected HeRC32, B50, 293RC21, and TERC21 cells, respectively.
Further analyses were conducted to determine if rep-cap amplification could also take place in other rep-cap stable cell clones derived from other cell backgrounds. For this purpose, two stable cell clones derived from low-passage-number 293 (293RC21) and TE671 cells (TERC21) and harboring integrated rep-cap genomes were similarly analyzed by Southern blotting. Following adenovirus infection, the endogenous rep-cap sequences were amplified only two- to threefold in the 293RC21 cells, a level much lower than that observed in HeRC32 and B50 cells (Fig. 2, lanes 8 and 9). In TERC21 cells, no rep-cap amplification was detected (Fig. 2, lanes 10 and 11). Overall, these analyses suggested that adenovirus-induced rep-cap amplification occurred preferentially in the HeLa-derived cell clones analyzed.
Amplified rep-cap sequences are extrachromosomal.
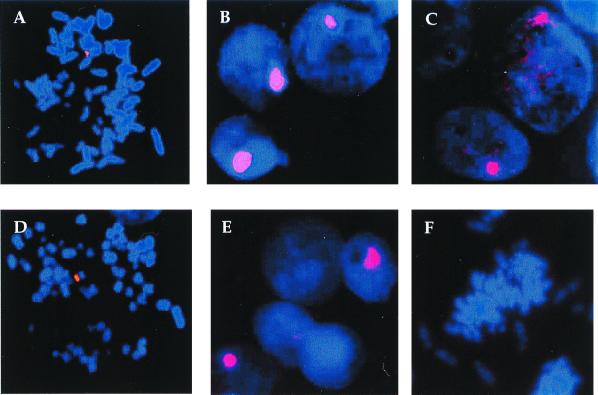
The next question concerned the status of the amplified rep-cap sequences. We wished to determine if the amplified rep-cap sequences are found in an integrated or in an extrachromosomal form. For this, rep-cap sequences present in control and adenovirus-infected HeRC32 and B50 cells were analyzed by FISH. Metaphase spreads of uninfected cells confirmed the presence of rep-cap sequences in an integrated state in both cell clones (Fig. 3A and D). The analysis performed 48 h following adenovirus infection showed an increase in the rep-cap signal, which appeared as a large dot (Fig. 3B and E). This result illustrated the amplification phenomenon previously detected by Southern blotting. However, because of the growth arrest induced by the adenovirus infection, it was not possible to visualize metaphases in these cells and thus to distinguish if the rep-cap signal following amplification colocalized with a chromosomal structure. To try to visualize intermediate forms of amplification, HeRC32 cells were infected with wild-type adenovirus at a suboptimal multiplicity of infection (MOI) of 1. In this case, different patterns could be observed. Particularly, some nuclei displayed a strong rep-cap signal, which was not concentrated in a single spot but was rather diffuse (Fig. 3C). This result suggested that amplified rep-cap sequences were present in an extrachromosomal form.
FIG. 3.
FISH analysis of noninfected and adenovirus-infected HeRC32 and B50 cells. Cells were prepared for FISH analysis as described in Materials and Methods, and were analyzed using a fluorescein-labeled rep-cap probe (4.5 kb) obtained by digesting pspRC with XbaI. (A) noninfected HeRC32 cells; (B) adenovirus-infected HeRC32 cells (MOI, 50); (C) adenovirus-infected HeRC32 cells (MOI, 1); (D) noninfected B50 cells; (E) adenovirus-infected B50 cells (MOI, 50); (F) noninfected control HeLa cells. Magnification, ×1,000.
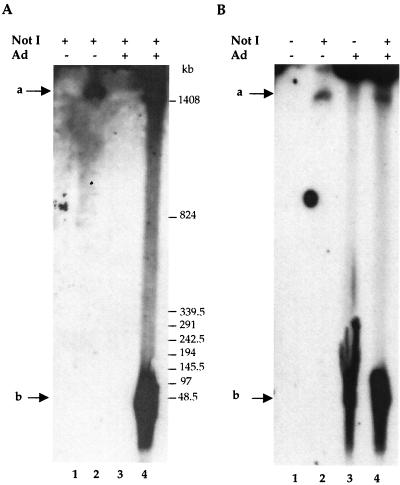
To confirm this observation, total genomic DNA extracted from infected and uninfected HeRC32 cells, was analyzed by pulsed-field gel electrophoresis followed by Southern blot analysis using a rep probe. Digestion of total DNA extracted from uninfected HeRC32 cells with NotI, which does not cut the rep-cap DNA, released a unique high-molecular-weight band presumably containing the integrated rep-cap copies (Fig. 4A, lane 2). Following adenovirus infection of HeRC32 cells, an additional, faster-migrating form was detected (Fig. 4A, lane 4). Neither of these signals was detected by using DNA from control or adenovirus-infected HeLa cells (Fig. 4A, lanes 1 and 3). The highest-molecular-weight band seen with DNA from adenovirus-infected HeRC32 cells was not detected by using undigested DNA (Fig. 4B, lane 3), highlighting the specificity of the probe. Conversely, the faster-migrating band was still detected using by undigested DNA (Fig. 4B, lane 3), suggesting that this form corresponded to an extrachromosomal molecule containing rep-cap sequences.
FIG. 4.
Analysis of rep-cap amplified DNA molecules by pulsed-field gel electrophoresis. Samples for pulsed-field gel electrophoresis were prepared from noninfected or adenovirus-infected HeRC32 cells (MOI, 50) as described in Materials and Methods and were analyzed using a rep probe (1.4 kb). Where indicated, DNA was digested with NotI, which does not cut in the rep-cap genome. (A) Lanes 1 and 2, noninfected HeLa and HeRC32 cells, respectively; lanes 3 and 4, adenovirus-infected (48 h) HeLa and HeRC32 cells, respectively. (B) Lanes 1 and 2, noninfected HeRC32 cells; lanes 3 and 4, adenovirus-infected (48 h) HeRC32 cells. The two arrows indicate the positions of the integrated (a) and extrachromosomal (b) rep-cap fragments.
Cellular but not adenovirus polymerases are involved in the amplification process.
The above results indicated that upon adenovirus infection, integrated rep-cap sequences were amplified and extruded from the chromosomal structure. To further elucidate this phenomenon, it was important to determine if the amplification of rep-cap sequences resulted from the activity of cellular or adenovirus polymerases. To answer this question, rep-cap amplification was analyzed after infection of HeRC32 cells with an adenovirus mutant harboring a thermosensitive mutation in the E2b gene encoding the viral polymerase (Ad.ts149). HeRC32 cells were infected with Ad.ts149 and maintained for 48 h at either 32°C (the permissive temperature) or 39°C (the nonpermissive temperature). Analysis of total DNA by Southern blotting and hybridization with a rep probe indicated that inactivation of the adenovirus polymerase at 39°C did not inhibit rep-cap amplification, which reached a level similar to that observed in cells infected at 32°C (Fig. 5A, lanes 6 and 7). This result indicated that the adenovirus polymerase was not involved in the rep-cap amplification and further suggested the involvement of cellular polymerases in this process.
FIG. 5.
(A) Effect of thermosensitive adenovirus mutants on rep-cap amplification. HeRC32 cells were infected with Ad.ts125 or Ad.ts149 at an MOI of 50 and incubated at either 32 or 39°C. Forty-eight hours later, total genomic DNA was extracted and analyzed using a rep probe as indicated in the legend to Fig. 1. Lanes 1 and 2, standard rep-cap genome copies; lane 3, DNA from noninfected HeRC32 cells; lanes 4 and 5, DNA from HeRC32 cells infected with Ad.ts125 at 32 and 39°C, respectively; lanes 6 and 7, DNA from HeRC32 cells infected with Ad.ts149 at 32 and 39°C, respectively. The position of the expected 1.4-kb rep band is indicated. (B) Effect of aphidicolin on adenovirus-induced rep-cap amplification. HeRC32 cells were infected with Ad5 (MOI, 50) for 2 h at 37°C and then either left untreated or incubated in the presence of aphidicolin at the final concentrations indicated. Two micrograms of total DNA extracted 48 h later was analyzed by dot blot using a rep (1.4-kb) or DBP (1.6-kb) probe. The DBP probe was obtained by digesting plasmid pMSG-DBP-EN (19) with HindIII and SfiI. Lane 1, DNA from noninfected HeRC32 cells; lane 2, DNA from adenovirus-infected HeRC32 cells; lanes 3 to 6, DNA from adenovirus-infected HeRC32 cells incubated in the presence of increasing concentrations of aphidicolin.
To confirm this hypothesis, rep-cap amplification was analyzed in the presence of an inhibitor of cellular polymerases. For this, HeRC32 cells were infected with wild-type adenovirus for 2 h. After this period, the medium was changed and cells were incubated with different concentrations of aphidicolin, a drug known to inhibit the activity of polymerases α, δ, and ɛ (16, 20). Two days later, DNA was analyzed by dot blot and hybridized either to a rep probe, to monitor rep-cap amplification, or to an E2a probe, to monitor the effect of the drug on adenovirus replication. As shown in Fig. 5B, the addition of aphidicolin strongly inhibited rep-cap amplification, with a maximum effect reached at a concentration of 2.5 μg/ml. In contrast, aphidicolin did not inhibit adenovirus replication. Overall, these results indicated that a cellular polymerase(s) was involved in the amplification process.
rep-cap amplification can be induced in the presence of DBP and Rep proteins.
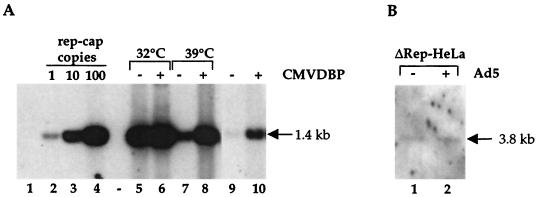
Previous results indicated that the adenovirus E2b gene was not necessary for rep-cap amplification. To further investigate the role of adenovirus, the same analysis was performed using another adenovirus mutant harboring a thermosensitive mutation in the E2a gene encoding the DBP (Ad.ts125). As previously described, HeRC32 cells were infected with Ad.ts125 and maintained for 48 h at either 32°C (the permissive temperature) or 39°C (the nonpermissive temperature). Analysis of the rep-cap copy number by Southern blotting indicated that amplification was severely reduced upon inactivation of the DBP (Fig. 5, lanes 4 and 5). This result suggested that this adenovirus factor might play a key role in the observed phenomenon. To confirm this hypothesis, a plasmid harboring the E2a gene under the control of the cytomegalovirus (CMV) promoter (CMVDBP) was transfected into HeRC32 cells 6 h prior to infection with Ad.ts125 at both the permissive and nonpermissive temperatures. Analysis of rep-cap DNA 48 h after infection revealed that rep-cap amplification could be restored to normal levels when cells were infected with Ad.ts125 at 39°C and transfected with CMVDBP (Fig. 6, lanes 7 and 8).
FIG. 6.
(A) Effect of the adenovirus DBP on rep-cap amplification. HeRC32 cells were infected with Ad.ts125 (MOI, 50) at the indicated temperature, and total DNA was analyzed by Southern blotting using a rep probe (1.4 kb) as described in the legend to Fig. 1. Where indicated, the CMVDBP plasmid (10 μg) was transfected into 4 × 106 HeRC32 cells using Exgen (EuroMedex), either alone or 6 h prior to adenovirus infection. In this case, the transfection was done at 37°C and the cells were switched to the indicated temperature immediately after adenovirus infection. Lane 1, DNA from noninfected HeLa cells; lanes 2, 3, and 4, standard rep-cap genome copies; lane 5, DNA from HeRC32 cells infected with Ad.ts125 at 32°C; lane 6, DNA from HeRC32 cells transfected with CMVDBP and infected with Ad.ts125 at 32°C; lane 7, DNA from HeRC32 cells infected with Ad.ts125 at 39°C; lane 8, DNA from HeRC32 cells transfected with CMVDBP and infected with Ad.ts125 at 39°C; lane 9, DNA from noninfected HeRC32 cells; lane 10, DNA from HeRC32 cells transfected with the CMVDBP plasmid. (B) Analysis of rep-cap amplification in ΔRep-HeLa cells. Total DNA was extracted from uninfected (lane 1) and adenovirus-infected (lane 2) ΔRep-HeLa cells, digested with PstI, and analyzed on a Southern blot as previously indicated. Since the deletion in the rep sequence removes one PstI site, the size of the expected band is 3.8 kb.
To further validate the role of DBP in the amplification process, HeRC32 cells were transfected with plasmid CMVDBP alone and analyzed for rep-cap copy number by Southern blotting. A detectable level of amplification was seen under this condition (Fig. 6A, lane 10). The relatively low level of amplification seen upon transfection of CMVDBP was likely due to the inefficient transfection of this plasmid in HeRC32 compared to the efficiency of adenovirus infection.
To verify this, HeRC32 cells transfected with the CMVDBP plasmid were analyzed by FISH to detect rep-cap amplification. As shown in Fig. 7A and B, an amplified rep-cap signal was detected in a small proportion of cells, reflecting the overall transfection efficiency (approximately 5%). As previously observed in adenovirus-infected HeRC32 cells, it was not possible to visualize metaphases in cells displaying an amplified rep-cap signal. No amplification was observed using a control plasmid (data not shown). These results indicated that among the adenovirus genes, the gene encoding the DBP was sufficient to support rep-cap amplification.
FIG. 7.
FISH analysis of HeRC32 cells transfected with the CMVDBP plasmid. A total of 4 × 106 HeRC32 cells were transfected with 10 μg of the CMVDBP plasmid using Exgen (EuroMedex). Forty-eight hours later, the cells were prepared for FISH analysis as indicated in Materials and Methods. The samples were analyzed using a fluorescein-labeled rep-cap probe. Two typical examples of rep-cap amplification are shown. (A) Untransfected HeRC32 cells; (B and C) transfected HeRC32 cells. Magnification, ×1,000.
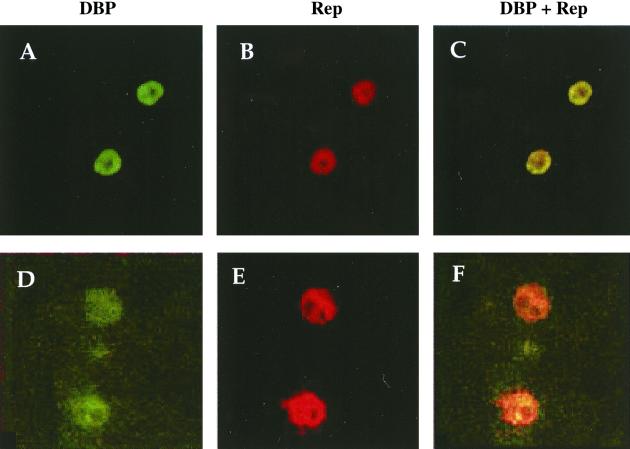
If these results clearly identified the DBP as the adenovirus factor able to induce the amplification process, they did not exclude the possibility that other proteins, and particularly the Rep proteins, participated in this phenomenon. To elucidate this point, HeRC32 cells transfected with the CMVDBP plasmid were first analyzed by immunofluorescence to detect Rep protein synthesis. As shown in Fig. 8, both spliced and unspliced Rep proteins were detected in cells transfected with the CMVDBP plasmid alone. This result indicated that Rep proteins were expressed in cells transfected with the CMVDBP plasmid and further suggested their involvement in the amplification process. To confirm this hypothesis, a stable cell clone harboring a mutated rep-cap genome (ΔRep-HeLa), unable to produce Rep proteins, was isolated. As expected, no amplification of integrated rep-cap sequences was detected following wild-type adenovirus infection (Fig. 6B). Overall, these results strongly suggested that the Rep proteins were implicated in the amplification process.
FIG. 8.
Detection of Rep and DBP proteins following transfection of the CMVDBP plasmid into HeRC32 cells. A total of 6 × 104 HeRC32 cells grown on glass slides were transfected with 0.4 μg of the CMVDBP plasmid. Forty-eight hours later, the cells were fixed and analyzed by immunofluorescence using an anti-DBP (26) and an anti-Rep 68/40 (A, B, and C) or anti-Rep 78/52 (D, E, and F) antibody (39). Cells were photographed with either a fluorescein (A and D) or a rhodamine (B and E) filter. In panels C and F, the two images are superimposed. Magnification, ×1,000.
DISCUSSION
rep-cap amplification, first mentioned by Liu et al. (21), was described using the HeRC32 cell line (3). Using Southern blot analysis we showed that 48 h after adenovirus infection, the rep-cap copy number was increased at least 100-fold. This increase in the number of rep-cap genome copies correlated with both a high level of Rep and Cap protein synthesis and rAAV assembly, thus supporting the idea that the newly amplified rep-cap copies were used as templates for rep and cap gene expression.
In this study, we further investigated the mechanisms underlying rep-cap amplification. First, by comparing different stable rep-cap cell lines, we found that among the various cell backgrounds examined, rep-cap amplification occurred preferentially in the HeLa-derived cell clones. rep-cap sequences integrated in the genome of 293 and TE671 cells were barely amplified (Fig. 2). This observation suggests that the HeLa cell background is critical for this phenomenon, and it can be related to the fact that, at least in our hands, this cell type is also optimal for rAAV production (3). Interestingly, cellular extracts from uninfected HeLa cells have been reported to be able to support in vitro AAV replication in the presence of Rep proteins (24, 37). These characteristics might be related to the presence in these cells of several copies of an HPV18 genome in which E2 is deleted (22). Indeed, HPV has also been reported to exert a helper activity for AAV replication (25, 34). Alternatively, these properties might be related to the presence of a cell type-specific factor. We are currently testing these hypotheses by examining if rep-cap amplification can also occur in stable rep-cap cell clones derived from SiHa cells which, like the HeLa cells, harbor the HPV genome (22).
Second, this study examined the status of the amplified rep-cap sequences. The data obtained by FISH analysis confirmed the tremendous increase in the rep-cap copy number detected by Southern blotting (Fig. 3). However, a clear-cut analysis of the status of these amplified sequences was obtained only after pulsed-field gel electrophoresis of the DNA. Using this method, it was found that amplified DNA is present in an extrachromosomal form 48 h after adenovirus infection (Fig. 4).
Amplification of endogenous cellular genes, and particularly oncogenes, has been extensively described as a common phenomenon occurring during tumor progression. Furthermore, cellular gene amplification can also occur as a response to various drugs such as DNA-damaging agents (30). Amplified sequences are found either integrated, under the form of homogeneously staining regions (HSR), or extrachromosomally. In this case, amplified sequences are usually identified as double-minute chromosomes (DMs). These high-molecular-weight circular DNA molecules autonomously replicate using a cellular replication origin, but, lacking centromeres, they do not segregate with chromosomes and as a consequence are usually lost upon cell division (33). A third class of amplified structures has also been described as submicroscopic circular DNA molecules termed “episomes”. Although the precise mechanism of gene amplification is still unclear, it has been proposed that DMs, which are the predominant cytogenic manifestation of gene amplification, are derived from smaller episomes which progressively enlarge and can lead to HSR by integrating back in the chromosomal structure (33). The extrachromosomal rep-cap sequences detected in our model might be defined as episomal structures resembling those leading to DMs. It should be noted that rep-cap amplification was not observed following treatment of the cells with DNA-damaging agents such as hydroxyurea, UV exposure, and the X-ray irradiation (data not shown). As such, rep-cap amplification could represent a unique model of gene amplification.
Third, this study aimed at identifying the minimal cellular and viral factors involved in rep-cap amplification. Using an adenovirus harboring a thermosensitive mutation in the E2b gene, we found that rep-cap amplification still occurred even in the absence of a functional adenovirus polymerase (Fig. 5A). This result also indicated that adenovirus replication per se was not required for rep-cap amplification. As shown in the case of wild-type AAV DNA replication (24), we further demonstrated that rep-cap amplification can be completely abolished by treating the cells with aphidicolin (Fig. 5B), a drug known to inhibit the activity of the cellular polymerases α, δ, and ɛ (16, 20). The similarity to wild-type AAV replication extends further to the requirement for a functional DBP. Indeed, by using an adenovirus harboring a thermosensitive mutation in the E2a gene, it was shown that the DBP was essential for rep-cap amplification (Fig. 5A and 6A). This protein is the only adenovirus factor directly implicated in AAV DNA replication. Ward et al. recently showed that DBP was essential in vitro, to increase processing of DNA replication, presumably by stabilizing single-stranded templates (35, 36). The involvement of DBP in rep-cap amplification was further demonstrated by transfecting a plasmid encoding this protein into HeRC32 cells and by showing that amplification events could be detected by Southern blotting and FISH analysis (Fig. 6A and 7). Although these results do not exclude the implication of other adenovirus factors in rep-cap amplification, they clearly demonstrated that the DBP alone is sufficient.
The last question concerned the role of the AAV gene products and particularly the Rep proteins. We found that, upon transfection of the CMVDBP plasmid, both spliced and unspliced Rep proteins were detected (Fig. 8). This observation, which is in agreement with a previous report by Chang and Shenk, who demonstrated that DBP was able to trans-activate the p5 promoter (4), suggested the possible involvement of Rep proteins in the amplification process. Abolishment of Rep proteins in adenovirus-infected stable HeLa cell clones (ΔRep-HeLa) harboring a rep-cap genome unable to produce Rep proteins also suggested that they are needed for amplification (Fig. 6B). Importantly, the fact that Rep 78 and Rep 52 were still expressed in Ad.ts125-infected HeRC32 cells at a nonpermissive temperature (data not shown), i.e., under conditions in which amplification no longer occurred (Fig. 5A), further confirmed that Rep proteins, and particularly Rep 78 and Rep 52, were not sufficient alone and that a functional DBP was also needed to induce rep-cap amplification. Finally, although the DBP is able to stimulate Rep protein synthesis alone (4), it is possible that a maximal level of amplification requires an optimal rate of rep gene expression that is obtained only in the presence of the E1a gene product (5).
Given these findings, we assume that rep-cap amplification is the result of the activity of at least three main factors: DBP, cellular polymerases, and Rep proteins. It remains to be seen if rep-cap amplification results from the presence of a cellular origin of replication or from one present in the viral genome. Analysis of stable rep-cap cell clones harboring critical deletions of the AAV rep-cap sequences will help resolve this issue. It is possible to envision that the combination of these trans (Rep, DBP, cellular polymerases, and presumably some unknown factor related to HeLa cells) and cis (a viral or cellular origin of replication) elements generate unscheduled overreplication of rep-cap sequences. The fact that the endogenous integrated rep-cap copies are still detected in adenovirus-infected HeRC32 cells (Fig. 4B) indicates that the original rep-cap sequences are not excised from the chromosome during rep-cap amplification. Further analysis of these extrachromosomal molecules together with the sequence of the integrated rep-cap genomes will help define the mechanism of amplification.
In conclusion, our observations constitute a first step toward the elucidation of the mechanism underlying rep-cap amplification in HeLa cells. These findings have important implications for the development of future generations of rep-cap cell lines able to produce optimal levels of Rep and Cap proteins upon adenovirus infection.
ACKNOWLEDGMENTS
We are grateful to Michael Linden and Matthew Weitzman for critical reading of the manuscript. We thank Marie-Claire Devilder and Jean-Paul Moisan for technical assistance.
This work was supported by the Association Française contre les Myopathies (AFM), Vaincre les Maladies Lysosomales (VML), the Association Nantaise de Thérapie Génique (ANTG), and the Fondation pour la Thérapie Génique en Pays de la Loire. P.M. was supported by a sponsored research agreement from Genopoietic Inc.
J. Tessier and G. Chadeuf contributed equally to this work.
REFERENCES
- 1.Berns K I, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Carter B J. Adeno-associated virus helper functions. In: Tijssen P, editor. Handbook of parvoviruses. Boca Raton, Fla: CRC Press; 1990. pp. 255–282. [Google Scholar]
- 3.Chadeuf G, Favre D, Tessier J, Provost N, Nony P, Kleinschmidt J, Moullier P, Salvetti A. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Chang L-S, Shenk T. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–2109. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang L-S, Shi Y, Shenk T. Adeno-associated virus p5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 7.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark K R, Liu X, McGrath J P, Johnson P R. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum Gene Ther. 1999;10:1031–1039. doi: 10.1089/10430349950018427. [DOI] [PubMed] [Google Scholar]
- 9.Clark K R, Voulgaropoulou F, Fraley D M, Johnson P R. Cell lines for the production of recombinant adeno-associated virus. Hum Gene Ther. 1995;6:1329–1341. doi: 10.1089/hum.1995.6.10-1329. [DOI] [PubMed] [Google Scholar]
- 10.Conway J E, ap Rhys C M J, Zolotukhin I, Zolotukhin S, Muzyczka N, Hayward G S, Byrne B J. High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I expressing AAV-2 rep and cap. Gene Ther. 1999;6:986–993. doi: 10.1038/sj.gt.3300937. [DOI] [PubMed] [Google Scholar]
- 11.Dubielzig R, King J A, Weger S, Kern A, Kleinschmidt J A. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J Virol. 1999;73:8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensinger M J, Ginsberg H S. Selection and preliminary characterization of temperature-sensitive mutants of type 5 adenovirus. J Virol. 1972;10:328–339. doi: 10.1128/jvi.10.3.328-339.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao G-P, Qu G, Faust L Z, Engdahl R K, Xiao W, Hughes J V, Zoltick P W, Wilson J M. High-titer adeno-associated viral vectors from a rep/cap cell line and hybrid shuttle virus. Hum Gene Ther. 1998;9:2353–2362. doi: 10.1089/hum.1998.9.16-2353. [DOI] [PubMed] [Google Scholar]
- 14.Graham F L, Prevec L. Manipulation of adenovirus vectors. In: Murray E J, editor. Gene transfer and protocol. Clifton, N.J: The Humana Press, Inc.; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 15.Grimm D, Kleinschmidt J A. Progress in adeno-associated virus type 2 vector production: promises and prospects for clinical use. Hum Gene Ther. 1999;10:2445–2450. doi: 10.1089/10430349950016799. [DOI] [PubMed] [Google Scholar]
- 16.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 17.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 18.Inoue N, Russel D W. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klessig D F, Brough D E, Cleghon V. Introduction, stable integration, and controlled expression of a chimeric adenovirus gene whose product is toxic to the recipient human cell. Mol Cell Biol. 1984;4:1354–1362. doi: 10.1128/mcb.4.7.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M Y W T, Tan C-K, Downey K M, So A G. Structural and functional properties of calf thymus DNA polymerase delta. Prog Nucleic Acids Res Mol Biol. 1981;26:83–96. doi: 10.1016/s0079-6603(08)60396-7. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Clark K R, Johnson P R. Production of recombinant adeno-associated virus vectors using a packaging cell line and a hybrid recombinant adenovirus. Gene Ther. 1999;6:293–299. doi: 10.1038/sj.gt.3300807. [DOI] [PubMed] [Google Scholar]
- 22.Meissner J D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 23.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 24.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogston P, Raj K, Beard P. Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J Virol. 2000;74:3494–3504. doi: 10.1128/jvi.74.8.3494-3504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 27.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvetti A, Chadeuf G, Orève S, Favre D, Cherel Y, Champion- Arnaud P, David-Ameline J, Moullier P. Factors influencing recombinant adeno-associated virus production. Hum Gene Ther. 1998;9:695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- 29.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimke R T. Gene amplification in cultured cells. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- 31.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder R O. Adeno-associated virus-mediated gene delivery. J Gene Med. 1999;1:166–175. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<166::AID-JGM34>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 33.Wahl G M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 34.Walz C, Deprez A, Dupressoir T, Dürst M, Rabreau M, Schlehofer J R. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J Gen Virol. 1997;78:1441–1452. doi: 10.1099/0022-1317-78-6-1441. [DOI] [PubMed] [Google Scholar]
- 35.Ward P, Dean F B, O'Donnell M E, Berns K I. Role of the adenovirus DNA-binding protein in in vitro adeno-associated virus DNA replication. J Virol. 1998;72:420–427. doi: 10.1128/jvi.72.1.420-427.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward P, Linden R M. A role for single-stranded templates in cell-free adeno-associated virus DNA replication. J Virol. 2000;74:744–754. doi: 10.1128/jvi.74.2.744-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wonderling R S, Kyostio S R, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Zolotukhin I, Im D S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]