Abstract
The Epstein-Barr virus (EBV) BamHI A mRNAs were originally identified in cDNA libraries from nasopharyngeal carcinoma, where they are expressed at high levels. The RNAs are differentially spliced to form several open reading frames and also contain the BARF0 open reading frame at the 3′ end. One cDNA, RK-BARF0, included a potential endoplasmic reticulum-targeting signal peptide sequence. The RK-BARF0 protein is shown here to interact with the Notch4 ligand binding domain, using yeast two-hybrid screening, coimmunoprecipitation, and confocal microscopy. This interaction induces translocation of a portion of the full-length unprocessed Notch4 to the nucleus by using the Notch nuclear localization signal. These effects of RK-BARF0 on Notch intracellular location indicate that EBV possibly modulates Notch signaling. Unprocessed Notch4 was also detected in immunoprecipitated complexes from EBV-infected cells by using a rabbit antiserum raised against a BARF0-specific peptide. This finding provides additional evidence for expression of RK-BARF0 and its interaction with Notch during EBV infection. In EBV-infected, EBNA2-negative cells, RK-BARF0 induced the expression of EBV latent membrane protein 1 (LMP1), and this induction was dependent on the RK-BARF0/Notch interaction domain. The activation of LMP1 expression by RK-BARF0 may be responsible for expression of LMP1 in EBV latent infections in the absence of EBNA2.
Epstein-Barr virus (EBV) is linked to the development of several malignancies including endemic Burkitt's lymphoma, posttransplantation lymphoma, Hodgkin's disease, and nasopharyngeal carcinoma (NPC) (35). Infection of B lymphocytes with EBV in vitro results in immortalized lymphoblastoid cells (23). Although virus is not produced in transformed lymphocytes, at least nine viral proteins, including six nuclear antigens (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNALP) and three membrane proteins (LMP1, LMP2A, and LMP2B), are expressed (23). Molecular genetic analyses have demonstrated that EBNA2, EBNA3A, EBNA3C, EBNALP, and LMP1 are critical for in vitro B-lymphocyte transformation (5, 22, 27, 46), while EBNA3B, LMP2A, and LMP2B are dispensable (26, 45). EBNA2 is the major regulator of viral transcription and transactivates the expression of LMP1 (50), in part through its interaction with the DNA binding protein, RBP-Jκ (CBF1) (14, 15, 19). RBP-Jκ is a component of the Notch signaling pathway and is activated through its interaction with the intracellular cytoplasmic domain of Notch (9, 44). These interactions convert RBP-Jκ from a repressor of transcription to an activator (18). EBNA2 seems to be a functional homologue of activated Notch in EBV-infected cells.
EBV gene expression varies in infected tissue in the distinct diseases and in different cell types. In NPC, most of the nuclear antigens are not detected and only EBNA1, LMP1, and LMP2A are consistently expressed (4, 7, 51). In addition, a family of rightward transcripts from the BamHI A region was originally identified in NPC, where they are abundantly and consistently expressed (3, 12, 13, 16). Structural analysis of clones obtained from a cDNA library of nude-mouse-passaged NPC tumor, C15, revealed that the BamHI A transcripts are differentially spliced, giving rise to a family of transcripts, which are 3′-end coterminal (37, 41). All of the transcripts include the open reading frame (ORF) BamHI A rightward frame 0 (BARF0) at their 3′ ends, which is predicted to encode a 174-amino-acid (aa) protein. Patients with NPC have antibodies to the in vitro translation product of this ORF, suggesting that the protein is expressed in vivo (13). Several additional ORFs are formed by the distinct splices. One cDNA, RK-BARF0, was obtained that extended the BARF0 ORF at the 5′ end and would potentially encode a 279-aa protein (37).
The N terminus of RK-BARF0 protein contains a highly hydrophobic region that resembles an endoplasmic reticulum (ER)-targeting signal peptide sequence; however, other potential motifs were not identified. To clarify the function of the RK-BARF0 protein, the yeast two-hybrid system was used to identify cellular proteins that potentially interact with RK-BARF0 in epithelial cells. Using an epithelial cell-specific cDNA library, the RK-BARF0 protein was found to interact with extracellular domain of the Notch family proteins Notch3 and Notch4.
Notch defines a family of transmembrane receptor proteins found in a variety of organisms including mammals (43). Notch is synthesized in the ER and further processed in the trans-Golgi network (TGN) to produce two fragments, which are then linked through disulfide bonds and transported to the plasma membrane (2, 34). The amino-terminal fragment is the extracellular subunit and contains multiple epidermal growth factor (EGF)-like repeats. Through direct contact with a cell that expresses a Notch ligand, such as Delta or Jagged, the cytoplasmic domain of Notch is proteolytically cleaved and released from the plasma membrane (38, 42). The cleaved form of Notch is then thought to interact with modulators and effectors of Notch signaling such as Deltex and the RBP-Jκ transcription factor, the mammalian homologue of the Drosophila Suppressor of Hairless protein (18). The relative abundance, subcellular location, and intermolecular interactions of Notch all contribute to the multiplicity of effects of Notch signaling. This study evaluates the effects of the RK-BARF0/Notch interaction on Notch and on viral gene expression.
MATERIALS AND METHODS
Plasmid construction.
The GAL4 DNA binding domain (GAL4BD) fusions with RK-BARF0 and deletion mutants were constructed in vector pAS2-1 (Clontech) by PCR amplification of the RK-BARF0 cDNA fragments encoding aa 12 to 279, 119 to 279, 12 to 128, 12 to 158, 12 to 179, 12 to 209 and 12 to 249 of RK-BARF0, followed by cloning into the NdeI-SalI or NdeI-SmaI sites of pAS2-1 to produce pASRK12–279, pASRK119–279, pASRK12–128, pASRK12–158, pASRK12–179, pASRK12–209, pASRK12–249, respectively. To express RK-BARF0 and its C-terminally truncated mutants with epitope tags, pRK-BARF0, pRK1–179, and pRK1–158 were amplified by PCR from the RK-BARF0 cDNA fragment, encoding aa 1 to 279, 1 to 179, or 1 to 158 of RK-BARF0 with FLAG and six-histidine (FLAG/His6) epitope tags in the carboxy terminus, respectively, and subcloned into the blunt-ended HindIII site of pMEP4 (Invitrogen). pBARF0 was constructed by PCR amplification of pRK-BARF0, encoding aa 106 to 279 of RK-BARF0 with a C-terminal FLAG epitope tag, and was also subcloned into pMEP4.
pNot4 containing full-length human Notch4 cDNA was constructed by fusing four clones from the human lung MATCHMAKER cDNA library (Clontech) and a DNA fragment, containing nucleotides 3011 to 6033 of Notch4 cDNA (kindly provided by K. Sugaya, National Institute of Radiological Sciences, Chiba, Japan). To express Notch4 protein with three myc epitope tags at the C terminus (Notch4-myc), pNot4M3 was constructed by PCR amplification of pNot4 and subcloning into pA3M (1), encoding three iterated copies of the myc epitope. To express intracellular functional domains of truncated Notch4 protein (TMNotch4) which is lacking the region of Notch4 from aa 1559 to 2010, two DNA fragments, including nt 1 to 3776 and 3770 to 4675, were isolated by EcoRI-ScaI digestion of pNot4 and PCR amplification of pNot4, respectively. These DNA fragments were subcloned into EcoRI-XbaI sites of pA3M to yield pNot4TM. An inducible Notch4 expression plasmid was synthesized by transferring an EcoRI-XbaI fragment from the myc-tagged Notch4 gene from pNot4M3 into the HindIII site of pMEP4 to yield pNot4(M). All of the DNA constructs were sequenced at the University of North Carolina—Chapel Hill (UNC) Automated DNA Sequencing Facility on a model 377 DNA Sequencer (Perkin-Elmer, Applied Biosystems Division).
Yeast two-hybrid screening and analysis.
Yeast transformation was performed by the method of Gietz et al. (11). The yeast strain Y190 (Clontech) was transformed with pASRK12-279, and transformants were plated on Trp− selection medium. GAL4BD-RK12-297 fusion protein expression was verified in single colonies by immunoblotting using the GAL4 DNA-BD antibody (Clontech). The pASRK12-279 transformant was subsequently transformed with the human lung MATCHMAKER cDNA library, and selection was done on Trp− Leu− His− selection medium in the presence of 25 mM 3-aminotriazole (Sigma). Colonies with moderate to intense growth were tested for β-galactosidase (β-gal) expression.
To verify the interaction of these clones with RK12-279, positive colonies were plated on Leu− selection medium containing 10 μg of cycloheximide per ml to lose pASRK12-279, and Y190 cells carrying only library-derived plasmid were mated with the Y187 transformant with pAS2–1, pASRK12–128, or pASRK119–279. Library-derived plasmids were recovered by transformation of competent DH5α cells with total DNA preparations from Y190 cells followed by selection for ampicillin resistance. To identify the interaction domain of RK-BARF0 with the cDNA clone from yeast two-hybrid screening, Y190 was transformed with pASRK12–128, pASRK12– 158, pASRK12–179, pASRK12–209, pASRK12–249, and pASRK12–279 and mated with library-derived plasmid-transformed Y187. Mated yeast cells were plated on Trp− Leu− His− selection medium containing 25 mM 3-aminotriatole and tested for β-gal expression.
Cell culture and transfection.
The H1299 cell line, derived from a human non-small-cell lung carcinoma, was maintained in Dulbecco's modified Eagle's medium containing penicillin and streptomycin and supplemented with 10% fetal bovine serum (Gibco BRL). To obtain H1299 cells expressing FLAG/His6 epitope-tagged RK-BARF0, RK1–179, RK1–158, BARF0, or pMEP4 and myc epitope-tagged Notch4 or pA3M, the DNA constructs were transfected using Lipofectin (Gibco BRL). To obtain stably expressing cell lines, transfected H1299 cells were selected in the presence of 200 μg of hygromycin B (Boehringer) per ml and 600 μg of G418 (Gibco BRL) per ml. BJAB and P3HR1 cell lines, derived from EBV-negative and -positive Burkitt's lymphoma, respectively, were maintained in RPMI 1640 medium containing penicillin and streptomycin and supplemented with 10% fetal bovine serum. Stable DG75 and Raji cell lines containing the inducible myc epitope-tagged Notch4 or pMEP4 and stable P3HR-1 cell lines containing the inducible FLAG/His6 epitope-tagged RK-BARF0, RK1–179, RK1–158, or pMEP4 were produced after electroporation and selection in the presence of 200 μg of hygromycin B.
Coimmunoprecipitation of RK-BARF0 and Notch4 from H1299 epithelial cells.
H1299 cells (5 × 105) were transiently transfected with 2.5 μg of FLAG/His6 epitope-tagged RK-BARF0, RK1–179, RK1–158, or pMEP4 and 2.5 μg of myc epitope-tagged Notch4 or pA3M plasmids and induced by addition of 5 μM of CdCl2 approximately 24 h after transfection. The cells were scraped from tissue culture dishes at 16 h postinduction and lysed for 30 min on ice in lysis buffer (20 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% 3-[(3-cholamidopropyl)-dimethylammonia]-1-propanesulfonate [CHAPS]) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM sodium orthovanadate, and 5 μg of aprotinin per ml. Cell lysates were clarified by centrifugation. The myc epitope-tagged Notch4 or FLAG/His6 epitope-tagged RK-BARF0 were immunoprecipitated with anti-FLAG M2 monoclonal antibody-conjugated beads (Sigma) or anti-myc monoclonal antibody-conjugated beads (Santa Cruz) for 8 h at 4°C. The beads were then washed, and protein complexes were recovered by boiling in sodium dodecyl sulfate (SDS) sample buffer. Protein samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to Immobilon-P (Millipore). Anti-OctA-probe polyclonal antibody (Santa Cruz) was used at 0.5 μg per ml for the detection of FLAG/His6 epitope-tagged RK-BARF0, RK1–179 or RK1–158. Anti-myc probe polyclonal antibody (Santa Cruz) was used at 0.5 μg per ml for the detection of myc epitope-tagged Notch4. Horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham) was used at a 1:2,000 dilution as the secondary antibody to detect bound primary antibody. Reactive proteins were detected by incubation of washed filters in SuperSignal substrate (Pierce) followed by exposure to X-ray film (Kodak).
Coimmunoprecipitation of Notch4 from Burkitt's lymphoma cell lines using rabbit antiserum raised against an RK-BARF0 peptide.
DG75 and Raji cell lines containing the inducible myc epitope-tagged Notch4 or pMEP4 were induced by addition of 5 μM CdCl2 approximately 36 h after plating. The cell lysates were prepared as described above and were immunoprecipitated using an affinity-purified rabbit antiserum raised against a BARF0-specific peptide and Gammabind Sepharose beads (Pharmacia). The beads were washed, and protein complexes were recovered by boiling in SDS sample buffer. To detect the myc-tagged Notch4, the protein samples were subjected to SDS-PAGE, transferred to Immobilon-P, and reacted with anti-myc epitope monoclonal antibody 9E10 (Santa Cruz) at 0.5 μg per ml.
Immunofluorescence analyses.
H1299 cells containing inducible FLAG/His6 epitope-tagged RK-BARF0, RK1–179, or RK1–158 and/or myc epitope-tagged Notch4 were induced and analyzed 24 h postinduction. The cells were fixed with 50% methanol–50% acetone at −20°C and incubated with 10 μg of anti-FLAG M2 monoclonal antibody per ml and 0.7 μg of anti-myc polyclonal rabbit antibody per ml. The cells were washed and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories) (1:200 dilution) and Lissamine rhodamine-conjugated (LSRC) goat anti-rabbit antibody (Jackson) (1:300 dilution). The cells were washed, mounted with VECTASHIELD mounting medium (Vector), and visualized with a Zeiss Axioplan fluorescence microscope. Confocal microscopy was performed with a Zeiss confocal microscopy system in the UNC Research Microscopy Laboratory. Confocal fluorescence images were collected and translated to TIFF format using NIH Image, and composite figures were created using Adobe Photoshop 5.0.2.
Preparation of nuclear lysate.
Stable H1299 clones (2 × 106) containing FLAG/His6 epitope-tagged RK-BARF0 or pMEP4 were plated, transiently transfected with 10 μg of myc-tagged Notch4-expressing plasmids using Lipofectin, and induced with 5 μM CdCl2 approximately 24 h after transfection. To purify the nucleus, the cells were lysed at 24 h postinduction in hypotonic lysis buffer (10 mM Tris-HCl [pH 7.8], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) containing 5 μg of aprotinin per ml. Nuclei were isolated by centrifugation at 1,500 × g for 5 min at 4°C and lysed for 30 min on ice in solubilization buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA, 0.5% SDS, 0.1% sodium deoxycholate, 1% NP-40) containing 1 mM PMSF and 5 μg of aprotinin per ml. To make whole-cell lysates, the cells were lysed in solubilization buffer. The protein concentrations of nuclear and whole-cell lysates were quantitated using the DC protein assay reagent (Bio-Rad). Equivalent amounts of protein were analyzed by immunoblotting for Notch4-myc, using 1 μg of monoclonal antibody 9E10 per ml, and detected with horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham). Reactive proteins were detected by incubation of washed filers in SuperSignal substrate followed by exposure to X-ray film.
Analysis of LMP1 induction and RK-BARF0 secretion in P3HR1.
Stable P3HR1 cell lines (107) containing FLAG/His6 epitope-tagged RK-BARF0, RK1–179, RK1–158, or pMEP4 were plated and induced by addition of 1.5 μM CdCl2. The cells were collected by centrifugation from tissue culture flasks at 48 h postinduction and lysed for 30 min on ice in RIPA buffer (20 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.2% SDS, 0.2% sodium deoxycholate) containing 1 mM PMSF, 0.5 mM sodium orthovanadate, and 5 μg of aprotinin per ml. Cell lysates were subjected to SDS-PAGE and transferred to Immobilon-P. RK-BARF0, RK1–179, and RK1–158 were detected with an anti-His probe at 0.5 μg per ml. LMP1 expression was detected by CS1-4 (DAKO) at a 1:200 dilution. The culture medium was recentrifuged at 3,000 × g for 30 min to remove debris. Supernatants were recovered and incubated with anti-FLAG M2 beads for 8 h at 4°C. The beads were then washed with lysis buffer, and protein complexes were recovered by boiling in SDS sample buffer. Protein samples were subjected to SDS-PAGE and transferred to Immobilon-P. Anti-His-probe polyclonal antibody (Santa Cruz Biotechnology) was used at 0.5 μg per ml for the detection of FLAG/His6 epitope-tagged RK-BARF0, RK1–179, and RK1–158.
RESULTS
A yeast two-hybrid screen reveals that the extracellular regions of Notch3 and Notch4 interact with a predicted hydrophobic helix domain of RK-BARF0.
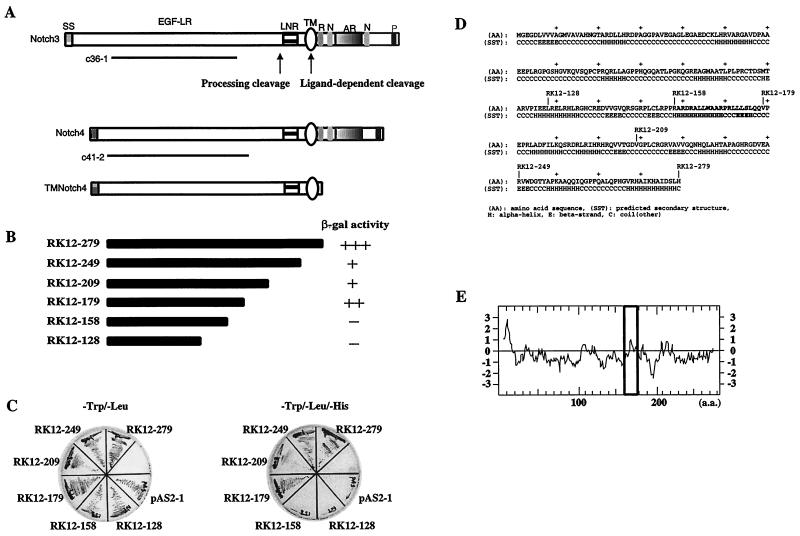
The RK-BARF0 protein contains a signal peptide-like sequence at its N terminus; however, other regions of similarity to known proteins were not identified (37). To further analyze the function of RK-BARF0, the yeast two-hybrid system was used to identify cellular proteins that interact with RK-BARF0. The highly hydrophobic region at the N terminus was deleted, and the DNA encoding aa 12 to 279 was fused in frame to the GAL4 DNA binding domain (GAL4BD). The GAL4-activating domain (GAL4AD) was fused to a human lung cDNA library. Approximately 8.5 × 105 transformants were tested for growth on selection medium. Sequence analysis of two positive clones, c36-1 and c41-2, revealed that the cDNAs represented the extracellular domains of human Notch3 and Notch4, respectively (Fig. 1A). The extracellular region of Notch proteins has multiple EGF-like repeats, and Notch3 and Notch4 contain 34 and 29 repeats, respectively (48). Other protein-interacting domains of Notch, including the RAM domain and the Notch or ankyrin repeats, were not contained in the c36-1 and c41-2 clones, suggesting that RK-BARF0 must interact with Notch through the EGF-like repeat elements.
FIG. 1.
(A) Schematic representation of the Notch clones obtained from two-hybrid screening. Clones containing the EGF-like repeat regions of Notch3 (c36-1) and Notch4 (c41-2) interacted with RK-BARF0 by yeast two-hybrid screening. A schematic representation of the Notch4 mutant lacking the entire functional intracellular region (TMNotch4) that is used this study is shown. The sites of the proteolytic cleavage for processing and ligand-dependent cleavage are indicated. Structural motifs within the Notch proteins are indicated as follows: SS, signal sequence; EGF-LR, EGF-like repeats; LNR, Notch/lin-12 like repeats; TM, transmembrane domain; R, RAM domain; N, NLS; AR, cdc10/ankyrin repeats; P, PEST sequence. (B) Schematic representation of C-terminal deletion mutants of RK-BARF0 and β-gal activity in the yeast mating assay. The full-length (aa 12 to 279) and five C-terminal deletion (aa 12 to 249, 12 to 209, 12 to 179, 12 to 158, and 12 to 128) constructs of RK-BARF0 fused to GAL4BD were transfected in yeast Y190 cells and mated with yeast Y187 cells containing the Notch4 EGF-like repeats region/GAL4AD fusion construct. Mated yeast cells were plated on Trp− Leu− medium plates, and the relative β-gal activity was determined. The time taken for more than 80% of the colonies to become blue is shown: +++, <1 h; ++, 1 to 3 h; +, 3 to 6 h; −, no blue colonies is >8 h. (C) Growth patterns of mated yeast strains on His− selective medium. Mated yeast cells were plated on Trp− Leu− medium plates and Trp− Leu− His− medium plates and incubated at 30°C for 5 days. (D) The predicted secondary structure of RK-BARF0 was determined using a 3D-1D compatibility algorithm. Bold letters indicate the sequence of the interaction-domain of RK-BARF0 with Notch. (E) The predicted hydrophobicity of the RK-BARF0 protein was determined by Kyte-Doolittle analysis. Bold square indicates the hydrophobicity of the interaction domain of RK-BARF0 with Notch.
To verify these interactions between RK-BARF0 and the two Notch clones, yeast Y190 cells containing the c41-2/GAL4AD fusion were mated with Y187 transformants carrying pAS2-1, encoding GAL4BD only, pASRK12–128, encoding the amino-terminal 12 to 128 aa of RK-BARF0 fused to GAL4BD, or pASRK119–279, encoding the carboxy terminal 119 to 279 aa of RK-BARF0 fused to GAL4BD. After mating, the yeast cells were plated on selective medium. Only yeast containing pASRK119–279, representing the carboxy terminus of RK-BARF0, could grow in the absence of histidine and also had high levels of β-gal activity (data not shown). These results revealed that aa 119 to 279 of RK-BARF0 interact with Notch4. To further define the Notch interaction domain of RK-BARF0, five C-terminal deletion constructs of RK-BARF0 fused to GAL4BD were tested for Notch4 interaction by mating and yeast two-hybrid analysis (Fig. 1B). The yeast strains carrying RK12–279, RK12–249, RK12–209, and RK12–179 could grow on the selective medium with moderate to high levels of β-gal activity, while the deletion constructs representing the amino terminus, RK12–128 and RK12–158 could not grow on selective media (Fig. 1B and 1C). Similar results were obtained from mating analyses with these mutants using yeast strains carrying human Notch3 clone c36-1 (data not shown). This analysis indicated that the region of RK-BARF0 between aa 159 and 179 is necessary for interaction with Notch. The hydrophobicity plot by Kyte-Doolittle analysis (25) and potential secondary structure predicted by 3D-1D compatibility algorithm (21) indicated that the region between aa 159 and 179 of RK-BARF0 has a hydrophobic helical structure, suggesting that RK-BARF0 interacts with the extracellular region of Notch protein through a hydrophobic helix (Fig. 1D and E).
RK-BARF0 associates with Notch4 in vivo.
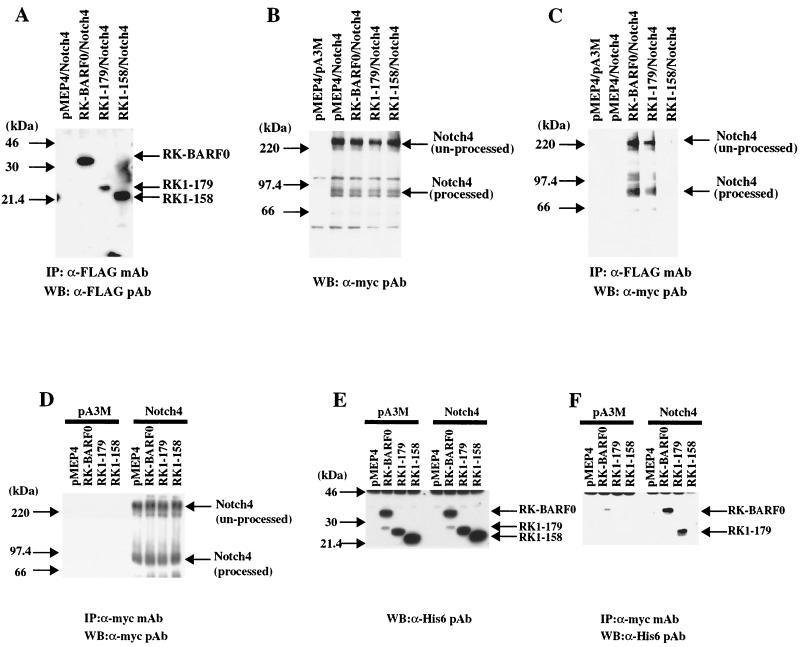
To examine the interaction of RK-BARF0 and Notch4 in vivo, a full-length Notch4 cDNA was synthesized and cloned in frame into a pA3M, which is a pcDNA3 plasmid containing three repeated c-myc epitopes at the carboxy terminus under the control of the human cytomegalovirus immediate-early promoter. Three RK-BARF0 constructs, RK-BARF0 (aa 1 to 279), RK1–179, and RK1–158, containing FLAG/His6 epitopes at the C terminus, were also prepared and cloned into pMEP4, which contains the inducible human metallothionein promoter. We have consistently achieved excellent protein expression in epithelial cells by using the metallothionein promoter, which enables expression of potentially toxic proteins in cell lines. The Notch4 and RK-BARF0 plasmids were transfected into the human epithelial cell line, H1299. After induction with CdCl2, lysates were prepared and complexes containing RK-BARF0 were immunoprecipitated using the anti-FLAG M2 monoclonal antibody. Western blot analyses of the immunoprecipitated complexes indicated that RK-BARF0 and the C-terminal deletion mutant, RK1–158, were expressed at similar levels, with somewhat lower expression of RK1–179 (Fig. 2A). Although equivalent amounts of transfected Notch4 were detected in the total-cell lysates, immunoblotting with an anti-myc polyclonal antibody to identify myc epitope-tagged Notch4 (Notch4-myc) detected Notch4 in precipitated complexes containing RK-BARF0 and RK1–179 but not RK1–158 (Fig. 2B and C). This observation is consistent with the results of yeast two-hybrid analysis and indicated that aa 159 to 179 of RK-BARF0 are also necessary for the interaction between RK-BARF0 and Notch in vivo. Interestingly, the unprocessed full-length form of Notch4 and also the intracellular C-terminal, processed form of Notch4, which lacks the EGF-like repeats, were detected in complexes with both RK-BARF0 and RK1–179. The presence of the intracellular fragment of processed Notch in the precipitated RK-BARF0 complexes indicates that RK-BARF0 also interacts with processed Notch4 containing both the extracellular and intracellular subfragments, joined by cysteine bonds. These bonds are denatured during SDS-PAGE such that only the intracellular Notch fragment which contains the myc epitope is detected by Western blotting. Comparison of the amount of Notch4 present in the total-cell lysate with that detected in the RK-BARF0 containing complexes indicated that approximately 15% of the transfected Notch4 was present in the complexes. However, RK-BARF0 also interacts with Notch1 (data not shown). This interaction with endogenously expressed Notch1 would decrease the amount available for interaction with transfected Notch4.
FIG. 2.
RK-BARF0 associates with processed and unprocessed forms of Notch4 in vivo. H1299 cells (5 × 105) were transiently transfected with both FLAG/His6-tagged RK-BARF0 (aa 1 to 279), RK1–179, RK1–158, or pMEP4 and Notch4myc or pA3M. RK-BARF0, RK1–179, and RK1–158 were induced with 5 μM CdCl2 for 16 h, immunoprecipitated, and analyzed on immunoblots. (A) Immunoblot with an anti-FLAG antibody to identify RK-BARF0, RK1–179, and RK1–158 in the immunoprecipitated RK-BARF0 complexes. (B) Immunoblot analysis with anti-myc antibody to identify Notch4 expression in the transfected cells. (C) Identification of processed and unprocessed forms of Notch4 in the immunoprecipitated (IP) RK-BARF0 complexes using anti-myc epitope polyclonal antibodies. (D) Immunoblot with anti-myc antibody to identify Notch4 in the immunoprecipitated Notch4 complexes. (E) Immunoblot analysis with antihistidine antiserum to determine the expression of RK-BARF0, RK1–179, and RK1–158 in the transfected cells. (F) Detection of RK-BARF0 and RK1–179, using antihistidine antibody in the Notch4 complexes immunoprecipitated with anti-myc antibody.
The interaction of Notch4 with RK-BARF0 was confirmed in the reciprocal immunoprecipitation. Immunoblot analysis of the immunoprecipitated Notch4-containing complexes detected equivalent amounts of Notch4 in the complexes (Fig. 2D). Using an anti-His polyclonal antibody, immunoblots of the total-cell lysates indicated that RK-BARF0 and RK1–179 were expressed at equal levels and that there was a somewhat greater expression of RK1–158 (Fig. 2E). In agreement with the data presented above, only RK-BARF0 and RK1–179 were detected in the Notch4-containing immunoprecipitated complexes (Fig. 2F).
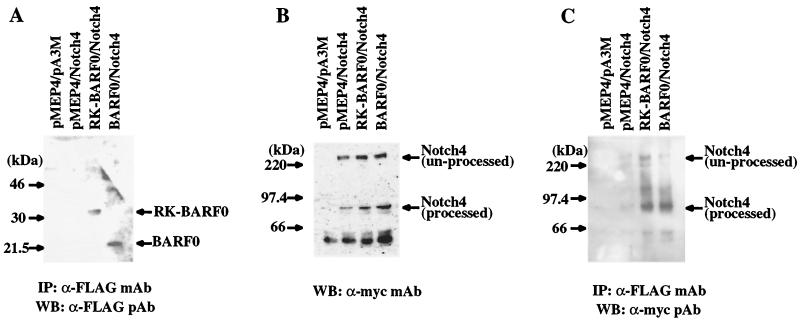
It is known that Notch has a signal sequence in the N terminus, is synthesized in the ER, and is processed in the TGN (2). Since the RK-BARF0 protein also has a signal peptide-like sequence at its amino terminus, it is likely to also be synthesized in the ER. The interaction of RK-BARF0 with unprocessed Notch suggests that the RK-BARF0/Notch interaction occurs in the ER prior to Notch processing. The BARF0 ORF, which is present at the 3′ end of all of the variably spliced BamHI A transcripts, lacks the N-terminal hydrophobic sequence of RK-BARF0 and contains aa 106 to 279 of RK-BARF0, including the Notch-interacting domain. BARF0, tagged with the FLAG epitope, was cloned into the pMEP4 vector, and RK-BARF0 and BARF0 were transfected with Notch4-myc into H1299 cells. After induction of expression and precipitation, immunoblot analysis indicated that both RK-BARF0 and BARF0 were present in the precipitated complexes (Fig. 3A). Notch4 was expressed at equivalent levels in each of the transfections (Fig. 3B). The processed form of Notch4 was present in the immunoprecipitated complexes with BARF0, but only trace levels of unprocessed Notch were present, comparable to that detected in precipitates prepared from cells containing the pMEP4 vector control (Fig. 3C). This observation indicates that the potential protein product of the BARF0 ORF also can interact with Notch and suggests that the N-terminal hydrophobic region of RK-BARF0 is necessary for the interaction with unprocessed Notch4.
FIG. 3.
BARF0 interacts with processed Notch. (A) Immunoblot of RK-BARF0 or BARF0 (aa 106 to 279) after induction and immunoprecipitation (IP) with an anti-FLAG polyclonal antibody. (B) Immunoblot analysis of whole cell lysates to identify Notch4 expression after transfection. (C) Identification of processed and unprocessed forms of Notch4 in the RK-BARF0 and BARF0 immunoprecipitated complexes. WB, Western blot.
RK-BARF0 induces the translocation of unprocessed Notch4 to the nucleus.
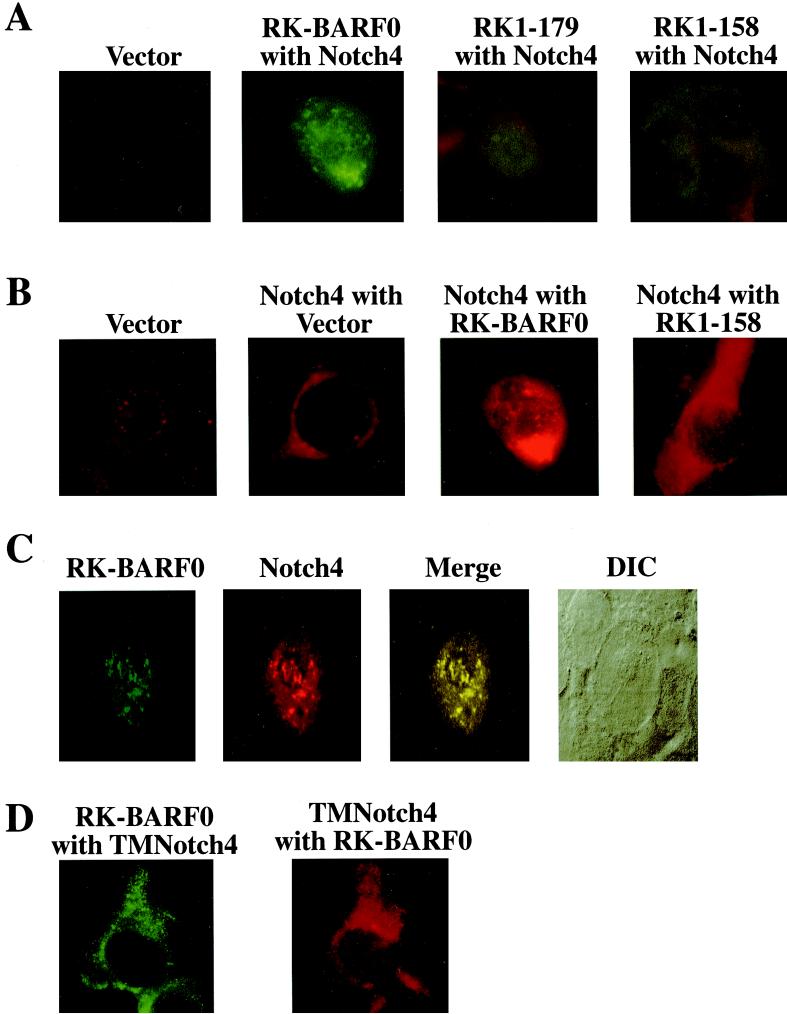
To further analyze the interaction between RK-BARF0 and Notch4, the intracellular location of RK-BARF0 and Notch was determined. Stable cell lines containing Notch4 and the FLAG/His6-tagged RK-BARF0 and deletion mutants in H1299 cells were prepared and analyzed for protein expression and intracellular location by using the anti-FLAG M2 monoclonal antibody and a fluorescein-conjugated anti-mouse secondary antibody to detect RK-BARF0. RK-BARF0 and RK1–179 were predominantly found in a punctate staining pattern throughout the nucleus and in the perinuclear region, possibly representing the ER (Fig. 4A). RK-BARF0 was also mainly detected in the nuclear pellet of biochemically fractionated RK-BARF0-expressing H1299 cells (data not shown). However RK1–158, which does not interact with Notch4 protein, was localized throughout the cytoplasm (Fig. 4A). These data indicate that the region of RK-BARF0 which is required for the interaction with Notch protein is also necessary for the nuclear localization of RK-BARF0.
FIG. 4.
RK-BARF0 Colocalizes with Notch4 in the nucleus. (A) Cell lines containing both Notch4-myc and FLAG/His6-tagged RK-BARF0, RK1–179, RK1–158 or pMEP4 expression plasmids were induced and stained using a monoclonal antibody to FLAG and FITC-conjugated anti-mouse immunoglobulin G (IgG) to identify RK-BARF0. (B) Cell lines containing Notch4-myc or both Notch4-myc and FLAG/His6-tagged RK-BARF0, RK1–158, or the pMEP4 vector were stained with anti-myc polyclonal antibody and LSRC-conjugated anti-rabbit IgG to identify Notch4. (C) A stable cell line containing Notch4-myc and FLAG/His6-tagged RK-BARF0 was stained with anti-FLAG M2 monoclonal antibody and anti-myc polyclonal antibody using FITC-conjugated anti-mouse IgG to detect RK-BARF0 and RISC-conjugated anti-rabbit IgG to detect Notch4. Confocal images were collected, and composite figures were created using Adobe Photoshop 5.0.2. The differential interference contrast microscopy image of the cell is presented. (D) A cell line containing the extracellular to transmembrane region of Notch4 (TMNotch4)-myc in pA3M and FLAG/His6-tagged RK-BARF0 was stained with a monoclonal FLAG and FITC-conjugated anti-mouse IgG to identify RK-BARF0 and anti-myc polyclonal antibody and LSRC-conjugated anti-rabbit IgG to identify TMNotch4.
To determine if the interaction with RK-BARF0 affected Notch localization, stable H1299 cell lines expressing the Notch4 protein alone or in the presence of RK-BARF0 were analyzed. In the absence of RK-BARF0, the Notch4-myc protein was found throughout the cytoplasm, with no staining of the nucleus (Fig. 4B, Notch4 with Vector). In contrast, in the presence of RK-BARF0, Notch4 was detected in the nucleus and perinuclear region (Fig. 4B, Notch4 with RK-BARF0). When Notch4 was coexpressed with the RK1–158 deletion mutant of RK-BARF0 which cannot interact with Notch, Notch4 remained in the cytoplasm (Fig. 4B, Notch4 with RK1–158). These data suggest that the interaction of RK-BARF0 with Notch4 protein induces the nuclear localization of Notch4. The staining for RK-BARF0 and Notch4 in the same cell in Fig. 4A, RK-BARF0 with Notch4, and Fig. 4B, Notch4 with RK-BARF0, suggested colocalization. The interaction and colocalization of RK-BARF0 and Notch4 in the nucleus were confirmed using confocal microscopy. RK-BARF0 and Notch4 protein were detected in punctate nuclear granules using fluorescein-conjugated secondary antiserum for detection of RK-BARF0 and rhodamine-conjugated secondary antiserum for detection of Notch4. The colocalized proteins are indicated in yellow (Fig. 4C). The cell and nuclear shape were revealed by differential interference contrast microscopy (Fig. 4C).
RK-BARF0 does not have a potential nuclear localization signal (NLS), and the deletion mutant of RK-BARF0 that lacks the Notch-interacting region does not translocate to the nucleus. However, an NLS is present in the intracellular portion of Notch4, and forms of Notch that lack the extracellular domain localize to the nucleus (1, 42). To determine if the localization of RK-BARF0 and Notch in the nucleus is dependent on the Notch NLS, a deleted form of Notch containing the extracellular domain and transmembrane region and lacking the entire intracellular region containing the NLS (TMNotch4) was coexpressed with RK-BARF0 (Fig. 4D). RK-BARF0 (Fig. 4D, RK-BARF0 with TMNotch4) and TMNotch4 (Fig. 4D, TMNotch4 with RK-BARF0) were both detected in the cytoplasm. These data indicate that the translocation of the RK-BARF0/Notch complex to the nucleus is dependent on the Notch NLS. The interaction of RK-BARF0 with the Notch extracellular domain apparently affects the tertiary structure of Notch such that the Notch/RK-BARF0 complex is transported to the nucleus through the Notch NLS.
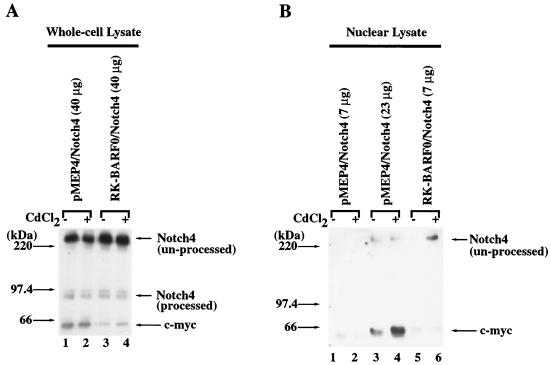
The translocation of Notch4 protein to the nucleus was also examined by transfection of Notch4 into the H1299 cell line containing RK-BARF0 in the pMEP4 inducible-expression plasmid. Analysis of the whole-cell lysates detected equivalent expression of unprocessed Notch4 in whole-cell lysates from the transfected cell lines with or without induction of the pMEP4 promoter (Fig. 5A). The presence of transfected Notch4 in the nuclear fraction was determined before and after induction of RK-BARF0 expression. Unprocessed Notch4 was not detected in equivalent amounts of nuclear lysate from cells that contained the pMEP4 vector or in cells that contained RK-BARF0 without induction. However, after induction of RK-BARF0 expression, a portion of unprocessed Notch4 was detected in the nuclear fraction (Fig. 5B). The amount of Notch4 detected in 7 μg of nuclear protein after induction of RK-BARF0 expression was two- to threefold greater than that detected in 23 μg of nuclear protein from the vector-containing cells. The detection of unprocessed Notch4 in the nuclear fraction only after induction of RK-BARF0 expression confirmed that the nuclear localization of the unprocessed form of Notch protein is dependent on of RK-BARF0.
FIG. 5.
RK-BARF0 induces the translocation of unprocessed Notch4 to the nucleus. Stable cell clones containing FLAG/His6 epitope-tagged RK-BARF0 or pMEP4 expression plasmids were transiently transfected with Notch4-myc and treated for 24 h with 8 μM CdCl2 or left untreated. Protein (40 μg) of whole-cell lysates (A) and 7 or 23 μg of nuclear lysates (B) were subjected to SDS-PAGE and analyzed for the presence of Notch4-myc by using anti-myc (9E10) antibody.
RK-BARF0 has an ER-targeting signal and can be secreted.
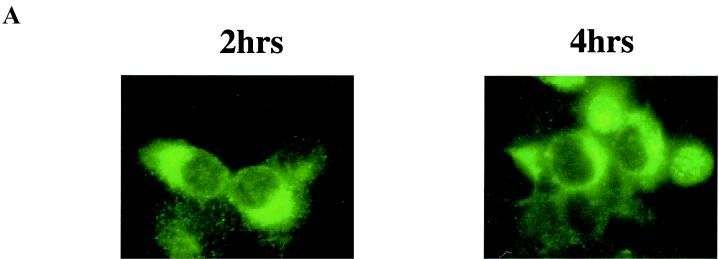
The interaction of RK-BARF0 with unprocessed Notch suggests that the amino-terminal hydrophobic region of RK-BARF0 is indeed an ER-targeting sequence and that RK-BARF0 and Notch are synthesized and interact in the ER prior to Notch processing in the Golgi. In H1299 cells, 2 h after induction, RK-BARF0 was detected in the perinuclear region, characteristic of the ER, and by 4 h after induction, RK-BARF0 was present in the nucleus in some cells (Fig. 6A).
FIG. 6.
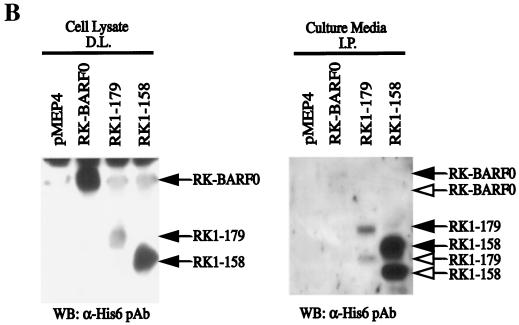
RK-BARF0 is expressed in the ER and can be secreted. (A) Stable H1299 cell lines containing FLAG/His6-tagged RK-BARF0 were induced by 5 μM CdCl2 to express RK-BARF0, and cells were stained 2 and 4 h postinduction with anti-FLAG antibody and FITC-conjugated anti-mouse antibody to determine RK-BARF0 localization. (B) Culture medium (8 ml/5 × 106 cells) of induced P3HR1 cells containing FLAG/His6-tagged RK-BARF0, RK1–179, RK1–158, or pMEP4 expression plasmids was incubated with anti-FLAG M2 beads. Immunoprecipitates (I.P.) from culture medium and 50 μg of protein of whole-cell lysates were subjected to SDS-PAGE. FLAG/His6-tagged RK-BARF0, RK1–179, or RK1–158 were detected with the anti-His- probe polyclonal antibody. Solid arrows indicate the protein bands which are identified in both whole-cell lysate and cell culture media; open arrows indicate the protein bands which are present only in the cell culture media. D.L., direct load; WB, Western blot.
A signal peptide sequence not only suggests that RK-BARF0 is targeted to the ER but also suggests that RK-BARF0 could be processed and secreted into the medium. To determine if RK-BARF0 can be secreted, cell lines stably expressing RK-BARF0, RK1–179, and RK1–158 were established in the P3HR1 cell line, which expresses a very low level of endogenous Notch1. The full-length proteins were detected in the whole-cell lysates; however, RK1–158, which does not interact with Notch, and a smaller form, probably representing RK1–158, after cleavage of the signal peptide, were abundant in the culture medium (Fig. 6B). RK1–179 and its cleaved form were also clearly evident; however, only trace levels of RK-BARF0 and a smaller form were detected. These data suggest that RK-BARF0 can be processed and secreted. The greatly increased secretion of RK1–158, which does not interact with Notch and does not translocate to the nucleus, suggests that the interaction with Notch may inhibit the secretion of RK-BARF0.
RK-BARF0 induces LMP1 expression in EBNA2-negative P3HR1 cells.
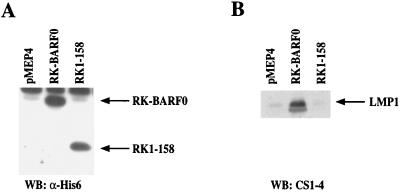
EBNA2 is the major viral transactivator of expression of the EBV oncogene LMP1. Transactivation of LMP1 by EBNA2 is complex; however, it is partially mediated through its interaction with RBP-Jκ. Interestingly, in EBV-infected NPC, LMP1 is expressed in the absence of EBNA2. P3HR1 is an EBV-positive Burkitt's lymphoma cell line in which the EBNA2-encoding region is deleted, and it expresses very low levels of LMP1. To determine if the RK-BARF0/Notch interaction positively affects expression of LMP1 in the absence of EBNA2, stable cell lines containing RK-BARF0 and the noninteracting deletion, RK1–158, were derived from P3HR1 cells and analyzed for LMP1 expression. After induction of expression of RK-BARF0 in P3HR1 cells, the amount of LMP1 was considerably increased in comparison with that in P3HR1 cells expressing RK1–158, the mutant that does not interact with Notch, or the vector control, pMEP4 (Fig. 7). These results indicate that RK-BARF0 can activate LMP1 expression in an EBNA2-independent manner through the Notch interaction domain.
FIG. 7.
RK-BARF0 induces LMP1 expression in EBNA2-negative P3HR1 cells. Stable cell lines containing FLAG/His6-tagged RK-BARF0, RK1–158, or pMEP4 expression plasmids were treated for 48 h with 1.5 μM CdCl2. (A) Immunoblot analysis of 80 μg of protein of whole-cell lysates with an anti-His-probe to identify RK-BARF0 and RK1–158. (B) Immunoblot analysis of 60 μg of protein of whole-cell lysates by anti-LMP1 (CS1-4) to identify LMP1. WB, Western blot.
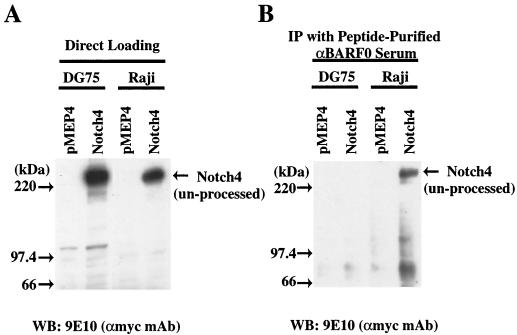
Coimmunoprecipitation of RK-BARF0 and Notch4 from the EBV-infected Raji cell line.
We have previously produced a rabbit antiserum to a BARF0-specific peptide representing aa 115 to 132 of RK-BARF0 (10). This antiserum specificially reacts with a glutathione transferase-BARF0 fusion protein, in vitro-translated BARF0, and transfected RK-BARF0 (10). The antiserum also cross-reacts with a cellular protein that is abundant in DG75 and other EBV-negative Burkitt's lymphoma cells (24). This protein has since been identified as major histocompatibility complex (MHC) class II (T. Sculley, personal communication). Since expression of class II is induced by EBV infection and the estimated molecular weight of RK-BARF0 is the same as that of MHC class II, the detection of endogenously expressed RK-BARF0 using this reagent is obscured. To use the RK-BARF0/Notch interaction to assess the expression of RK-BARF0 expression in EBV-infected cells, stable cell lines containing myc-tagged Notch4 in pMEP4 were established from the EBV-positive cell line Raji and the EBV-negative cell line DG75. Expression of Notch 4 was induced by treatment with 5 μM CdCl2 for 36 h prior to preparation of cell lysates. Immunoblot analysis indicated that Notch4 expression was induced in both cell lines with increased expression in DG75 cells (Fig. 8A). The RK-BARF0 antiserum was enriched by affinity purification with the BARF0 specific peptide and used for immunoprecipitation from the Notch4-expressing Raji and DG75 cells. Immunoblot analysis using the anti-myc monoclonal antibody detected Notch4 in the immunoprecipitated complexes from Raji cells but not from DG75, despite the higher level of Notch4 expression in these cells (Fig. 8B). In addition, Notch4 was not detected after immunoprecipitation of MHC class II from induced Raji or DG75 cells (data not shown). Immunoprecipitation of unprocessed Notch4 from the EBV-infected Raji cell line, using the BARF0-specific antibody, provides indirect evidence that the RK-BARF0 protein is expressed in EBV-positive Raji cells.
FIG. 8.
RK-BARF0 peptide rabbit antiserum precipitates transfected Notch4 protein from Raji cells but not from DG75. Stable cell lines containing myc-tagged Notch4 or pMEP4 expression plasmids were treated for 36 h with 5 μM CdCl2. (A) Protein (100 μg) from cell lysates was subjected to SDS-PAGE, and myc-tagged Notch4 was identified using the 9E10 monoclonal antibody. (B) Protein from cell lysates (1,250 μg) was precipitated using peptide affinity-purified rabbit antiserum against RK-BARF0 (10). Immunoprecipitates (IP) were subjected to SDS-PAGE, and myc-tagged Notch4 was identified using 9E10 monoclonal antibody. WB, Western blot.
DISCUSSION
Notch4 (also called Int3) is a member of the Notch receptor family and is thought to be involved in endothelial and epithelial differentiation in mammals (48, 49). Notch was originally identified as a Drosophila neurogenic gene required for correct segregation of epidermal cells from neuronal cell precursors during embryogenesis, but subsequent studies demonstrated that Notch is widely expressed during embryonic and adult development and mediates many cell-cell interactions during fly development (8). Notch directly activates gene expression through its interaction with the DNA binding protein RBP-Jκ, the mammalian homologue of the Drosophila Suppressor of Hairless protein. The major transcriptional regulator of EBV during latent infection in B lymphocytes, EBNA2, also directly interacts with RBP-Jκ in a manner analogous to that of Notch and activates cellular and viral promoters through RBP-Jκ (14, 15, 19). The activated form of mouse Notch1 can substitute for EBNA2 and can transactivate the EBV LMP1 promoter (17). The interaction of RBP-Jκ with EBNA2 is apparently carefully regulated and is thought to be affected by at least three other EBV proteins, EBNA3A, EBNA3B, and EBNA3C, which also interact with RBP-Jκ and possibly negatively regulate EBNA2-mediated transactivation (36, 52).
The data presented here indicate that the potential EBV protein, RK-BARF0, interacts with the Notch in the ER prior to processing in the TGN. The biogenesis of Notch is complex, with expression in the ER, processing in the TGN, and cleavage at the plasma membrane to release the cytoplasmic fragment, which activates cellular gene expression through its interaction with RBP-Jκ (18). The interaction of RK-BARF0 with the Notch ligand binding domain apparently unmasks the Notch NLS and induces translocation of unprocessed Notch to the nucleus. This interaction also results in a decrease in the levels of total Notch (data not shown). These effects on Notch may inhibit Notch activation and signaling and also reduce the amount of RBP-Jκ that is bound to Notch. This effect might facilitate the interaction of EBNA2 with RBP-Jκ to activate the expression of LMP1.
However, in most of the malignancies associated with EBV, including NPC, LMP1 is expressed in the absence of EBNA2 (35). The BamHI A RNAs are abundantly expressed in NPC and were originally identified in cDNA libraries from NPC RNA (12, 16). The levels of these RNAs are considerably lower in transformed lymphocytes, and it is difficult to detect the mRNAs on Northern blots (12, 37). The difference in the relative abundance of the RNAs may indicate that the proteins potentially encoded by these RNAs are predominantly expressed and function in infections where the EBNA2 and EBNA3 proteins are not expressed.
The data presented here indicate that the RK-BARF0/Notch interaction can induce the expression of LMP1 in the absence of EBNA2. The regulation of LMP1 expression is complex, and EBNA2-dependent and -independent elements have been identified (47). An ATF/CRE element in the promoter mediates EBNA2-independent activation of the LMP1 promoter and binds ATF1, ATF2, and c-Jun (39). The Notch/RK-BARF0 interaction may activate LMP1 expression through effects on RBP-Jκ or through other pathways affected by Notch. Notch interacts with the cytoplasmic protein Deltex, which inhibits activation of the c-Jun terminal kinase (JNK), and recent studies also implicate Notch in activation of NF-κB p50-containing complexes (6, 28–30, 32, 33). Notch can also affect gene expression independently of its interaction with RBP-Jκ. Dominant negative forms of RBP-Jκ do not block the effects of Notch on myogenic differentiation of C2C12 cells, and Notch mutants lacking the RBP-Jκ binding site and the Notch NLS can still activate transcription (31). Notch4 is frequently rearranged by mouse mammary tumor virus integration, and Notch gene family members are activated through chromosomal translocations. In human tumors, novel Notch4 mRNAs have also been identified that contain the intracellular domain and lack the RBP-Jκ binding site (20).
Multiple potential ORFs are formed by the differentially spliced BamHI A mRNAs; however, the proteins encoded by the RNAs remain unidentified. The most abundant 4.8-kb mRNA was originally predicted to have the structure found in the RB2 cDNA, and the two largest ORFs identified in this cDNA are the RK103 ORF and the RB2 ORF (37). A recent report has confirmed the splice donor and acceptor sites described in the RB2 cDNA. The data indicate that the RB2 ORF is identical to an ORF identified in the A73 cDNA and that the RPMS1 ORF, originally described in the C22.2 cDNA, is identical to the RK103 ORF (37, 40). These ORFs encode proteins when expressed with a recombinant epitope tag, and potential interacting proteins have been identified using the yeast two hybrid. Interestingly, RPMS1 also interacts with RBP-Jκ and negatively regulates a synthetic promoter containing four copies of the EBNA2-responsive element of the BamHI C promoter (40).
Clearly, these ORFs can encode proteins that have interesting biologic properties; however, antisera that specifically recognize the RPMS1 or RB2 protein products have not yet been produced. A previously described rabbit antiserum produced using a BARF0-specific peptide was shown to identify a glutathione S-transferase–BARF0 fusion protein, in vitro-translated RK-BARF0, and transfected RK-BARF0 (10). This antiserum was subsequently shown to also react with a cellular protein that was expressed at high levels in the EBV-negative Burkitt's lymphoma cell line DG75 and other EBV-negative B-cell lines (24). This cellular protein was identical in size to transfected RK-BARF0 and obscured the detection of the authentic protein in EBV-infected cells or tissues. In this study, the antiserum was affinity purified using the BARF0 peptide to reduce reactivity with the cellular protein and the interaction between Notch and RK-BARF0 was analyzed. The detection of unprocessed Notch4 in the complexes immunoprecipitated from the EBV-positive Raji cell line but not from the-EBV negative DG75 cell line provides indirect evidence for the expression of RK-BARF0.
The direct interaction of RK-BARF0 with Notch provides another mechanism through which EBV affects or usurps the Notch signaling pathway. The protein product of the RK-BARF0 ORF is the first viral protein that has been shown to interact with Notch and affect its intracellular location. The interaction of several EBV proteins with components of the Notch signaling pathway highlights the multiple mechanisms that regulate this pathway and affect cellular and viral gene expression and growth control.
ACKNOWLEDGMENTS
We thank S. Milgram and K. Sugaya for kindly providing the amplified human lung cDNA library and partial human Notch4 clone, respectively.
This work was supported by NIH grant CA32979 (N.R.-T.) and by the Yamanouchi Foundation for Research on Metabolic Disorders and The Ryoichi Naito Foundation for Medical Research (S.K.).
REFERENCES
- 1.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 2.Blaumueller C M, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 3.Brooks L A, Lear A L, Young L S, Rickinson A B. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol. 1993;67:3182–3190. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3257–3262. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diederich R J, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994;120:473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- 7.Fahraeus R, Fu H L, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadav M, Busson P, et al. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 8.Fortini M E, Artavanis-Tsakonas S. Notch: neurogenesis is only part of the picture. Cell. 1993;75:1245–1247. doi: 10.1016/0092-8674(93)90611-s. [DOI] [PubMed] [Google Scholar]
- 9.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 10.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 12.Gilligan K, Sato H, Rajadurai P, Busson P, Young L, Rickinson A, Tursz T, Raab-Traub N. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J Virol. 1990;64:4948–4956. doi: 10.1128/jvi.64.10.4948-4956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilligan K J, Rajadurai P, Lin J C, Busson P, Abdel-Hamid M, Prasad U, Tursz T, Raab-Traub N. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 16.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofelmayr H, Strobl L J, Stein C, Laux G, Marschall G, Bornkamm G W, Zimber-Strobl U. Activated mouse Notch 1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J Virol. 1999;73:2770–2780. doi: 10.1128/jvi.73.4.2770-2780.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 20.Imatani A, Callahan R. Identification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell lines. Oncogene. 2000;19:223–231. doi: 10.1038/sj.onc.1203295. [DOI] [PubMed] [Google Scholar]
- 21.Ito M, Matsuo Y, Nishikawa K. Prediction of protein secondary structure using the 3D–1D compatibility algorithm. Comput Appl Biosci. 1997;13:415–424. doi: 10.1093/bioinformatics/13.4.415. [DOI] [PubMed] [Google Scholar]
- 22.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieff E. Epstein-Barr virus and its replication. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 24.Kienzle N, Buck M, Greco S, Krauer K, Sculley T B. Epstein-Barr virus-encoded RK-BARF0 protein expression. J Virol. 1999;73:8902–8906. doi: 10.1128/jvi.73.10.8902-8906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Longnecker R, Miller C L, Tomkinson B, Miao X Q, Kieff E. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J Virol. 1993;67:5068–5074. doi: 10.1128/jvi.67.8.5068-5074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuno K, Diederich R J, Go M J, Blaumueller C M, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 29.Matsuno K, Eastman D, Mitsiades T, Quinn A M, Carcanciu M L, Ordentlich P, Kadesch T, Artavanis-Tsakonas S. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- 30.Matsuno K, Go M J, Sun X, Eastman D S, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- 31.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 32.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oswald F, Liptay S, Adler G, Schmid R M. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol. 1998;18:2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan D, Rubin G M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 35.Raab-Traub N. Pathogenesis of Epstein-Barr virus and its associated malignancies. Semin Virol. 1996;7:315–323. [Google Scholar]
- 36.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadler R H, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 39.Sjoblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith P R, de Jesus O, Turner D, Hollyoake M, Karstegl C E, Griffin B E, Karran L, Wang Y, Hayward S D, Farrell P J. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J Virol. 2000;74:3082–3092. doi: 10.1128/jvi.74.7.3082-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith P R, Gao Y, Karran L, Jones M D, Snudden D, Griffin B E. Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors. J Virol. 1993;67:3217–3225. doi: 10.1128/jvi.67.6.3217-3225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 43.Sugaya K, Sasanuma S, Nohata J, Kimura T, Fukagawa T, Nakamura Y, Ando A, Inoko H, Ikemura T, Mita K. Gene organization of human NOTCH4 and (CTG)n polymorphism in this human counterpart gene of mouse proto-oncogene Int3. Gene. 1997;189:235–244. doi: 10.1016/s0378-1119(96)00857-8. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 45.Tomkinson B, Kieff E. Use of second-site homologous recombination to demonstrate that Epstein- Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 49.Uyttendaele H, Soriano J V, Montesano R, Kitajewski J. Notch4 and Wnt-1 proteins function to regulate branching morphogenesis of mammary epithelial cells in an opposing fashion. Dev Biol. 1998;196:204–217. doi: 10.1006/dbio.1998.8863. [DOI] [PubMed] [Google Scholar]
- 50.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young L, Alfieri C, Hennessy K, Evans H, O'Hara C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 52.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]