Abstract
Background
Dyslipidemia is a well-known risk factor for cardiovascular disease, the leading cause of mortality worldwide. Although habitual intake of fish oil is associated with cardioprotective effects through triglyceride reduction, the interactions of fish oil with the genetic predisposition to dysregulated lipids remain elusive.
Objectives
We examined whether fish oil supplementation modifies the association between genetically predicted and observed concentrations of total cholesterol, low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, and triglycerides.
Methods
A total of 441,985 participants with complete genetic and phenotypic data from the UK Biobank were included. Polygenic scores (PGS) of the 4 lipids were calculated in participants of diverse ancestries. For each lipid, multivariable linear regression models were used to assess if fish oil supplementation modified the association between PGS and the observed circulating concentration, with adjustment for relevant covariates.
Results
Fish oil supplementation attenuates the associations between genetically predicted and observed circulating concentrations of total cholesterol, LDL cholesterol, and triglycerides while accentuating the corresponding association for HDL cholesterol among 424,090 participants of European ancestry. Consistent significant findings were obtained using PGS calculated based on multiple genome-wide association studies or alternative PGS methods. For triglycerides, each standard deviation (SD) increment in PGS is associated with 0.254 [95% confidence interval (CI): 0.248, 0.259] SD increase in the observed concentration among European-ancestry participants who reported fish oil usage. In contrast, a stronger association was observed in nonusers (0.267; 95% CI: 0.263, 0.270). Consistently, we showed that fish oil significantly attenuates the association between genetically predicted and observed concentrations of triglycerides in African-ancestry participants.
Conclusions
Fish oil supplementation attenuates the association between genetically predicted and observed circulating concentrations of total cholesterol, LDL cholesterol, and triglycerides while accentuating the corresponding association for HDL cholesterol in individuals of European ancestry. Further research is needed to understand the clinical implications of these findings.
Keywords: fish oil supplementation, lipids, polygenic scores, gene-diet interaction, triglycerides.
Introduction
Dyslipidemia is well-known to be a major risk factor for cardiovascular disease (CVD), the leading cause of death globally [1]. Elevated concentrations of total cholesterol, low-density lipoprotein (LDL) cholesterol, and triglycerides serve as major risk factors for increased risks of ischemic events, as shown in epidemiologic and Mendelian randomization studies [[2], [3], [4], [5], [6], [7]]. Conversely, a raised concentration of high-density lipoprotein (HDL) cholesterol may have protective roles against CVD, autoimmune disease, and cancers [8,9]. Numerous studies have found that fish oil supplementation, predominantly eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), exerts cardioprotective effects by reducing the triglyceride concentration [[10], [11], [12]]. However, the role of fish oil supplementation in modulating genetic susceptibility to elevated lipid concentrations remains insufficiently investigated.
In addition to environmental factors, studies have highlighted the essential role of genetic factors in dyslipidemia [13]. Genetic effects have been estimated to explain > 50% of the phenotypic variance in blood lipid concentrations [14]. The differential effect of a genotype in individuals with different environmental exposures, known as gene-environment interaction, also plays a crucial role in determining outcomes [15,16]. A prior genome-wide interaction study identified 4 novel loci whose associations with blood lipids were modified by fish oil supplementation [17]. Expanding the previous single-variant analysis to a genome scale, we hypothesized that the genome-wide genetic effects on blood lipid concentrations, captured by polygenic scores (PGS), could be modified by habitual fish oil supplementation.
In the current study, we aimed to explore the modifying effects of fish oil supplementation on the associations between the genetically predicted and the observed concentrations of total cholesterol, LDL cholesterol, HDL cholesterol, and triglycerides. Large-scale genome-wide association studies (GWAS) have identified numerous single nucleotide polymorphisms (SNPs) significantly associated with circulating lipid concentrations [18,19]. Leveraging this information and employing novel PGS derivation approaches, we aggregated individual lipid-associated SNPs into genome-scale PGS for estimating the genetically predicted circulating lipid concentrations among participants in UK Biobank (UKB). Then, for each of the 4 lipids, we examined the effect of interaction between fish oil supplementation and the PGS on the observed lipid concentration. Although our primary analysis focused on participants of European (EUR) ancestry, we also investigated the interaction effects in African (AFR), Central/South Asian (CSA), and East Asian (EAS) populations.
Methods
Study population
This study was conducted using data from UKB (application number 48818), which is a prospective population-based study of over 0.5 million participants aged 37–73 y at recruitment from 2006 to 2010 across the United Kingdom [20]. The UKB study received ethical approval from the National Health Service North West Centre for Research Ethics Committee, and all participants provided electronically signed consent before joining the study. Participants provided information on sociodemographic, lifestyle, and medical records via questionnaires at baseline. Blood samples for genotyping and biomarker measurements were also collected at recruitment. Of 502,365 individuals who attended baseline assessment, we excluded 8176 individuals from the analysis due to missing fish oil supplementation data, mismatched information between phenotypic and genetic sex, sex chromosome aneuploidy, outliers for heterogeneity and missing genotype rate, and having a high degree of genetic kinship (≥ 10 third-degree relatives identified). The genetic ancestry of participants has been previously defined in the Pan-ancestry genetic analysis of the UK Biobank (Pan-UKBB). Our analysis primarily focused on EUR ancestry participants, as they comprised the largest ancestral group in this resource. We also conducted analyses in participants of AFR, CSA, and EAS ancestries.
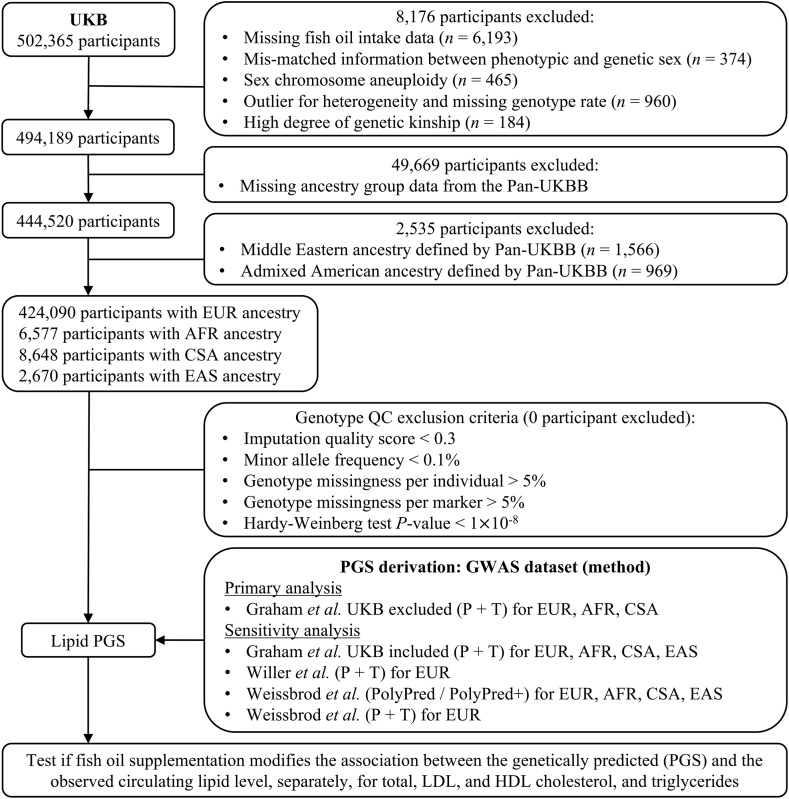
Genetic data were generated through genotyping by either the UKB Axiom Array or the United Kingdom BiLEVE Array. Genetic imputation was further performed with the Haplotype Reference Consortium and the UK10K haplotype reference panel [21]. Quality control for genotypic data was based on the following inclusion criteria: 1) imputation quality score > 0.3, 2) minor allele frequency > 0.1%, 3) genotype missingness per individual < 5%, 4) genotype missingness per marker < 5%, and 5) Hardy-Weinberg test P value > 1 × 10−8. Figure 1 presents a flowchart outlining the inclusion and exclusion of participants throughout our study.
FIGURE 1.
Overview of the study design and participant flow. Our study participants were drawn from UK Biobank, while the calculation of polygenic scores leveraged previous genome-wide association studies of blood lipids. AFR, African; CSA, Central/South Asian; EAS, East Asian; EUR, European; GWAS, genome-wide association study; Pan-UKBB, Pan-ancestry genetic analysis of the UK Biobank; PGS, polygenic scores; UKB, UK Biobank.
Assessment of fish oil supplementation status
The fish oil supplementation status of study participants was determined during their initial assessment center visit using a touchscreen questionnaire. Specifically, participants were asked about their regular consumption of certain supplements, such as fish oil intake (including cod liver oil). Of note, the touchscreen questionnaire did not collect information regarding the dose, frequency, or duration of the supplementation.
Assessment of blood lipids and dyslipidemia
Our primary outcomes of interest were 4 serum lipid parameters, including total cholesterol, LDL cholesterol, HDL cholesterol, and triglycerides. Their concentrations in baseline serum samples were ascertained using the Beckman Coulter AU5800 chemistry analyzer. The serum concentrations were standardized so that the effects could be interpreted as per 1 SD increase in the corresponding blood lipid.
We also explored secondary outcomes related to dyslipidemia, including hypercholesterolemia, hyperlipidemia, and hypertriglyceridemia. Hypercholesterolemia was defined as LDL cholesterol ≥130 mg/dL (≥3.4 mmol/L) or non-HDL cholesterol ≥160 mg/dL (≥4.1 mmol/L). Hyperlipidemia was defined as non-HDL cholesterol ≥220 mg/dL (≥5.7 mmol/L) and triglycerides ≥150 mg/dL (≥1.7 mmol/L). Hypertriglyceridemia was defined as triglycerides ≥150 mg/dL (≥1.7 mmol/L) [22].
Polygenic score calculation
We computed PGS for participants of EUR, AFR, CSA, and EAS ancestries in the UKB. PGS measures the aggregated effects of genetic variants on the circulating concentrations of total cholesterol, LDL cholesterol, HDL cholesterol, and triglycerides. In addition to the individual-level genetic data from the UKB participants, the calculation of PGS has 2 important considerations: 1) a previous GWAS of the trait of interest and 2) an algorithm to select variants and determine their corresponding weights. In our primary analysis, we used summary statistics from the largest and most recent GWAS of blood lipids to date, which excluded UKB and included up to 930,672 EUR, 92,555 AFR, and 34,135 CSA ancestry participants [19]. We applied the pruning and thresholding (P + T) algorithm to select variants for PGS calculation. Genome-wide significant variants were identified with a P value threshold of < 5 × 10−8. Independent variants were obtained by linkage disequilibrium (LD)-based pruning (r2 = 0.1, window size = 250 kb) with in-sample LD in UKB ancestry groups. The PGS of UKB participants was calculated with the weighted method: weighted PGS = β1 × SNP1 + β2 × SNP2 + … + βn × SNPn, where SNPn is the number of effect alleles of the nth SNP, and the β-coefficient is the effect size of the corresponding allele, which was directly extracted from GWAS summary statistics. Weighted PGS was further standardized to enable interpretation of per 1 SD change in PGS.
To evaluate the robustness of our findings to different GWAS datasets (e.g., inclusion vs. exclusion of UKB; GWAS sample sizes) and PGS derivation algorithms (including different parameters in P + T), we performed 4 sets of sensitivity analysis. The first 2 sets of sensitivity analysis applied the same P + T algorithms as in the primary analysis on 2 different GWAS datasets. The first was the GWAS meta-analysis of UKB and other cohorts, which was also released by Graham et al. [19], containing up to 1,320,016 EUR, 99,432 AFR, 40,963 CSA, and 146,492 EAS ancestry participants. The second was an early GWAS that included up to 187,365 EUR individuals, as previously reported by Willer et al. [18]. Our third and fourth sets of sensitivity analysis utilized PGS previously developed by Weissbrod et al. [23], who provided the selected variants and their corresponding weights. The PGS in the third sensitivity analysis was developed using novel algorithms, PolyPred for EUR ancestry and PloyPred+ for AFR, CSA, and EAS ancestries, and the main GWAS summary statistics were from UKB. PolyPred linearly combines 2 complementary predictors derived from GWAS in EUR-ancestry participants. The first predictor applies functionally informed fine-mapping to estimate the causal effects of genome-wide variants, whereas the second predictor jointly analyzes genome-wide variants to capture all signals in extremely polygenic loci. PolyPred+ was developed for non-EUR ancestries. In addition to the 2 EUR-based predictors, it has a third predictor that is trained in samples from the target non-EUR population. The PGS in our fourth set of sensitivity analysis was developed using the P + T algorithm but with different parameters (pruning r2 = 0.5, window size = 250 kb), and the GWAS was from EUR-ancestry UKB participants. The predictive power of these PGS in our primary and sensitivity analyses was evaluated with Pearson correlation analysis between the genetically predicted (i.e., PGS) and the observed circulating concentrations in UKB participants, stratified by ancestry groups and fish oil supplementation status (Supplementary Table 1). Notably, our primary analysis utilized PGS that were developed using GWAS that excluded UKB. This was to avoid overfitting bias in our gene-diet interaction analysis in UKB.
Covariates
A series of sociodemographic, behavioral, and genetic covariates were included in our analysis with 2 statistical models. Age, sex, and BMI (kg/m2) were obtained from baseline assessments at recruitment. The 22 baseline assessment center locations were situated throughout the United Kingdom. Socioeconomic status, as calculated by the Townsend deprivation index, was assigned to each participant corresponding to their home postcode at recruitment [24]. Smoking and alcohol drinking status were defined as never, former, and current. Physical activity concentrations were categorized as low, moderate, and high following the International Physical Activity Questionnaire protocol [25]. Statin use was ascertained through self-report during verbal interviews. Genotyping array was included as a covariate to control for possible batch effects. The top 20 ancestry principal components derived from the Pan-UKBB were employed as covariates to minimize the confounding effects of population stratification.
Statistical analysis
Multivariable linear regression models were conducted to assess the modifying impacts of fish oil supplementation on the associations between the genetically predicted (i.e., PGS) and the observed concentrations of blood lipids. The analysis was done separately for each of the 4 blood lipids and for each of the 4 ancestry groups. In Model 1, we adjusted for sex, age, age2, assessment centers, genotyping array, and the top 20 genetic principal components. Model 2 added additional covariates, including BMI, Townsend deprivation index, smoking status, alcohol status, physical activity, and statin use. These covariates were selected based on previous genetic and epidemiological studies of fish oil supplementation and blood lipids [[17], [18], [19],26]. For each model, we performed stratified analysis by fish oil supplementation status to estimate the effect of the PGS () on the observed concentration of the corresponding lipid in the 2 stratified groups. The model was as follows: observed lipid concentration = β0 + × PGSlipid + . We further evaluated the interaction effect by applying the same model to the combined sample and added an interaction term (PGS fish oil): observed lipid concentration = β0 + × PGSlipid + × fish oil + × PGSlipid × fish oil + . To further account for the potential modifying effects of sex and age, we repeated the above analyses with Model 2 in stratified groups by sex (male or female) and age at recruitment (median cut-off). The assumptions underlying multivariable linear regression, including linearity, normality of residuals, homoscedasticity, and no multicollinearity, were evaluated with diagnostic plots and residual analysis [27]. Only the model for observed triglyceride concentrations showed apparent violations of assumptions for the normality of residuals and homoscedasticity. Following previous studies of circulating triglycerides [19,28], we performed sensitivity analysis with natural log transformation of the phenotype, and the interaction between fish oil and the triglyceride PRS remained statistically significant.
Additionally, we applied multivariable logistic regression with Model 2 to perform exploratory analyses for 3 dyslipidemia-related outcomes, including hypercholesterolemia, hyperlipidemia, and hypertriglyceridemia. For each of the 3 outcomes and each of the 4 lipid PGS, the following model was applied: logit (probability of the dyslipidemia outcome) = β0 + × PGSlipid + × fish oil + × PGSlipid × fish oil + . The same covariates as the primary analysis of observed lipid concentrations were included: sex, age, age2, assessment centers, genotyping array, the top 20 genetic principal components, BMI, Townsend deprivation index, smoking status, alcohol status, physical activity, and statin use. The assumptions underlying multivariable logistic regression, including linearity in the logit for the continuous independent variable (i.e., the lipid PRS) and absence of multicollinearity, were evaluated with diagnostic plots and variance inflation factor analysis [29]. No violations of assumptions were found. All analyses were conducted using R (version 4.2.1).
Results
Study cohort
A total of 441,985 participants from the UKB with complete genetic and phenotypic data were included in this study, consisting mostly of individuals of EUR ancestry and small proportions of other ancestries (Figure 1). We present the baseline characteristics of participants of EUR ancestry according to their fish oil intake status in Table 1. After quality control, 134,720 (31.8%) of study participants reported habitual use of fish oil supplements, whereas the remaining 289,370 participants were grouped as nonconsumers of fish oil. The majority of fish oil users were female (56%), with a mean age of 59 years. Those in the fish oil intake group were more likely to be older, female, current alcohol drinkers, statin users, engaging in high physical activity, and having higher concentrations of total cholesterol, LDL cholesterol, and HDL cholesterol, but less likely to be current smokers or have an elevated BMI or triglyceride concentration. Among other ancestry groups, 2,289 (34.8%) AFR, 1,942 (22.5%) CSA, and 845 (31.6%) EAS participants took fish oil supplementation. Their baseline characteristics are displayed in Supplementary Table 2.
TABLE 1.
Baseline characteristics of participants of European ancestry in the UK Biobank1
| Fish oil intake (n = 134,720) | No fish oil intake (n = 289,370) | |
|---|---|---|
| Age, y (SD) | 59 (7.3) | 56 (8.1) |
| Sex, female (%) | 75,726 (56) | 153,758 (53) |
| BMI, kg/m2 (SD) | 27 (4.5) | 28 (4.9) |
| Total cholesterol, mmol/L (SD) | 5.78 (1.16) | 5.68 (1.14) |
| LDL cholesterol, mmol/L (SD) | 3.61 (0.88) | 3.55 (0.87) |
| HDL cholesterol, mmol/L (SD) | 1.48 (0.39) | 1.44 (0.38) |
| Triglycerides, mmol/L (SD) | 1.75 (1.00) | 1.76 (1.04) |
| Smoking status (%) | ||
| Never | 71,756 (53) | 157,498 (54) |
| Previous | 51,940 (39) | 97,930 (34) |
| Current | 10,527 (8) | 32,968 (11) |
| Alcohol status (%) | ||
| Never | 4075 (3) | 9220 (3) |
| Previous | 4302 (3) | 10,411 (4) |
| Current | 126,232 (94) | 269,500 (93) |
| Physical activity (%) | ||
| Low | 16,930 (13) | 47,061 (16) |
| Moderate | 43,674 (32) | 96,758 (33) |
| High | 48,761 (36) | 90,921 (31) |
| Statin use, yes (%) | 24,121 (18) | 45,463 (16) |
Values are numbers (%) for categorical variables and mean (SD) for continuous variables.
Fish oil supplementation modifies the associations between genetically predicted and observed concentrations of blood lipids in EUR-ancestry participants
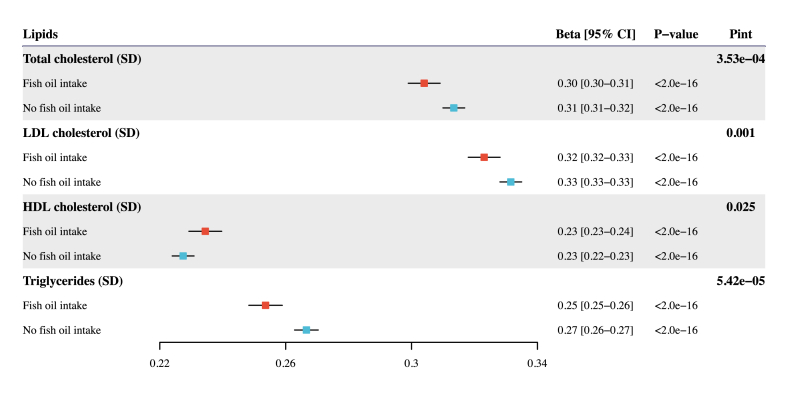
In general, fish oil supplementation attenuates the association between genetically predicted and observed circulating concentrations of total cholesterol, LDL cholesterol, and triglycerides while accentuating the corresponding association for HDL cholesterol. For the primary analysis using PGS from Graham et al. [19] with UKB excluded, there was strong evidence that fish oil supplementation modifies the associations between PGS and the corresponding observed concentration for 4 lipids in either the partially or fully adjusted model (Figure 2, Table 2) [19]. We performed 4 sets of sensitivity analysis to evaluate the robustness of these findings to different GWAS and methods for PGS derivation. When we generated PGS using 2 other GWAS summary statistics from Graham et al. [19] with UKB included and Willer et al. [18], consistently significant interaction results were observed for the 4 lipids (Supplementary Figure 1, Supplementary Table 3). We also calculated 2 more sets of PGS based on the β-coefficients previously generated by Weissbrod et al. [23] employing 2 methods, PolyPred and P + T. Similarly, we discovered that fish oil supplementation significantly modifies the associations between genetically predicted and observed concentrations of blood lipids (Supplementary Figure 2, Supplementary Table 3).
FIGURE 2.
Association of lipid polygenic scores with the corresponding observed lipid concentrations stratified by the fish oil intake status. This analysis was performed for each of the 4 lipids, separately, in UKB participants of European ancestry and utilized PGS from Graham et al. [19] with UKB excluded and the P + T method. Effect estimates and 95% confidence intervals were scaled to per 1 SD increase in PGS measured in SD units. GWAS, genome-wide association study; PGS, polygenic scores; Pint, P value for interaction; UKB, UK Biobank.
TABLE 2.
Association of lipid polygenic scores with the corresponding observed lipid concentrations stratified by the fish oil intake status in UKB participants of European ancestry1
| Fish oil intake |
No fish oil intake |
Interaction2 |
|||||
|---|---|---|---|---|---|---|---|
| Lipids | n | (95% CI) | n | (95% CI) | n | (95% CI) | P value |
| Total cholesterol | |||||||
| Model 13 | 128,634 | 0.244 (0.239, 0.249) | 276,134 | 0.265 (0.261, 0.268) | 404,768 | −0.022 (−0.028, −0.015) | 9.98×10-12 |
| Model 24 | 103,738 | 0.304 (0.299, 0.309) | 222,391 | 0.313 (0.310, 0.317) | 326,129 | −0.011 (−0.017, −0.005) | 3.53×10-4 |
| LDL cholesterol | |||||||
| Model 1 | 128,393 | 0.262 (0.257, 0.267) | 275,625 | 0.280 (0.277, 0.284) | 404,018 | −0.019 (−0.025, −0.012) | 6.27×10-9 |
| Model 2 | 103,538 | 0.323 (0.318, 0.328) | 221,994 | 0.332 (0.328, 0.335) | 325,532 | −0.010 (−0.016, −0.004) | 0.001 |
| HDL cholesterol | |||||||
| Model 1 | 117,709 | 0.235 (0.230, 0.240) | 252,786 | 0.230 (0.227, 0.234) | 370,495 | 0.005 (−0.001, 0.011) | 0.139 |
| Model 2 | 94,880 | 0.234 (0.229, 0.240) | 203,514 | 0.227 (0.224, 0.231) | 298,394 | 0.007 (0.001, 0.013) | 0.025 |
| Triglycerides | |||||||
| Model 1 | 128,544 | 0.252 (0.247, 0.257) | 275,897 | 0.265 (0.262, 0.269) | 404,441 | −0.013 (−0.020, −0.007) | 2.45×10-5 |
| Model 2 | 103,674 | 0.254 (0.248, 0.259) | 222,215 | 0.267 (0.263, 0.270) | 325,889 | −0.013 (−0.020, −0.007) | 5.42×10-5 |
The lipid polygenic scores were calculated by utilizing the P + T algorithm and the Graham et al. [19] GWAS with UKB excluded. refers to the change in the observed lipid concentration (in the unit of phenotypic SD) per SD increment in the PGS of the corresponding lipid. PGS, polygenic score; UKB, UK Biobank.
The interaction P values were obtained from the interaction term between lipid PGS and fish oil supplementation in corresponding models that further adjusted for lipid PGS and fish oil supplementation.
Model 1 adjusted for sex, age, age2, assessment centers, genotyping array, and the top 20 genetic principal components.
Model 2 additionally adjusted for BMI, Townsend deprivation index, smoking status, alcohol status, physical activity, and statin use.
In the stratified analysis with the fully adjusted model (i.e., Model 2), fish oil supplementation significantly attenuates the association between the genetically predicted and the observed concentration of total cholesterol: each SD increase in PGS was associated with a 0.304 SD increase (95% CI: 0.299, 0.309) in the observed concentration of total cholesterol among fish oil supplement users, compared with a 0.313 SD increase (95% CI: 0.310, 0.317) among nonusers (Figure 2, Table 2) [19]. Similarly, for LDL cholesterol, each SD increment in PGS resulted in an increase of 0.323 SD (95% CI: 0.318, 0.328) in LDL cholesterol concentration among fish oil users, in comparison with 0.332 SD increase (95% CI: 0.328, 0.335) among nonusers. The corresponding association effect for triglycerides was 0.254 (95% CI: 0.248, 0.259) in the fish oil group and 0.267 (95% CI: 0.263, 0.270) in the group not taking fish oil. Lastly, the impact of PGS was more pronounced in the fish oil group (0.234; 95% CI: 0.229, 0.240) for HDL cholesterol compared to participants without fish oil intake (0.227; 95% CI: 0.224, 0.231). Consistent patterns were observed in the 4 sets of sensitivity analysis (Supplementary Table 3). In summary, fish oil supplementation attenuates the associations between genetically predicted and observed concentrations of total cholesterol, LDL cholesterol, and triglycerides while amplifying the association for HDL cholesterol in EUR-ancestry participants.
PGS-fish oil interaction effects in other ancestry groups
In participants of AFR ancestry from UKB, we found that fish oil supplementation consistently attenuates the association between the genetically predicted and the observed concentration of triglycerides (Supplementary Table 4). When stratified by fish oil supplementation status, each SD increase in PGS was associated with a 0.099 SD (95% CI: 0.070, 0.128) rise in triglyceride concentration among fish oil users, compared with a 0.150 SD (95% CI: 0.125, 0.174) increase among those not taking fish oil. However, this modification effect of fish oil supplementation was not observed in participants of CSA and EAS ancestries (Supplementary Tables 5, 6). Moreover, analyses involving participants from other ancestry groups did not indicate significant interaction effects on total cholesterol, LDL cholesterol, and HDL cholesterol. In these diverse populations, the influence of PGS on lipid concentrations remained statistically significant even after adjusting for fish oil supplementation and multiple covariates (Supplementary Table 7). In summary, our interaction analyses in 3 non-EUR ancestry groups revealed that fish oil supplementation attenuates the association between the genetically predicted and the observed concentration of triglycerides in participants of AFR ancestry.
PGS-fish oil interaction effects by sex and age stratification
To examine the possible modifying impacts of sex and age on the PGS-fish oil interaction effects on observed blood lipids, we repeated the interaction analysis in sex and age-stratified subgroups of EUR-ancestry participants. In sex-stratified subgroups, PGS-fish oil interaction effects on total cholesterol, LDL cholesterol, and triglycerides were more pronounced in males than in females. By contrast, the interaction effect on HDL cholesterol was stronger in females than in males (Table 3, Supplementary Table 8) [19]. In the age-stratified analysis, the PGS-fish oil interaction effects were statistically significant in both the younger and the older groups for triglycerides, only significant in the older group for HDL cholesterol, and not significant in either group for total cholesterol and LDL cholesterol (Table 3, Supplementary Tables 9, 10) [19].
TABLE 3.
Association of lipid polygenic scores with the corresponding observed lipid concentrations stratified by the fish oil intake status and sex (or age) in UKB participants of European ancestry1
| Fish oil intake |
No fish oil intake |
Interaction2 |
||||
|---|---|---|---|---|---|---|
| Lipids | n | (95% CI) | n | (95% CI) | (95% CI) | P value |
| Total cholesterol | ||||||
| Females | 55,969 | 0.314 (0.307, 0.321) | 113,526 | 0.318 (0.314, 0.323) | −0.007 (−0.015, 0.002) | 0.114 |
| Males | 47,769 | 0.290 (0.282, 0.297) | 108,865 | 0.306 (0.301, 0.311) | −0.018 (−0.027, −0.010) | 3.36×10−5 |
| LDL cholesterol | ||||||
| Females | 55,879 | 0.342 (0.335, 0.349) | 113,351 | 0.346 (0.342, 0.351) | −0.006 (−0.015, 0.002) | 0.133 |
| Males | 47,659 | 0.298 (0.291, 0.305) | 108,643 | 0.314 (0.309, 0.319) | −0.018 (−0.027, −0.009) | 4.84×10−5 |
| HDL cholesterol | ||||||
| Females | 50,787 | 0.260 (0.253, 0.268) | 103,067 | 0.250 (0.244, 0.255) | 0.011 (0.001, 0.020) | 0.023 |
| Males | 44,093 | 0.205 (0.198, 0.212) | 100,447 | 0.205 (0.200, 0.209) | 0.000 (−0.008, 0.008) | 0.979 |
| Triglycerides | ||||||
| Females | 55,944 | 0.211 (0.205, 0.217) | 113,463 | 0.218 (0.214, 0.222) | −0.007 (−0.014, 0.000) | 0.067 |
| Males | 47,730 | 0.305 (0.296, 0.314) | 108,752 | 0.318 (0.312, 0.324) | −0.014 (−0.025, −0.003) | 0.011 |
| Total cholesterol | ||||||
| Younger3 | 38,019 | 0.320 (0.312, 0.328) | 120,497 | 0.325 (0.320, 0.329) | −0.006 (−0.015, 0.003) | 0.206 |
| Older3 | 65,719 | 0.292 (0.286, 0.298) | 101,894 | 0.296 (0.291, 0.301) | −0.007 (−0.015, 0.001) | 0.095 |
| LDL cholesterol | ||||||
| Younger | 37,941 | 0.339 (0.331, 0.348) | 120,266 | 0.346 (0.341, 0.351) | −0.007 (−0.017, 0.002) | 0.121 |
| Older | 65,597 | 0.312 (0.305, 0.318) | 101,728 | 0.311 (0.306, 0.316) | −0.002 (−0.010, 0.006) | 0.583 |
| HDL cholesterol | ||||||
| Younger | 34,650 | 0.228 (0.220, 0.236) | 110,062 | 0.226 (0.222, 0.231) | 0.002 (−0.008, 0.011) | 0.736 |
| Older | 60,230 | 0.238 (0.232, 0.245) | 93,452 | 0.229 (0.223, 0.234) | 0.010 (0.001, 0.018) | 0.027 |
| Triglycerides | ||||||
| Younger | 37,985 | 0.253 (0.244, 0.262) | 120,391 | 0.265 (0.260, 0.270) | −0.013 (−0.023, −0.003) | 0.014 |
| Older | 65,689 | 0.253 (0.247, 0.260) | 101,824 | 0.268 (0.262, 0.273) | −0.015 (−0.024, −0.006) | 0.001 |
PGS, polygenic score; UKB, UK Biobank.
The lipid polygenic scores were calculated by utilizing the P + T algorithm and the Graham et al. [19] GWAS with UKB excluded. refers to the change in the observed lipid concentration (in the unit of phenotypic SD) per SD increment in the PGS of the corresponding lipid. These regression analyses were performed with Model 2, which adjusted for sex, age, age2, assessment centers, genotyping array, the top 20 genetic principal components, BMI, Townsend deprivation index, smoking status, alcohol status, physical activity, and statin use.
The interaction P values were obtained from the interaction term between lipid PGS and fish oil supplementation in corresponding models that further adjusted for lipid PGS and fish oil supplementation.
The sample were dichotomized into the younger and the older groups based on the median age.
Fish oil supplementation modifies the associations between genetically predicted lipid concentrations and dyslipidemia in EUR-ancestry participants
We examined if fish oil supplementation modifies the associations between genetically predicted lipid concentrations and 3 dyslipidemia-related outcomes, including hypercholesterolemia, hyperlipidemia, and hypertriglyceridemia. For hypercholesterolemia, significant interaction effects were observed for 3 blood lipids. Genetically predicted total cholesterol (odds ratio [OR] 1.122; 95% CI: 1.119, 1.125), LDL cholesterol (OR 1.131; 95% CI: 1.129, 1.134), and triglycerides (OR 1.037; 95% CI: 1.034, 1.040) were associated with higher odds of hypercholesterolemia in fish oil users. By contrast, these associations were accentuated in those not taking fish oil: 1.133 (95% CI: 1.131, 1.135) for total cholesterol, 1.141 (95% CI: 1.139, 1.142) for LDL cholesterol, and 1.044 (95% CI: 1.042, 1.046) for triglycerides (Supplementary Table 11). The associations of the 4 lipid PGS with hyperlipidemia were not significantly altered by fish oil intake status (Supplementary Table 12). For hypertriglyceridemia, significant interaction effects were observed for 2 blood lipids. Genetically predicted total cholesterol (OR 1.038; 95% CI: 1.035, 1.041) and LDL cholesterol (OR 1.012; 95% CI: 1.009, 1.015) were positively associated with the odds of hypertriglyceridemia in the fish oil group. These associations were accentuated in nonconsumers, with OR of 1.043 (95% CI: 1.041, 1.045) for total cholesterol and 1.017 (95% CI: 1.015, 1.019) for LDL cholesterol, respectively (Supplementary Table 13). In summary, we observed a general trend that fish oil supplementation attenuates the associations of genetically predicted blood lipids with hypercholesterolemia and hypertriglyceridemia.
Discussion
In this cross-sectional study involving 424,090 participants of EUR ancestry, our results indicated that fish oil supplementation modifies the associations between genetically predicted and observed concentrations of 4 blood lipids. Specifically, fish oil supplementation attenuates the associations for total cholesterol, LDL cholesterol, and triglycerides while accentuating the association for HDL cholesterol. Analyzing 6,577 AFR, 8,648 CSA, and 2,670 EAS ancestry participants, we only found significant interactions for triglycerides in participants of AFR ancestry, showing that fish oil supplementation attenuates the association between the genetically predicted and the observed concentration of triglycerides. Some of these significant PGS-fish oil interactions could be further modified by sex and age. Moreover, our exploratory analysis with dyslipidemia-related outcomes revealed that fish oil supplementation attenuates the associations of genetically predicted lipid concentrations with hypercholesterolemia and hypertriglyceridemia. Collectively, our findings supported the hypothesis that genome-wide genetic effects on blood lipid concentrations captured by PGS could be modified by habitual fish oil supplementation.
Although substantial studies have highlighted the effects of fish oil supplements in improving the lipid profile in hyperlipidemic patients, their influence on genetic susceptibility to dyslipidemia remains unclear [2,11,[30], [31], [32], [33]]. A recent dose-response meta-analysis of 90 randomized controlled trials with 72,598 participants demonstrated that DHA + EPA supplementation (>2 g/d) reduced triglyceride concentration [34]. Skulas-Ray et al. [35] concluded that pharmacological doses of omega-3 fatty acids (>3 g/d total EPA + DHA) effectively decrease triglycerides and can be safely combined with other lipid-lowering agents, particularly statins. The Japan EPA Lipid Intervention Study, involving 14,981 hypercholesterolemia Japanese patients, found that 1.8 g/d EPA in combination with a statin led to lower triglycerides compared with statin therapy alone [36]. In the current study of non-EUR ancestry groups, we also observed that fish oil attenuates the association between the genetically predicted and observed concentrations of triglycerides among participants of AFR ancestry, but no significant interaction was detected among participants of CSA and EAS ancestries, which may be due to insufficient sample sizes. Large-scale studies covering more representative samples from diverse ancestries are needed to further validate our findings. Regarding genetic factors, some studies have investigated the interactions between fish oil and lipid-related genetic variants [17, [37], [38], [39], [40]]. Instead of examining specific genetic variants, our study leveraged the cumulative effects of many genetic variants across the genome to assess the overall genetically predicted concentrations of circulating lipids. Our findings affirmed the possible protective role of fish oil against genetic susceptibility to an unhealthy lipid profile, including a decreased concentration of HDL cholesterol and elevated concentrations of total cholesterol, LDL cholesterol, and triglycerides. Additionally, we directly evaluated the modifying effects of fish oil on the associations of the 4 lipid PGS with 3 dyslipidemia-related outcomes, including hypercholesterolemia (LDL cholesterol ≥3.4 mmol/L or non-HDL cholesterol ≥4.1 mmol/L), hyperlipidemia (non-HDL cholesterol ≥5.7 mmol/L and triglycerides ≥1.7 mmol/L), and hypertriglyceridemia (triglycerides ≥1.7 mmol/L). We observed a general trend that fish oil attenuates the associations of genetically predicted blood lipids with hypercholesterolemia and hypertriglyceridemia. However, no significant modifying effects were observed for hyperlipidemia or on the associations of the PGS of 2 lipids (i.e., HDL cholesterol and triglycerides) with hypertriglyceridemia. It is known that quantitative traits offer better statistical power over binary traits in regression analysis [41]. So, the lack of significant findings may reflect the loss of statistical power, especially for the much smaller case number of hyperlipidemia. Another possible explanation is that reverse causation obscures the interaction signals. Because of the well-known benefit of fish oil in reducing triglycerides [11,12], individuals with hyperlipidemia or hypertriglyceridemia are more likely to take fish oil in addition to other lipid-lowering treatments. We emphasize that our findings need to be replicated in the context of randomized controlled trials.
The precise mechanisms underlying the observed interactions between fish oil supplementation and the genetically predicted lipid concentrations are not yet fully elucidated. These modifying effects could potentially be attributed to the multifaceted benefits derived from fish oil supplements. The putative mechanisms through which fish oil supplementation influences human lipoprotein metabolism include inhibition of VLDL triglyceride synthesis, decreased apoprotein B synthesis, enhancement of VLDL turnover via an increased fractional catabolic rate of VLDL, repression of LDL synthesis, and reduction of postprandial lipemia [42]. Additionally, fish oil intake has been found to have beneficial associations with several factors implicated in cardiovascular health, including inflammatory, oxidative, thrombotic, vascular, and arrhythmogenic parameters [11,[43], [44], [45], [46], [47], [48]]. However, it is plausible that other mechanisms may also be involved in these modifying effects. Future functional studies are warranted to discern the underlying biological mechanisms.
To the best of our knowledge, this is the first study to evaluate the modifying effects of fish oil supplementation on the associations between genetically predicted and observed concentrations of blood lipids. The major strength of our study lies in the large number of participants of EUR ancestry, providing sufficient statistical power to detect significant interactions. Another strength of our study is the inclusion of other ancestry groups, where we confirmed that fish oil also attenuates the association of the genetically predicted and the observed concentration of triglycerides in the AFR population. We used various GWAS summary statistics and PGS methodologies to calculate PGS and applied a fully adjusted model with a wealth of data on covariates, including socioeconomic characteristics, lifestyle factors, and statin use. Notably, the PGS for the primary analysis incorporated multiple SNPs from the largest and most recent GWAS across different cohorts, excluding any sample overlap with the UKB, thereby circumventing the potential issue of spurious candidate gene interactions [49]. Population structure can lead to inflated estimates of PGS prediction [49]. To address this, we adjusted all models for the top 20 genetic principal components to minimize potential bias. This comprehensive study design ensured the robustness and consistency of our findings.
Several potential limitations should also be considered. First, the self-reported fish oil intake is vulnerable to both measurement error and recall bias. Second, since fish oil supplementation status was assessed only at baseline, we were unable to evaluate the effects of longitudinal changes or extended duration of use. Third, our data lacked details on specific doses of fish oil supplementation, and future research is needed to discern the independent roles of DHA and EPA at specific doses. Fourth, UKB recruited people aged between 37 and 73 years at baseline, and our findings may not be generalizable to other age groups, such as children and adolescents [50]. Fifth, though our study included multiple ancestries, caution is advised when extrapolating our findings to non-EUR populations, and future investigations with larger sample sizes are necessary. Sixth, it is important to note that there are other dietary factors not controlled in our models, such as intake of saturated fat or seafood, which may also provide the same active nutrients in fish oil (e.g., EPA and DHA) and may also influence blood lipid concentrations. Seventh, some dietary or lifestyle factors may be correlated with the tendency to take fish oil supplements. As a result, our observed PRS-fish oil interaction effects on blood lipids may not be interpreted as the causal effect of fish oil. Lastly, given the observational design of our study, we cannot completely rule out reverse causation or residual confounding.
In conclusion, our findings reveal that fish oil supplementation attenuates the associations between genetically predicted and observed circulating concentrations of total cholesterol, LDL cholesterol, and triglycerides while accentuating the corresponding association for HDL cholesterol. Moreover, we detected that fish oil attenuates the corresponding association for triglycerides among participants of AFR ancestry. These findings support the hypothesis that genome-wide genetic effects on blood lipid concentrations captured by PGS could be modified by habitual fish oil supplementation. Our study has at least 2 notable public health and clinical implications. First, it supports the habitual use of fish oil supplements to improve the blood lipid profile. Second, it cautions against the direct comparison of lipid PGS between individuals with and without fish oil supplementation. Further studies with larger sample sizes from diverse ancestry and accurate dose information of fish oil supplements, especially those in the context of randomized controlled trials, are warranted to corroborate and expand our findings and to illuminate their clinical implications.
Acknowledgments
We would like to express our heartfelt gratitude to the UK Biobank participants and administrative staff. This research was conducted using the UK Biobank resource under Application Number 48818. This work uses data provided by patients and collected by the NHS as part of their care and support. These data are copyrighted, 2022, NHS England. Reused with the permission of the NHS England and UK Biobank. All rights reserved. This research used data assets made available by National Safe Haven as part of the Data and Connectivity National Core Study, led by Health Data Research United Kingdom in partnership with the Office for National Statistics and funded by United Kingdom Research and Innovation (grant ref MC_PC_20058).
Author contributions
The authors’ responsibilities were as follows – YS and KY: designed the study; YS: performed data analysis, prepared visualizations, and wrote the original draft of the manuscript; TM, AB, HX, and NBB: contributed to the data analysis; YS, YS (yeshen@uga.edu), CL, and KY: provided statistical advice; YS and KY: interpreted the results; KY: critically revised the paper; all authors read and approved the final manuscript and took responsibility for the integrity of the work as a whole.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was funded by the University of Georgia Research Foundation and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM143060. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data availability
Access to the UK Biobank resource is available via an approved application (http://www.ukbiobank.ac.uk). GWAS summary statistics generated by Graham et al. [19] are publicly available at http://csg.sph.umich.edu/willer/public/glgc-lipids2021/results/ancestry_specific/. GWAS summary statistics generated by Willer et al. [18] are available for public download at http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST002001-GCST003000/GCST002221/harmonised/. PGS coefficients generated by Weissbrod et al. [23] are available for public download at https://alkesgroup.broadinstitute.org/polypred_results.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajcnut.2024.07.009.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Roth G.A., Mensah G.A., Johnson C.O., Addolorato G., Ammirati E., Baddour L.M., et al. Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J. Am. Coll. Cardiol. 2020;76(25):2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt D.L., Steg P.G., Miller M., Brinton E.A., Jacobson T.A., Ketchum S.B., et al. Cardiovascular risk reduction with icosapent ethyl for hypertriglyceridemia. N. Engl. J. Med. 2019;380(1):11–22. doi: 10.1056/NEJMoa1812792. [DOI] [PubMed] [Google Scholar]
- 3.Libby P. Triglycerides on the rise: should we swap seats on the seesaw? Eur. Heart J. 2015;36(13):774–776. doi: 10.1093/eurheartj/ehu500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klempfner R., Erez A., Sagit B.Z., Goldenberg I., Fisman E., Kopel E., et al. Elevated triglyceride level is independently associated with increased all-cause mortality in patients with established coronary heart disease: twenty-two-year follow-up of the bezafibrate infarction prevention study and registry. Circ. Cardiovasc. Qual. Outcomes. 2016;9(2):100–108. doi: 10.1161/CIRCOUTCOMES.115.002104. [DOI] [PubMed] [Google Scholar]
- 5.Ference B.A., Ginsberg H.N., Graham I., Ray K.K., Packard C.J., Bruckert E., et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2017;38(32):2459–2472. doi: 10.1093/eurheartj/ehx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toth P.P., Granowitz C., Hull M., Liassou D., Anderson A., Philip S. High triglycerides are associated with increased cardiovascular events, medical costs, and resource use: a real-world administrative claims analysis of statin-treated patients with high residual cardiovascular risk. J. Am. Heart Assoc. 2018;7(15) doi: 10.1161/JAHA.118.008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris W.S., Ginsberg H.N., Arunakul N., Shachter N.S., Windsor S.L., Adams M., et al. Safety and efficacy of omacor in severe hypertriglyceridemia. J. Cardiovasc. Risk. 1997;4(5–6):385–391. [PubMed] [Google Scholar]
- 8.von Eckardstein A., Nordestgaard B.G., Remaley A.T., Catapano A.L. High-density lipoprotein revisited: biological functions and clinical relevance. Eur. Heart J. 2023;44(16):1394–1407. doi: 10.1093/eurheartj/ehac605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manson J.E., Bassuk S.S., Cook N.R., Lee I.M., Mora S., Albert C.M., et al. Vitamin D, marine n-3 fatty acids, and primary prevention of cardiovascular disease current evidence. Circ. Res. 2020;26(1):1112–1128. doi: 10.1161/CIRCRESAHA.119.314541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Sun Y., Yu Q., Song S., Brenna J.T., Shen Y., et al. Higher ratio of plasma omega-6/omega-3 fatty acids is associated with greater risk of all-cause, cancer, and cardiovascular mortality: a population-based cohort study in UK Biobank. eLife. 2024;12 doi: 10.7554/eLife.90132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholls S.J., Lincoff A.M., Garcia M., Bash D., Ballantyne C.M., Barter P.J., et al. Effect of high-dose omega-3 fatty acids vs corn oil on major adverse cardiovascular events in patients at high cardiovascular risk: the STRENGTH randomized clinical trial. JAMA. 2020;324(22):2268–2280. doi: 10.1001/jama.2020.22258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grevengoed T.J., Trammell S.A., Svenningsen J.S., Makarov M.V., Nielsen T.S, Jacobsen J.C.B., et al. An abundant biliary metabolite derived from dietary omega-3 polyunsaturated fatty acids regulates triglycerides. J. Clin. Invest. 2021;131(6) doi: 10.1172/JCI143861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H., Wang W., Zhang C., Xu C., Duan H., Tian X., et al. Heritability and genome-wide association study of plasma cholesterol in Chinese adult twins. Front. Endocrinol. (Lausanne) 2018;9:677. doi: 10.3389/fendo.2018.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corraini P., Olsen M., Pedersen L., Dekkers O.M., Vandenbroucke J.P. Effect modification, interaction and mediation: an overview of theoretical insights for clinical investigators. Clin. Epidemiol. 2017;9:331–338. doi: 10.2147/CLEP.S129728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ottman R. Gene-environment interaction: definitions and study designs. Prev. Med. 1996;25(6):764–770. doi: 10.1006/pmed.1996.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis M., Li C., Sun Y., Zhou J., Li X., Brenna J.T., et al. Genome-wide association study of fish oil supplementation on lipid traits in 81,246 individuals reveals new gene-diet interaction loci. PLoS Genet. 2021;17(3) doi: 10.1371/journal.pgen.1009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham S.E., Clarke S.L., Wu K.H., Kanoni S., Zajac G.J.M., Ramdas S., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600(7890):675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civeira F., Arca M., Cenarro A., Hegele R.A. A mechanism-based operational definition and classification of hypercholesterolemia. J. Clin. Lipidol. 2022;16(6):813–821. doi: 10.1016/j.jacl.2022.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W.J., Khera A.V., et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54(4):450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan H., Roderick P., Martin D. The index of multiple deprivation 2000 and accessibility effects on health. J. Epidemiol. Community Health. 2004;58(3):250–257. doi: 10.1136/jech.2003.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleland C., Ferguson S., Ellis G., Hunter R.F. Validity of the International Physical Activity Questionnaire (IPAQ) for assessing moderate-to-vigorous physical activity and sedentary behaviour of older adults in the United Kingdom. BMC Med. Res. Methodol. 2018;18(1):176. doi: 10.1186/s12874-018-0642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z.H., Zhong W.F., Liu S., Kraus V.B., Zhang Y.J., Gao X., et al. Associations of habitual fish oil supplementation with cardiovascular outcomes and all cause mortality: evidence from a large population based cohort study. BMJ. 2020;368:m456. doi: 10.1136/bmj.m456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H.Y. Statistical notes for clinical researchers: simple linear regression 3 - residual analysis. Restor. Dent. Endod. 2019;44(1):e11. doi: 10.5395/rde.2019.44.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying S., Heung T., Thiruvahindrapuram B., Engchuan W., Yin Y., Blagojevic C., et al. Polygenic risk for triglyceride levels in the presence of a high impact rare variant. BMC Med. Genomics. 2023;16(1):281. doi: 10.1186/s12920-023-01717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoltzfus J.C. Logistic regression: a brief primer, Acad. Emerg. Med. 2011;18(10):1099–1104. doi: 10.1111/j.1553-2712.2011.01185.x. 10.1111/j.1553-2712.2011.01185.x [DOI] [PubMed] [Google Scholar]
- 30.The ORIGIN Trial Investigators. Bosch J., Gerstein H.C., Dagenais G.R., Diaz R., Dyal L., et al. n-3 fatty acids and cardiovascular outcomes in patients with dysglycemia. N. Engl. J. Med. 2012;367(4):309–318. doi: 10.1056/NEJMoa1203859. [DOI] [PubMed] [Google Scholar]
- 31.Ganda O.P., Bhatt D.L., Mason R.P., Miller M., Boden W.E. Unmet need for adjunctive dyslipidemia therapy in hypertriglyceridemia management. J. Am. Coll. Cardiol. 2018;2(3):7330–7343. doi: 10.1016/j.jacc.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 32.Abdelhamid A.S., Martin N., Bridges C., Brainard J.S., Wang X., Brown T.J., et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018;7(7):CD012345. doi: 10.1002/14651858.CD012345.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marston N.A., Giugliano R.P., Im K., Silverman M.G., O'Donoghue M.L., Wiviott S.D., et al. Association between triglyceride lowering and reduction of cardiovascular risk across multiple lipid-lowering therapeutic classes: a systematic review and meta-regression analysis of randomized controlled trials. Circulation. 2019;140(16):1308–1317. doi: 10.1161/CIRCULATIONAHA.119.041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T., Zhang X., Zhou N., Shen Y., Li B., Chen B.E., et al. Association between omega-3 fatty acid intake and dyslipidemia: a continuous dose-response meta-analysis of randomized controlled trials. J. Am. Heart Assoc. 2023;12(11):12e029512. doi: 10.1161/JAHA.123.029512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skulas-Ray A.C., Wilson P.W.F., Harris W.S., Brinton E.A., Kris-Etherton P.M., Richter C.K., et al. Omega-3 fatty acids for the management of hypertriglyceridemia: a science advisory from the American Heart Association. Circulation. 2019;140(12):e673–e691. doi: 10.1161/CIR.0000000000000709. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y., Yokoyama M., Origasa H., Matsuzaki M., Matsuzawa Y., Ishikawa Y., et al. Effects of EPA on coronary artery disease in hypercholesterolemic patients with multiple risk factors: sub-analysis of primary prevention cases from the Japan EPA Lipid Intervention Study (JELIS) Atherosclerosis. 2008;200(1):135–140. doi: 10.1016/j.atherosclerosis.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Madden J., Williams C.M., Calder P.C., Lietz G., Miles E.A., Cordell H., et al. The impact of common gene variants on the response of biomarkers of cardiovascular disease (CVD) risk to increased fish oil fatty acids intakes. Annu. Rev. Nutr. 2011;31:203–234. doi: 10.1146/annurev-nutr-010411-095239. [DOI] [PubMed] [Google Scholar]
- 38.Ferguson J.F., Phillips C.M., McMonagle J., Perez-Martinez P., Shaw D.I., Lovegrove J.A., et al. NOS3 gene polymorphisms are associated with risk markers of cardiovascular disease, and interact with omega-3 polyunsaturated fatty acids. Atherosclerosis. 2010;211(2):539–544. doi: 10.1016/j.atherosclerosis.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 39.Lindi V., Schwab U., Louheranta A., Laakso M., Vessby B., Hermansen K., et al. Impact of the Pro12Ala polymorphism of the PPAR-gamma2 gene on serum triacylglycerol response to n-3 fatty acid supplementation. Mol. Genet. Metab. 2003;79(1):52–60. doi: 10.1016/s1096-7192(03)00065-9. [DOI] [PubMed] [Google Scholar]
- 40.Madden J., Carrero J.J., Brunner A., Dastur N., Shearman C.P., Calder P.C., et al. Polymorphisms in the CD36 gene modulate the ability of fish oil supplements to lower fasting plasma triacyl glycerol and raise HDL cholesterol concentrations in healthy middle-aged men. Prostaglandins Leukot. Essent. Fatty Acids. 2008;78(6):327–335. doi: 10.1016/j.plefa.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Yang J., Wray N.R., Visscher P.M.. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet. Epidemiol. 2010;34(3):254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 42.Stone N.J. Fish consumption, fish oil, lipids, and coronary heart disease. Circulation. 1996;94(9):2337–2340. doi: 10.1161/01.cir.94.9.2337. [DOI] [PubMed] [Google Scholar]
- 43.Duda M.K., O'Shea K.M., Lei B., Barrows B.R., Azimzadeh A.M., McElfresh T.E., et al. Dietary supplementation with omega-3 PUFA increases adiponectin and attenuates ventricular remodeling and dysfunction with pressure overload. Cardiovasc. Res. 2007;76(2):303–310. doi: 10.1016/j.cardiores.2007.07.002. 10.1016/j.cardiores.2007.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavie C.J., Milani R.V., Mehra M.R., Ventura H.O. Omega-3 polyunsaturated fatty acids and cardiovascular diseases. J. Am. Coll. Cardiol. 2009;54(7):585–594. doi: 10.1016/j.jacc.2009.02.084. [DOI] [PubMed] [Google Scholar]
- 45.Mozaffarian D., Wu J.H. Omega-3 fatty acids and cardiovascular disease: effects on risk factors, molecular pathways, and clinical events. J. Am. Coll. Cardiol. 2011;58(20):2047–2067. doi: 10.1016/j.jacc.2011.06.063. [DOI] [PubMed] [Google Scholar]
- 46.Tribulova N., Szeiffova Bacova B., Benova T.E., Knezl V., Barancik M., Slezak J. Omega-3 index and anti-arrhythmic potential of omega-3 PUFAs. Nutrients. 2017;9(11):1191. doi: 10.3390/nu9111191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirban A., Nandrean S., Gotting C., Tamler R., Pop A., Negrean M., et al. Effects of n-3 fatty acids on macro- and microvascular function in subjects with type 2 diabetes mellitus. Am. J. Clin. Nutr. 2010;91(3):808–813. doi: 10.3945/ajcn.2009.28374. [DOI] [PubMed] [Google Scholar]
- 48.Lee J.H., O'Keefe J.H., Lavie C.J., Harris W.S. Omega-3 fatty acids: cardiovascular benefits, sources and sustainability. Nat. Rev. Cardiol. 2009;6(12):753–758. doi: 10.1038/nrcardio.2009.188. [DOI] [PubMed] [Google Scholar]
- 49.Choi S.W., Mak T.S., O'Reilly P.F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 2020;15(9):2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frensham L.J., Bryan J., Parletta N. Influences of micronutrient and omega-3 fatty acid supplementation on cognition, learning, and behavior: methodological considerations and implications for children and adolescents in developed societies, Nutr. Rev. 2012;70(10):594–610. doi: 10.1111/j.1753-4887.2012.00516.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to the UK Biobank resource is available via an approved application (http://www.ukbiobank.ac.uk). GWAS summary statistics generated by Graham et al. [19] are publicly available at http://csg.sph.umich.edu/willer/public/glgc-lipids2021/results/ancestry_specific/. GWAS summary statistics generated by Willer et al. [18] are available for public download at http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST002001-GCST003000/GCST002221/harmonised/. PGS coefficients generated by Weissbrod et al. [23] are available for public download at https://alkesgroup.broadinstitute.org/polypred_results.