Abstract
The precise prediction of major histocompatibility complex (MHC)-peptide complex structures is pivotal for understanding cellular immune responses and advancing vaccine design. In this study, we enhanced AlphaFold’s capabilities by fine-tuning it with a specialized dataset consisting of exclusively high-resolution class I MHC-peptide crystal structures. This tailored approach aimed to address the generalist nature of AlphaFold’s original training, which, while broad-ranging, lacked the granularity necessary for the high-precision demands of class I MHC-peptide interaction prediction. A comparative analysis was conducted against the homology-modeling-based method Pandora as well as the AlphaFold multimer model. Our results demonstrate that our fine-tuned model outperforms others in terms of root-mean-square deviation (median value for Cα atoms for peptides is 0.66 Å) and also provides enhanced predicted local distance difference test scores, offering a more reliable assessment of the predicted structures. These advances have substantial implications for computational immunology, potentially accelerating the development of novel therapeutics and vaccines by providing a more precise computational lens through which to view MHC-peptide interactions.
Significance
Major histocompatibility complex (MHC) molecules are central to the immune system, enabling disease recognition and response by presenting diverse peptides to T cells. By fine-tuning the AlphaFold model with specialized high-resolution data, we offer a novel tool that surpasses existing methods in its ability to accurately predict the structure of class I MHC-peptide complexes. This enhanced predictive model has important implications for both infectious disease and cancer immunotherapy and has the potential to facilitate the development of targeted therapeutic strategies.
Introduction
MHC class I (MHC-I) molecules play a crucial role in the immune system and are found on the surface of most cells in the body. They present intracellular specific antigens, such as viral, bacterial, or cancerous peptides, to cytotoxic T cells, enabling T cells to recognize and respond to these threats (1).
MHC-I molecules are important to the functioning of the immune system. By understanding how they bind and present peptides, we can gain insights into disease mechanisms such as autoimmunity (2). This knowledge would also empower us to prevent certain infectious diseases through MHC-based vaccination (3). In the context of cancer immunotherapy, this understanding would allow the design of neoantigen vaccines that enhance the immune system’s ability to selectively target cancer cells (4).
To ensure that the immune system effectively detects and responds to a wide range of infections, each MHC-I molecule presents a variety of peptides to T cells. To achieve this, each MHC-I molecule has the capability to bind a broad class of different peptide sequences. Although each person presents only a small number of different MHC-I molecules (two alleles for each of the three MHC-I genes), many different MHC-I alleles are present in the population (5), leading to individual differences in MHC-I specificity. The diversity of MHC-I molecules and peptides allows the immune system to respond effectively to different threats and adapt to new challenges. However, this diversity also poses a significant challenge when it comes to predicting class I MHC-peptide complexes.
There are different approaches for predicting class I MHC-peptide complex structures, including molecular docking (6), molecular dynamics simulations (7,8), homology modeling (9), and machine-learning methods (10,11,12). The accuracy of their predictions can vary.
One of the advanced tools for predicting class I MHC-peptide complex structures is the Peptide ANchoreD modeling fRAmework (Pandora) (13). Pandora uses a database of known MHC structures as templates with anchors-restrained loop modeling for peptide conformation. However, Pandora has some limitations: rare alleles may lack suitable MHC templates, low sequence similarity in the peptide-binding groove can cause alignment issues, accurate anchor residues are needed, and ranking output models can be challenging due to different scoring functions.
Deep-learning methodologies have significantly enhanced the prediction of protein structural information across various degrees of complexity (14). These methodologies vary in the nature of the input data they require and in their architectural designs, as evidenced by recent studies (15,16,17,18). Among these innovative methods, AlphaFold stands out by employing a novel equivariant attention architecture to predict protein structures with remarkable accuracy. This approach leverages an extensive dataset of structural information to train its algorithm, enabling accurate prediction of protein structures even without closely related known structures. However, despite AlphaFold’s successes, its utility for MHC-peptide predictions has been somewhat limited by its generalized training across diverse protein types. This broad approach, while comprehensive, may not always capture the intricate nuances necessary for high-fidelity predictions within specific domains such as MHC-peptide interactions. Therefore, by fine-tuning AlphaFold with a dataset curated explicitly from high-resolution class I MHC-peptide crystal structures, we aim to enhance the model’s specificity and accuracy in this critical area, thereby overcoming one of the main limitations faced by practitioners utilizing this tool for specialized applications in immunological research.
We present an approach that leverages the robust capabilities of AlphaFold, fine-tuning it to significantly improve the prediction accuracy for class I MHC-peptide complex structures. Our model outperforms Pandora in terms of Cα root-mean-square deviation (RMSD) (median value for Cα atoms for peptides is 0.66 Å) and also provides enhanced predicted local distance difference test (lDDT) scores, offering a more reliable assessment of the predicted structures. Our model also does not require any input information about the anchor residues.
Related work
Structural overview of class I MHC-peptide complexes
MHC-I is a transmembrane protein composed of two non-covalently connected chains, α and -microglobulin (19). The α chain consists of three domains followed by a transmembrane part and a cytoplasmic tail. Each α domain is approximately 90 residues long.
and domains form a symmetric structure composed of curved α-helices. Together they form a peptide-binding groove between them. The most variability in MHC-I sequence is found in this groove region to create a variety for peptide specificity. and -microglobulin domains do not interact with the peptide.
The number of different peptides that a particular MHC-I molecule can bind varies depending on the specific MHC molecule, but in general peptide-binding groove geometry can accommodate short peptides of length 8–11 residues (20). However, slightly longer peptides have also been observed (21,22,23). Several peptide positions, typically at the N- and C-termini of the peptide, contribute significantly to the binding. These specific residues are known as anchors.
Homology modeling (Pandora)
One of the state-of-the-art tools for class I MHC-peptide complex structure prediction is the homology-based Pandora (13), which uses homology modeling for the MHC protein and performs anchor-restrained peptide modeling using MODELLER (24).
For the homology modeling of MHC-I, Pandora selects a single template from the custom-made database of known MHC structures. This approach requires proper template selection, which depends on the availability of the same allele type, group, or gene within the database.
After selecting the template, the alignment between the target MHC and the template is carried out. The sequence similarity between the target and the template may be low in the peptide-binding groove due to MHC sequence variability, which can increase the likelihood of alignment issues. However, this groove region is of utmost importance for modeling because it is the area where peptide binds to MHC-I.
One of the crucial requirements for Pandora is the inclusion of peptide anchors. Pandora utilizes this information for anchors-restrained loop modeling of the peptide. This information can be provided by the user or predicted by netMHCpan 4.1, although challenges may arise for non-canonical anchors.
Pandora provides 20 models for a single peptide-MHC pair and evaluates them using MODELLER’s internal scoring functions molpdf and DOPE. In some cases, the best-scored DOPE and molpdf structures are different, which can present a challenging decision for the user in selecting the best model. The authors suggest using molpdf scoring ranking.
AlphaFold
AlphaFold has been a transformative force in computational biology, redefining the landscape of protein structure prediction with its transformer-based architecture. This architecture is particularly well suited to processing sequential biological data, such as amino acid chains, due to its ability to capture long-distance interactions between amino acids—a fundamental factor in predicting how proteins fold.
The model’s capabilities, already proven in the Critical Assessment of Structure Prediction (CASP) competitions, saw further advancements as evidenced in CASP15. Recent research (25) demonstrated AlphaFold’s enhanced proficiency in the structure prediction of protein complexes, marking a significant step forward in the CASP15-CAPRI experiment. Furthermore, the scientific community has recognized the transformative impact of AI-based modeling, such as AlphaFold, on the accuracy of protein assembly prediction (26). These developments underscore AlphaFold’s broad potential, extending from structure prediction to advancing our understanding of disease pathology, drug discovery, and enzyme engineering.
However, the generalist nature of AlphaFold, while powerful, reveals limitations when tasked with highly specialized predictions, such as modeling the MHC-peptide complexes. The complexity inherent in these biological structures, along with their variability, necessitates tailored versions of the model. Recent advancements have seen the use of specific datasets and modified architectures to enhance the prediction of such complex interactions, exemplifying the need for specialized approaches in computational biology (27).
In the pursuit of enhancing AlphaFold’s predictions for MHC-peptide complexes, researchers have embarked on a variety of methodologies. One prominent approach involves the development of an AlphaFold-based pipeline which involves additional steps for multiple sequence alignment (MSA) or template selection (11). In addition, efforts have been made to fine-tune AlphaFold’s parameters on peptide-MHC class I and II structural and binding data, with the fine-tuned model achieving state-of-the-art classification accuracy (28). These advancements, while in some cases requiring more complex pipelines, reflect the nuanced balance between achieving broad predictive capabilities and the pursuit of granular structural details.
Materials and methods
Implementation of AlphaFold in PyTorch
In this study, we faced the challenge of AlphaFold’s unavailability for training code and its original implementation in JAX (29), which presents complexities in modification and testing. Inspired by the OpenFold model (30), we developed a custom version of AlphaFold using the PyTorch library (31). This approach not only allowed us to utilize the pre-trained weights of AlphaFold but also introduced significant flexibility in modifying the code as per our research needs.
One of the primary enhancements in our PyTorch-based AlphaFold implementation is the integration of checkpoints for optimal memory management. Given the extensive size of the AlphaFold network, these checkpoints are crucial for efficient memory usage, ensuring stable and effective processing even on large-scale data.
Furthermore, we leveraged the PyTorch-Lightning library (32), which significantly streamlined our workflow. PyTorch-Lightning abstracts and automates many routine tasks, enabling us to focus on the core aspects of our model. It facilitated effective training and easy distribution of computational workload across multiple GPUs and nodes. PyTorch-Lightning also brought additional advantages, such as simplified implementation of advanced optimization techniques and streamlined model-validation processes. It enhanced our model’s reproducibility and scalability, allowing us to efficiently experiment with various configurations and settings.
Dataset
We downloaded class I MHC-peptide complex structures from the RCSB Protein Data Bank (33), selecting X-ray structures with a resolution finer than 3.5 Å. We excluded structures containing non-standard amino acids or a significant number of unresolved residues. Additionally, we limited our selection to samples with peptide lengths ranging from 8 to 11 amino acids. From each MHC protein only and domains were used. Our final dataset consisted of 919 structures from various species, including humans (Homo sapiens), mice (Mus musculus), and other species. Specifically, the human dataset consisted of samples that encompassed at least eight distinct allele groups for HLA-A, 20 for HLA-B, and five for HLA-C. Additionally, the dataset included samples containing alleles from at least one of each of the HLA-E, HLA-F, and HLA-G genes. For mice, the dataset reflected allelic variation across five H2-K genes, two H2-D genes, and one H2-L gene. Furthermore, our dataset incorporated samples from other species, including chickens, swine, and macaques, to reflect genetic variability in one gene for each respective species. In total, the dataset included 618 unique peptides. The detailed breakdown of alleles can be found in the supporting material. We divided the dataset into three parts based on the release date, approximately a 60/20/20 split for training, validation, and testing. This allows the model to learn patterns and trends from historical data, and during the testing the model it is presented with unseen data. Additionally, cases with a 98% identity to any MHC sequence from the test, while sharing the same peptide, were removed from the test set. This precaution was taken to prevent data leakage from the training set to the testing set. The training set (releases from October 15, 1992 to December 7, 2016) and the validation set (releases from December 7, 2016 to June 17, 2020) were used for fine-tuning and hyperparameter optimization, while the test set (releases from June 17, 2020 to August 23, 2023) was reserved solely for final comparison.
To prepare the necessary input for AlphaFold, we generated MSAs using MMseqs2 (34), a software suite designed for fast and sensitive protein sequence searching. Our searches were conducted against the ColabFoldDB (35), which is a comprehensive and regularly updated database tailored for such analyses.
Metrics
In protein structure prediction, the RMSD is a crucial metric used to measure the average atomic distance between a predicted protein structure and a reference structure, typically comparing backbone atoms. For an accurate prediction, the RMSD should ideally be below 2.0 Å.
In our study, we compute several RMSD metrics to thoroughly assess the accuracy of predicted class I MHC-peptide interactions. All RMSD calculations are performed using the PyMol library (36). The primary metric we utilize is termed the MHC-peptide RMSD. To compute this, we first align the predicted and true structures based on the MHC chain. Following this alignment, the RMSD is calculated specifically for the Cα atoms of the peptide, providing a focused measure of the peptide’s positional accuracy relative to the MHC.
While the main and most informative metric for our analysis is the MHC-peptide RMSD, we also report additional metrics such as MHC-MHC RMSD, peptide-peptide RMSD, and the side-chain RMSD of anchor residues to provide a more comprehensive analysis of our prediction results, allowing for detailed evaluation of the separate components and their interaction within the complex. Detailed information can be found in the supporting material.
The lDDT (37) is another metric that evaluates the quality of a protein model at the local residue level. Unlike RMSD, lDDT is a superposition-free metric, meaning it does not require alignment of the structures and is insensitive to domain movements. It measures the local conformational similarity of each residue’s environment by comparing inter-atomic distances. The lDDT score ranges from 0 to 1, with higher values indicating better model quality.
AlphaFold’s predicted lDDT (plDDT) scores offer a valuable confidence measure for the predicted positions of residues within a protein structure. These scores are instrumental for researchers to gauge the reliability of specific regions within the predicted structural model, particularly when experimental structures are absent for validation. In our study, we have enhanced the original AlphaFold’s confidence predictions, enabling a more precise estimation of structure reliability.
Evaluating plDDT against lDDT can be done through comparison with known structures or through experimental validation. High plDDT values in regions that align with high lDDT scores from experimental data can indicate a successful prediction. To measure the quality of predicted lDDT scores, researchers often use statistical methods such as mean absolute error and the Pearson correlation coefficient.
Similar to RMSD, we would like to focus specifically on the peptide part of the complex, which is why we computed the lDDT and plDDT scores separately for the peptide portions of the MHC-peptide complexes.
Fine-tuning techniques
To refine the prediction capabilities of AlphaFold for class I MHC-peptide complexes, we started with the foundational AlphaFold multimer model v2.2. We explored several refinement techniques, focusing on both architectural modifications and training strategies.
Architectural enhancements
Our initial approach involved augmenting the existing AlphaFold model by adding extra Evoformer blocks, ranging from 1 to 10, to the pre-existing 48. The design of AlphaFold, with its inherent residual connections, allows for such integrations without compromising existing functionalities, potentially enriching the model’s learning capacity.
Template usage optimization
Another significant adjustment pertained to the use of templates. While the standard AlphaFold architecture processes protein chains individually, we innovated by incorporating information regarding the interactions between protein chains and peptides. This modification is crucial for our focus on MHC-peptide interactions, aiming to capture the subtleties of these complex molecular interplays.
Focused loss function
To enhance the precision of structural prediction specifically for peptide residues within MHC-peptide complexes, our study incorporated a refined approach to the AlphaFold model’s loss function. Recognizing the intricate nature of AlphaFold’s loss function, which comprises multiple components each calculating loss on an individual or pairwise residue basis, we devised a strategy to recalibrate this mechanism to favor our targeted peptide structures.
This recalibration was achieved through the implementation of a differential weighting system within the loss function. By constructing a binary mask for peptide residues and designating weights to all residues in the complex, each MHC residue was assigned a baseline weight of 1, whereas peptide residues received a baseline weight incremented by a . The serves as a hyperparameter with evaluated values ranging from 0 (indicating no additional emphasis) to 3.0 (signifying that the penalty for inaccuracies in peptide residue predictions is quadrupled relative to MHC residues). The integration of this weighting scheme is important for modulating the loss function, where it facilitates the computation of a weighted average for the components constituting the final loss.
Optimizing training parameters
On the training front, we experimented with various hyperparameters to optimize the model’s performance. This included testing different learning-rate schedulers, such as CyclicLR, StepLR, and CosineAnnealingLR, with learning rates spanning from 0.001 to 0.000001. We also employed , serving as an analog to batch size, to manage the model’s learning process more effectively. This broad range of hyperparameter experimentation allowed us to finely calibrate the model for optimal performance in predicting MHC-peptide structures.
Results and discussion
Optimal parameter settings: MHC-Fine model
After extensive experimentation, we have identified an optimal set of parameters for our AlphaFold-based model, now termed MHC-Fine, tailored for predicting class I MHC-peptide complex structures. These parameters were determined as the most effective in balancing computational efficiency with predictive accuracy.
Additional Evoformer blocks
We treated the number of additional Evoformer blocks as a hyperparameter and found it had a positive correlation with the lDDT scores for peptides and was one of the most important parameters. This indicates that more blocks could improve the model’s accuracy in predicting peptide structures. However, through hyperparameter optimization, we determined that adding exactly two additional Evoformer blocks was the best choice. This process involved testing different combinations of model parameters to find the one that worked best for our dataset. The inclusion of two blocks achieves the optimal balance, potentially allowing for the capture of additional information about MHC-peptide interactions without unnecessarily complicating the model.
Template utilization
The introduction of high-quality templates emerged as a crucial factor. Although the model was capable of learning independently of templates, their inclusion significantly accelerated the training convergence, underscoring their value in our fine-tuning process.
Focused loss function
The optimal value for the weighting factor was determined to be 0.4 through hyperparameter optimization, indicating that the loss function for peptide residues was adjusted to 1.4 times that of MHC residues. This outcome was lower than initially anticipated. Further investigation revealed that similar levels of accuracy could be achieved without increasing the loss for peptide residues. Nonetheless, the implementation of a peptide-specific mask remained beneficial for refining the training process. It provided enhanced visibility into the model’s learning progress, particularly allowing for the precise monitoring of peptide loss improvement even after the overall loss for the MHC-peptide complex had stabilized. This approach underscores the utility of the peptide mask in facilitating nuanced control over the model’s training dynamics.
Optimizing training parameters
We employed the CosineAnnealingLR scheduler with a learning rate of 0.0003. This configuration facilitated a more dynamic adjustment of the learning rate, aiding in finer convergence. We set to 1, with the model being trained across eight GPUs. This effectively meant a real batch size of eight, allowing for a more precise gradient update, which proved beneficial at this stage of the model’s training. The MHC-Fine model, with these refined parameters, has shown the capability to outperform established benchmarks. Its single-model framework simplifies the prediction process while maintaining high accuracy, marking a significant step forward in computational immunology.
Benchmarking baseline performances
To assess the baseline performance of AlphaFold on our test set prior to fine-tuning, we utilized parameters of all five multimer models of AlphaFold version 2.2 using our script. For each sample in the test set, we systematically evaluated the predicted structures generated by each model. The best result for each sample was selected based on the highest plDDT scores.
To ensure a comprehensive and fair comparison, we similarly evaluated the performance of Pandora. We meticulously removed any instances where our test set overlapped with Pandora’s template database to avoid any potential bias from Pandora having prior knowledge of the structures in our test set. For each sample, the most probable structure predicted by Pandora was selected, using the molecular probability density function (molpdf) as the scoring function.
Employing this approach allowed us to establish a robust benchmark for the original performance of both AlphaFold and Pandora. This benchmark serves as a critical reference point against which we could measure the enhancements achieved through our fine-tuning process.
Comparative analysis
Our fine-tuning of AlphaFold led to a model with enhanced predictive performance for MHC-peptide complexes, showing a statistically significant reduction in median RMSD values compared to both the original AlphaFold and the homology-modeling-based Pandora approach. Fig. 1 illustrates the distribution of RMSD values for predicted MHC-peptide complex structures across three different computational methods: the original AlphaFold (median RMSD calculated for Cα atoms of peptides 1.59 Å), Pandora (1.31 Å), and our MHC-Fine model (0.66 Å). The MHC-Fine model shows not only lower median RMSD but also a significantly narrower interquartile range, indicating higher accuracy and consistency in structure prediction. This suggests a closer approximation to the high-resolution crystal structures in our test dataset and indicates a marked improvement in the spatial accuracy of our model’s predictions. Further metrics and information concerning peptide length are available in the supporting material.
Figure 1.

Comparative analysis of prediction accuracy for class I MHC-peptide complexes using RMSD calculated for Cα atoms of peptides: performance of AlphaFold, Pandora, and MHC-Fine.
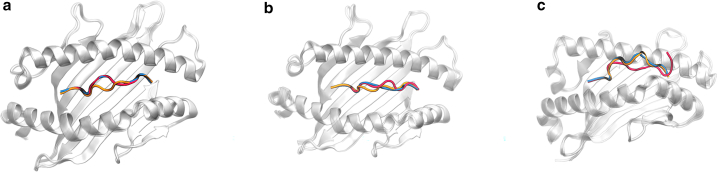
Our analysis is demonstrated through three case studies, each with varying prediction accuracy. Fig. 2 illustrates these differences: Fig. 2 a shows high accuracy with the fine-tuned AlphaFold, Fig. 2 b depicts moderate accuracy for both models, and Fig. 2 c highlights significant discrepancies in predictions.
Figure 2.
Comparative visualization of class I MHC-peptide complex prediction accuracy, reporting MHC-peptide RMSD calculated for Cα atoms. True peptide structure is shown in red, Pandora model in orange, and MHC-Fine in blue. (a) PDB: 6vb3: high precision of MHC-Fine. MHC-peptide RMSD for Cα atoms is 0.25 Å for MHC-Fine and 1.44 Å for Pandora. (b) PDB: 7n2o: moderate accuracy for both models. MHC-peptide RMSD for Cα atoms is 1.23 Å for MHC-Fine and 1.19 Å for Pandora. (c) PDB: 7mj7: significant deviations in predictions for both models. MHC-peptide RMSD for Cα atoms is 3.73 Å for MHC-Fine and 4.30 Å for Pandora. To see this figure in color, go online.
In addition, our fine-tuned model exhibited enhanced confidence in prediction. We evaluated the average plDDT values for peptide residues across all test samples, ensuring accurate and reliable lDDT score predictions. The mean absolute error between the true lDDT values and the predicted ones for peptide residues was 3.0, with a Pearson correlation coefficient of 0.62, showcasing good performance. Fig. 3 presents the distribution of the deviation in plDDT scores for peptide residues, revealing that the MHC-Fine model generates a significantly narrower error distribution compared to the original model. This reduced spread indicates more consistent predictions, suggesting that our fine-tuning process achieves a more dependable and precise measure of structural confidence with significantly fewer large deviations. Additionally, Fig. 4 illustrates that within our test dataset, a mean plDDT score for peptide residues above 90 is linked with an MHC-peptide RMSD of less than 1.5 Å, and scores above 95 correlate with RMSD of less than 1.0 Å. These findings highlight the refined accuracy of our model in structural prediction, particularly for peptide residues.
Figure 3.
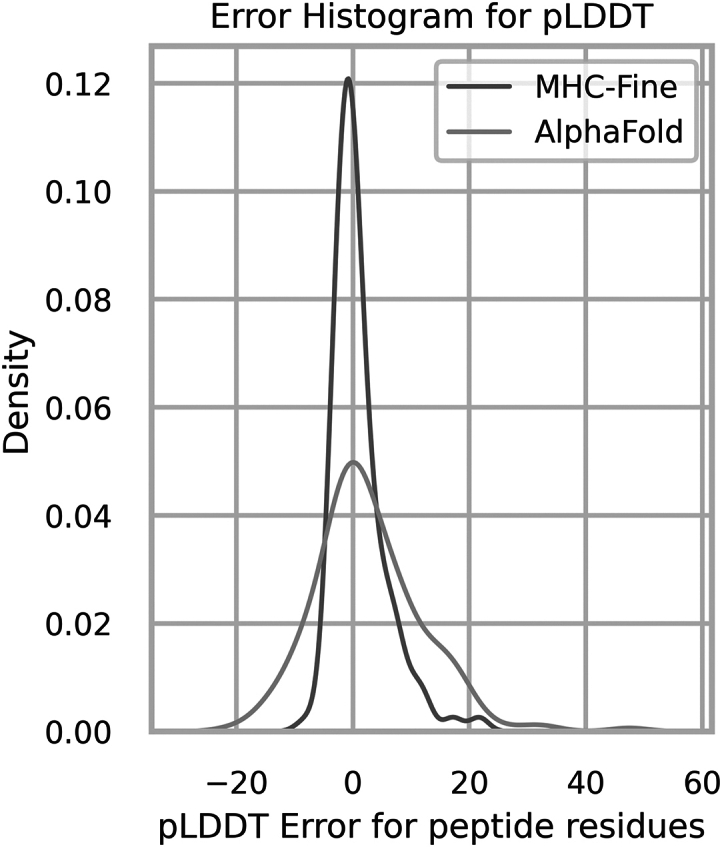
Error distribution for plDDT values of peptide residues. The red distribution represents the original AlphaFold model with a standard deviation of 9.2, whereas the blue distribution highlights our MHC-Fine model, which achieves a reduced standard deviation of 4.6, indicating a more precise confidence in structural predictions for peptide residues.
Figure 4.
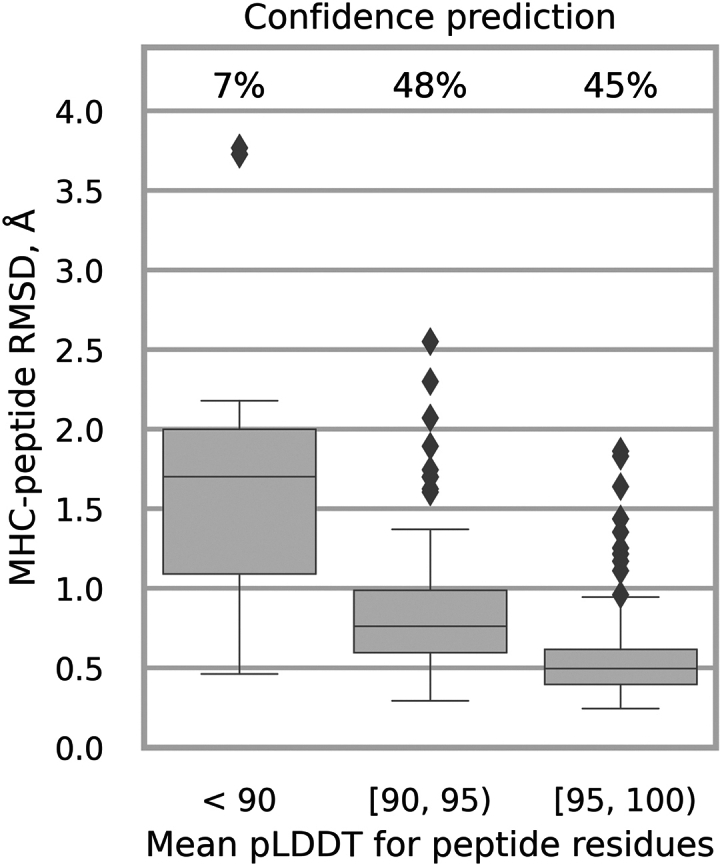
MHC-Fine confidence prediction. Samples are grouped by mean plDDT values for peptide residues. The chart displays the distribution of MHC-peptide RMSD alongside the percentage of samples within each specific plDDT range.
Conclusion
In conclusion, our study presents a refined AlphaFold model tailored for the intricate task of class I MHC-peptide complex structure prediction. By fine-tuning with high-resolution domain-specific data, we have achieved superior performance compared to both the original AlphaFold and traditional homology-modeling approaches. Our focused metrics on the peptide regions have yielded RMSD values indicative of high-precision predictions, while improvements in the plDDT scores reflect an enhanced confidence in the structural assessments provided by our model. These advancements hold promising implications for computational immunology, potentially expediting the discovery and design of novel therapeutics and vaccines.
Data and code availability
The inference code and datasets utilized in this study are publicly available to facilitate further research. To enhance accessibility and ease of use, we have developed a comprehensive Jupyter Notebook within Google Colaboratory. This interactive environment allows researchers to seamlessly execute the code and analyze the datasets featured in our study. The code, datasets, and the link to Notebook can be accessed via the following path: https://bitbucket.org/abc-group/mhc-fine/src/main/.
Author contributions
E.G., C.S., J.C.M., S.V., K.A.D., D.P., D.S., T.P., and D. Kozakov designed the research. E.G., D. Kalitin, D.S., Y.Z., T.N., and G.J. performed the research. E.G. and D.S. wrote the article. E.G., D. Kalitin, D.S., Y.Z., T.N., G.J., T.P., C.S., J.C.M., S.V., K.A.D., D.P., and D. Kozakov reviewed and edited the manuscript.
Acknowledgments
This work was supported in part by the National Institutes of Health grants RM1135136 and R01GM140098, and by National Science Foundation grants DMS-1664644 and DMS-2054251. This research used resources of the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science User Facility supported under contract DE-AC05-00OR22725.
Declaration of interests
The authors declare no competing interests.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used ChatGPT to check grammar and spelling and to improve readability and language. After using this service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Editor: Tamar Schlick.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2024.05.011.
Contributor Information
Dzmitry Padhorny, Email: dzmitry.padhorny@stonybrook.edu.
Dima Kozakov, Email: midas@laufercenter.org.
Supporting material
References
- 1.Neefjes J., Jongsma M.L.M., et al. Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Deng L., Mariuzza R.A. Recognition of self-peptide–MHC complexes by autoimmune T-cell receptors. Trends Biochem. Sci. 2007;32:500–508. doi: 10.1016/j.tibs.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosendahl Huber S., van Beek J., et al. van Baarle D. T cell responses to viral infections - opportunities for peptide vaccination. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott P.A., Hu Z., et al. Wu C.J. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson J., Barker D.J., et al. Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigo M.M., Antunes D.A., et al. Vieira G.F. Docktope: A web-based tool for automated PMHC-I modelling. Sci. Rep. 2015;5 doi: 10.1038/srep18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narzi D., Becker C.M., et al. Böckmann R.A. Dynamical Characterization of Two Differentially Disease Associated MHC Class I Proteins in Complex with Viral and Self-Peptides. J. Mol. Biol. 2012;415:429–442. doi: 10.1016/j.jmb.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Knapp B., Demharter S., et al. Deane C.M. Current status and future challenges in T-cell receptor/peptide/MHC molecular dynamics simulations. Briefings Bioinf. 2015;16:1035–1044. doi: 10.1093/bib/bbv005. [DOI] [PubMed] [Google Scholar]
- 9.Abella J.R., Antunes D.A., et al. Kavraki L.E. APE-Gen: A Fast Method for Generating Ensembles of Bound Peptide-MHC Conformations. Molecules. 2019;24:881. doi: 10.3390/molecules24050881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans R., O’Neill M., et al. Hassabis D. Protein complex prediction with AlphaFold-multimer. BioRxiv. 2021 doi: 10.1101/2021.10.04.463034. Preprint at. [DOI] [Google Scholar]
- 11.Mikhaylov V., Brambley C.A., et al. Levine A.J. Accurate modeling of peptide-MHC structures with AlphaFold. Structure. 2023;32:228–241. doi: 10.1016/j.str.2023.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley P. Structure-based prediction of T cell receptor:peptide-MHC interactions. Elife. 2023;12 doi: 10.7554/eLife.82813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzella D.F., Parizi F.M., et al. Xue L.C. PANDORA: A Fast, Anchor-Restrained Modelling Protocol for Peptide: MHC Complexes. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.878762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrisi M., Pollastri G., Le Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020;18:1301–1310. doi: 10.1016/j.csbj.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek M., DiMaio F., et al. Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury R., Bouatta N., et al. AlQuraishi M. Single-sequence protein structure prediction using a language model and deep learning. Nat. Biotechnol. 2022;40:1617–1623. doi: 10.1038/s41587-022-01432-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Z., Akin H., et al. Rives A. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023;379:1123–1130. doi: 10.1126/science.ade2574. [DOI] [PubMed] [Google Scholar]
- 18.Weissenow K., Heinzinger M., Rost B. Protein language-model embeddings for fast, accurate, and alignment-free protein structure prediction. Structure. 2022;30:1169–1177.e4. doi: 10.1016/j.str.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Madden D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 20.Momburg F., Roelse J., et al. Neefjes J.J. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J. Exp. Med. 1994;179:1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rist M.J., Theodossis A., et al. Burrows S.R. HLA Peptide Length Preferences Control CD8+ T Cell Responses. J. Immunol. 2013;191:561–571. doi: 10.4049/jimmunol.1300292. [DOI] [PubMed] [Google Scholar]
- 22.Tynan F.E., Burrows S.R., et al. Rossjohn J. T cell receptor recognition of a “super-bulged” major histocompatibility complex class I–bound peptide. Nat. Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 23.Li L., Peng X., et al. Bouvier M. Crystal structures of MHC class I complexes reveal the elusive intermediate conformations explored during peptide editing. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-40736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sali A. Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today. 1995;1:270–277. doi: 10.1016/s1357-4310(95)91170-7. [DOI] [PubMed] [Google Scholar]
- 25.Lensink M.F., Brysbaert G., et al. Wodak S.J. Impact of AlphaFold on structure prediction of protein complexes: The CASP15-CAPRI experiment. Proteins. 2023;91:1658–1683. doi: 10.1002/prot.26609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozden B., Kryshtafovych A., Karaca E. The impact of AI-based modeling on the accuracy of protein assembly prediction: Insights from CASP15. Proteins. 2023;91:1636–1657. doi: 10.1002/prot.26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Verburgt J., et al. Kihara D. Improved Peptide Docking with Privileged Knowledge Distillation using Deep Learning. bioRxiv. 2023 doi: 10.1101/2023.12.01.569671. Preprint at. [DOI] [Google Scholar]
- 28.Motmaen A., Dauparas J., et al. Bradley P. Peptide-binding specificity prediction using fine-tuned protein structure prediction networks. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2216697120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradbury J., Frostig R., et al. Zhang Q. JAX: composable transformations of Python+NumPy programs. 2018. http://github.com/google/jax
- 30.Ahdritz G., Bouatta N., et al. Alquraishi M. OpenFold: Retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization. bioRxiv. 2023 doi: 10.1101/2022.11.20.517210. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paszke A., Gross S., et al. Chintala S. Vol. 32. Curran Associates, Inc; 2019. PyTorch: an imperative style, high-performance deep learning library; pp. 8024–8035. (Advances in Neural Information Processing Systems). [Google Scholar]
- 32.Falcon W., The PyTorch Lightning team . 2019. PyTorch Lightning.https://github.com/Lightning-AI/lightning [Google Scholar]
- 33.Berman H.M., Westbrook J., et al. Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. http://www.rcsb.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 35.Mirdita M., Schütze K., et al. Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrödinger, L., and W. DeLano. PyMOL. http://www.pymol.org/pymol.
- 37.Mariani V., Biasini M., et al. Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The inference code and datasets utilized in this study are publicly available to facilitate further research. To enhance accessibility and ease of use, we have developed a comprehensive Jupyter Notebook within Google Colaboratory. This interactive environment allows researchers to seamlessly execute the code and analyze the datasets featured in our study. The code, datasets, and the link to Notebook can be accessed via the following path: https://bitbucket.org/abc-group/mhc-fine/src/main/.