Abstract
Purpose:
The purpose of this study was to assess the predictive capability of HER2DX assay following (neo)adjuvant trastuzumab–pertuzumab (HP)–based therapy in HER2-positive (HER2+) early breast cancer.
Experimental Design:
HER2DX was analyzed in baseline pretreatment tumors from the PHERGain trial. Patients with stage I–IIIA HER2+ early breast cancer were randomized to group A [docetaxel, carboplatin, and HP (TCHP)] and group B (HP ± endocrine therapy). PET response was evaluated after two cycles. Group A received TCHP for six cycles regardless of PET response. Group B continued with HP ± endocrine therapy for six cycles (PET responders) or with TCHP for six cycles (PET nonresponders). The primary objective of this retrospective study was to associate the HER2DX pathologic complete response (pCR) score with pCR. The secondary objective was the association of the HER2DX risk score with 3-year invasive disease-free survival (iDFS).
Results:
HER2DX was performed on 292 (82.0%) tumors. The overall pCR rate was 38.0%, with pCR rates of 56.4% in group A and 33.8% in group B. In multivariable analysis including treatment and clinicopathologic factors, the HER2DX pCR score (continuous variable) significantly correlated with pCR [OR, 1.29; 95% confidence interval (CI), 1.10–1.54; P < 0.001]. HER2DX-defined pCR-high, -med, and -low groups exhibited pCR rates of 50.4%, 35.8%, and 23.2%, respectively (pCR-high vs. pCR-low OR, 3.27; 95% CI, 1.54–7.09; P < 0.001). In patients with residual disease, the HER2DX high-risk group demonstrated numerically worse 3-year iDFS than the low-risk group (89.8% vs. 100%; HR, 2.70; 95% CI, 0.60–12.18; P = 0.197).
Conclusions:
HER2DX predicts pCR in the context of neoadjuvant HP-based therapy, regardless of chemotherapy addition, and might identify patients at higher risk of recurrence among patients with residual disease.
Translational Relevance.
In HER2-positive (HER2+) early breast cancer, there is a critical need to identify patients eligible for dual HER2 blockade with trastuzumab–pertuzumab (HP) without chemotherapy. The PHERGain trial revealed the HER2DX genomic assay’s robust ability to predict pathologic complete response (pCR) during neoadjuvant HP therapy, independent of chemotherapy, hormone receptor status, and early imaging response. HER2DX pCR score significantly correlated with pCR, offering a quantitative measure. Notably, HER2DX’s low-risk designation demonstrated a promising trend toward improved 3-year invasive disease-free survival, establishing its importance as a prognostic indicator. The assay provides insights into tumor responsiveness to HP and recurrence risk, potentially guiding personalized HP-based therapies, possibly with reduced or without chemotherapy, for HER2+ early breast cancer.
Introduction
Breast cancer is a heterogeneous disease, with HER2 overexpression accounting for approximately 15% to 20% of cases (1). Overexpression of HER2 is a well-established independent predictor of disease recurrence and cancer-related mortality (2, 3). Advances in the development of HER2-targeted agents have significantly improved the prognosis of patients with HER2-positive (HER2+) early breast cancer. This progress has led to the exploration of de-escalation strategies, such as less intensive chemotherapy regimens, particularly in the neoadjuvant setting (4–7).
One notable approach is the neoadjuvant dual HER2 blockade without chemotherapy (6), which has demonstrated remarkable pathological complete response (pCR) rates ranging from 20.5% to 36.3% (4, 8, 9). However, it is important to acknowledge that these rates are based on phase II trials with small sample sizes and may not be fully representative. Nonetheless, achieving pCR is a validated surrogate endpoint associated with improved long-term survival outcomes (10–13). These findings have prompted investigations into chemotherapy-sparing approaches for patients with HER2+ early breast cancer.
The PHERGain phase II trial represents a significant effort in evaluating the feasibility of a chemotherapy-free treatment strategy for HER2+ early breast cancer (9). This trial employs a neoadjuvant approach based on dual HER2 blockade with trastuzumab–pertuzumab (HP) and utilizes an 18-fluorine-fluorodeoxyglucose-PET (18F-FDG-PET) and pCR-adapted strategy for treatment decision-making. In this trial, patients with HER2+ early breast cancer with at least one breast lesion evaluable by 18F-FDG-PET were randomized to receive either docetaxel, carboplatin, trastuzumab, and pertuzumab (TCHP, group A) or HP ± endocrine therapy (group B). Notably, PHERGain demonstrated promising results, with a high percentage of 18F-FDG-PET responders in group B achieving a pCR, and a noteworthy 3-year invasive disease-free survival (iDFS) rate of 94.8% in group B despite sparing chemotherapy in around one third of patients (13). These findings support exploring 18F-FDG-PET-based and pCR-adapted strategies to identify patients with HER2+ early breast cancer who could safely omit chemotherapy.
Given the evolving landscape of de-escalation strategies, understanding predictive factors for treatment response becomes imperative. Among these factors, the HER2DX genomic assay has emerged as a promising tool (14–19). This assay, a 27-gene expression and clinical feature-based classifier, provides two independent scores predicting long-term prognosis and the likelihood of pCR in HER2+ early breast cancer (14). It integrates biological information related to immune response, luminal differentiation, tumor cell proliferation, and the expression of HER2 17q12-21 chromosomal amplicon, including the ERBB2 gene, along with clinical information.
HER2DX genomic assay has previously demonstrated prognostic value and the ability to predict pCR following trastuzumab-based therapy in various datasets (16–21). However, its performance in the context of (neo)adjuvant HP-based therapy, as investigated in the PHERGain trial, remains to be determined. In this study, we aim to evaluate the ability of HER2DX assay to predict outcomes in patients with HER2+ early breast cancer undergoing HP-based therapy, contributing valuable insights to the ongoing efforts in tailoring treatment strategies for this specific patient population.
Materials and Methods
Study design and patient population
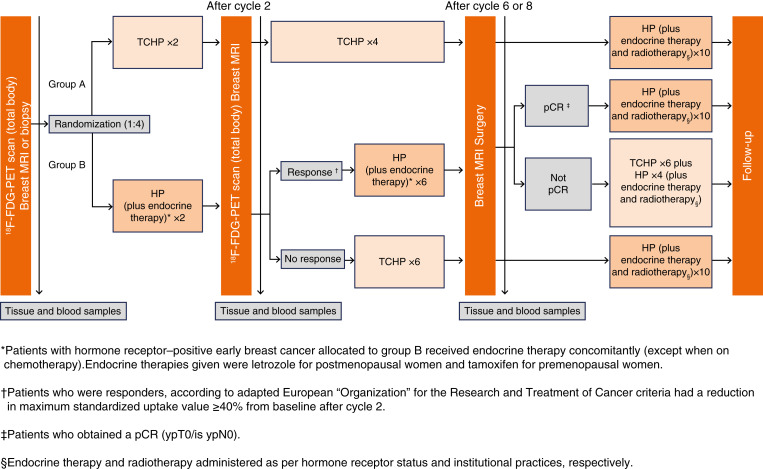
The PHERGain trial is a strategy-based, multicenter, randomized, noncomparative, open-label phase II study conducted across 45 hospitals in seven European countries (Fig. 1; refs. 9, 13). The trial targeted women aged 18 years or older with previously untreated, centrally confirmed, HER2+, stage I–IIIA, invasive, operable breast cancer, having at least one breast lesion assessable by 18F-FDG-PET. Additional inclusion criteria encompassed hormone receptor status assessment, Eastern Cooperative Oncology Group performance status of 0 or 1, left ventricular ejection fraction of at least 55%, and adequate organ function. Exclusion criteria included metastatic disease (stage IV), bilateral breast cancer, and prior systemic therapy for invasive breast cancer.
Figure 1.
PHERGain study design.
Patients were randomized at a 1:4 ratio and underwent an initial 18F-FDG-PET scan before being allocated to either TCHP [docetaxel (75 mg/m2, intravenous), carboplatin (area under the curve 6 mg/mL per min, intravenous), trastuzumab (600 mg fixed dose, subcutaneous), and pertuzumab (840 mg loading dose followed by 420 mg maintenance doses, intravenous)] (group A) or HP ± endocrine therapy (group B). The study’s protocol entailed an open-label approach to the administration of the investigational medications every 3 weeks. This included a loading dose of pertuzumab, a fixed dose of trastuzumab, and allowed for dose adjustments of docetaxel and carboplatin in response to toxicity. Following two cycles of HP ± endocrine therapy in group B, participants underwent another 18F-FDG-PET scan. Those showing a 18F-FDG-PET response continued with six more cycles of HP ± endocrine therapy before undergoing surgery. Patients were classified as 18F-FDG-PET responders if all target lesions exhibited a reduction from baseline of at least 40% of the maximum standardized uptake value on 18F-FDG-PET scans conducted after two cycles of study treatment, with no metabolic progression of non-target lesions. Those not meeting these criteria were classified as 18F-FDG-PET nonresponders.
Patients who achieved pCR were treated with adjuvant HP ± endocrine therapy, excluding chemotherapy, to complete a year of anti-HER2 treatment. Conversely, if there was no 18F-FDG-PET response after the initial two cycles of HP ± endocrine therapy, patients were treated with six cycles of TCHP before surgery. In instances in which a pCR was not achieved and chemotherapy was not previously administered in the neoadjuvant phase, adjuvant TCHP was provided. It is important to note that T-DM1 was not utilized throughout the study period.
Baseline assessments included core biopsies, hormone receptor status, and 18F-FDG-PET scans, with response evaluations based on both 18F-FDG-PET and magnetic resonance imaging. Patients proceeded to surgery 2 to 6 weeks after completing neoadjuvant therapy, followed by adjuvant HP for 1 year. The trial’s primary endpoints included the proportion of 18F-FDG-PET responders achieving a pCR in group B and 3-year iDFS in group B. Secondary endpoints involved other efficacy and surgical outcomes, safety evaluations, and health-related quality-of-life assessments.
The trial employed rigorous statistical analyses, including exact binomial tests for the primary endpoints and logistic regression models for secondary endpoints, with adjustments for hormonal receptor status. The study is registered with EudraCT (2016-002676-27) and ClinicalTrials.gov (NCT03161353).
HER2DX evaluation
This retrospective analysis utilized baseline pretreatment formalin-fixed, paraffin-embedded core tumor biopsies. HER2DX was performed at a central lab (Barcelona, Spain) in 292 tumor tissue samples (Supplementary Fig. S1). The HER2DX assay integrates clinical data (i.e., tumor stage and nodal status) and the expression of 27 genes implicated in four biological processes (immune infiltration, luminal differentiation, tumor cell proliferation, and HER2 amplicon expression) and provides a risk score, a pCR likelihood score, and the ERBB2 score (14).
Primary objective
The primary objective of this analysis was to investigate the association of HER2DX with pCR, defined as ypT0/is ypN0 per local assessment.
Secondary objective
In addition to the primary objective, an exploratory analysis was conducted to assess the association of the HER2DX risk score, defined using the original 50-point threshold of HER2DX, and correlates with 3-year iDFS. This analysis aimed to explore the potential prognostic value of HER2DX in predicting 3-year outcomes following treatment.
Statistical analysis
The validation of HER2DX in the PHERGain trial was an unplanned exploratory analysis. However, we assessed the statistical power of the study to determine the feasibility of the analysis. With a sample size of 292 patients, and assuming an odds ratio (OR) of 1.25 for the association between a 10-unit increase in HER2DX (continuous variable) and the pCR endpoint, the study had a statistical power >85% using a 5% two-sided alpha error. For the primary objective, univariable and multivariable logistic regression models were used to evaluate the association between HER2DX and pCR in terms of ORs with 95% confidence interval (95% CI). For the secondary objective, the Kaplan–Meier method was used to estimate iDFS at 3 years, and univariable Cox proportional-hazard models were used to obtain HRs with 95% CI. The C-statistic with 95% CI was estimated to evaluate the discrimination performance. No data imputation was performed. All tests were two-sided with a value of P < 0.05 considered statistically significant. All statistical analyses were conducted using R software 4.0.3.
Ethical considerations
This study adhered to ethical standards and was conducted in accordance with the principles outlined in the Declaration of Helsinki. The use of retrospective data from the PHERGain trial was approved by the relevant ethics committees. Informed written consent was obtained from each subject.
Data availability
Data are not publicly available, as participants of this study did not agree for their data to be shared publicly. Data can be requested from the corresponding author for academic use only, subject to a data transfer agreement and ethics committee approval.
Results
HER2DX genomic assay was successfully evaluated in 292 of 356 tumors, representing an 82.0% inclusion rate (Table 1). The median patient follow-up period extended to 3.6 years.
Table 1.
Patient characteristics.
| Trial population | HER2DX study population | |||
|---|---|---|---|---|
| Treatment group | n | % | n | % |
| Overall | 356 | 100 | 292 | 82 |
| Group A | 71 | 20 | 55 | 19 |
| Group B | 285 | 80 | 237 | 81 |
| Group B PET responder | 227 | 80 | 187 | 79 |
| Group B PET nonresponder | 58 | 20 | 50 | 21 |
| Tumor stage | ||||
| cT1 | 49 | 14 | 45 | 15 |
| cT2 | 245 | 69 | 199 | 68 |
| cT3 | 62 | 17 | 48 | 17 |
| Nodal status | ||||
| cN0 | 184 | 52 | 144 | 49 |
| cN1 | 151 | 42 | 130 | 45 |
| cN2 | 21 | 6 | 18 | 6 |
| Hormone receptor status | ||||
| Negative | 120 | 34 | 98 | 34 |
| Positive | 236 | 66 | 194 | 66 |
| pCR rates based on treatment | ||||
| Overall | 151 | 42 | 111 | 38 |
| Group A | 50 | 70 | 31 | 56 |
| Group B | 101 | 35 | 80 | 34 |
| Group B PET responder | 86 | 38 | 66 | 35 |
| Group B PET nonresponder | 15 | 26 | 14 | 28 |
HER2DX pCR score predictive capacity for pCR
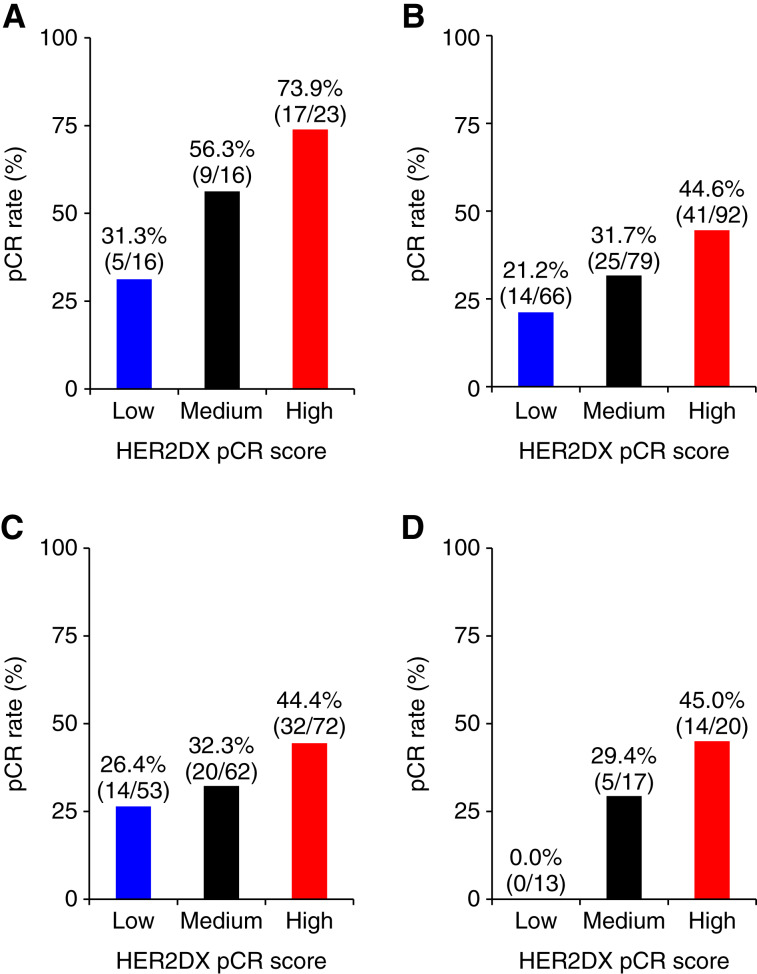
The overall pCR rate was 38.0%, with pCR rates of 56.4% in group A and 33.8% in group B. HER2DX pCR score, assessed both as a continuous and categorical variable, demonstrated a significant association with pCR in both univariate and multivariate logistic regression analyses (after adjusting for treatment and clinicopathologic factors such as hormone receptor status, treatment group, and 18F-FDG-PET response; Table 2). In multivariable analysis, HER2DX pCR score, as a continuous variable, significantly correlated with pCR (OR, per 10-unit increase, 1.29; 95% CI, 1.10–1.54; P < 0.001), adjusting for treatment and clinicopathologic factors. HER2DX-defined pCR-high, -med, and -low groups exhibited pCR rates of 50.4% (58/115), 35.8% (34/95), and 23.2% (19/82), respectively (pCR-high vs. pCR-low OR, 3.27; 95% CI, 1.54–7.09; P < 0.001). In group A (n = 55), the pCR rates of HER2DX pCR-high, -medium, and -low disease were 73.9% (17/23), 56.3% (9/16), and 31.3% (5/16), respectively (Fig. 2A; OR HER2DX pCR-high vs. -low, 6.23; 95% CI, 1.6–27.8; P = 0.011).
Table 2.
HER2DX pCR score association with pCR.
| HER2DX pCR score | OR per 10-unit increase | P value | |
|---|---|---|---|
| Univariate | Continuous variable | 1.31 (1.15–1.50) | <0.001 |
| Multivariatea | Continuous variable | 1.29 (1.10–1.54) | <0.001 |
| Univariate | Low | Reference | — |
| Medium | 1.85 (0.96–3.63) | 0.069 | |
| High | 3.37 (1.82–6.45) | <0.001 | |
| Multivariatea | Low | Reference | — |
| Medium | 1.84 (0.95–3.63) | 0.080 | |
| High | 3.27 (1.54–7.09) | <0.001 | |
Adjusted for the treatment arm, PET response, and hormone receptor status.
Figure 2.
pCR rates across HER2DX pCR score groups. A, All patients in group A; B, all patients in group B; C, patients in group B with a 18F-FDG-PET response; D, patients in group B without a 18F-FDG-PET response.
In group B (n = 237), the pCR rates of HER2DX pCR-high, -medium, and -low disease were 44.6% (41/92), 31.7% (25/79), and 21.2% (14/66), respectively (Fig. 2B; OR HER2DX pCR-high vs. -low, 2.99; 95% CI, 1.5–6.3; P = 0.003). Moreover, among patients in group B with an 18F-FDG-PET response and who did not receive neoadjuvant chemotherapy (n = 185), the pCR rates of HER2DX pCR-high, -medium, and -low disease were 44.4% (32/72), 32.3% (20/62), and 26.4% (14/53), respectively (Fig. 2C; OR HER2DX pCR-high vs. -low, 2.23; 95% CI, 1.1–4.9; P = 0.040). Finally, among group B patients without an 18F-FDG-PET response, which received neoadjuvant chemotherapy (n = 46), the pCR rates of HER2DX pCR-high, -medium, and -low disease were 45.0% (9/20), 29.4% (5/17), and 0.0% (0/13), respectively (Fig. 2D).
In the multivariable analysis, HER2DX pCR score was significantly associated with pCR rates regardless of hormone receptor status (Table 2). HER2DX pCR score distribution in hormone receptor–positive and hormone receptor–negative status and pCR rates in each group are shown in Supplementary Fig. S2.
Additionally, we performed three multivariable models for pCR prediction. Model 1 included clinicopathologic data but did not include HER2DX. Model 2 included clinicopathologic data and HER2DX pCR score as a categorical variable. Model 3 included clinicopathologic data and HER2DX pCR score as a continuous variable (Supplementary Table S1). In terms of the discrimination performance, the C-statistics for model 2 (0.71; 95% CI, 0.65–0.77) and model 3 (0.70; 95% CI, 0.64–0.77) were numerically better than those obtained with model 1 (0.69; 95% CI, 0.63–0.76), although not statistically significant (P = 0.13; Supplementary Table S2).
Effect of upfront versus delayed chemotherapy based on HER2DX pCR score
The pCR rate was 56.4% (31/55) in patients who underwent six cycles of upfront TCHP (group A). Conversely, for patients in group B lacking a PET response after two cycles of HP and subsequently receiving six cycles of TCHP, the pCR rate was 28.0% (14/50), a statistically significant difference (OR, 3.32; 95% CI, 1.5–7.5; P = 0.004). This distinction was clearer within the subset of patients with HER2DX pCR-high disease: a 74.0% pCR rate (17/23) was observed in group A, compared with a 45.0% pCR rate (9/20) in group B patients who, after not responding to two cycles of HP, were treated with six cycles of TCHP (OR, 3.47; 95% CI, 1.0–12.5; P = 0.053). Conversely, no significant difference in pCR rates was observed for HER2DX pCR-low disease between patients in group A and those in group B without a PET response and then treated with TCHP [26.0% (7/27) vs. 21.2% (14/66); OR, 1.3; 95% CI, 0.5–3.7; P = 0.626].
HER2DX ERBB2 score predictive capacity for pCR
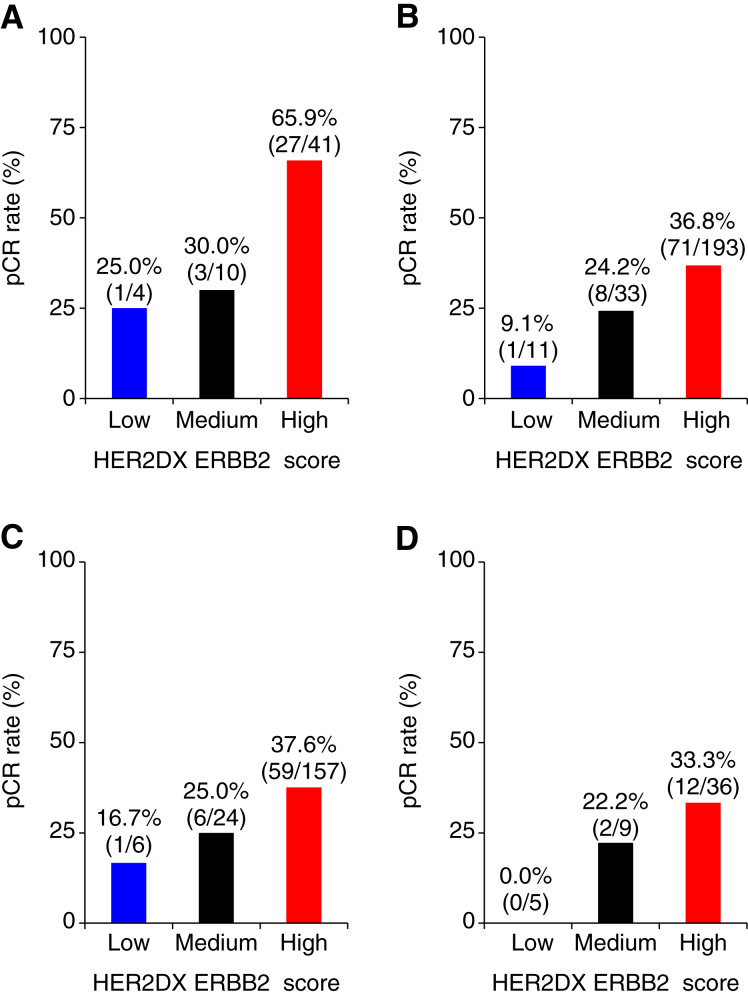
The ERBB2 score, assessed both as a continuous and as a predefined categorical variable (i.e., 80.1% ERBB2-high, 14.7% ERBB2-medium, and 5.2% ERBB2-low), demonstrated a significant association with pCR in both univariate and multivariate logistic regression analyses (after adjusting for treatment and clinicopathologic factors such as hormone receptor status, treatment group, and 18F-FDG-PET response; Table 3). In group A (n = 55), the pCR rate of HER2DX ERBB2-high disease was 65.9% (27/41) versus 25.0% (1/4) in ERBB2-low disease (OR, 5.8; 95% CI, 0.7–123.1; P = 0.144; Fig. 3A). In group B (n = 237), the pCR rate of HER2DX ERBB2-high disease was 36.8% (71/193) versus 9.1% (1/11) in ERBB2-low disease (OR, 5.82; 95% CI, 1.1–107.9; P = 0.096; Fig. 3B). Moreover, among group B patients with an 18F-FDG-PET response who did not receive neoadjuvant chemotherapy (n = 185), the pCR rate of HER2DX ERBB2-high disease was 37.6% (59/157) versus 16.7% (1/6) in pCR-low disease (OR, 3.01; 95% CI, 0.5–58.4; P = 0.320; Fig. 3C). Lastly, among group B patients without an 18F-FDG-PET response and, therefore, who also received neoadjuvant chemotherapy (n = 46), the pCR rate of HER2DX ERBB2-high disease was 33.3% (12/36) versus 0.0% (0/5) in ERBB2-low disease (Fig. 3D).
Table 3.
HER2DX ERBB2 score association with pCR.
| HER2DX ERBB2 score | OR per 10-unit increase | P value | |
|---|---|---|---|
| Univariable | Continuous variable | 1.38 (1.18–1.64) | <0.001 |
| Multivariablea | Continuous variable | 1.35 (1.14–1.61) | <0.001 |
| Univariable | Low | Reference | — |
| Medium | 2.23 (0.51–15.74) | 0.337 | |
| High | 4.68 (1.26–30.39) | 0.045 | |
| Multivariablea | Low | Reference | — |
| Medium | 2.30 (0.48–18.06) | 0.301 | |
| High | 4.90 (1.22–33.24) | 0.048 | |
Adjusted for the treatment arm, PET response, and hormone receptor status.
Figure 3.
pCR rates across HER2DX ERBB2 score groups. A, All patients in group A; B, all patients in group B; C, patients in group B with a 18F-FDG-PET response; D, patients in group B without a 18F-FDG-PET response.
HER2DX risk score association with 3-year iDFS
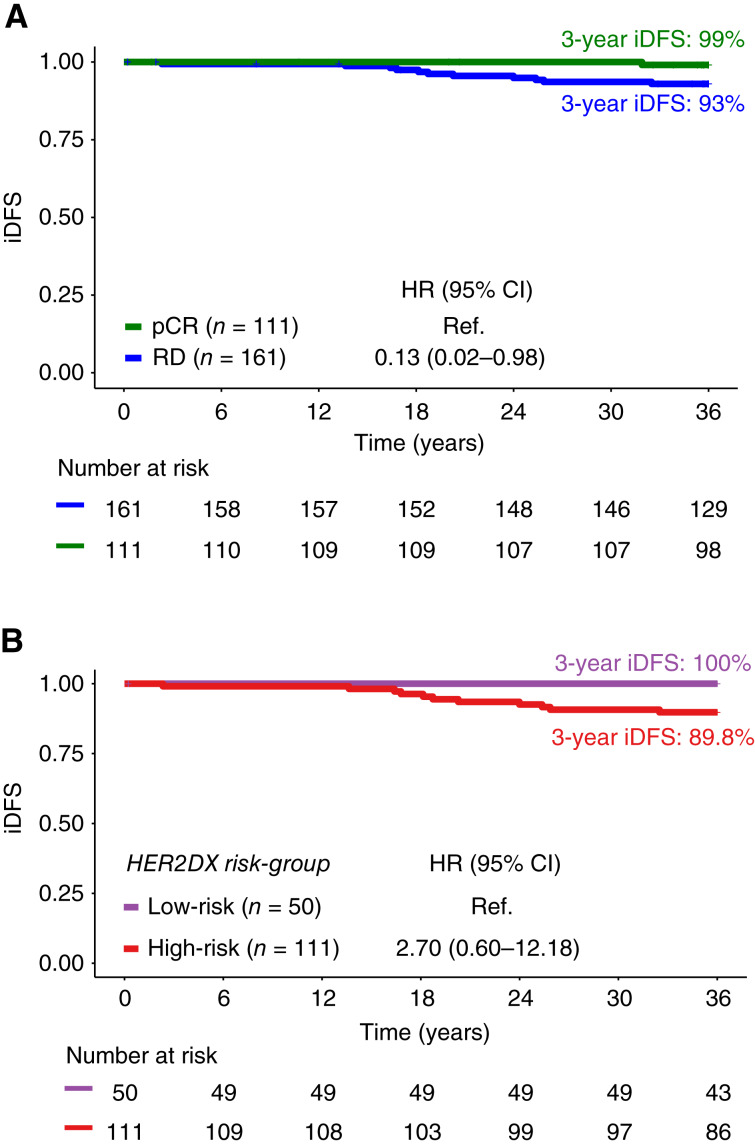
Data on 3-year iDFS were available for 272 patients with a total of 12 iDFS events. In this subgroup, pCR was significantly associated with better iDFS compared with residual disease (HR, 0.13; 0.02–0.98, P = 0.048). HER2DX low-risk and high-risk represented 36.8% and 63.2% of cases, respectively. Notably, 92% (11/12) of iDFS events occurred in patients with residual disease (Fig. 4A). According to the HER2DX risk score, all 12 patients experiencing an iDFS event had HER2DX high-risk disease. Moreover, among patients with residual disease, HER2DX high-risk had a 3-year iDFS of 89.8% compared with 100.0% in HER2DX low-risk (HR, 2.70; 95% CI, 0.6–12.2; P = 0.197; Fig. 4B).
Figure 4.
Three-year iDFS in the PHERGain trial with HER2DX results. A, According to pCR status; B, according to HER2DX risk score in patients with residual disease (RD).
Discussion
This study, which was retrospectively designed and embedded within the PHERGain trial, was undertaken to explore the predictive potential of the HER2DX genomic assay for neoadjuvant dual HER2 blockade using HP in patients with HER2+ early breast cancer. Briefly, our results confirm a significant association between HER2DX pCR score and the likelihood of achieving pCR, regardless of hormone receptor status, 18F-FDG-PET response, and treatment regimen. In addition, the HER2DX risk score shows promise in classifying patients with residual disease with distinct 3-year survival outcomes.
In the analysis of HER2DX-defined pCR risk groups, our study revealed distinct pCR rates in each subgroup. This prognostic capability remained consistent across diverse treatment groups, irrespective of the incorporation of chemotherapy. Particularly noteworthy were the findings observed in group A, in which patients receiving TCHP exhibited a striking pCR rate of 74.0% in the HER2DX pCR-high category, in stark contrast to the 31.0% rate observed in the pCR-low category. Importantly, these rates were notably diminished in the absence of chemotherapy. Accordingly, in group B with a 18F-FDG-PET response receiving a total of eight cycles of HP without chemotherapy, and selected by, the pCR rate in the HER2DX pCR-high category was 44.4%. These findings underscore the intrinsic link between anti-HER2 sensitivity and chemotherapy responsiveness (22). Moreover, these results emphasize that the administration of chemotherapy might not salvage pCR rates in patients lacking a 18F-FDG-PET response after two cycles of HP, thereby highlighting the interplay between anti-HER2 sensitivity and the role of chemotherapy in achieving optimal treatment outcomes, and the need to select the right treatment upfront at diagnosis.
The analysis of 3-year iDFS reinforces the clinical relevance of HER2DX in guiding treatment decisions. As expected, patients with residual disease exhibited a markedly inferior iDFS compared with those achieving pCR, affirming the prognostic impact of treatment response. Within the residual disease subgroup, a particularly noteworthy observation was that all patients experiencing iDFS events had HER2DX high-risk disease, underscoring the assay’s adeptness in identifying individuals at elevated risk of recurrence. Consequently, HER2DX risk stratification within the residual disease subgroup unveiled a numerically superior 3-year iDFS in low-risk patients (100%) as opposed to high-risk patients (89.8%), demonstrating a HR of 2.70. Of note, patients with residual disease following neoadjuvant treatment did not receive adjuvant T-DM1, in alignment with the treatment guidelines and drug approval status applicable at the time the PHERGain study was conducted. In summary, individuals characterized by HER2DX pCR high- and HER2DX low-risk disease profiles may emerge as optimal candidates for de-escalation treatment strategies, specifically with HP without chemotherapy, presenting a prospect for more tailored and targeted therapeutic interventions.
The findings from the HER2DX in the PHERGain study align with outcomes from previous validation studies (16–18, 21). A comprehensive patient-level meta-analysis involving 765 patients across seven studies has demonstrated that the HER2DX pCR score significantly correlates with pCR across various trastuzumab-based therapies, including dual HER2 blockade with lapatinib and trastuzumab, even in the absence of chemotherapy (16). It is noteworthy that the distribution of HER2DX pCR score categories within the PHERGain study mirrors the proportions observed in other studies, with approximately one third of patients falling into each of the three categories. Furthermore, the HER2DX risk score has consistently shown an association with survival outcomes, independent of clinicopathologic factors, including pCR status. In a subset of 150 patients who did not achieve pCR, those classified as low-risk by HER2DX experienced better event-free survival outcomes compared with their high-risk counterparts, with event-free survival rates at 6 years being 93.5% versus 78.8%, respectively (16). It is noteworthy that 9% of these patients were treated with adjuvant T-DM1 (16). Currently, the assay is under prospective evaluation in the CompassHER2-pCR trial (NCT04266249) and the DEFINITIVE trial (23), aiming to fine-tune the chemotherapy regimen in the neoadjuvant setting.
Although our study provides valuable insights into the predictive capacity of the HER2DX genomic assay in the context of neoadjuvant dual HER2 blockade with HP, it is important to acknowledge certain caveats and limitations that may impact the interpretation and generalizability of our findings. First, our analysis relied on retrospective data from the PHERGain trial, introducing inherent limitations associated with this study design. Although efforts were made to minimize biases and ensure data accuracy, the retrospective nature of the study warrants careful consideration of potential confounding variables. Second, certain subgroup analyses, particularly within the context of treatment groups (group A and group B with or without 18F-FDG-PET response), involved relatively small sample sizes with limited statistical power and precision. Therefore, findings from subgroup analyses should be interpreted with caution. Third, the addition of HER2DX did not statistically improve the discrimination of the model, although the analysis was underpowered to detect these differences. Fourth, the short follow-up of patients and low number of events preclude evaluating associations of the HER2DX risk score with patients’ outcomes, regardless of the pCR status. In a recent retrospective study, the HER2DX risk score was found significantly associated with long-term survival outcome in patients with residual disease (16). Despite these caveats and limitations, our study contributes valuable information about the potential clinical utility of the HER2DX assay in guiding treatment decisions for early-stage HER2+ breast cancer, and prospective validation is warranted.
To conclude, the ability of HER2DX to predict treatment response and 3-year outcomes, especially in the context of HP-based therapy, holds promise for individualizing treatment decisions. As the field progresses, incorporating genomic assays like HER2DX into routine clinical practice may facilitate a more precise and tailored approach to the management of HER2+ early breast cancer. Future research should focus on validating these findings in larger cohorts and exploring the integration of HER2DX into broader clinical algorithms for treatment decision-making.
Supplementary Material
Supplementary Tables 1 and 2, and Supplementary Figures 1 and 2.
Acknowledgments
This work was funded by MEDSIR and Reveal Genomics. F. Hoffmann-La Roche funded the PHERGain study and provided trastuzumab and pertuzumab for the study. F. Brasó-Maristany received funding from Fundación científica AECC Ayudas Investigador AECC 2021 (INVES21943BRAS).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
A. Llombart-Cussac reports grants and nonfinancial support from Roche Genentech during the conduct of the study, as well as personal fees from Pfizer, grants from Novartis, and grants and personal fees from Eli Lilly and Company, AstraZeneca, Daiichi Sankyo, and Gilead outside the submitted work. J. Pérez-García reports personal fees from Roche and other support from MEDSIR during the conduct of the study, as well as personal fees from AstraZeneca, Daiichi Sankyo, Seattle Genetics, Gilead, Eli Lilly and Company, and Eisai outside the submitted work. F. Brasó-Maristany reports a patent for PCT/EP2022/086493 pending, a patent for PCT/EP2023/060810 pending, a patent for EP23382703 pending, and a patent for EP23383369 pending. L. Paré reports a patent for PCT/EP2021/070788 pending and a patent for PCT/EP2021/086493 pending. G. Villacampa reports personal fees from Merck Sharp & Dohme, Pfizer, GlaxoSmithKline, Pierre Fabre, and Reveal Genomics outside the submitted work. M. Gion reports grants from Gilead, AstraZeneca, Pfizer, and Novartis outside the submitted work. P. Schmid reports grants and personal fees from AstraZeneca, Merck, Novartis, and Roche; personal fees from Bayer, Boehringer Ingelheim, Pfizer, Puma, Eisai, Celgene, and Medivation; and grants from Astellas, Genentech, and OncoGenex during the conduct of the study as well as from Astellas, Genentech, and OncoGenex outside the submitted work. M. Colleoni reports grants from Roche outside the submitted work. P. Galván reports other support from Reveal Genomics during the conduct of the study. J.S. Parker reports personal fees from Reveal Genomics during the conduct of the study, other support from GeneCentric outside the submitted work, and a patent for PAM50-Prosigna with royalties paid from Veracyte. W. Buckingham reports other support from Reveal Genomics outside the submitted work, reports being a shareholder of Everything Genetic Ltd., and reports that the wife is an employee and shareholder of Natera Inc. C.M. Perou reports personal fees from Reveal Genomics outside the submitted work. P. Villagrasa reports other support from Reveal Genomics SL outside the submitted work, as well as a patent for PCT/EP2023/060810 pending and a patent for PCT/EP2021/086493 pending. M. Sampayo-Cordero reports other support from MEDSIR during the conduct of the study, as well as other support from MEDSIR and personal fees from Optimapharm outside the submitted work. A. Prat reports grants and personal fees from Reveal Genomics during the conduct of the study, as well as grants and personal fees from Roche, AstraZeneca, Daiichi Sankyo, and Novartis; personal fees from Pfizer and Ona Therapeutics outside the submitted work; in addition, A. Prat reports a patent for HER2DX pending and licensed to Reveal Genomics and a patent for ERBB2 issued and licensed to Reveal Genomics. J. Cortés reports grants and personal fees from Roche during the conduct of the study as well as grants and personal fees from Roche, AstraZeneca, Merck Sharp & Dohme, Eisai, and Pfizer; other support from Leuko and MAJ3 Capital; personal fees from Seattle Genetics, Daiichi Sankyo, Eli Lilly and Company, Bioasis, Clovis Oncology, Boehringer Ingelheim, Ellipses, HiberCell, BioInvent, GEMoaB, Gilead, Menarini, Zymeworks, Reveal Genomics, Scorpion Therapeutics, ExpreS2ion Biotechnologies, Jazz Pharmaceuticals, AbbVie, Novartis, Stemline Therapeutics, BridgeBio, BioNTech, Biocon, and Circle Pharma; grants from Queen Mary University of London, IQVIA, ARIAD Pharmaceuticals, Guardant Health, Bayer Healthcare, Piqur Therapeutics, and Baxalta GmbH outside the submitted work; in addition, J. Cortés reports a patent for Pharmaceutical Combinations of a Pi3k Inhibitor and a Microtubule Destabilizing Agent, a patent for WO 2014/199294 A issued, and a patent for HER2 as a Predictor of Response to Dual HER2 Blockade in the Absence of Cytotoxic Therapy. A. Prat, A. Llombart-Cussac, and J. Cortés report a patent for US 2019/0338368 A1 licensed. No disclosures were reported by the other authors.
Authors’ Contributions
A. Llombart-Cussac: Conceptualization, investigation, writing–original draft, writing–review and editing. J. Pérez-García: Conceptualization, resources, investigation, writing–original draft, writing–review and editing. F. Brasó-Maristany: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft. L. Paré: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. G. Villacampa: Data curation, validation, investigation, visualization, writing–review and editing. M. Gion: Resources, writing–review and editing. P. Schmid: Resources, writing–review and editing. M. Colleoni: Resources, writing–review and editing. M.R. Borrego: Resources. P. Galván: Data curation, methodology. J.S. Parker: Validation, methodology, writing–review and editing. W. Buckingham: Writing–review and editing. C.M. Perou: Writing–review and editing. P. Villagrasa: Writing–review and editing. J.A. Guerrero: Writing–review and editing. M. Sampayo-Cordero: Writing–review and editing. M. Mancino: Data curation, writing–review and editing. A. Prat: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing. J. Cortés: Conceptualization, resources, supervision, investigation, writing–original draft.
References
- 1. Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. J Clin Oncol 2018;36:2105–22. [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez-Angulo AM, Litton JK, Broglio KR, Meric-Bernstam F, Rakkhit R, Cardoso F, et al. High risk of recurrence for patients with breast cancer who have human epidermal growth factor receptor 2–positive, node-negative tumors 1 cm or smaller. J Clin Oncol 2009;27:5700–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prat A, Carey LA, Adamo B, Vidal M, Tabernero J, Cortés J, et al. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst 2014;106:dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Llombart-Cussac A, Cortés J, Paré L, Galván P, Bermejo B, Martínez N, et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol 2017;18:545–54. [DOI] [PubMed] [Google Scholar]
- 5. Chang JCN, Mayer IA, Forero-Torres A, Nanda R, Goetz MP, Rodriguez AA, et al. Tbcrc 006: a multicenter phase II study of neoadjuvant lapatinib and trastuzumab in patients with HER2-overexpressing breast cancer. J Clin Oncol 2011;29:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prat A, Baselga J. Dual human epidermal growth factor receptor 2 (HER2) blockade and hormonal therapy for the treatment of primary HER2-positive breast cancer: one more step toward chemotherapy-free therapy. J Clin Oncol 2013;31:1703–6. [DOI] [PubMed] [Google Scholar]
- 7. Waks AG, Desai NV, Li T, Poorvu PD, Partridge AH, Sinclair N, et al. A prospective trial of treatment de-escalation following neoadjuvant paclitaxel/trastuzumab/pertuzumab in HER2-positive breast cancer. NPJ Breast Cancer 2022;8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gianni L, Pienkowski T, Im Y-H, Roman L, Tseng LM, Liu MC, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol 2012;13:25–32. [DOI] [PubMed] [Google Scholar]
- 9. Pérez-García JM, Gebhart G, Ruiz Borrego M, Stradella A, Bermejo B, Schmid P, et al. Chemotherapy de-escalation using an 18F-FDG-PET-based pathological response-adapted strategy in patients with HER2-positive early breast cancer (PHERGain): a multicentre, randomised, open-label, non-comparative, phase 2 trial. Lancet Oncol 2021;22:858–71. [DOI] [PubMed] [Google Scholar]
- 10. Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164–72. [DOI] [PubMed] [Google Scholar]
- 11. Harbeck N, Nitz UA, Christgen M, Kümmel S, Braun M, Schumacher C, et al. De-escalated neoadjuvant trastuzumab-emtansine with or without endocrine therapy versus trastuzumab with endocrine therapy in HR+/HER2+ early breast cancer: 5-year survival in the WSG-ADAPT-TP trial. J Clin Oncol 2023;41:3796–804. [DOI] [PubMed] [Google Scholar]
- 12. Nitz U, Gluz O, Graeser M, Christgen M, Kuemmel S, Grischke EM, et al. De-escalated neoadjuvant pertuzumab plus trastuzumab therapy with or without weekly paclitaxel in HER2-positive, hormone receptor-negative, early breast cancer (WSG-ADAPT-HER2+/HR−): survival outcomes from a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 2022;23:625–35. [DOI] [PubMed] [Google Scholar]
- 13. Pérez-García JM, Cortés J, Ruiz-Borrego M, Colleoni M, Stradella A, Bermejo B, et al. 3-year invasive disease-free survival with chemotherapy de-escalation using an 18F-FDG-PET-based, pathological complete response-adapted strategy in HER2-positive early breast cancer (PHERGain): a randomised, open-label, phase 2 trial. Lancet 2024;403:1649–59. [DOI] [PubMed] [Google Scholar]
- 14. Prat A, Guarneri V, Pascual T, Brasó-Maristany F, Sanfeliu E, Paré L, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine 2022;75:103801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prat A, Guarneri V, Paré L, Griguolo G, Pascual T, Dieci MV, et al. A multivariable prognostic score to guide systemic therapy in early-stage HER2-positive breast cancer: a retrospective study with an external evaluation. Lancet Oncol 2020;21:1455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villacampa G, Tung NM, Pernas S, Paré L, Bueno-Muiño C, Echavarría I, et al. Association of HER2DX with pathological complete response and survival outcomes in HER2-positive breast cancer. Ann Oncol 2023;34:783–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waks AG, Ogayo ER, Paré L, Marín-Aguilera M, Brasó-Maristany F, Galván P, et al. Assessment of the HER2DX assay in patients with ERBB2-positive breast cancer treated with neoadjuvant paclitaxel, trastuzumab, and pertuzumab. JAMA Oncol 2023;9:835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bueno-Muiño C, Echavarría I, López-Tarruella S, Roche-Molina M, Del Monte-Millán M, Massarrah T, et al. Assessment of a genomic assay in patients with ERBB2-positive breast cancer following neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab. JAMA Oncol 2023;9:841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guarneri V, Brasó-Maristany F, Dieci MV, Griguolo G, Paré L, Marín-Aguilera M, et al. HER2DX genomic test in HER2-positive/hormone receptor-positive breast cancer treated with neoadjuvant trastuzumab and pertuzumab: a correlative analysis from the PerELISA trial. EBioMedicine 2022;85:104320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marín-Aguilera M, Jares P, Sanfeliu E, Villacampa G, Hernández-Lllán E, Martínez-Puchol AI, et al. Analytical validation of HER2DX genomic test for early-stage HER2-positive breast cancer. ESMO Open 2024;9:102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Villacampa G, Pascual T, Brasó-Maristany F, Paré L, Martínez-Sáez O, Cortés J, et al. Prognostic value of HER2DX in early-stage HER2-positive breast cancer: a comprehensive analysis of 757 patients in the Sweden Cancerome Analysis Network-Breast dataset (SCAN-B). ESMO Open 2024;9:102388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pascual T, Fernandez-Martinez A, Tanioka M, Dieci MV, Pernas S, Gavila J, et al. Independent validation of the PAM50-based chemo-endocrine score (CES) in hormone receptor–positive HER2-positive breast cancer treated with neoadjuvant anti–HER2-based therapy. Clin Cancer Res 2021;27:3116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. European Commission . Diagnostic HER2DX-guided treatment for patients with early-stage HER2-positive breast cancer. Available from:https://cordis.europa.eu/project/id/101136953. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1 and 2, and Supplementary Figures 1 and 2.
Data Availability Statement
Data are not publicly available, as participants of this study did not agree for their data to be shared publicly. Data can be requested from the corresponding author for academic use only, subject to a data transfer agreement and ethics committee approval.