Abstract
Durable response to chimeric antigen receptor T (CART) cell therapy remains limited in part due to CART cell exhaustion. Here, we investigate the regulation of CART cell exhaustion with three independent approaches including: a genome-wide CRISPR knockout screen using an in vitro model for exhaustion, RNA and ATAC sequencing on baseline and exhausted CART cells, and RNA and ATAC sequencing on pre-infusion CART cell products from responders and non-responders in the ZUMA-1 clinical trial. Each of these approaches identify interleukin (IL)-4 as a regulator of CART cell dysfunction. Further, IL-4-treated CD8+ CART cells develop signs of exhaustion independently of the presence of CD4+ CART cells. Conversely, IL-4 pathway editing or the combination of CART cells with an IL-4 monoclonal antibody improves antitumor efficacy and reduces signs of CART cell exhaustion in mantle cell lymphoma xenograft mouse models. Therefore, we identify both a role for IL-4 in inducing CART exhaustion and translatable approaches to improve CART cell therapy.
Subject terms: Translational immunology, Interleukins, Epigenetics in immune cells, Translational research
The application and therapeutic success of CAR-T cell approaches are limited by the development of T cell exhaustion. Here, Stewart et al discover a role for IL-4 in driving CD8+ CAR-T cell exhaustion and demonstrate the improvement of CAR-T cell effectivity with interruption of IL-4 signalling.
Introduction
Chimeric antigen receptor T (CART) cell therapy has evolved as a potentially curative therapy in a subset of patients with hematological malignancies1. While CART cell therapy results in impressive overall response rates over 70%, durable response rates remain limited to 30–40%, and most patients relapse within the first year of therapy2–4. Several mechanisms of CART cell failure have been identified, including the limited in vivo expansion and persistence of CART cells5,6.
It has become increasingly evident that T cell exhaustion contributes to CART cell failure in the clinic7,8. T cell exhaustion is an epigenetically regulated state of dysfunction that results from chronic stimulation through either the T cell receptor (TCR) in CD8+ T cells or through the CAR in CART cells9–12. Exhaustion is characterized by phenotypic, functional, transcriptional, and epigenetic changes. Phenotypic alterations include the upregulation of multiple inhibitory receptors on a cell such as programmed cell death protein 1 (PD-1), T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), cytotoxic T-lymphocyte associated protein 4 (CTLA-4), and lymphocyte-activation gene 3 (LAG-3)9. Functional alterations include a decreased ability to proliferate and to produce effector cytokines such as interleukin (IL)-2 and tumor necrosis factor (TNF)-α, followed by losing the ability to produce interferon (IFN)-γ at later stages9. In addition, exhausted T cells experience metabolic changes such as impaired glycolysis13. Transcriptional and epigenetic changes include alterations in the activity of several transcription factors including: TCF-7, TOX, T-BET, EOMES, PRDM1, NR4A3, BATF, and EGR2, as well as AP-1 and RUNX family members14–26. In an effort to control the epigenetic response to chronic stimulation, several studies have used genetic engineering tools to overexpress or knockout individual molecules. Some examples include the overexpression of the AP-1 family member c-Jun and the deletion of molecules such as the methyltransferase DNMT3A, the inflammatory regulators REGNASE-1 and ROQUIN-1, and the transcription factors PRDM1 and NR4A311,25,27,28. While these approaches have both enhanced the field’s understanding of CART cell exhaustion as well as established a framework to prevent its development, a complete understanding of molecular pathways and potential targets for therapeutic intervention has not been achieved thus far.
It is likely that the occurrence of CART cell exhaustion varies by CAR construct and disease type. For example, independent studies have demonstrated that CD28-costimulated CART cells are more susceptible to a state of exhaustion as compared to 41BB-costimulated CART cells5,29,30. In addition, CAR constructs that experience a higher occurrence of tonic signaling have also been positively associated with the development of CART cell exhaustion29. Further, current literature supports a higher risk of exhaustion in CART cells used for the treatment of solid tumors due to the added challenges of an immunosuppressive tumor microenvironment31.
We focused our studies on the epigenetic regulation of exhaustion in CART cells targeting the CD19 antigen (CART19) and containing a CD28 costimulatory domain (CART19-28ζ) as a model. To do so, we employed the following three independent strategies: (1) a genome-wide clustered regularly interspaced short palindromic repeat (CRISPR) knockout screen in healthy donor CART19-28ζ cells using an in vitro model for exhaustion, (2) RNA and assay for transposase-accessible chromatin (ATAC) sequencing on baseline and chronically stimulated CART19-28ζ cells from healthy donors using an in vitro model for exhaustion, and (3) RNA and ATAC sequencing on pre-infusion axicabtagene ciloleucel (axi-cel) products from patient responders and non-responders in the pivotal ZUMA-1 clinical trial that led to the initial FDA approval of axi-cel32. Collectively, our independent approaches identified IL-4 as a key regulator of CART cell exhaustion. Subsequently, we performed validation studies to confirm the role of IL-4 on CART cell function using in vitro and in vivo models, ultimately proposing IL-4 neutralization as a strategy to improve CART cell anti-tumor activity.
Results
Establishing an in vitro model for CART19 cell exhaustion
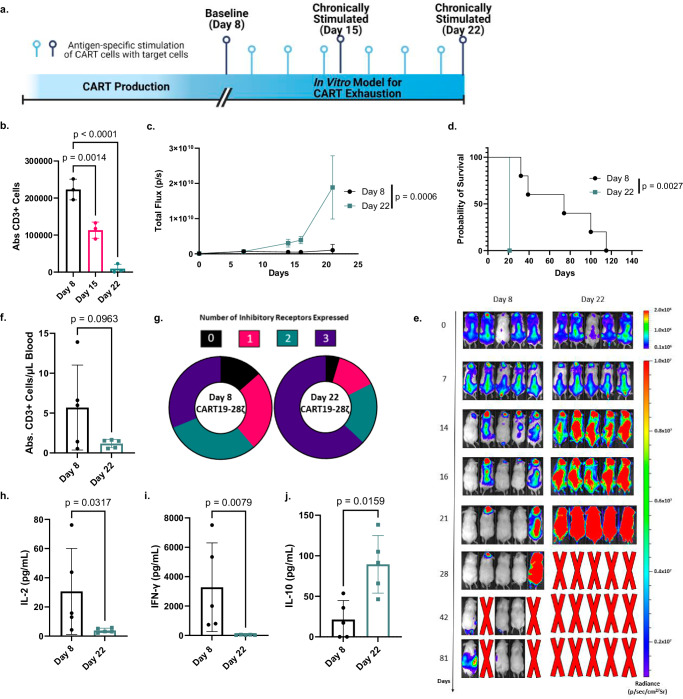
To better understand the development of CART cell exhaustion, we first designed an in vitro model to induce exhaustion in CART19 cells generated from healthy donor T cells (Fig. 1a and Supplementary Fig. S1). This model was designed both to be scalable and to focus specifically on the development of exhaustion through chronic stimulation of the T cells. In CD8+ T cells, exhaustion has been modeled in vitro by the chronic stimulation of the T cells through the T cell receptor (TCR)9,33. To model this phenomenon in CART19-28ζ cells, we chronically stimulated CART cells through the CAR with the addition of fresh CD19+ target cells to the culture every other day. When CART19-28ζ cells were chronically stimulated with the CD19+ mantle cell lymphoma cell line, JeKo-1, they became progressively dysfunctional as evident by reduced CART cell antigen specific expansion in vitro (Fig. 1b). Additionally, chronically stimulated CART cells exhibited phenotypical and functional signs of exhaustion such as the increased co-expression of multiple inhibitory receptors and the decreased production of effector cytokines such as IL-2 and TNF-α (Supplementary Fig. S2a–d). Furthermore, these cells exhibited reduced polyfunctionality as determined by the number of T cells secreting three or more cytokines (Supplementary Fig. S2e). Importantly, changes in antigen specific expansion, the co-expression of inhibitory receptors, and the production of effector cytokines appear to be a result of chronic stimulation and not due to a change in the percentage of CAR+ T cells (Supplementary Fig. S2f) or due to long-term co-culture (Supplementary Fig. S3).
Fig. 1. An in vitro model for chronic stimulation induces phenotypic and functional signs of exhaustion in CART19-28ζ cells.
a Schematic depicting an in vitro model for exhaustion in CART cells (Fig. 1a was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license). b Absolute CD3+ cell count, as determined with flow cytometry, after Day 8 (baseline), Day 15 (1 week of chronic stimulation), and Day 22 (2 weeks of chronic stimulation) CART19-28ζ cells were co-cultured with JeKo-1 target cells at a 1:1 ratio for 5 days (One-way ANOVA with average of two technical replicates for three biological replicates, mean ± standard deviation (SD)). c In vivo antitumor activity of Day 22 and Day 8 CART19-28ζ cells in a JeKo-1 xenograft model. NOD-SCID-IL2rγ−/− (NSG) mice were engrafted with the CD19+ luciferase+ JeKo-1 cells (1 x 106 cells I.V.). Mice underwent bioluminescent imaging weekly to confirm engraftment and to monitor tumor burden. Total flux is depicted over time following treatment with CART19-28ζ (0.9 x 106 cells I.V.) on Day 0 (Two-way ANOVA, n = 5 mice per group, mean ± SD). d Overall survival curve based on JeKo-1 xenograft mouse model comparing treatment with Day 8 or Day 22 CART19-28ζ cells (Log-rank (Mantel–Cox) test, n = 5 mice per group) e Bioluminescent imaging of the tumor growth in the JeKo-1 xenograft mouse model. f In vivo CART cell expansion as determined by absolute count of hCD45+CD3+ cells per μL of blood by flow cytometry on Day 15 of the JeKo-1 xenograft model. (two-sided t test, n = 5 mice per group, mean ± SD) g Circle graphs showing the average portion of CART cells expressing multiple inhibitory receptors (0—black, 1—pink, 2—green, 3—dark purple) based on flow cytometry detection of PD-1, CTLA-4, and TIM-3 on human CD3+ cells in the peripheral blood of mice on Day 15 of the JeKo-1 xenograft model. (Average portion from n = 5 mice per group) h–j Human cytokine levels for IL-2, IFN-γ, and IL-10 as determined by Multiplex bead assay of mouse serum collected from peripheral blood on Day 15 of the JeKo-1 xenograft model. (two-sided t test, n = 5 mice per group, mean ± SD). Source data for (b–d) and (f–j) are provided in the Source Data file.
To further examine the function and phenotype of chronically stimulated CART cells, we utilized a mantle cell lymphoma xenograft mouse model that stress tests CART19 cells (Supplementary Fig. S4a). In this model, mice are allowed to develop high disease burden so that standard CART19 doses fail to induce a complete remission of the tumor. Mice were then randomized to receive treatment with either baseline (Day 8) or chronically stimulated (Day 22) CART19-28ζ cells. Treatment with chronically stimulated CART19-28ζ cells resulted in a significant reduction in anti-tumor activity (Fig. 1c and Supplementary Fig. S4b) and overall survival (Fig. 1d, e and Supplementary Fig. S4c), compared to treatment with baseline CART19-28ζ. In addition, there was a trend for decreased CART expansion (Fig. 1f and Supplementary Fig. S4d) and a significant upregulation of multiple inhibitory receptors (Fig. 1g and Supplementary Fig. S4e, f) on CART cells in the peripheral blood of mice treated with chronically stimulated CART19-28ζ cells. Further, peripheral blood cytokine analysis two weeks following CART cell injection showed significantly decreased levels of several effector cytokines (Supplementary Fig. S4g), including IL-2 (Fig. 1h) and IFN-γ (Fig. 1i), in mice treated with chronically stimulated CART cells. These mice also showed significantly elevated levels of the inhibitory cytokine IL-10 (Fig. 1j). These findings corroborate our in vitro data and align with an exhausted phenotype.
Having demonstrated that our in vitro model for CART cell exhaustion leads to phenotypic and functional changes associated with T cell exhaustion, we next tested whether this model is applicable to other tumor models and CAR constructs. Consistent with our initial data, chronic stimulation of CART19-28ζ cells with the CD19+ acute lymphoblastic leukemia cell line, NALM-6 (Supplementary Fig. S5), or chronic stimulation of 4-1BB-costimulated CART19 (CART19-BBζ) cells with JeKo-1 cells (Supplementary Fig. S6) resulted in functional and phenotypical signs of T cell exhaustion. As such, our in vitro model is a representative model for inducing CART cell exhaustion in different tumor models and different CAR constructs.
CRISPR screen identifies a role for the IL-4 pathway in CART failure
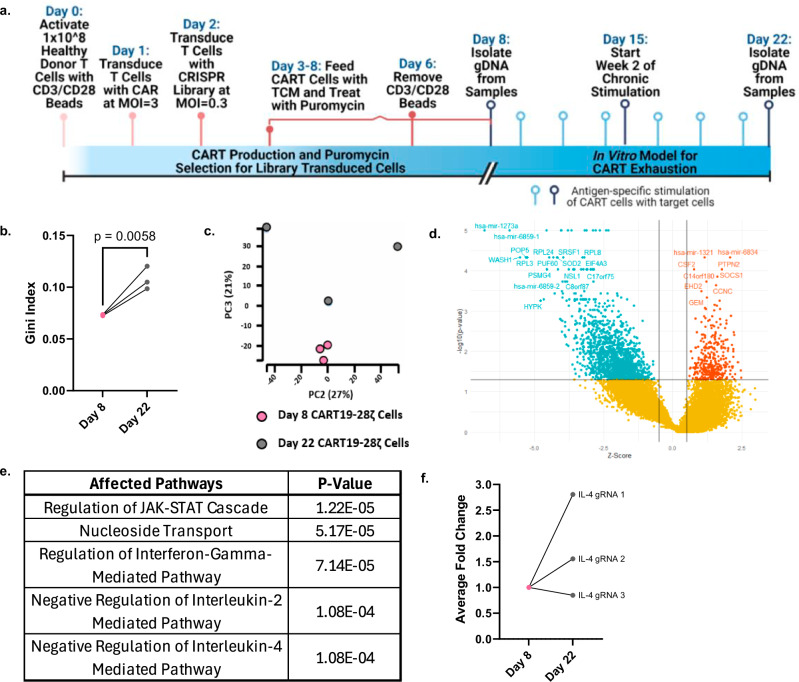
To investigate genes and pathways that can be altered to protect CART cells from exhaustion, we conducted a genome-wide CRISPR knockout screen using healthy donor CART19-28ζ cells. To do this, we scaled our in vitro model for CART cell exhaustion by transducing 1 × 108 CART cells with the GeCKO v2 library A on Day 2 at a multiplicity of infection (MOI) of 0.3 (Fig. 2a)34. Then, we selected for transduced cells through treatment with puromycin from Day 3 to Day 8.
Fig. 2. A genome-wide CRISPR knockout screen identifies a role for the IL-4 pathway in the development of CART cell dysfunction resulting from chronic stimulation.
a Schematic depicting the in vitro genome-wide CRISPR knockout screen conducted in healthy donor CART19-28ζ cells (Fig. 2a was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license). b The Gini index on Day 8 and Day 22 of the CRISPR screen (Gini index was calculated with MAGeCK-VISPR and compared with a two-sided t test, three biological replicates). c Principal component analysis plot of gRNA representation in the CRISPR screen at Day 8 and Day 22 (MAGeCK-VISPR maximum likelihood estimation (MLE) analysis with three biological replicates). d Volcano plot showing genes that were positively (red) or negatively (green) selected by Day 22 of the CRISPR screen as compared to Day 8 (Results from MAGeCK-VISPR MLE analysis with three biological replicates). e Top pathways identified by gene ontology enrichment analysis of the positively selected genes (FDR < 0.25 as determined with MAGeCK-VISPR analysis with three biological replicates). f Average fold-change of IL-4 gRNA representation from Day 8 to Day 22 of the CRISPR screen from three biological replicates (fold change calculated with normalized counts of IL-4 targeting gRNAs as determined with MAGeCK-MLE with three biological replicates). Source data for (b) and (f) are provided in the Source Data file.
By Day 22 of the chronic stimulation assay, we observed positive selection of guide RNAs (gRNAs) as seen by an increase in the Gini index from Day 8 to Day 22 (Fig. 2b) and principal component analysis (PCA) (Fig. 2c). Additionally, there were little changes in the presence of non-targeting gRNAs from Day 8 to Day 22 of the CRISPR screen (Supplementary Fig. S7a). However, to adequately assess changes in gRNA representation throughout the screen, we normalized all gRNA counts to counts from the list of 1000 non-targeting gRNAs. To further verify the quality of the CRISPR screen, we examined the top negatively and positively selected genes. Top results from gene set enrichment analysis of the negatively selected genes include pathways associated with ribosomal genes and translational processes (Supplementary Fig. S7b). This is expected as knockout of these pathways leads to a strong negative selection phenotype35. Among the top positively selected genes, there are several genes that have previously been associated with CART cell dysfunction (CSF2, SOCS1, and PTPN2) (Fig. 2d). Knockout of these genes with CRISPR Cas9 in previously published studies has resulted in increased proliferative ability and decreased signs of CART cell dysfunction36–39. Together, these data indicate that the genome-wide CRISPR knockout screen effectively identified positively, and negatively selected genes associated with CART cell dysfunction.
To investigate key pathways associated with CART cell exhaustion, we performed gene ontology enrichment analysis on the list of positively selected genes. This analysis revealed a role for several cytokine signaling pathways in CART cell exhaustion including the IFN-γ, IL-2, and IL-4 pathways (Fig. 2e). In an independent causal network analysis of the positively selected genes with QIAGEN ingenuity pathway analysis (IPA), the IL-4 receptor (IL4R) was identified as a top regulator (Supplementary Fig. S8). Upon closer investigation of the gRNAs targeting IL-4 in the screen, two out of the three gRNAs targeting IL-4 had enhanced representation in the chronically stimulated (Day 22) samples as compared to baseline (Day 8) samples (Fig. 2f). This suggests that the IL-4 pathway is involved in CART cell exhaustion.
IL-4 is involved in regulation of exhaustion
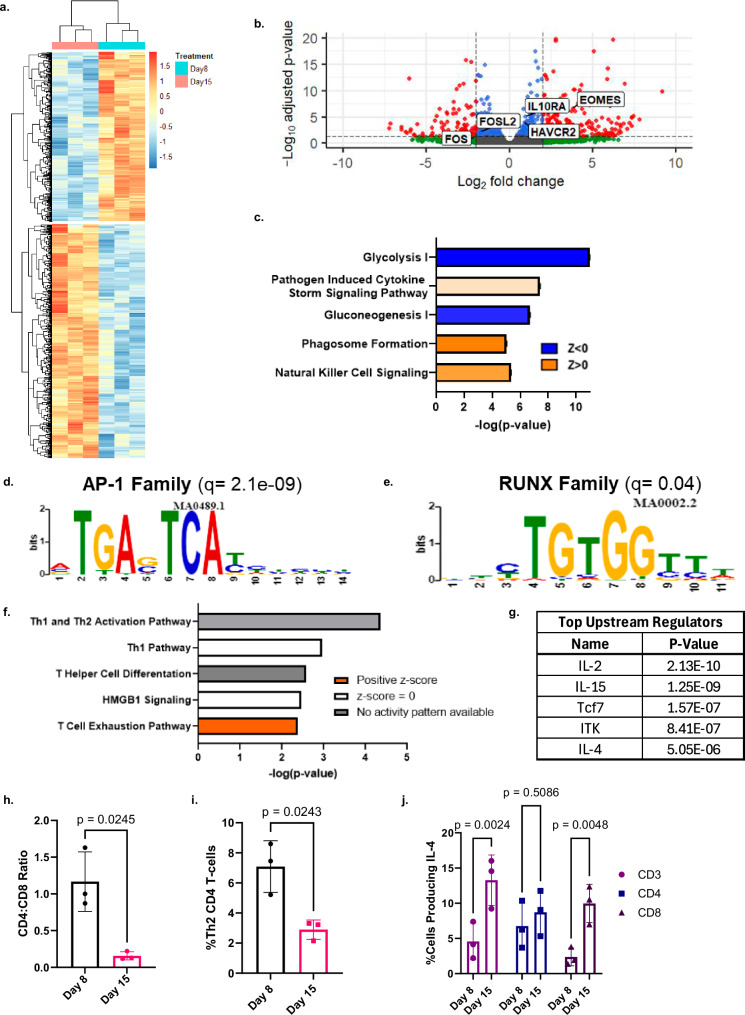
To further interrogate the top pathways identified with the genome-wide CRISPR knockout screen, and to enhance the field’s understanding of the epigenetic regulation of CART cell exhaustion, we interrogated the transcriptome and chromatin accessibility pattern of baseline and chronically stimulated CART19-28ζ cells from our in vitro model for exhaustion. RNA sequencing of baseline (Day 8) and chronically stimulated (Day 15) CART19-28ζ cells revealed the development of a distinct transcriptomic profile (Fig. 3a). Several genes that have previously been correlated with an exhausted phenotype, such as EOMES, IL10RA, and HAVCR2 (TIM-3), were confirmed to be upregulated in chronically stimulated CART19-28ζ cells (Fig. 3b). Additionally, the transcription of AP-1 family members that have previously been negatively correlated with the development of exhaustion, such as FOS and FOSL2, were downregulated (Fig. 3b)27. These findings further validate the development of CART exhaustion in our model.
Fig. 3. Transcriptomic and chromatin accessibility interrogation reveals a role for IL-4 in the development of CART cell exhaustion that is independent of its classical role in Th2 polarization.
a, b Heat map and volcano plot showing differentially expressed genes when comparing chronically stimulated (Day 15) to baseline (Day 8) CART19-28ζ cells (DESEQ2 with three biological replicates, padj < 0.05). In the volcano plot, significantly differentially expressed genes (padj < 0.05) are colored red. c Top differentially regulated pathways as determined by QIAGEN IPA of differentially expressed genes (DESEQ2 with three biological replicates, padj < 0.05). d, e Motifs enriched in chronically stimulated as compared with baseline CART19-28ζ cells (MEME/TOMTOM analysis with three biological replicates). f, g Top canonical pathways and upstream regulators as identified by QIAGEN IPA of genes that were both differentially expressed (DESEQ2 with three biological replicates, padj < 0.05) and differentially accessible (DiffBind with DESEQ2 using three biological replicates, padj < 0.05). h Ratio of CD4+ to CD8+ CART19-28ζ cells in baseline (Day 8) and chronically stimulated (Day 15) cell populations from the in vitro model for exhaustion (Paired two-sided t test, average of two technical replicates for three biological replicates, mean ± SD). i The percent of CD4+ CART19-28ζ cells that are Th2 polarized as determined by CCR6-CCR4+CXCR3- via flow cytometry in baseline (Day 8) and chronically (Day 15) stimulated CART19-28ζ cell populations (Paired two-sided t test, average of two technical replicates for three biological replicates, mean ± SD). j The percent of either CD3+, CD4+, or CD8+ CART19-28ζ cells producing IL-4 as determined by intracellular staining for IL-4 by flow cytometry following four hours of antigen-specific CAR stimulation through co-culturing Day 8 or Day 15 CART19-28ζ cells with JeKo-1 cells at a 1:5 effector-to-target (E:T) cell ratio (Two-way ANOVA, average of two technical replicates for three biological replicates, mean ± SD). Source data for (h–j) are provided in the Source Data file.
To determine the activity of signaling pathways during the development of exhaustion, we performed pathway analysis using both the list of differentially expressed genes and their respective fold changes. This analysis revealed a strong downregulation in pathways associated with metabolism such as glycolysis and gluconeogenesis and highlighted the importance of cytokine signaling in the development of exhaustion (Fig. 3c).
Next, we performed ATAC sequencing on baseline (Day 8) and chronically stimulated (Day 15) CART19-28ζ cells to investigate the epigenetic changes responsible for the development of exhaustion. Consistent with the transcriptomic changes and existing literature, there was enhanced chromatin accessibility at exhaustion-related gene loci (e.g., PDCD1 and ENTPD1) in our chronically stimulated samples (Supplementary Fig. S9a, b) as well as enhanced motif accessibility for AP-1 and RUNX family members (Fig. 3d, e)27,33.
After verifying that known transcriptomic and epigenetic changes associated with exhaustion are seen following chronic stimulation of CART cells in our in vitro model for exhaustion, we next asked how the development of exhaustion was being regulated. To uncover epigenetic regulators of CART cell exhaustion, we overlapped the genes that were both differentially expressed and differentially accessible. Then, using QIAGEN IPA, we evaluated top affected pathways and upstream regulators by using both the list of overlapped genes and their respective fold changes, as determined with RNA sequencing. The top enriched pathway was the T cell exhaustion pathway (Fig. 3f). Other pathways with indeterminant enrichment statuses include pathways involved with Th1, Th2 or T helper cell activation or differentiation (Fig. 3f). Top predicted upstream regulators include IL-2, IL-15, TCF-7, ITK, and IL-4 (Fig. 3g). IL-2, IL-15, TCF-7 and ITK have previously been linked to the development of T cell exhaustion9,16,40–42. However, while IL-4 has classically been associated with Th2 polarization in CD4+ T cells, its role in the development of CART cell exhaustion has not been well-studied.
IL-4 production in CD8+ CART cells increases upon chronic stimulation
Given both the identification of IL-4 as a top upstream regulator in the development of exhaustion and the inclusion of pathways related to T helper cell differentiation in the top affected pathways, we asked whether IL-4 was identified as a top upstream regulator as a result of changes in the T helper cell polarization or as a result of the development of exhaustion.
To approach this question, we utilized multiple independent approaches to differentiate T cell exhaustion from T helper cell polarization in our in vitro model for CART cell exhaustion. First, we observed a sharp decrease in the CD4+ population of CART19-28ζ cells following chronic stimulation, an established finding during the development of exhaustion (Fig. 3h)9. Second, we inspected the Th1 and Th2 populations of cells within the declining CD4+ population and observed a significant decrease in the Th2 population within the CD4+ cells (Fig. 3i) and an increase in the Th1 population (Supplementary Fig. S10a). Third, we evaluated the serum cytokine levels of the Th2 cytokines IL-5 and IL-13 in JeKo-1 xenograft mice treated with either baseline (Day 8) or chronically stimulated (Day 22) CART19-28ζ cells and observed a reduction in the levels of these cytokines in mice treated with chronically stimulated CART19-28ζ cells (Supplementary Fig. S10b, c). Fourth, we observed an increase in IL-4 production in CART19-28ζ cells following chronic stimulation with JeKo-1 cells (Fig. 3j). Notably, there was a significant increase in IL-4 production in CD8+ CART19-28ζ cells, but not in CD4+ CART19-28ζ cells (Fig. 3j).
Overall, IL-4 was identified as a top upstream regulator of exhaustion based on epigenetic changes observed from Day 8 to Day 15 of our in vitro model for exhaustion. While this model also produces a shift in the T helper cell population, the shift is towards a Th1 population. This is unexpected given the role of IL-4 in polarizing CD4+ T cells towards a Th2 phenotype. However, we believe IL-4 is mainly identified as a top upstream regulator as a result of its impact on the CD8+ population of CART cells. This is supported by both an increase in the production of IL-4 by CD8+ CART cells and a significant reduction in CD4+ CART cells following chronic stimulation.
IL-4 is enriched in CART cell products from non-responders
Next, we aimed to determine the significance of our findings in patients treated with CART19-28ζ cells. We interrogated the transcriptome and chromatin accessibility pattern of pre-infusion axi-cel products from responders and non-responders in the pivotal ZUMA-1 clinical trial that led to the initial FDA approval of axi-cel32. In this analysis, responders were defined as patients who achieved complete remission as best response while non-responders were defined as patients who experienced stable or progressive disease. There was no observed difference in the percent of T cells expressing CAR between responders and non-responders (Supplementary Fig. S11a).
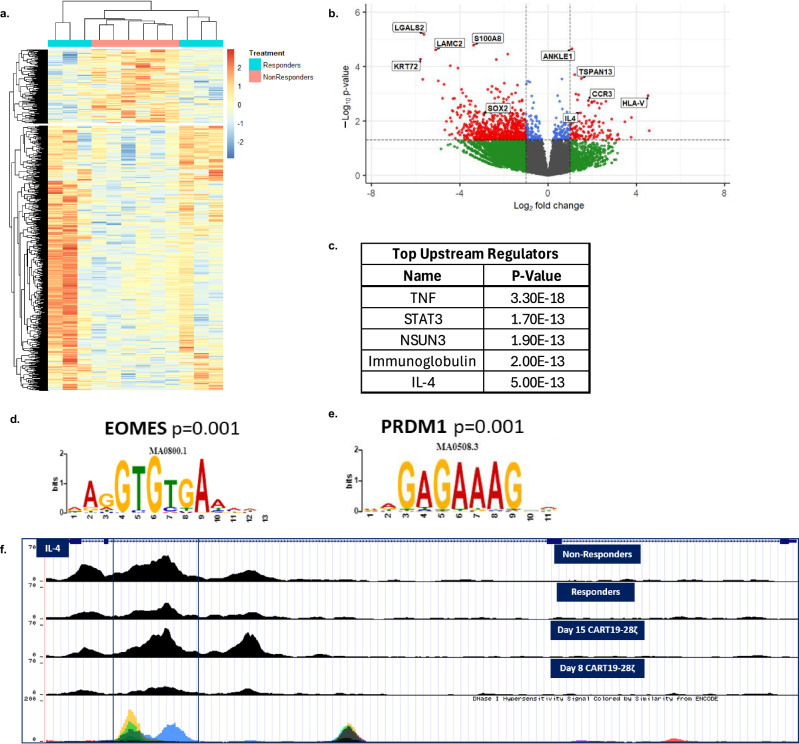
Interrogation of the transcriptome with RNA sequencing of pre-infusion axi-cel products from 6 responders and 6 non-responders showed clustering of non-responder samples (Fig. 4a). The top upregulated genes in non-responders include IL-4 and CCR3, a chemokine receptor that is known to be induced by IL-4 and IL-2 (Fig. 4b)43. Additionally, analysis of the differentially expressed genes with QIAGEN IPA identified IL-4 as one of the upstream regulators, along with other genes such as TNF and STAT3 (Fig. 4c). Interestingly, when we evaluate changes in chromatin accessibility between baseline axi-cel products from responders and non-responders, we see many similarities to the findings observed following chronic stimulation of healthy donor CART19-28ζ cells in our in vitro model for exhaustion. Motif analysis revealed an enrichment of motif binding sites for EOMES and PRDM1 (Fig. 4d, e). Both of these transcription factors have previously been associated with the development of exhaustion22,23. Additionally, non-responders showed similar enhancement of chromatin accessibility to exhausted cell populations at exhaustion-related gene loci such as PDCD1, HAVCR2 (TIM-3), EOMES, and IL-10 (Supplementary Fig. S12a–d). These findings indicate that epigenetic changes in baseline CART cell products contribute to CART cell exhaustion and failure in the clinic.
Fig. 4. Transcriptomic and chromatin accessibility interrogation of pre-infusion axi-cel products from responders and non-responders in the ZUMA-1 clinical trial identifies IL-4 as a regulator of response.
a, b Heat map and volcano plot showing differentially expressed genes when comparing pre-infusion axi-cel products from non-responders to pre-infusion axi-cel products from responders (DESEQ2, six biological replicates per condition, p < 0.05). In the volcano plot, significantly differentially expressed genes (p < 0.05) are colored red. c Top upstream regulators as determined by QIAGEN IPA of differentially expressed genes between non-responder and responder samples (DESEQ2, six biological replicates per condition, p < 0.05). d, e Enriched motifs in pre-infusion products from non-responders as compared to pre-infusion products from responders (MEME/TOMTOM analysis with six biological replicates per condition). f ATAC signal track of IL-4 gene locus from averaged signal for each experimental condition (axi-cel products from non-responders (n = 6), axi-cel products from responders (n = 6), Day 15 CART19-28ζ cells (n = 3), and Day 8 CART19-28ζ cells (n = 3)) as visualized with the UCSC genome browser.
Looking more specifically at the IL-4 locus, we see enhanced chromatin accessibility in non-responder samples as compared to responders (Fig. 4f). The chromatin accessibility pattern mirrors the change in accessibility observed when healthy donor CART19-28ζ cells are chronically stimulated. In particular, there is enhanced accessibility in both non-responder and chronically stimulated samples at a hypersensitivity site in intron 2 (Fig. 4f). This site correlates with an enhancer locus for IL-4, HS2, that specifically regulates IL-4 without impacting other Th2 cytokines44. Collectively, these data indicate that IL-4 regulation in non-responders is consistent with changes seen following the development of CART cell exhaustion in our in vitro model for exhaustion.
IL-4 induces exhaustion independently of the presence of CD4+ CART cells
Due to our identification of IL-4 as a key regulator of CART cell exhaustion through three independent approaches, we performed in vitro studies to directly evaluate phenotypic and functional changes that occur when CART19-28ζ cells are treated with human recombinant IL-4 (hrIL-4). In addition, we also performed experiments to determine if IL-4 regulates CART cell function independently of CD4+ CART cells.
Upon stimulating either bulk (CD3+) or CD8+ CART19-28ζ cells once with JeKo-1 target cells in the presence of 20 ng/mL hrIL-4 (vs. diluent), CART19-28ζ cells showed signs of dysfunction such as reduced cytotoxicity (Fig. 5a) and reduced proliferative ability as determined through CFSE staining (Supplementary Fig. S13a). These changes appear to be independent of a direct impact of hrIL-4 on JeKo-1 cells as treatment of JeKo-1 cells with hrIL-4 alone did not affect their growth or survival (Supplementary Fig. S13b). Additionally, these changes appear to be independent of a change in the percentage of CAR+ T cells as treatment of CART cells with hrIL-4 does not change CAR expression (Supplementary Fig. S13c).
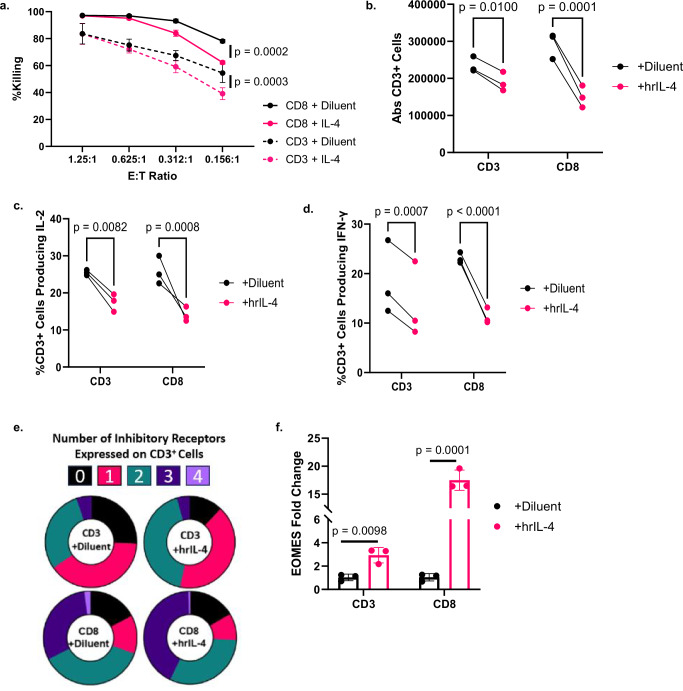
Fig. 5. Treatment of CART19-28ζ cells with hrIL-4 leads to phenotypical and functional signs of exhaustion in a CD4-independent manner.
a Percent killing as measured with bioluminescent imaging after Day 8 CD3+ or CD8+ CART19-28ζ cells were co-cultured with luciferase+ JeKo-1 cells at various E:T cell ratios for 48 hours in the presence of either 20ng/mL human recombinant IL-4 (hrIL-4) or diluent (Two-way ANOVA, average of two technical replicates for three biological replicates, mean ± SD). b, f CD3+ and CD8+ CART19-28ζ cells were kept in media supplemented with 100 IU/mL hrIL-2 and chronically stimulated from Day 8 to Day 15 of the in vitro model for exhaustion in the presence of either 20ng/mL hrIL-4 or diluent control. b Absolute CD3+ cell count as measured with flow cytometry after Day 15 CART19-28ζ cells were co-cultured with JeKo-1 cells at a 1:1 E:T cell ratio for five days. (Two-way ANOVA, average of two technical replicates for three biological replicates). c, d The percent of CD3+ cells producing IL-2 and IFN-γ as determined with intracellular staining and flow cytometry after Day 15 CART19-28ζ cells were co-cultured with JeKo-1 cells at a 1:5 E:T cell ratio for four hours (Two-way ANOVA, average of two technical replicates for three biological replicates). e The percent of CART19-28ζ cells co-expressing multiple inhibitory receptors (0—black, 1—pink, 2—green, 3—dark purple, 4—light purple) on Day 15 as determined by flow cytometric detection of PD-1, CTLA-4, TIM-3, and LAG-3 on CD3+ cells (Circle plots from one representative biological replicate). f The change in the transcription of EOMES as determined with RT-qPCR of Day 15 CD3+ or Day 15 CD8+ CART19-28ζ cells (Paired two-sided t-tests average of two technical replicates for three biological replicates, mean ± SD). Source data are provided as a Source Data file.
Next, we sought to evaluate the impact of IL-4 on CART19-28ζ cells in the presence of chronic stimulation using our in vitro model for CART cell exhaustion. Upon chronic stimulation of either bulk or CD8+ CART19-28ζ cells in the presence of 20 ng/mL hrIL-4 (vs. diluent), CART19-28ζ cells showed enhanced functional and phenotypic signs of exhaustion such as (1) reduced expansion (Fig. 5b), (2) reduced production of effector cytokines such as IL-2 and IFN-γ (Fig. 5c, d and Supplementary Fig. S13d, e), and (3) an increase in the percent of cells coexpressing inhibitory receptors (Fig. 5e and Supplementary Fig. S13f, g). The increase in the coexpression of inhibitory receptors appears to be dose-dependent with the greatest increase seen at the highest dose of hrIL-4 tested, 20 ng/mL (Supplementary Fig. S13h).
With evidence that IL-4 induces CART cell exhaustion independently of CD4+ CART cells, we next investigated the transcriptional changes responsible for IL-4-induced CART cell dysfunction. Given that 1) a prior study in CD8+ T cells showed the induction of the transcription factor EOMES by IL-445 and that 2) EOMES is enriched in both chronically stimulated healthy donor CART cells and non-responder axi-cel products, we asked whether treatment of CART19-28ζ cells with hrIL-4 would induce the expression of EOMES. After chronically stimulating either bulk or CD8+ CART19-28ζ cells in the presence of hrIL-4, EOMES transcription is induced as compared to the diluent-treated group (Fig. 5f).
Next, to evaluate if IL-4-induced CART cell dysfunction is dependent on additional interactions between the CART19-28ζ cells and target cells, we evaluated functional and phenotypical changes when CART19-28ζ cells are chronically stimulated with CD19-conjugated beads in the presence of either hrIL-4 or diluent. In this tumor-free assay, chronic stimulation of CART19-28ζ in the presence of hrIL-4 enhances the exhaustion profile as seen by (1) decreased production of IL-2, (2) increased coexpression of inhibitory receptors, and (3) decreased CART cell expansion (Supplementary Fig. S14a–d). This suggests that IL-4-induced CART cell exhaustion is due to a direct effect on CART cells and is independent of CART-tumor interactions.
Given the direct effect of hrIL-4 on CART19-28ζ cells, we next assessed if hrIL-4 induces CART cell exhaustion in different CAR constructs and tumor models. To test this, we generated a variety of CART cells by using CAR constructs that are similar to the constructs used in the clinic, including 1) CART19-BBζ, 2) BCMA-targeting 4-1BB-costimulated CART cells (BCMA CART-BBζ), and 3) CS1-targeting CD28-costimulated CART cells (CS1 CART-28ζ) (Supplementary Fig. S1b)46–49. Chronic stimulation of CART19-BBζ with JeKo-1 cells (Supplementary Fig. S15), BCMA CART-BBζ with OPM-2 multiple myeloma cells (Supplementary Fig. S16), or CS1 CART-28ζ with OPM-2 cells (Supplementary Fig. S17) in the presence of hrIL-4 enhanced the exhaustion phenotype. These data indicate that IL-4 supplementation can drive CART cell exhaustion independent of construct design or tumor models.
IL-4 neutralization improves the longevity and efficacy of CART cells
Given that IL-4 induces signs of CART cell exhaustion, we next examined whether disrupting the IL-4 pathway in CART cells with gene editing could reduce the prevalence of exhaustion. To start, we used CRISPR Cas9 to create IL-4 knockdown CART cells. Using two separate gRNAs, we were able to create targeted mutations to reduce CART19-28ζ production of IL-4 upon stimulation with CD19+ JeKo-1 cells (Supplementary Fig. S18a, b). At baseline, IL-4 knockdown CART19-28ζ cells showed enhanced cytotoxicity (Supplementary Fig. S18c, d) as compared with control gRNA CART19-28ζ cells. IL-4 knockdown CART cells also showed reduced phenotypical and functional signs of exhaustion following chronic stimulation such as (1) increased CART expansion (Supplementary Fig. S18e, f), (2) increased production of effector cytokines such as IL-2 and IFN-γ (Supplementary Fig. S18g–i), and (3) reduced co-expression of multiple inhibitory receptors (Supplementary Fig. S18j).
Importantly, IL-4 knockdown did not significantly alter the CD4-to-CD8 ratio at baseline or following chronic stimulation (Supplementary Fig. S19a–c). This indicates that IL-4’s role in driving CART cell exhaustion is not due to a change in the CD4-to-CD8 ratio. Further, given our previous finding that IL-4 supplementation can drive an exhausted phenotype in CD8+ CART cells, we looked at functional and phenotypical changes in CD8+ CART cells following chronic stimulation of IL-4 knockdown CART19-28ζ cells with JeKo-1 cells. IL-4 knockdown appears to reduce the exhausted phenotype of CD8+ CART19-28ζ cells as seen by an increase in the production of effector cytokines such as IL-2 and IFN-γ (Supplementary Fig. S20a–c) and a reduction in the co-expression of multiple inhibitory receptors (Supplementary Fig. S20d).
Next, to further evaluate if IL-4 reception by CART cells can drive an exhausted phenotype, we generated IL4R knockdown CART19-28ζ cells by using CRISPR Cas9 (Supplementary Fig. S21a). Following chronic stimulation with JeKo-1 cells, IL4R knockdown CART cells showed reduced phenotypical and functional signs of exhaustion such as (1) increased proliferative ability (Supplementary Fig. S21b), (2) decreased co-expression of multiple inhibitory receptors (Supplementary Fig. S21c), and (3) increased production of effector cytokines (Supplementary Fig. S21d, e). Further, the CD8+ population of IL4R knockdown CART cells also showed reduced phenotypical and functional signs of exhaustion (Supplementary Fig. S22). This further supports the notion that IL-4 can drive an exhausted phenotype in CD8+ CART cells.
To validate our in vitro findings that IL-4 pathway editing can improve the activity of CART19-28ζ cells, we also tested IL-4 and IL4R knockdown CART cells in vivo. We utilized a CD19+ JeKo-1 stress xenograft mouse model, similar to the one previously described (Supplementary Fig. S23a). In this model, both IL-4 and IL4R knockdown CART cells showed improved antitumor activity as compared with control gRNA CART19-28ζ cells (Supplementary Fig. S23b). Additionally, IL-4 and IL4R knockdown CART cells appeared to expand more in vivo (Supplementary Fig. S23c). This indicates that editing of the IL-4 pathway in CART cells can improve the activity of CART cells both in vitro and in vivo.
While CRISPR editing of the IL-4 pathway showed promise both in vitro and in vivo, we also decided to explore the use of an IL-4 monoclonal antibody (mAb) to neutralize IL-4. This represents a translatable strategy to improve CART cell outcomes in the clinic, as there are currently available, FDA-approved IL-4 mAbs50–52.
The addition of 10 μg/mL of a commercially available IL-4 mAb (clone MP4-25D2) in co-cultures of CART19 and JeKo-1 cells showed complete neutralization of IL-4 (Supplementary Fig. S24a) and an increase in cytotoxicity (Supplementary Fig. S24b) as compared to treatment with an IgG control antibody. Further, when CART19-28ζ cells are chronically stimulated in the presence of 10 μg/mL IL-4 mAb, there is a significant decrease in the percent of CART19-28ζ cells that express the inhibitory receptor PD-1 (Supplementary Fig. S24c). Chronic stimulation of CART19-28ζ cells in the presence of 10 µg/mL IL-4 mAb does not significantly alter the percentage of CAR+ T cells (Supplementary Fig. S24d).
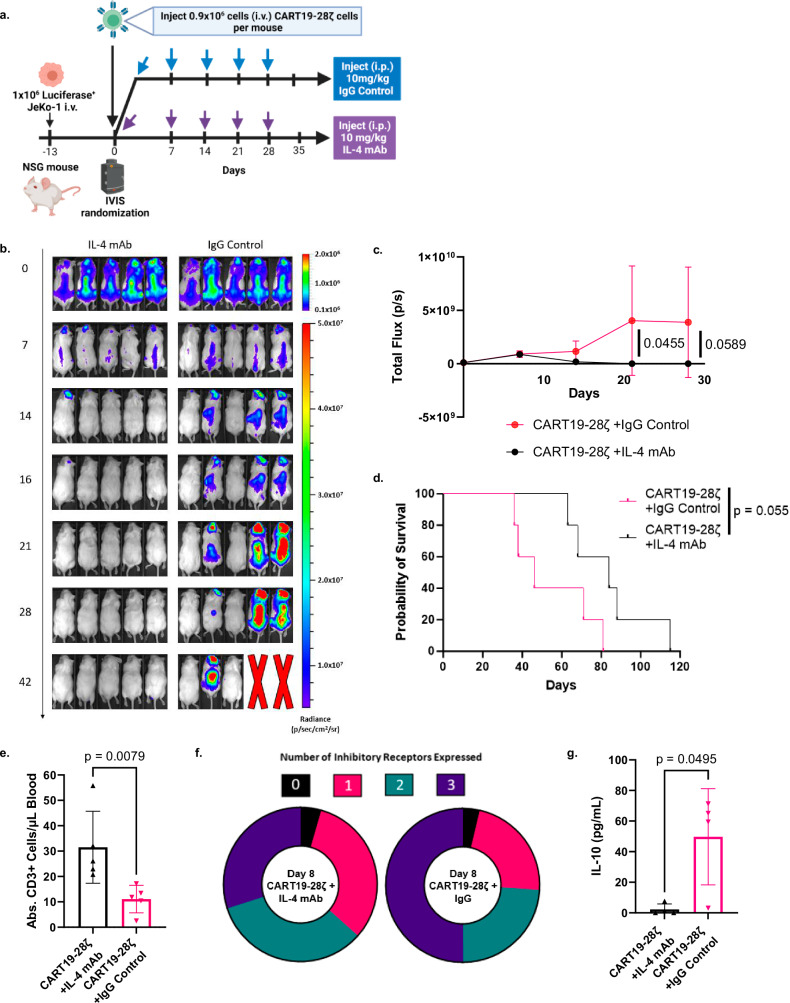
To further investigate the combination of CART19-28ζ cells with an IL-4 mAb, we used our CD19+ JeKo-1 stress xenograft mouse model (Fig. 6a), similar to the one depicted in Supplementary Fig. S3a. Following CART cell injection, mice were randomized based on tumor burden to weekly intraperitoneal (i.p.) injections of either 10 mg/kg IL-4 mAb or an IgG control antibody for 5 weeks. In this model, the majority of mice treated with CART19-28ζ cells and an IgG control antibody were unable to effectively clear their tumor burden (Fig. 6b, c). In contrast, all mice treated with a combination of CART19-28ζ cells and an IL-4 mAb were able to effectively clear their tumor burden and delay tumor relapse (Fig. 6b, c). Thus, combining CART19-28ζ cells with an IL-4 mAb resulted in enhanced antitumor activity (Fig. 6b, c) and a trend for prolonged overall survival (Fig. 6d).
Fig. 6. Combination of CART19-28ζ cells with an IL-4 monoclonal antibody improves overall treatment efficacy and CART cell function in an in vivo mouse model for mantle cell lymphoma.
a Schema for mantle cell lymphoma xenograft mouse model in NSG mice used to test the treatment efficacy of CART19-28ζ cells combined with 10mg/kg IL-4 monoclonal antibody (mAb) as compared with CART19-28ζ combined with 10mg/kg IgG control antibody (Fig. 6a was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license). b, c Tumor progression as monitored by bioluminescence imaging over time following injection of CART cells on Day 0 (Two-way ANOVA, n = 5 mice per group, mean ± SD). d Overall survival curve comparing CART19-28ζ cell treatment combined with either an IL-4 mAb or IgG control antibody (Log-rank (Mantel–Cox) test, n = 5 mice per group). e Absolute hCD45+CD3+ cells per μL of blood on Day 15 of the in vivo study as determined by flow cytometric measurement of cells that are human CD45+ and human CD3+ after collecting peripheral blood via tail vein bleeding (two-sided t test, n = 5 mice per group, mean ± SD). f Circle graph showing the average portion of CART cells positive for multiple inhibitory receptors (0—black, 1—pink, 2—green, 3—dark purple) as determined with flow cytometric detection of human CD3+ cells positive for PD-1, TIM-3, and/or CTLA-4 in the peripheral blood of mice on Day 15 of the in vivo study (Average value from n = 5 mice per group). g The concentration of IL-10 in the serum of mice in the in vivo model two weeks after the injection of CART cells. Serum was collected through tail vein bleeding of the mice, and cytokine concentration was determined with the use of the Milliplex MAP Human High Sensitivity T Cell Panel Premixed 13-plex (two-sided t test, n = 5 mice per group, mean ± SD). Source data for (c–g) are provided in the Source Data file.
Within this model, we evaluated CART19-28ζ cell function and phenotype through peripheral blood sampling two weeks following CART cell injection. The combination of the IL-4 mAb with CART19-28ζ cells improved CART cell expansion (Fig. 6e), reduced the percent of CART cells expressing multiple inhibitory receptors (Fig. 6f), and reduced the secretion of the inhibitory cytokine IL-10 (Fig. 6g and Supplementary Fig. S25a). Consistent results were observed in a low-tumor burden CD19+ JeKo-1 xenograft mouse model, where all mice were able to clear tumor load regardless of combination therapy (Supplementary Fig. S25b–d). Together, our data demonstrates that IL-4 neutralization with a mAb reduces exhaustion and improves anti-tumor activity of CART19-28ζ cells.
Discussion
Our study started with the goal of investigating the development of CART cell exhaustion due to its known role in negative therapeutic outcomes in CART cell therapy8. We therefore developed and utilized an in vitro model for CART cell exhaustion that resulted in phenotypic, functional, transcriptional, and epigenetic changes associated with exhaustion. Using this in vitro model for exhaustion, we performed a genome-wide CRISPR knockout screen that helped us determine genes and pathways that can be altered to protect CART cells from exhaustion, including the IL-4 pathway. In a second approach, we identified IL-4 as a top upstream regulator of CART cell exhaustion by performing RNA and ATAC sequencing on baseline and chronically stimulated CART19-28ζ cells. Finally, in a third approach, we evaluated clinically relevant determinants of CART cell response by performing RNA and ATAC sequencing on pre-infusion axi-cel products from responders and non-responders in the pivotal ZUMA-1 clinical trial. This independent approach not only showed epigenetic changes associated with exhaustion in pre-infusion CART cells from non-responders, but it also identified IL-4 as a top regulator of CART cell dysfunction. Thus, three independent approaches highlighted the involvement of IL-4 in CART cell exhaustion using both preclinical models and clinical trial samples.
Our results demonstrated that IL-4 induces a state of exhaustion, as evident by reduced T cell effector functions, increased expression of inhibitory receptors, as well as a transcriptional and epigenetic signature of exhaustion. Collectively, these studies showed that 1) IL-4 has a direct impact on CART cells that is independent of the presence of tumor cells, 2) IL-4 supplementation drives CART cell exhaustion in various CAR constructs with both CD28 and 4-1BB costimulatory domains, and in lymphoma or myeloma models, 3) IL-4 regulates CART cell exhaustion in both retrovirally (as seen in our RNA and ATAC sequencing studies of preinfusion products from the ZUMA-1 clinical trial) and lentivirally (as seen in our in vitro and in vivo experiments) transduced CART cells and 4) IL-4 drives exhaustion in CD8+ CART cells even in the absence of CD4+ CART cells. However, interestingly, and consistent with data showing a higher level of exhaustion with CD28-costimulated CART cells, IL-4 appears to have a more dramatic impact on CART19-28ζ cells (Fig. 5) than CART19-BBζ cells (Supplementary Fig. S15). Together, our data indicate a role for IL-4 in the development of exhaustion regardless of CAR construct, tumor model, transduction method, or the presence of CD4+ T cells53.
Importantly, our results indicate that IL-4-mediated CART cell exhaustion is associated with lack of response in the clinic. RNA and ATAC sequencing of axi-cel products from the ZUMA-1 clinical trial demonstrated IL-4 to be significantly upregulated in CART19 cells from non-responders prior to their infusion. This finding is different from what has been reported for how Th2 CART cells are associated with response to CART19 cell therapy in the clinic54. In one report of single-cell RNA sequencing of 41BB co-stimulated CART19 cells (tisagenlecleucel products), it was found that CART19 cells with a Th2 phenotype are associated with durable remission after therapy54. In another study for outcomes post-TCR therapies, CD4+ Th2 cell cytokine profiles were associated with inferior outcomes55.
Our study also validated three approaches to mitigate CART cell exhaustion through IL-4 pathway interruption. In one approach, we generated IL-4 knockdown CART19-28ζ cells which showed improved antitumor activity both in vitro and in vivo and reduced phenotypical and functional signs of exhaustion following chronic stimulation in vitro. In a second approach, we generated IL4R knockdown CART19-28ζ cells which showed improved antitumor activity in vivo and reduced signs of exhaustion in vitro. Finally, in a third approach, we used a mAb to neutralize IL-4’s activity. IL-4 neutralization with a mAb resulted in improved CART cell antitumor efficacy both in vitro and in vivo while also reducing signs of exhaustion. While the first two approaches were limited due to incomplete knockout, all three of these approaches appear to be promising therapeutic strategies to improve response to CART cell therapy through IL-4 depletion.
In particular, we believe the combination of CART cell therapy with an IL-4 mAb is an actionable approach to improve durable response. This strategy is highly clinically translatable, as IL-4-targeted therapies are in various stages of clinical development and are generally well-tolerated. The IL-4 mAb, pascolizumab, was well-tolerated in a phase II clinical trial for the treatment of asthma50; dupilumab, a mAb blocking IL-4 and IL-13, is FDA-approved for multiple allergic diseases, including eczema and asthma51; and pitrakinra, a small molecule inhibitor of IL-4 signaling, has been shown to be safe in a phase II clinical trial for the treatment of asthma52. The combination of FDA-approved CART19 cells with existing therapeutics to neutralize or antagonize IL-4 is an attractive approach because it avoids further genetic editing of CART cells, which carries additional risks and complicates the already lengthy and expensive CART production process.
In addition, IL-4 neutralization with a mAb would also account for IL-4 production by other components of the tumor microenvironment. In previous studies, it has been shown that IL-4 is upregulated in the tumor microenvironment due in part to cancer-associated fibroblasts and M2 macrophages56–58. Additionally, IL-4 is said to promote the progression of solid tumors, enhance the generation of immunosuppressive myeloid cells, and dampen antitumor immune responses57. Thus, the combination of CART cell therapy with an IL-4 neutralizing antibody in the treatment of solid tumors holds the potential to not only directly improve the activity of the CART cells, but also to modulate the tumor microenvironment to promote tumor killing.
In summary, our findings utilized relevant preclinical models as well as patient samples from a pivotal clinical trial to identify a role for IL-4 in CART cell exhaustion that is independent of the known role of IL-4 in polarizing CD4+ CART cells. This study also presents IL-4 neutralization as a clinically feasible strategy to prevent CART exhaustion and to enhance CART antitumor efficacy. CART intrinsic dysfunction remains a major challenge to clinical efficacy in both hematological and solid tumors. This study illuminates a mechanism for preventing CART exhaustion and proposes a strategy to develop more effective CART cell therapies.
Methods
The research described in this article complies with all relevant ethical regulations. The use of recombinant DNA in the laboratory was approved by the Mayo Clinic Institutional Biosafety Committee (IBC), IBC number HIP00000252.43. De-identified healthy donor T cells were isolated from blood samples that were obtained from apheresis donor cones collected during platelet apheresis at Mayo Clinic. Specific experiments to study resistance to CART cell therapy were approved by Mayo Clinic Institutional Review Board (IRB 18-005745). Mice were purchased from Jackson Laboratories and were cared for within the Department of Comparative Medicine at the Mayo Clinic under an approved Institutional Animal Care and Use Committee protocol (A00001767-16-R22).
Cell Lines
The mantle cell lymphoma cell line, JeKo-1, the acute lymphoblastic leukemia cell line, NALM6, and the epithelial-like cell line, 293T, were purchased from ATCC, Manassas, VA, USA (cat. #CRL-3006, cat. #CRL-3273, and cat. #CRL-3216). The multiple myeloma cell line, OPM-2, was purchased from DSMZ, Braunschweig, Germany (cat. #ACC 50). For cytotoxicity and in vivo experiments, JeKo-1 cells were transduced with luciferase-ZsGreen lentivirus (Addgene, Cambridge, MA, USA) and sorted to 100% purity prior to use. JeKo-1 and OPM-2 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Gaithersburg, MD, USA) with 1% penicillin-streptomycin-glutamine (Gibco, Gaithersburg, MD, USA) and 20% fetal bovine serum (FBS, Sigma, St. Louis, MO, USA). NALM6 cells were cultured in RPMI 1640 medium with 1% penicillin-streptomycin-glutamine and 10% FBS. 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning, Glendale, Arizona, USA) with 1% penicillin-streptomycin-glutamine and 10% FBS. These cell lines were passaged less than 20 times, and they were tested monthly to confirm negative mycoplasma contamination.
CART cell production for in vitro and in vivo studies
The use of recombinant DNA in the laboratory was approved by the Mayo Clinic Institutional Biosafety Committee (IBC), IBC number HIP00000252.43. Healthy donor CART cells used in vitro and in vivo validation studies were generated as shown in Supplementary Fig. S1a. Briefly, peripheral blood mononuclear cells (PBMCs) were isolated using SepMate PBMC Isolation Tubes (STEMCELL Technologies, Vancouver, Canada) from blood samples that were obtained from apheresis donor cones collected during platelet apheresis at the Mayo Clinic. Next, T cells were isolated from the PBMCs through negative selection with an EasySep Human T Cell Isolation Kit with a RoboSep machine (STEMCELL Technologies, Vancouver, Canada). Then, the T cells were activated through the addition of CTSTM (Cell Therapy Systems) DynabeadsTM CD3/CD28 beads (Life Technologies, Oslo, Norway) at a 3:1 bead-to-cell ratio. After 24 hours of bead stimulation, T cells were transduced with lentiviral particles encoding the specific CAR at a multiplicity of infection (MOI) of 3. To generate lentiviral particles, we transiently transfected 293T cells with a CAR expression plasmid with Lipofectamine 3000 transfection reagent (cat. # L3000001, Invitrogen, Carlsbad, CA, USA). The studies included in this paper utilized the following CARs: (1) a second-generation CAR targeting CD19 through an scFv derived from an anti-human CD19 antibody clone FMC63 with a CD28 costimulatory domain (CART19-28ζ), (2) a second-generation CAR targeting CD19 through an scFv derived from an anti-human CD19 antibody clone FMC63 with a 4-1BB costimulatory domain (CART19-BBζ), (3) a second-generation CAR targeting CS1 through an scFv targeting CS1 through antibody clone Luc90 with a CD28 costimulatory domain, and 4) a second-generation CAR targeting BCMA through C11D5.3 clone with a 4-1BB costimulatory domain (Supplementary Fig. S1b)49.
CART cells were maintained at a concentration of 1x106 cells/mL throughout the production period with T cell media made with X-Vivo15 (Lonza, Walkersville, MD, USA) supplemented with 10% human serum albumin (Corning, NY, USA) and 1% penicillin-streptomycin-glutamine (Gibco, Gaithersburg, MD, USA). On Day 6 of the production period, the CD3/CD28 beads were removed from the cell suspension using magnetic separation before CAR expression was evaluated via flow cytometry as shown in Supplementary Fig. S26. Then, the CART cells were rested in T cell media until Day 8. On Day 8, CART cells were either cryopreserved for future experiments or used fresh for experiments, as indicated. Before use in functional experiments, the cells were thawed in T cell medium and rested overnight.
Similar to the above protocol, to generate IL-4 or IL4R knockdown CART19-28ζ cells, T cells were isolated from PBMCs originating from de-identified healthy donor and activated with CTSTM (Cell Therapy Systems) DynabeadsTM CD3/CD28 beads (Life Technologies, Oslo, Norway) at a 3:1 bead-to-cell ratio. After 24 hours of stimulation, on Day 1, T cells were transduced with lentiviral particles encoding the CD19 CAR with a CD28 costimulatory domain at a multiplicity of infection (MOI) of 3. On day 2, CART cells were transduced with an IL-4, IL4R, or non-targeting control gRNA lentiCRISPRv2 lentivirus. To make lentivirus for these studies, we transiently transfected 293T cells with a lentiCRISPRv2 plasmid that contained a specified gRNA. Two separate gRNAs targeting IL-4 were tested (IL-4 gRNA1: TGATATCGCACTTGTGTCCG and IL-4 gRNA2: CAAGTGCGATATCACCTTAC) and compared to a non-targeting control gRNA (GTATTACTGATATTGGTGGG). One gRNA targeting IL4R (TGAGCATCTCTACTTGCGAG) was tested and compared to the non-targeting control gRNA. The lentiCRISPRv2 plasmids for each gRNA are publicly available and were ordered through GenScript. Starting on Day 3, CART cells transduced with the CRISPR lentivirus were selected by supplementing the T cell media with 1μg/mL puromycin. On Day 6, the CD3/28 beads were removed from the cell suspension using magnetic separation before CAR expression was evaluated via flow cytometry as described above. Then, the CART cells were rested in T cell media supplemented with 1μg/mL puromycin until Day 8. On Day 8, CART cells were washed three times to remove the puromycin before use in functional assays.
CART cell production for RNA and ATAC sequencing
Healthy donor and pre-infusion patient CART cells used for RNA and ATAC sequencing (Figs. 3a–g, 4) were generated by KITE Pharma and sent to the Kenderian laboratory for sequencing2,3,32.
Multi-parametric flow cytometry
Antibodies were purchased from BioLegend, eBioscience, or BD Biosciences (Table S1). Flow cytometry was performed with a three-laser CytoFLEX (Beckman Coulter, Chaska, MN, USA). Analyses were performed with Kaluza Analysis 2.1 software (Beckman Coulter) or FlowJo X10.0.7r2 software (Becton Dickenson). More specifically, we performed the following:
CAR detection
Protein L staining with a Biotin Protein L primary antibody (cat. #M00097, GenScript, Piscataway, NJ, USA) and a secondary antibody for streptavidin (cat. #405203, BioLegend, San Diego, CA, USA) were used to detect CAR on lentivirally generated CART cells, and an anti-Whitlow linker antibody was used to detect CAR expression on retrovirally generated CART cells used for RNA and ATAC sequencing experiments. Positive staining was determined based on staining of a T cell from the same biological replicate that was not transduced with CAR lentivirus (Supplementary Fig. S26)
Intracellular staining of cytokines
A degranulation and intracellular cytokine assay was used to determine changes in the production of cytokines. Briefly, CART cells were co-cultured with JeKo-1, NALM6, or OPM-2 target cells at a 1:5 effector-to-target cell ratio for four hours with the addition of CD28 (clone L293, cat #348040, BD Biosciences, San Diego, CA, USA), CD49d (clone L25, cat #340976, BD Biosciences, San Diego, CA, USA), monesin (cat #420701, BioLegend, San Diego, CA, USA), and CD107a (clone H4A3) FITC (cat #555800, BD Biosciences, San Diego, CA, USA). After four hours of co-culture, intracellular staining of T cells was performed by first staining with Live/Dead Aqua (cat #L34966, Invitrogen, Carlsbad, CA, USA) before using the FIX & PERMTM Cell Permeabilization Kit (cat #GAS001S100 and cat #GAS002S100, Life Technologies, Oslo, Norway). Then, intracellular staining was performed with the following antibodies: IL-2 (clone 5344.111) PE-CF594 (cat #562384, BD Biosciences, San Diego, CA, USA), IL-4 (clone MP4-25D2) APC (cat #554486, BD Biosciences, San Diego, CA, USA), IFN-γ (clone 4S.B3) APC-efluor 780 (cat #47-7319-42, eBioScience, San Diego, CA, USA), GM-CSF (clone BVD2-21C11) BV421 (cat #562930, BD Biosciences, San Diego, CA, USA), and TNF-α (clone Mab11) AF700 (cat #502928, BioLegend, San Diego, CA, USA). Positive gating for each cytokine was determined based on unstained controls (Supplementary Fig. S28).
Surface staining for inhibitory receptors
To determine changes in the expression of inhibitory receptors from in vitro assays, cells were stained with the following antibodies: CD3 (clone SK7) APC-Cy7 (cat #344818, BioLegend, San Diego, CA, USA), CD8 (clone SK1) PerCP (cat #344708, BioLegend, San Diego, CA, USA), PD-1 (clone EH12.2H7) BV-421 (cat #329920, BioLegend, San Diego, CA, USA), TIM-3 (clone F38-2E2) PE (cat #345006, BioLegend, San Diego, CA, USA), CTLA-4 (clone BNI3) PE-Cy7 (cat #369614, BioLegend, San Diego, CA, USA), and LAG-3 (clone 3DS223H) FITC (cat #11-2239-42, eBioscience, San Diego, CA, USA). Positive gating for each inhibitory receptor was determined based on unstained controls (Supplementary Fig. S29).
Evaluating changes in the Th1/Th2 phenotype by flow cytometry
To determine changes in the Th1/Th2 phenotype of the CART cells, T cells were stained with the following antibodies: CD4 (clone OKT4) FITC (cat #11-0048-42, eBioscience, San Diego, CA, USA), CCR4 (clone L291H4) PerCP (cat #L291H4, BioLegend, San Diego, CA, USA), CCR6 (clone 11A9) APC (cat #560619, BD Biosciences, San Diego, CA, USA), and CXCR3 (clone G02H7) APC-Cy7 (cat #353722, BioLegend, San Diego, CA, USA). Th2 cells were identified as CD4+CCR6-CCR4+CXCR3- and Th1 cells were identified as CD4+CCR6-CCR4-CXCR3+ (Supplementary Fig. S30).
In vitro model for CART cell exhaustion
Following CART cell production, baseline (Day 8) CART cells were co-cultured with target cells at a 1:1 effector-to-target cell ratio. Every other day for a week, CART cells were restimulated with target cells by adding the same number of target cells that was added to baseline CART cells (Fig. 1a). Following one week of chronic stimulation (Day 15), CART cells were isolated through the combined use of CD4 and CD8 microbeads (cat #130-045-101 and cat #130-045-201, Miltenyi Biotec, Auburn, CA, USA). Briefly, CD4 and CD8 beads were added to the washed cell pellet at a 1:1 ratio according to product instructions. Following magnetic separation with LS columns (cat #130-042-401, Miltenyi Biotec, Auburn, CA, USA), purity was verified through staining with anti-human CD3 (clone SK7) APC-Cy7 (cat #344818, BioLegend, San Diego, CA, USA) and Live/Dead Aqua (cat #L34966, Invitrogen, Carlsbad, CA, USA). Then, the function and phenotype of the CART cells were interrogated by evaluating inhibitory receptor expression, cytokine production, and proliferative ability. The remaining Day 15 CART cells were further chronically stimulated by continuing the in vitro model for exhaustion for an additional week to generate two-week chronically stimulated CART cells (Day 22). During this additional week, CART cells were re-stimulated every other day with the same number of target cells that was added on Day 15. Bead separation of the CART cells from the co-culture did not impair the activity of the CART cells (Supplementary Fig. S27).
Experiments utilizing the in vitro model for exhaustion for CD8+ CART cell exhaustion (Fig. 5) followed the same protocol as described above, but the media was supplemented with 100 IU/mL hrIL-2 (cat #78145, STEMCELL Technologies, Vancouver, Canada).
Tumor-free in vitro model for exhaustion
Similar to our in vitro model for exhaustion that utilizes target cells, CART cells were chronically stimulated with CD19-coupled magnetic beads (cat #MBS-K005, Acro Biosystems, Newark, DE, USA). To do this, CART19-28ζ cells were stimulated on Day 8 by adding 10μg/mL CD19-coated beads to the co-culture. Then, every other day for one or two weeks (Fig. 1a), CART cells were restimulated with CD19-coated beads by adding an additional 10μg/mL CD19-coated beads to the cell suspension. Finally, on Days 8, 15, and 22, the function and phenotype of the CART cells were interrogated by evaluating inhibitory receptor expression, cytokine production, and proliferative ability.
T cell functional experiments
To determine changes in a CART cells ability to expand, CART cells were plated at a 1:1 effector-to-target cell ratio on Day 0 of the proliferation assay. On Day 3, 100μL of media was removed from each well and cryopreserved for cytokine release experiments as described in a subsequent section. Cells were then fed with 100μL of T cell media and incubated until Day 5. On Day 5, the absolute CD3+ cell count was determined via flow cytometry with CD3 (clone SK7) APC-Cy7 (cat #344818, BioLegend, San Diego, CA, USA). For proliferation assays that tested the effects of hrIL-4 on baseline CART cells, the 100μL of media added on Day 3 contained 20ng/mL hrIL-4 (cat #78045, STEMCELL Technologies, Vancouver, Canada).
For proliferation assays that utilized CFSE staining, CART19 cells were stained with the CellTrace CFSE Cell Proliferation Kit (cat #C34554, Invitrogen, Carlsbad, CA, USA) on Day 0 according to manufacturer instructions. Then, the cells were washed and counted prior to being plated at a 1:1 effector-to-target cell ratio with JeKo-1 target cells. On Day 3, 100µL of media was removed from each well, and 100µL of fresh T cell media was added to each well. On Day 5, the percent of CFSE- CART cells was determined via flow cytometry after staining for CD3 (clone SK7) APC-Cy7 (cat #344818, BioLegend, San Diego, CA, USA). Negative gating was determined by CFSE stained CART cells that were kept in T cell media for 5 days without stimulation.
Briefly, for cytotoxicity assays, luciferase+ target cells (JeKo-1) were incubated at the indicated effector-to-target cell ratios for 48 hours as listed in the specific experiment. Killing was calculated by bioluminescence imaging on a GloMax Explorer (Promega, Madison, WI, USA) after treating samples with 1μL D-luciferin (30μg/mL) per 100μL sample volume (Gold Biotechnology, St. Louis, MO, USA) for 5 minutes prior to imaging.
Real time-quantitative polymerase chain reaction
Total RNA was extracted with QIAzol lysis reagent (Qiagen, Gaithersburg, MD, USA), RNeasy Plus Mini Kit (Qiagen, Gaithersburg, MD, USA), and RNase-Free DNase Set (Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s protocol. cDNA was generated using iScript Advanced cDNA Kit for RT-qPCR (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Real Time-Quantitative Polymerase Chain Reaction (RT-qPCR) was completed according to the manufacturer’s instructions for RT-qPCR SsoAdvanced Universal SYBR Green Supermix (cat # S7563, Invitrogen, Carlsbad, CA, USA). The primer sequences used were as follows: forward primer EOMES (5’-GGCCTCTGTGGCTCAAATTC-3’), reverse primer EOMES (5’-GCAGTGGGATTGAGTCCGTT-3’), forward primer GAPDH (5’-GGAGCGAGATCCCTCCAAAAT-3’)59, reverse primer GAPDH (5’-GGCTGTTGTCATACTTCTCATGG-3’)59, forward primer TBP (5’-CTCACAGGTCAAAGGTTTAC-3’)60, reverse primer TBP (5’-GCTGAGGTTGCAGGAATTGA-3’)60.
Mantle cell lymphoma xenograft mouse model
Male and female 6- to 8-week-old NOD-SCID-IL2rγ−/− (NSG) mice were purchased from Jackson Laboratories and were cared for within the Department of Comparative Medicine at the Mayo Clinic under an approved Institutional Animal Care and Use Committee protocol (A00001767-16-R22). Mice were allowed to acclimate for two weeks before being included in experiments. The mice were housed in a pathogen-free, biosafety level 2+ animal facility in social housing (2-5 mice per cage). As part of their housing, the mice were exposed to a 12-hour dark/12-hour light cycle with an ambient temperature at 21 °C ± 1 °C and a humidity of 50% ± 10%. After tumor inoculation, mice were monitored regularly to assess weight changes and body condition. As part of our IACUC approved protocol, mice were euthanized following weight loss of greater than or equal to 20% weight loss, worsening body condition as seen by an inability to ambulate and reach food or water, or a body condition score of 1 or less.
In the mantle cell lymphoma xenograft mouse model which stress-tested CART19-28ζ cells, 0.8–1 × 106 luciferase+ JeKo-1 cells were engrafted via tail vein injection. Tumor burden was monitored through bioluminescent imaging with a Xenogen IVIS-200 Spectrum camera (PerkinElmer, Hopkinton, MA, USA). 10 min prior to imaging, mice were injected with 3mg D-luciferin (Gold Biotechnology, St. Louis, MO, USA) through an intraperitoneal injection. Once the tumor burden reached approximately 1x108 photons/second, mice were weighed and randomized based on tumor burden into treatment groups. CART cells were administered via tail vein injection as indicated in each specific experiment. For the mice receiving a combination of CART19-28ζ cells and either an IL-4 mAb (MP4-25D2, cat #BE0240, BioXCell, Lebanon, NH, USA) or an IgG control antibody (cat #BE0088, BioXCell, Lebanon, NH, USA), mice received weekly intraperitoneal injections of 10mg/kg antibody.
Following the start of treatment, mice were followed for tumor burden as determined with bioluminescence imaging, CART cell expansion and cytokine profile of peripheral blood, and overall survival. Following peripheral blood sampling, 50μL of blood was lysed of red blood cells with FACSTM Lysing Solution (cat #349202, BD Biosciences, San Diego, CA, USA) prior to antibody staining with anti-human CD45 (clone HI30) BV421 (cat #304032, BioLegend, San Diego, CA, USA), anti-mouse CD45 (clone 30-F11) APC-eFluor780 (cat #47-0451-82, eBioscience, San Diego, CA, USA), anti-human CD3 (clone SK7) BV605 (cat #344836, BioLegend, San Diego, CA, USA), anti-human CD20 (clone 2H7) APC (cat #302310, BioLegend, San Diego, CA, USA), anti-human CD8 (clone SK1) PerCP (cat #344708, BioLegend, San Diego, CA, USA), anti-human PD-1 (clone MIH4) FITC (cat #11-9969-42, eBioscience, San Diego, CA, USA, anti-human TIM-3 (clone F38-2E2) PE (cat #345006, BioLegend, San Diego, CA, USA), and anti-human CTLA-4 (clone BNI3) PE-Cy7 (cat #369614, BioLegend, San Diego, CA, USA). Flow cytometry was then performed to determine the absolute CD3+ T cell count and the expression of inhibitory receptors (Supplementary Fig. S31). Serum from peripheral blood was then used for multiplex analysis of cytokines as described below.
Serum and supernatant analysis of cytokine concentration
Cytokine concentration in the supernatant of in vitro assays and in the serum of mice was determined with MILLIPLEX MAP Human High Sensitivity T Cell Panel Premixed 13-plex (cat #HSTCMAG28PMX13BK, Millipore Sigma, Ontario, Canada) according to the kit’s instructions with 25μL of either serum or supernatant per sample. Analysis was completed with Belysa software after running the samples on a Luminex 200 (Millipore Sigma, Ontario, Canada).
RNA sequencing
For RNA sequencing of healthy donor CART cells, CART cells were first isolated from co-culture using combined CD4 and CD8 microbeads (cat #130-045-101 and cat #130-045-201, Miltenyi Biotec, Auburn, CA, USA) as described above. Then, RNA was isolated from 1 × 106 cells by using the miRNeasy Micro kit (Qiagen, Gaithersburg, MD, USA) and treated with RNase-Free DNase Set (Qiagen, Gaithersburg, MD, USA). RNA quality was assessed by High Sensitivity RNA Tapestation (Agilent Technologies Inc., California, USA) before library construction with the SMARTer Stranded Total RNA-Seq Kit v2—Pico Input Mammalian (Takara Bio USA INC., California, USA). Next, RNA sequencing was performed on an Illumina NovaSeq S4 (Illumina, California, USA). CD Genomics provided the output as fastq files through a file transfer protocol (ftp). Fastq files were first evaluated for quality using FASTQC. Then, Cutadapt was used to remove Illumina adapters before FASTQC was run again to evaluate quality after adapter removal. Next, the paired-end reads for each condition were aligned with STAR using the genome reference consortium human build 38 patch release 13 (GRCh38.p13) downloaded from NCBI61. HTSeq was used to generate expression counts for each gene, and DESeq2 was used to normalize the data and calculate differential expression62,63. Differentially expressed genes were determined based on a false discovery rate (FDR) less than 0.05. Volcano plots were generated with the package EnhancedVolcano64. Heatmaps were generated with the R package pheatmap. Pathway analysis was conducted through the use of Qiagen IPA (Qiagen Inc., https://digitalinsights.qiagen.com/IPA)65.
RNA sequencing of pre-infusion axi-cel samples was completed by Kite Pharma and the sequencing files were shared with the Kenderian Laboratory. Briefly, frozen cell pellets were used for RNA isolation and library preparation. Then, strand-specific RNA sequencing (2 × 150 bp) with Poly-A selected RNA was performed. Following sequencing, the latest human genome (GRCh38.84) was downloaded from the Ensembl database and STAR was used to align the paired-end reads for each sample to the genome. HTSeq was used to generate expression counts for each gene, and DESeq2 was used to normalize the data and calculate differential expression62,63. Differential expression was determined based on a p-value less than 0.05. Volcano plots were generated with the package EnhancedVolcano64. Heatmaps were generated with the R package pheatmap. Pathway analysis was conducted through the use of Qiagen IPA (Qiagen Inc., https://digitalinsights.qiagen.com/IPA).
ATAC sequencing
For all samples, 1 × 105 cells were washed in PBS and pelleted. The cell pellet was resuspended in 100 μL freezing medium, composed of 10% dimethyl sulfoxide (Sigma, St. Louis, MO, USA) and 90% FBS (Sigma, St. Louis, MO, USA). Then, it was stored at −80 °C prior to shipment to CD Genomics for library preparation with the Nextera kit and sequencing services with the HISEQ 4000. CD Genomics provided the output as fastq files through ftp. Quality check was performed first with FASTQC. Then, the Nextera adapter sequences were trimmed and a minimum length of 45 base-pairs was employed per ENCODE recommendations using Cutadapt. FASTQC was then re-run to ensure quality. Next, paired-end reads for each sample were aligned to the UCSC reference genome for human genome 38 (hg38), patch release 13 with Bowtie2. The resulting bam file was sorted with samtools and mitochondrial reads were removed with the BAMQC program. Peak calling was performed on the sorted and cleaned bam file using the MACS2 package with broad peak calling. Differential peak accessibility analysis was conducted with DESeq2 in the DiffBind package after removing the blacklisted regions associated with hg38 and normalizing the data based on library size. Next, the differentially accessible peaks were annotated using the R package “ChIPSeeker”66. Differentially accessible genes were determined with using FDR less than 0.05 for healthy donor CART samples and with p-value less than 0.05 for patient CART samples. Motif analysis was conducted using MEME suites. Chromatin accessibility profiles were created with the UCSC genome browser with averaged BigWig files for each condition. Briefly, BigWig files were created from sorted BAM files using the bamCoverage function. Then, averaged BigWig files for each experimental condition were generated with the mean function of Wiggletools.
CRISPR screen considerations
The protocol used for this CRISPR screen was adapted from a previously published article67. Briefly, the library A of the Human CRISPR Knockout Pooled Library (GeCKO v2) (cat #1000000048, AddGene, Cambridge, MA, USA) was amplified through the use of Endura Electrocompetent Cells (cat # 60242-1, Biosearch Technologies, Middlesex, UK). An even sgRNA distribution of the amplified library was verified through next-generation sequencing and the use of MAGeCK analysis based on the calculated Gini index and the number of gRNAs in the library with zero counts68.
After verifying a low Gini index and a low number of unrepresented gRNAs in the amplified library, lentivirus was generated as described above. Lentivirus for the library was titered using a puromycin selection method. On Day 2 of the CRISPR screen, CART cells were transduced with library lentivirus at an MOI of 0.3. From Days 3-8 of the screen, puromycin selection was conducted to purify the population of CART cells successfully transduced with the GeCKO library A lentivirus. Then, on Days 8 and 22 of the screen, CART cells were isolated from co-culture using combined CD4 and CD8 microbeads (cat #130-045-101 and cat #130-045-201, Miltenyi Biotec, Auburn, CA, USA) as described above and a pellet of 3.5x107 cells CART cells were cryopreserved.
CRISPR screen analysis
To prepare for sequencing, 3.5 × 107 cells from baseline (Day 8) and chronically stimulated (Day 22) timepoints were washed and pelleted before storage at −20 °C. Next, genomic deoxyribonucleic acid (gDNA) was isolated from each cell pellet with the Quick-DNA Midiprep Plus Kit (cat #D4075, Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. An ethanol precipitation protocol was performed to enhance the purity of the gDNA following isolation. Next, the gDNA was prepared for sequencing through library PCR. For the library PCR, the NEBNext® High-Fidelity 2X PCR Master Mix (cat #M0541S, New England BioLabs, Ipswich, MA, USA) was used with the primers (Supplementary Table S2) and the cycling conditions (Supplementary Table S3) that were developed and described in the referenced protocol paper67. Finally, the PCR reactions for each sample were pooled and the product was purified by running it on a 2% (wt/vol) agarose gel before extracting it with the QIAquick Gel Extraction Kit (cat #28704, Qiagen, Gaithersburg, MD, US).
Next, amplicon deep sequencing was performed by CD Genomics using the PE150 (Illumina, San Diego, CA, USA). CD Genomics provided the output as paired end fastq files through ftp. Paired-end read files for each sample were merged with bbmerge before using MAGeCK-VISPR to perform quality and differential expression analysis68. The maximum likelihood estimation method within MAGeCK-VISPR was used to determine selection of gRNAs in the screen as normalized to the list of 1000 non-targeting gRNAs included in the GeCKO library A. Volcano plots depicting the positively and negatively selected gRNAs was generated with ggplot and gene set enrichment analysis was performed on both the top negatively and the top positively-selected genes (FDR < 0.25) to identify the top affected pathways using Enrichr and QIAGEN IPA65,69,70.
Statistics
In vitro and in vivo experiments were performed using technical and biological replicates for appropriate statistical analyses. The method of p-value calculation is indicated in the respective figure legends. The pairing of biological replicates was used for statistical tests. GraphPad Prism (La Jolla, CA, USA) and Microsoft Excel (Redmond, WA, USA) were used to analyze the experimental data. Fold change calculations for RT-qPCR results were calculated with the delta-delta Ct method71.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This study was partly funded by Kite, a Gilead company (S.S.K.), Mayo Clinic Center for Individualized Medicine (S.S.K.), Mayo Clinic Comprehensive Cancer Center (S.S.K.), Mayo Clinic Center for Regenerative Biotherapeutics (SSK), National Institutes of Health K12CA090628 (S.S.K.) and R37CA266344-01 (S.S.K.), Department of Defense grant CA201127 (S.S.K.), Minnesota Partnership for Biotechnology and Medical Genomics, and Predolin Foundation (R.L.S. and S.S.K.). C.M.S., K.Y., O.S., and J.H.G. are supported by the Mayo Clinic Graduate School of Biomedical Sciences. Schematics are created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Author contributions
C.M.S. and S.S.K. conceptualized the project and designed experiments; C.M.S., E.L.S., R.L.S., M.J.C., T.H., B.K., and L.M., and performed experiments; C.M.S. analyzed data and prepared manuscript figures; C.M.S., E.L.S., and S.S.K. wrote the manuscript; C.M.S., E.L.S., R.L.S., M.J.C., T.H., B.K., L.M., I.C., C.M.R., K.Y., O.S., J.H.G., E.O., W.I., A.G.M., J.B., J.K., N.S., M.M., S.F., and S.K. edited and approved the final manuscript; C.M.S., M.J.C., and J.B. performed the bioinformatic analyses, with consultation from W.I. and A.G.M.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw and processed files from the in vitro genome-wide CRISPR screen are available in the Gene Expression Omnibus (GEO) repository under accession code GSE273299. The raw and processed files from RNA sequencing of baseline and chronically stimulated healthy donor CART19-28ζ cells are available in GEO under accession code GSE273294. The raw and processed files from ATAC sequencing of baseline and chronically stimulated healthy donor CART19-28ζ cells are available in GEO under accession code GSE273297. Due to patient privacy, we have provided processed files from RNA (Supplementary Data S1) and ATAC sequencing (Supplementary Data S2-S13) of pre-infusion patient CART cells. For RNA sequencing, we have provided a supplementary file with non-normalized counts, and for ATAC sequencing we have provided supplementary peak files. Source data from in vitro and in vivo experiments are provided as a supplementary file. Source data are provided with this paper.
Competing interests
S.S.K. is an inventor on patents in the field of CAR immunotherapy that are licensed to Novartis (through an agreement between Mayo Clinic, University of Pennsylvania, and Novartis). R.L.S., M.J.C., and S.S.K. are inventors on patents in the field of CAR immunotherapy that are licensed to Humanigen (through Mayo Clinic). S.S.K. is an inventor on patents in the field of CAR immunotherapy that are licensed to Mettaforge (through Mayo Clinic). S.S.K. receives research funding from Kite, Gilead, Juno, BMS, Novartis, Humanigen, MorphoSys, Tolero, Sunesis/Viracta, LifEngine Animal Health Laboratories Inc, and Lentigen. S.S.K. has participated in advisory meetings with Kite/Gilead, Calibr, Luminary Therapeutics, Humanigen, Juno/BMS, Capstan Bio, and Novartis. SSK has served on the data safety and monitoring board with Humanigen. S.S.K. has severed a consultant for Torque, Calibr, Novartis, Capstan Bio, and Humanigen. J.B., J.K., M.M., N.S., and S.F. are employed by Gilead. C.M.S., M.M., S.F., and S.S.K. are inventors on intellectual property related to this work. All other authors do not have competing interests to disclose at this time.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51978-3.
References
- 1.Sakemura, R., Cox, M. J., Hefazi, M., Siegler, E. L. & Kenderian, S. S. Resistance to CART cell therapy: lessons learned from the treatment of hematological malignancies. Leuk Lymphoma62, 2052–2063 (2021). 10.1080/10428194.2021.1894648 [DOI] [PubMed] [Google Scholar]
- 2.Neelapu, S. S. et al. Five-year follow-up of ZUMA-1 supports the curative potential of axicabtagene ciloleucel in refractory large B-cell lymphoma. Blood141, 2307–2315 (2023). [DOI] [PMC free article] [PubMed]
- 3.Locke, F. L. et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol.20, 31–42 (2019). 10.1016/S1470-2045(18)30864-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuster, S. J. et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. 380, 45–56 (2018). [DOI] [PubMed]
- 5.Wittibschlager, V. et al. CAR T-cell persistence correlates with improved outcome in patients with B-cell lymphoma. Int. J. Mol. Sci.24, 5688 (2023). [DOI] [PMC free article] [PubMed]
- 6.Shah, N. N. & Fry, T. J. Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol.16, 372–385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zebley, C. C. et al. CD19-CAR T cells undergo exhaustion DNA methylation programming in patients with acute lymphoblastic leukemia. Cell Rep.37, 110079 (2021). 10.1016/j.celrep.2021.110079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beider, K. et al. Molecular and functional signatures associated with CAR T cell exhaustion and impaired clinical response in patients with B cell malignancies. Cells11, 1140 (2022). [DOI] [PMC free article] [PubMed]
- 9.Wherry, E. J. T cell exhaustion12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Weber, E. W. et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science372, eaba1786 (2021). 10.1126/science.aba1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prinzing, B. et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med.13, eabh0272 (2021). 10.1126/scitranslmed.abh0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu, B. et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol.18, 573-582 (2017). [DOI] [PMC free article] [PubMed]
- 13.Li, F., Liu, H., Zhang, D., Ma, Y. & Zhu, B. Metabolic plasticity and regulation of T cell exhaustion. Immunology167, 482–494 (2022). 10.1111/imm.13575 [DOI] [PubMed] [Google Scholar]
- 14.Beltra, J.-C. et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity.52, 825–841 (2020). 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J. et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature567, 530–534 (2019). 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Z. et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity51, 840–855.e845 (2019). 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, P. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature.490, 543–546 (2012). 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLane, L. M. et al. Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells. Cell Rep.35, 109120 (2021). 10.1016/j.celrep.2021.109120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci USA.116, 12410–12415 (2019). 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo, H. et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol.22, 983–995 (2021). 10.1038/s41590-021-00964-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yigit, B. et al. SLAMF6 as a regulator of exhausted CD8+ T cells in cancer. Cancer Immunol Res.7, 1485–1496 (2019). 10.1158/2326-6066.CIR-18-0664 [DOI] [PubMed] [Google Scholar]
- 22.Li, J., He, Y., Hao, J., Ni, L. & Dong, C. High levels of eomes promote exhaustion of anti-tumor CD8(+) T cells. Front. Immunol.9, 2981 (2018). 10.3389/fimmu.2018.02981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, H. et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity31, 309–320 (2009). 10.1016/j.immuni.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung, I. Y. et al. Type I Interferon signaling via the EGR2 transcriptional regulator potentiates CAR T cell-intrinsic dysfunction. Cancer Discov.13, 1636–1655 (2023). [DOI] [PMC free article] [PubMed]
- 25.Jung, I. Y. et al. BLIMP1 and NR4A3 transcription factors reciprocally regulate antitumor CAR T cell stemness and exhaustion. Sci .Transl. Med.14, eabn7336 (2022). 10.1126/scitranslmed.abn7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, X. et al. Depletion of BATF in CAR-T cells enhances antitumor activity by inducing resistance against exhaustion and formation of central memory cells. Cancer Cell40, 1407–1422 e1407 (2022). 10.1016/j.ccell.2022.09.013 [DOI] [PubMed] [Google Scholar]
- 27.Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature.576, 293–300 (2019). 10.1038/s41586-019-1805-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mai, D. et al. Combined disruption of T cell inflammatory regulators Regnase-1 and Roquin-1 enhances antitumor activity of engineered human T cells. Proc Natl Acad Sci USA120, e2218632120 (2023). 10.1073/pnas.2218632120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long, A. H. et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med.21, 581–590 (2015). 10.1038/nm.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harush, O. et al. Preclinical evaluation and structural optimization of anti-BCMA CAR to target multiple myeloma. Haematologica, 107, 2395–2407 (2022). [DOI] [PMC free article] [PubMed]
- 31.Hou, A. J., Chen, L. C. & Chen, Y. Y. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov.20, 531–550 (2021). 10.1038/s41573-021-00189-2 [DOI] [PubMed] [Google Scholar]
- 32.Neelapu, S. S. et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med.377, 2531–2544 (2017). 10.1056/NEJMoa1707447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belk, J. A. et al. Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell40, 768–786.e767 (2022). 10.1016/j.ccell.2022.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods11, 783–784 (2014). 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, W. et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol.16, 281 (2015). 10.1186/s13059-015-0843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox, M. J. et al. GM-CSF disruption in CART cells modulates T cell activation and enhances CART cell anti-tumor activity. Leukemia36, 1635–1645 (2022). 10.1038/s41375-022-01572-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutra Del Galy, A. et al. In vivo genome-wide CRISPR screens identify SOCS1 as intrinsic checkpoint of CD4(+) T(H)1 cell response. Sci. Immunol.6, eabe8219 (2021). 10.1126/sciimmunol.abe8219 [DOI] [PubMed] [Google Scholar]
- 38.Wiede, F. et al. PTPN2 phosphatase deletion in T cells promotes anti-tumour immunity and CAR T-cell efficacy in solid tumours. EMBO J39, e103637 (2020). 10.15252/embj.2019103637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaFleur, M. W. et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol.20, 1335–1347 (2019). 10.1038/s41590-019-0480-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y. et al. IL-2 regulates tumor-reactive CD8(+) T cell exhaustion by activating the aryl hydrocarbon receptor. Nat. Immunol.22, 358–369 (2021). 10.1038/s41590-020-00850-9 [DOI] [PubMed] [Google Scholar]
- 41.Parry, H. M. et al. Long-term ibrutinib therapy reverses CD8(+) T cell exhaustion in b cell chronic lymphocytic leukaemia. Front. Immunol.10, 2832 (2019). 10.3389/fimmu.2019.02832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y. et al. Co-expression IL-15 receptor alpha with IL-15 reduces toxicity via limiting IL-15 systemic exposure during CAR-T immunotherapy. J. Transl. Med.20, 432 (2022). 10.1186/s12967-022-03626-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jinquan, T., Quan, S., Feili, G., Larsen, C. G. & Thestrup-Pedersen, K. Eotaxin activates T cells to chemotaxis and adhesion only if induced to express CCR3 by IL-2 together with IL-4. J. Immunol.162, 4285–4292 (1999). 10.4049/jimmunol.162.7.4285 [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, S. et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat. Immunol.12, 77–85 (2011). 10.1038/ni.1966 [DOI] [PubMed] [Google Scholar]
- 45.Carty, S. A., Koretzky, G. A. & Jordan, M. S. Interleukin-4 regulates eomesodermin in CD8+ T cell development and differentiation. PLoS ONE9, e106659 (2014). 10.1371/journal.pone.0106659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parikh, R. H. & Lonial, S. Chimeric antigen receptor T-cell therapy in multiple myeloma: A comprehensive review of current data and implications for clinical practice. CA Cancer J. Clin.73, 275–285 (2023). 10.3322/caac.21771 [DOI] [PubMed] [Google Scholar]
- 47.Raje, N. et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med.380, 1726–1737 (2019). 10.1056/NEJMoa1817226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakemura, R. et al. Targeting cancer-associated fibroblasts in the bone marrow prevents resistance to CART-cell therapy in multiple myeloma. Blood139, 3708–3721 (2022). 10.1182/blood.2021012811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterner, R. M. et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood133, 697–709 (2019). 10.1182/blood-2018-10-881722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bagnasco, D., Ferrando, M., Varricchi, G., Passalacqua, G. & Canonica, G. W. A critical evaluation of anti-il-13 and anti-il-4 strategies in severe asthma. Int. Arch. Allergy Immunol.170, 122–131 (2016). 10.1159/000447692 [DOI] [PubMed] [Google Scholar]
- 51.Thibodeaux, Q. et al. A review of dupilumab in the treatment of atopic diseases. Hum. Vaccin. Immunother.15, 2129–2139 (2019). 10.1080/21645515.2019.1582403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slager, R. E. et al. IL-4 receptor polymorphisms predict reduction in asthma exacerbations during response to an anti-IL-4 receptor alpha antagonist. J. Allergy Clin. Immunol.130, 516–522.e514 (2012). 10.1016/j.jaci.2012.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva-Filho, J. L., Caruso-Neves, C. & Pinheiro, A. A. S. IL-4: an important cytokine in determining the fate of T cells. Biophys. Rev.6, 111–118 (2014). 10.1007/s12551-013-0133-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai, Z. et al. Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci. Adv.8, eabj2820 (2022). 10.1126/sciadv.abj2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowicki, T. S. et al. Infusion product TNFalpha, Th2, and STAT3 activities are associated with clinical responses to transgenic T-Cell Receptor Cell therapy. Cancer Immunol. Res.11, 1589–1597 (2023). 10.1158/2326-6066.CIR-23-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakemura, R. et al. Targeting cancer associated fibroblasts in the bone marrow prevents resistance to chimeric antigen receptor T cell therapy in multiple myeloma. Blood134, 865–865 (2019). 10.1182/blood-2019-123277 [DOI] [Google Scholar]
- 57.Ito, S. E., Shirota, H., Kasahara, Y., Saijo, K. & Ishioka, C. IL-4 blockade alters the tumor microenvironment and augments the response to cancer immunotherapy in a mouse model. Cancer Immunol. Immunother.66, 1485–1496 (2017). 10.1007/s00262-017-2043-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirlekar, B. Tumor promoting roles of IL-10, TGF-beta, IL-4, and IL-35: Its implications in cancer immunotherapy. SAGE Open Med.10, 20503121211069012 (2022). 10.1177/20503121211069012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai, W. et al. PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nat. Commun.10, 5800 (2019). 10.1038/s41467-019-13608-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sturmlechner, I. et al. p21 produces a bioactive secretome that places stressed cells under immunosurveillance. Science374, eabb3420 (2021). 10.1126/science.abb3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Putri, G. H., Anders, S., Pyl, P. T., Pimanda, J. E. & Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics38, 2943–2945 (2022). 10.1093/bioinformatics/btac166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blighe, K., Rana, S. & Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labelinghttps://github.com/kevinblighe/EnhancedVolcano (2021).
- 65.Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics30, 523–530 (2014). 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics31, 2382–2383 (2015). 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- 67.Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc.12, 828–863 (2017). 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li, W. et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol.16, 1–13 (2015). 10.1186/s13059-015-0843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics14, 128 (2013). 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr Protoc1, e90 (2021). 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408 (2001). 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw and processed files from the in vitro genome-wide CRISPR screen are available in the Gene Expression Omnibus (GEO) repository under accession code GSE273299. The raw and processed files from RNA sequencing of baseline and chronically stimulated healthy donor CART19-28ζ cells are available in GEO under accession code GSE273294. The raw and processed files from ATAC sequencing of baseline and chronically stimulated healthy donor CART19-28ζ cells are available in GEO under accession code GSE273297. Due to patient privacy, we have provided processed files from RNA (Supplementary Data S1) and ATAC sequencing (Supplementary Data S2-S13) of pre-infusion patient CART cells. For RNA sequencing, we have provided a supplementary file with non-normalized counts, and for ATAC sequencing we have provided supplementary peak files. Source data from in vitro and in vivo experiments are provided as a supplementary file. Source data are provided with this paper.