Abstract
5‐Fluorouracil (5‐FU) has been used for chemotherapy for colorectal and other cancers for over 50 years. The prevailing view of its mechanism of action is inhibition of thymidine synthase leading to defects in DNA replication and repair. However, 5‐FU is also incorporated into RNA causing defects in RNA metabolism, inhibition of pseudouridine modification, and altered ribosome function. We examined the impact of 5‐FU on post‐transcriptional small RNA modifications (PTxMs) and the expression and export of RNA into small extracellular vesicles (sEVs). EVs are secreted by all cells and contain a variety of proteins and RNAs that can function in cell‐cell communication. We found that treatment of colorectal cancer (CRC) cells with 5‐FU represses sEV export of miRNA and snRNA‐derived RNAs, but promotes export of snoRNA‐derived RNAs. Strikingly, 5‐FU treatment significantly decreased the levels of pseudouridine on both cellular and sEV small RNA profiles. In contrast, 5‐FU exposure led to increased levels of cellular small RNAs containing a variety of methyl‐modified bases. These unexpected findings show that 5‐FU exposure leads to altered RNA expression, base modification, and aberrant trafficking and localization of small RNAs.
Keywords: 5‐FU, EV export, extracellular vesicles, miRNA, pseudourine, RNA modification
5‐FU treatment blocks miRNA export in small extracellular vesicles. Uracil residues (red) in normal cellular and small extracellular vesicle (sEV) RNA are frequently modified to pseudouridine (Ψ; blue) across multiple RNA subtypes. Exposure to 5‐FU inhibits pseudouridine formation and blocks miRNA export.
1. INTRODUCTION
Extracellular vesicles (EVs) are a diverse class of membrane‐bound particles that vary in size, cargo composition, biogenesis pathways, and delivery mechanisms (Couch et al., 2021; Dixson et al., 2023). The demonstration that EVs released from B cells could functionally activate T cells accelerated interest in the EV field, which was further increased by findings that EVs contain an array of protein and RNA cargo that can be transferred between donor and recipient cells (Raposo et al., 1996; Ratajczak et al., 2006; Skog et al., 2008; Thery et al., 2002; Valadi et al., 2007). It is now appreciated that all cells release EVs and that EVs constitute a unique form of cell‐cell communication, from near‐neighbour interactions to communication between distant cells and organs (Buzas, 2023; Couch et al., 2021; Dixson et al., 2023; Maas et al., 2017; van Niel et al., 2018; Wortzel et al., 2019).
Heterogeneous populations of EVs are released from cells with current work focused on precise characterization of different classes of vesicles based on size and biogenesis pathways (Abels & Breakefield, 2016; Colombo et al., 2014; Jeppesen et al., 2023; Thery et al., 2018). As it is difficult to identify a specific biogenesis pathway from standard EV preparations, we will use the term small EVs (sEV) to describe our vesicle preparations. Non‐vesicular particles and lipoproteins are also released from cells that can transfer protein and RNA cargo between cells (Allen et al., 2022; Jeppesen et al., 2019; Vickers et al., 2011; Zhang et al., 2018, 2019, 2021). Here, we report our investigation into the role(s) of PTxMs on small RNA selection and sEV export in response to 5‐FU treatments of CRC cells.
Protein, RNA and lipid cargo associated with sEVs varies in a cell‐context dependent manner (Dixson et al., 2023; Maas et al., 2017; Tkach & Thery, 2016). Almost all known classes of coding and noncoding RNA transcripts (mRNA, rRNA, tRNA, miRNA, snRNA, snoRNA, Y RNAs, ncRNAs) have been detected in sEVs (Cha et al., 2015; Dellar et al., 2022; Fritz et al., 2016; Nolte‐’t Hoen et al., 2012; Turchinovich et al., 2019; Veziroglu & Mias, 2020). The majority of RNAs within sEVs correspond to small RNAs and fragments of larger transcripts (<200 nt) (Mateescu et al., 2017; Turchinovich et al., 2019). Among small RNAs, miRNAs constitute one of the best characterized RNA components in sEVs because they exist as full length RNAs within sEVs and because their transfer between cells can be assayed using standard reporter assays (Cha et al., 2015). Numerous studies have proposed that miRNA export into sEVs is not a random sampling of cellular miRNAs, but rather a regulated pathway of selection, sorting, and export, thus providing a new layer of gene regulation (Cha et al., 2015; McKenzie et al., 2016; Santangelo et al., 2016; Shurtleff et al., 2016, 2017; Squadrito et al., 2014; Villarroya‐Beltri et al., 2013). Regulatory hypotheses have proposed that specific sequence motifs promote selective export due to recognition by candidate RNA binding proteins, but no universal targeting sequence or binding protein has thus far been identified (Bolukbasi et al., 2012; Garcia‐Martin et al., 2022; Santangelo et al., 2016; Shurtleff et al., 2016; Temoche‐Diaz et al., 2019; Villarroya‐Beltri et al., 2013).
Over 300 hundred post‐transcriptional base modifications have been identified with accompanying writers, readers, and erasers for some of these modifications (Roundtree et al., 2017; Schaefer et al., 2017; Zaccara et al., 2019). We previously found that knockdown of Mettl3, a writer of N6‐methyladenosine (m6A) modification, reduced sEV levels of a subset of miRNAs that contain consensus sequences for m6A leading us to propose that PTxMs serve as part of small RNA export signals (Abner et al., 2021). Here, we identify additional PTxMs on small RNAs in CRC cells and their exported sEVs, with pseudouridine being the most abundant PTxM. To determine the effect of altered pseudouridine modification on sEV small RNA export, we treated CRC cells with 5‐FU which is incorporated into RNA and inhibits pseudouridine modification (Gu et al., 1999; Karijolich et al., 2010; Kufe & Major, 1981; Liang et al., 2022; Noordhuis et al., 2004; Samuelsson, 1991; Zhao & Yu, 2007). 5‐FU has long been used in cancer treatment with the presumed mechanism of action being inhibition of thymidine synthesis to block DNA replication (Longley et al., 2003). However, recent work has shown that the effects of 5‐FU are complex, with effects on a variety of RNAs, some of which lead to altered ribosome function and translational recoding (Chalabi‐Dchar et al., 2021; Ge et al., 2017; Mojardin et al., 2013; Therizols et al., 2022). We found that 5‐FU treatment dramatically decreases the levels of pseudouridine in RNA isolated from both cells and sEVs. Remarkably, 5‐FU treatment causes decreased export of miRNAs and spliceosomal snRNA‐derived RNAs, but has the opposite effect on snoRNA‐derived RNA export. We also discovered that base methylation of cellular RNA is unexpectedly increased after 5‐FU exposure. Together, our results show that 5‐FU treatment alters cellular gene expression patterns, causing massive changes in PTxM status on cellular small RNAs, decreased sEV export of miRNAs and snRNAs, and increased export snoRNA‐derived fragments.
2. MATERIALS AND METHODS
2.1. Cell culture
DLD‐1 cells are a CRC line isolated from a patient tumour (Tibbetts et al., 1977; Shirasawa et al., 1993). We validated that our line is a heterozygous KRAS mutant (G13D) line by PCR amplification of genomic DNA and DNA sequencing.
2.2. Extracellular vesicle isolation
Cells were seeded at 6.5 × 106 in T175 flasks and cultured in DMEM medium with 10% foetal bovine serum (FBS), 2 mM l‐glutamine, 1× MEM non‐essential amino acids, 100 U/mL of penicillin, and 100 U/mL of streptomycin. When the cells reached 70% confluence, they were washed three times with phosphate‐buffered saline (PBS) and cultured in a serum‐free medium with or without 10 µM 5‐FU for 48 hrs to avoid small RNA contamination from FBS. Media from at least three T175 flasks were pooled for sEV isolation as in described (Abner et al., 2021; Hinger et al., 2020). Briefly, conditioned media was centrifuged at 1000 × g for 10 min at room temperature. Supernatants were transferred to a new centrifuge tube and spun at 2000 × g for 20 min at 4°C after which the supernatant was again transferred to a new centrifuge tube and spun at 10,000 × g for 30 min at 4°C. The supernatants were collected, transferred to clean ultracentrifuge tubes and spun at 167,000 × g for 17 h at 4°C. The resulting pellets were washed twice by resuspension 1 mL sterile Dulbecco's PBS and pelleting at 167,000 × g for 70 min at 4°C. After the final wash, pellets were resuspended in 50 µL of sterile 1× DPBS.
2.3. RNA isolation
Cells were collected by scraping, washed in PBS, and resuspended in PBS prior to isolation of total RNA using the Quick‐RNA Miniprep kit (Zymo Research). Total RNA from pelleted sEVs in PBS was isolated using the same kit.
2.4. Small RNA sequencing (small RNA‐seq)
RNA libraries were generated using the NEXTFlex Small RNA Library Preparation Kits v3 (Perkin) with the following modifications: (1) 3′‐ and 5′‐adaptors were diluted 1:8, (2) 3′‐adaptor ligations were performed overnight in multiple steps −25°C for 2 h, 20°C for 4 h and 16°C overnight, (3) following cDNA synthesis and before barcoding PCR, step F protocol was followed, and (4) PCR amplification was 20 cycles. Following PCR amplification, individual libraries were size‐selected (136–200 bp product) using Pippin Prep (Sage Sciences). Size‐selected libraries were quantified using Qubit Fluorometer. Libraries were checked for quality and sequenced using Illumina short‐read technology. Libraries were pooled (equimolar multiplexed libraries) and sequenced (PE150) using the NovaSeq6000 (Illumina) platform at the VANTAGE Core (Vanderbilt University). Demultiplexing and bioinformatic analyses was performed using the TIGER pipeline (Allen et al., 2018). Briefly, Cutadapt (v1.16) was used to trim 3′ adaptors and all reads with <16 nucleotides (nts) were removed (Martin, 2011). Quality control on both raw reads and adaptor‐trimmed reads was performed using FastQC (v0.11.9)(www.bioinformatics.babraham.ac.uk/projects/fastqc). The adaptor‐trimmed reads were mapped to the hg19 genome, with additional rRNA and tRNA reference sequences, by Bowtie1 (v1.1.2) allowing only one mismatch (Langmead & Salzberg, 2012).
2.5. Total RNA sequencing
Bulk RNA sequencing libraries were prepared using Universal RNAseq kits (Tecan), as per manufacturer's instructions. Libraries were cleaned (Zymo), checked for quality using the Agilent bioanalyzer, quantified (Qubit), and pooled based on equimolar concentrations of libraries. Pooled libraries were sequenced using Illumina short‐read technology based on PE150 on the NovaSeq6000 (Illumina). After sequencing, samples (libraries) were demultiplexed and analysed. Briefly, reads were trimmed to remove adapter sequences using Cutadapt v2.10)(Martin, 2011) and aligned to the Gencode GRCh38.p13 genome using STAR (v2.7.8a)(Dobin et al., 2013). Gencode v38 gene annotations were provided to STAR to improve the accuracy of mapping. Quality control on both raw reads and adaptor‐trimmed reads was performed using FastQC (v0.11.9). FeatureCounts (v2.0.2)(Liao et al., 2014) was used to count the number of mapped reads to each gene. Heatmap3 (Zhao et al., 2014) was used for cluster analysis and visualization. Significant differential expressed genes with absolute fold change >2 or <0.5, and adjusted p value (padj) < 0.05 were determined using DESeq2 (v1.30.1) (Love et al., 2014). Genome Ontology and KEGG pathway over‐representation analyses were performed on differentially expressed genes using the WebGestaltR package (NULL) (Wang et al., 2017). Gene set enrichment analysis (GSEA) was performed using GSEA package (v4.3.2) (Subramanian et al., 2005) on database v2022.1.Hs.
2.6. Mass spectrometry
Liquid chromatography tandem mass spectrometry (LC–MS/MS) was used to quantify PTxMs based on standard curves. Main nucleosides–adenosine (A), cytidine (C), guanosine (G) and uridine (U)–were obtained from Sigma‐Aldrich. 1‐methyladenosine (m1A), 2‐methyladenosine (m2A), 6‐methyladenosine (m6A), 8‐methyladenosine (m8A), 3‐methylcytidine (m3C), pseudouridine, 5‐methylcytidine (m5C), 5‐methyluridine (m5U), 1‐methylguanosine (m1G), 2‐methylguanosine (m2G), 2‐dimethylguanosine (m2 2G), 7‐methylguanosine (m7G) were purchased from Carbosynth. [15N]5‐2‐deoxyadenosine was obtained from Cambridge Isotope Laboratories. Cellular and sEV RNAs were enzymatically digested to single nucleosides under neutral conditions using the Nucleoside Digestion Mix (BioLabs). RNA nucleosides were combined with 1 nM of [15N]‐dA internal standard immediately prior to analysis. Digested nucleosides were separated using a Hypersil GOLD aQ C18 column (100‐mm length × 2.1‐mm inner diameter, pore size 175 Å, particle size 1.9 µm)(ThermoFisher) at a flow rate of 0.4 mL/min using 0.1% (v/v) formic acid in water (solvent A) and acetonitrile with 0.1% (v/v) formic acid (solvent B). The gradient profile applied to each sample was as follows: 0–6 min, 0% B; 6−7.5 min, 1% B; 7.5‐9.5 min, 6% B; 9.5–10.5 min, 10.5–12 min, 50% B; 12–14 min, 75% B; 14–16 min, 75% B. Columns were equilibrated prior to subsequent injections (10 µL) and maintained at 40°C. MS analysis was performed on a Shimadzu Nexera system in‐line with a QTRAP 6500 with an electrospray ionization source (Applied Biosystems). Multiple reaction monitoring (MRM) in positive ion mode was used to survey known modifications. Data acquisition and sample processing were performed using AB SCIEX Analyst Software 1.6.2 (Applied Biosystems). Nucleosides were quantified by converting peak area under the curve to nmol using standard curves of candidate nucleosides. To compare modification levels across conditions, modified nucleoside values were normalized to total nucleosides analysed.
2.7. Statistics
To compare between two groups, two‐tailed unpaired t‐tests were used with p < 0.05 considered significant. The Wilcox‐rank sum test was used to compare U percentages between miRNAs with significant interaction effect and those without.
3. RESULTS
To determine the effects of 5‐FU treatment on gene expression and small RNA trafficking, we exposed the patient‐derived CRC cell line DLD‐1 (Shirasawa et al., 1993; Tibbetts et al., 1977) to 10µM 5‐FU (Ge et al., 2017) for 48 h in serum‐free media to avoid sEV contamination. While serum depletion can affect growth, both control and 5‐FU treated cells were deprived of serum for 48 h so that differential gene expression could be directly compared. We chose 10µM of 5‐FU based on previous experiments examining the effect of 5‐FU on translational recoding of mRNA in the CRC cell line SW‐480 (Ge et al., 2017). Like SW‐480 cells, DLD‐1 cells are relatively resistant to high concentrations of 5‐FU and remained healthy after exposure to 10µΜ 5‐FU for 48 h (Bracht et al., 2010; Ge et al., 2017). Total RNA was purified from both cell lysates and sEVs and RNA sequencing was performed on 5‐FU treated cells and controls.
3.1. 5‐FU induces upregulation of nucleoside and RNA metabolism genes
We first analysed bulk total RNAseq data from DLD‐1 cells in the presence or absence of 5‐FU. 5‐FU treatment caused significant (adjusted p < 0.05) differential (absolute fold change > 2.0) expression of 807 transcripts with 365 up‐ and 442 down‐regulated RNAs when comparing treated to untreated cells (Figure 1a, b, Table S1).
FIGURE 1.
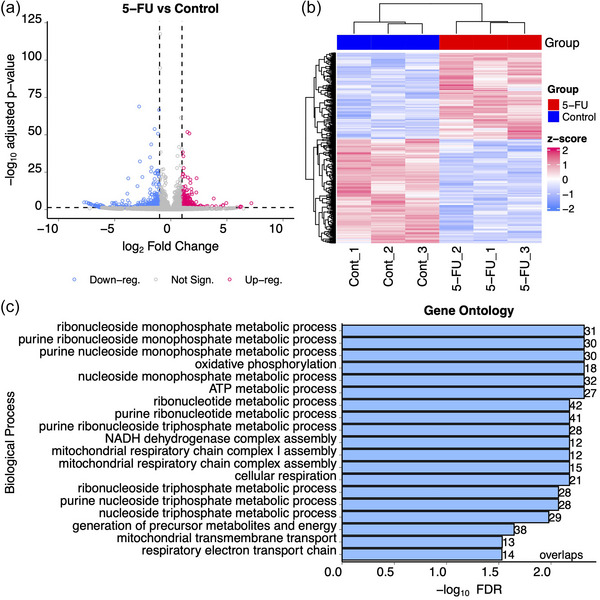
Differential gene expression in DLD‐1 cells after exposure to 5‐FU. DLD‐1 cells were exposed or not to 10 uM 5‐FU for 48 h in the absence of serum after which RNA was isolated and RNAseq was performed on transcripts > 200 nt. (a) Volcano plot showing up‐ and down‐regulated genes after treatment with 5‐FU. Blue dots show downregulated genes and red dots show upregulated genes with at least 2‐fold changes in expression and adjusted p values < 0.05. Grey dots represent transcripts whose expression did not significantly change after 5‐FU treatment. (b) Hierarchical clustering analysis was performed on triplicate RNAseq samples prepared from DLD‐1 cells treated (red) or not (blue) with 5‐FU. The heat map shows up‐ (red) and down‐ (blue) regulated genes. (c) Gene ontology analysis was performed using the WebGestaltR package to identify overlaps between gene annotation sets after exposure to 5‐FU (Wang et al., 2017).
To identify specific pathways and processes (Gene Ontology) that were affected by 5‐FU treatment, over‐representation analyses were performed using WebGestalt with DESeq2 pair‐wise comparisons. This approach showed significant enrichment of nucleoside metabolism pathways (Figure 1c). Also, 136 RNA metabolism pathway genes (GO:0016070) were found to be significantly increased (FDR = 1.5 × 10−5) by 5‐FU treatment (Table S1). These include several PTxM enzymes: tRNA methyltransferase 61 (TRM61, FC = 2.17, p = 0.004), dihydrouridine synthase 1 like 1 (DUSL1, FC = 2.25, p = 0.0000073), c16orf42 ribosome maturation factor (TSR3M, FC = 2.17, p = 0.011), and pseudouridine synthase 7 (PUS7, FC = 2.15, p = 0.0023). These observations are consistent with previous reports of thymidine synthase inhibition and DNA replication defects by 5‐FU, but are also consistent with RNA‐based defects (Chalabi‐Dchar et al., 2021; Ge et al., 2017; Liang et al., 2022; Mojardin et al., 2013; Therizols et al., 2022).
3.2. Effects of 5‐FU on expression and export of small RNAs
To investigate the impact of 5‐FU on small RNA expression and export in sEVs, high‐throughput small RNA‐seq was completed on RNA isolated from DLD‐1 cells and sEVs in control and 5‐FU treated conditions (Table S2). Cellular and sEV small RNAs were quantified using short‐read Illumina sequencing with degenerate base adapters for library generation. The data met quality control metrics as analysed using the TIGER small RNA pipeline (Allen et al., 2018) (Figures 2a, S1A, B). The most abundant class of host small RNAs detected in cells were miRNAs, followed by small RNAs derived from parental rRNA (rDRs) (Figure 2a). In contrast, the most abundant host small RNA classes in sEVs were tRNA‐derived RNAs (tDRs) and Y RNA‐derived RNAs (yDRs) (Figure 2a). Hierarchical clustering and correlation analyses showed that the cellular and sEV profiles were distinct with no association between the two profiles (Figure 2b).
FIGURE 2.
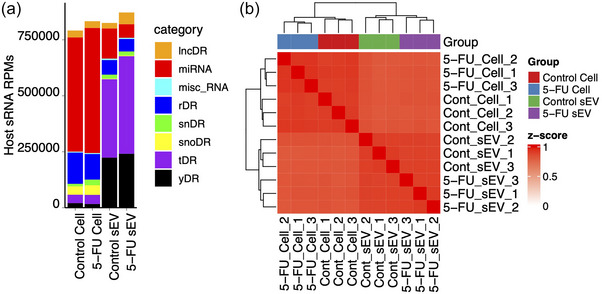
Small RNAseq of DLD‐1 cells and sEVs treated with 5‐FU. Small RNAseq was performed on RNA (<200 nt) isolated from DLD‐1 cells and sEVs treated or not with 5‐FU. (a) Small RNA read totals normalized to reads per million (RPM) from the indicated RNA subclasses after RNAseq on cellular and sEV RNA isolated in the presence or absence of 5‐FU. RNA subclasses include long noncoding RNA‐derived RNAs (lncDRs), miRNAs, miscellaneous small RNAs, rRNA‐derived RNAs (rDRs), spliceosomal snRNA‐derived RNAs (snDRs), snoRNA‐derived RNAs (snoDRs), tRNA‐derived RNAs (tDRs) and Y RNA‐derived RNAs (yDRs). (b) Distinct clustering of miRNA expression patterns across triplicate RNAseq samples in cells and sEVs after treatment with 5‐FU.
3.3. Minimal effects of 5‐FU on tDR, rDR, and YDR expression and sEV export
The functional roles of PTxMs on small RNAs are only beginning to emerge; however, this new level of gene regulation holds great potential for cellular small RNA selection and trafficking for degradation or export. Due to inhibition of pseudouridine modification by incorporation of 5‐FU (Karijolich et al., 2010), we expected differential cellular and sEV expression profiles for highly modified rDRs and tDRs (Borchardt et al., 2020; Taoka et al., 2018). However, when we examined tDRs in cells treated or not with 5‐FU, we found very few differentially expressed tDRs with only 4 up‐regulated and none down‐regulated (padj < 0.05 and fold‐change > 1.5 or <0.67) (Figure S2, Table S2). When we examined tDRs in sEVs treated or not with 5‐FU, we detected 8 up‐ and 14 down‐regulated tDRs (padj < 0.05 and fold‐change > 1.5 or <0.67) (Figure S2, Table S2). For rDRs in cells treated with 5‐FU, we failed to identify up‐regulation of rDRs and only found 2 rDRs to be down‐regulated in cells (Figure S2, Table S2). For rDRs from sEVs purified from cells either treated or not with 5‐FU, we did not detect any up‐regulated rDRs and found only four to be down‐regulated in sEVs (padj < 0.05 and fold‐change >1.5 or <0.67) (Figure S2, Table S2). For YDRs in cells treated with 5‐FU, 3 YDRs were up‐regulated and 1 was down‐regulated (padj < 0.05 and fold‐change >1.5 or <0.67) (Figure S2, Table S2). In sEVs purified from cells either treated or not with 5‐FU, 7 YDRs were up‐regulated and two were down‐regulated (padj < 0.05 and fold‐change >1.5 or <0.67) (Figure S2, Table S2). Overall, the effects of 5‐FU on these classes of RNA were minimal.
3.4. 5‐FU represses miRNA and snDR export and promotes snoDR export to sEVs
In contrast to modest effects on tDRs, rDRs and YDRs, we observed significant changes in expression and sEV localization for miRNAs, snRNA‐derived RNAs (snDRs), and snoRNA‐derived RNAs (snoDRs) (Figure 3, Table S2). At the cellular level, small subsets of miRNAs were altered (Figure 3a), with 22 up‐regulated miRNAs and 13 down‐regulated miRNAs after 5‐FU treatment (padj < 0.05 and fold‐change >1.5 or <0.67) (Table S2). In contrast, we observed significant down‐regulation of miRNA reads in sEVs in the presence of 5‐FU (Figure 3a), with 276 down‐ and only six up‐regulated miRNAs (padj < 0.05 and fold‐change > 1.5 or < 0.67) (Table S2). Combining the cellular and sEV data, treatment with 5‐FU caused a massive reduction of miRNA reads in sEVs, suggesting an export defect. To determine if any of the observed decreases in miRNA expression in sEVs were simply a result of decreased expression in cells exposed to 5‐FU, we determined interaction effects between the datasets. We found that 248 miRNAs were significantly downregulated in sEVs but did not show significant downregulation in cells (Table S3, columns E and H, lines 3–280), 13 miRNAs showed significant downregulation in cellular expression levels but not in sEVs (Table S3, columns E and H, lines 281–293), and 15 miRNAs were downregulated in both cells and sEVs (Table S3, columns E and H, lines 294–308). The fact that the great majority of miRNAs showed decreased levels in sEVs without corresponding decreases in cellular expression implies that treatment with 5‐FU causes a trafficking defect.
FIGURE 3.
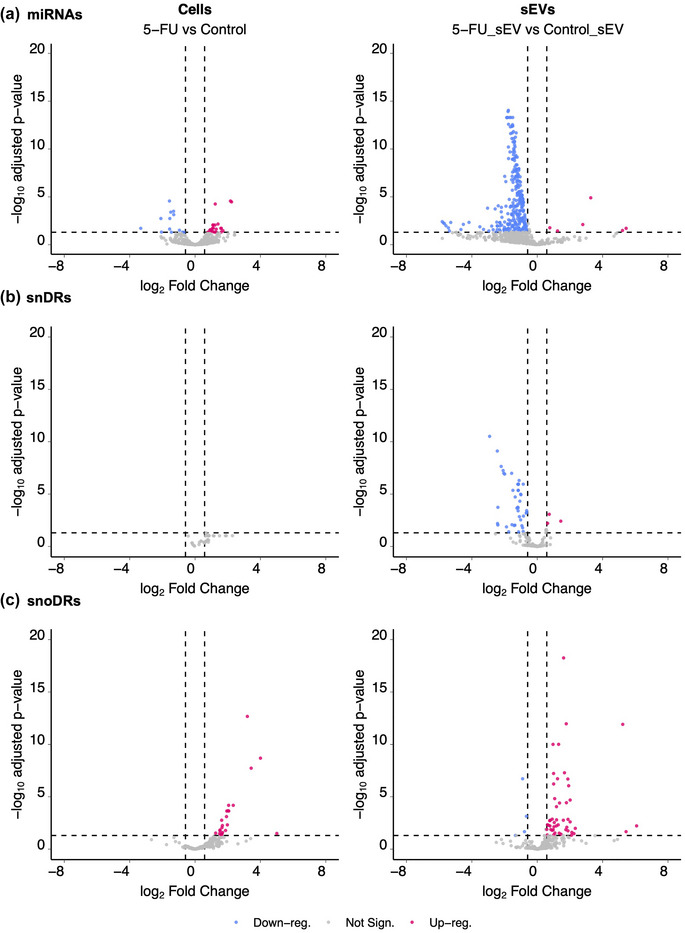
Differentially expressed small RNAs comparing DLD‐1 cells and sEVs grown in the presence or absence of 5‐FU. Volcano plots showing up‐ and down‐regulated miRNAs (a), snDRs (b) and snoDRs (c) in cells and sEVs treated or not with 5‐FU. Blue (downregulated) and red (upregulated) dots represent individual RNAs with significant changes in gene expression after treatment with 5‐FU (padj < 0.05 and fold‐change >1.5 or < 0.67).
For snRNAs, we found that no snDRs were differentially expressed in cells after treatment with 5‐FU; however, in sEVs, 43 snDRs were downregulated and 3 snDRs were upregulated (Figure 3b; Table S2). This largely mirrors the miRNA data, with significant down‐regulation of reads in sEVs suggesting an export defect for snDRs in the presence of 5‐FU.
For snoRNAs, 23 snoDRs were up‐regulated and none were downregulated in cells after treatment with 5‐FU. In sEVs, 50 snoDRs were upregulated and 4 were downregulated (Figure 3c; Table S2). Compared to the effects of 5‐FU on RNA export for miRNAs and snRNAs, the opposite was observed for snoRNAs with mostly upregulation observed in sEVs.
3.5. Differential effects of 5‐FU on uridine‐containing small RNA export
We next examined RNA export and uridine content focusing on the dramatic changes in miRNA export into sEVs in the presence of 5‐FU. For this, we re‐examined differential miRNA expression after 5‐FU treatment to identify miRNAs that show contrasting coordinate regulation between cells and sEVs meaning either down‐regulation in cells and up‐regulation in sEVs or up‐regulation in cells and down‐regulation in sEVs. We found six miRNAs that were co‐ordinately up‐regulated in cells and down‐regulated in sEVs (Figure 4a). Interestingly, when we examined the base composition of these miRNAs, all 6 miRNAs contained at least seven uridine residues across the short mature miRNA sequence. Two of these (miR‐340‐5p and miR‐92a‐1‐5p) not only have increased uridine content in the mature sequence, but also contain uridine‐rich precursor loops. Given that 5‐FU inhbits pseudouridine formation, these data suggest that pseudouridine modification could be important for miRNA export since the most affected miRNAs contain multiple uridines that could be modified. While we currently lack the tools to determine which specific bases within a given miRNA might be modified, we performed a Wilcoxon Rank Sum test to compare the overall percentage of U content when comparing miRNAs that are either retained in cells or exported to sEVs in the presence of 5‐FU. We found a statistically significant increase in the percentage of uridines in retained versus exported miRNAs (Figure 4b). This supports the hypothesis that pseudouridine modification is important for RNA export.
FIGURE 4.
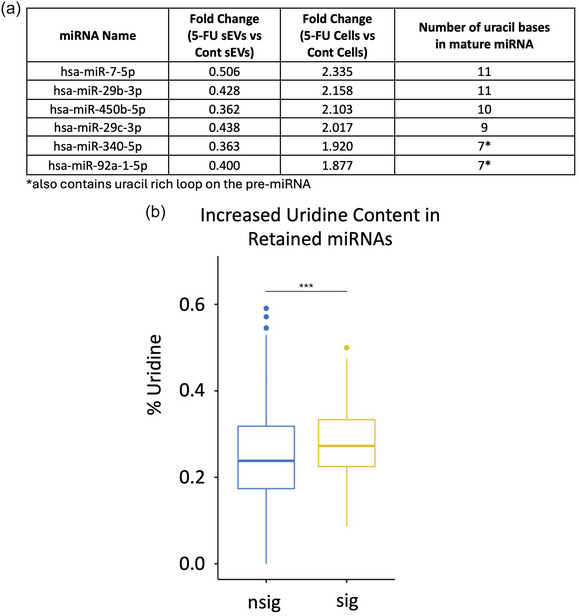
Increased uridine content in cellular‐retained miRNAs after 5‐FU treatment. (a) miRNAs that display coordinate up‐regulation in DLD‐1 cells and down‐regulation in sEVs after treatment with 5‐FU are shown with the number of uridine residues in each mature miRNA. miR‐340 and miR‐92a also contain uridine rich precursor loop sequences. (b) Overall enrichment of uridine residues in miRNAs retained in cells after 5‐FU treatment. The percentage of U residues in cellular retained miRNAs was compared between those that were significantly enriched (sig) or not (nsig) in cells after 5‐FU exposure using the Wilcoxon rank sum test. A significant increase (p < 0.001) in uridine content was detected in cellular‐retained RNAs with the average and standard deviation as indicated.
3.6. 5‐FU treatment reduces pseudouridine levels in cells and sEVs
To determine if 5‐FU treatment alters the levels of pseudouridine or other PTxMs, LC‐MS/MS approaches were used to quantify different PTxM levels in total RNA from cells and sEVs. This allowed precise quantitation of the levels of the four main nucleosides and 14 PTxMs based on internal and external standards. In DLD‐1 cells, 5‐FU treatment was found to slightly increase uridine (U) levels in cells, but did not affect the other three nucleosides in sEVs when normalized to total nucleoside levels (Figure 5a,b). For uridine modification, pseudouridine detection was significantly decreased in RNA isolated from both cells and sEVs after 5‐FU treatment (Figure 5c). Conversely, dihydrouridine (DHU) levels were found to be significantly increased in 5‐FU treated DLD‐1 cells with no observable changes in sEVs (Figure 5d). Also, 3‐methyluridine (m3U) and 5‐methyluridine (m5U) levels were not altered in 5‐FU‐treated cells or sEVs (Figure S3A,B). For cytosine modification, the cellular levels of 5‐methylcytosine (m5C) and 3‐methylcytosine (m3C) were found to be significantly increased in 5‐FU treated cells without changes in sEVs (Figure 5e,f). For adenosine modification, N1‐methyladenosine (m1A) levels were significantly increased in 5‐FU treated cells, however, N6‐methyladenosine (m6A) levels were not affected by 5‐FU treatment (Figures 5g and S3C). For inosine (deaminated adenosine) modification, 1‐methylinosine (m1I) levels were significantly increased with 5‐FU treatment, but m1I levels were undetectable in sEVs (Figure 5h). Inosine (I) levels were detected in both cells and sEVs, however, the levels of I were not affected by 5‐FU treatment (Figure S3D). For guanosine modification, 1‐methylguanosine (m1G), 7‐methylguanosine (m7G), 2‐methylguanosine (m2G), N2, N2‐dimethylguanosine (m2 2G), and N2, 7‐dimethylguanosine (m2 7G) levels were all significantly increased after treatment with 5‐FU (Figures 5i–l, S3E). With the exception of m2 7G, these guanosine PTxMs were also detected in sEVs, however, their levels were not altered by 5‐FU (Figures 5i–l, S3E). Together, the LC‐MS/MS data show that while post‐transcriptional methylation of nucleosides is significantly increased in cells treated with 5‐FU, pseudouridine levels are significantly decreased in both treated cells and sEVs.
FIGURE 5.
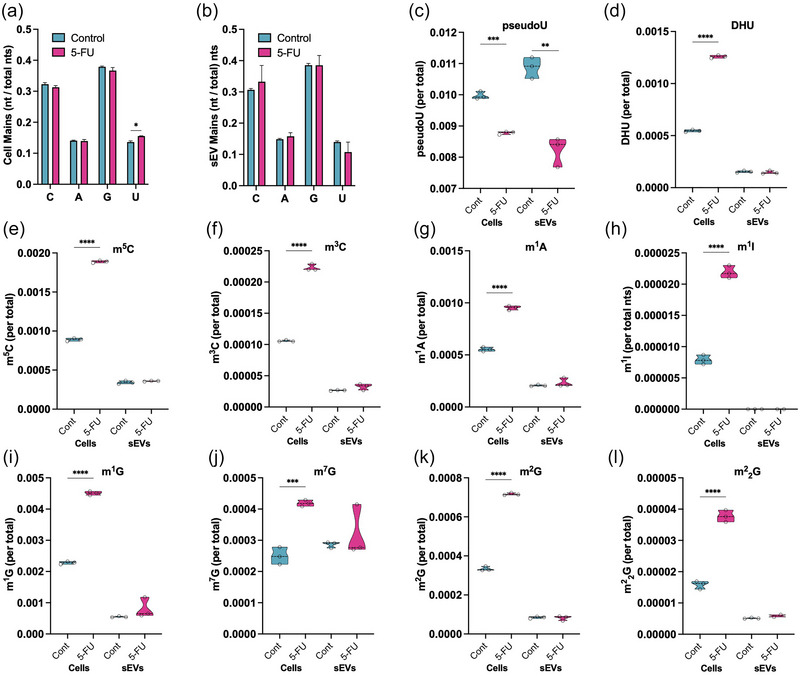
Reduced pseudouridine modification in cells and sEVs after 5‐FU treatment. LC–MS/MS approaches were used to quantify RNA base modifications in RNA from cells and sEVs. (a) Nucleoside content in 5‐FU treated DLD‐1 cells showed a modest but significant increase (p < 0.05) in uridine content when compared to untreated cells. No changes were observed for the other RNA nucleosides. (b) No changes in nucleoside levels were detected in sEVs purified from cells treated or not with 5‐FU. (c) Pseudouridine levels were significantly decreased in both cells and sEVs after 5‐FU treatment. (d−l) Quantitation of the effect of the indicated base modifications in cells and sEVs treated or not with 5‐FU. Base modifications include dihydrouridine (DHU), methylated cytosine (m5C and m3C), methylated adenosine (m1A), methylated inosine (m1I), and methylated guanosine (m1G, m7G, m2G) and demethylated guanosine (m2 2G). To compare modification levels across conditions, modified nucleoside values were normalized to the total levels for each nucleoside. For all graphs, the error bars indicate average and standard deviation.
4. DISCUSSION
Here, we report the first analysis of RNA profiles in sEVs from CRC cells treated with 5‐FU. Historically, the mechanism of action of 5‐FU has been viewed as primarily affecting DNA synthesis due to misincorporation and inhibition of thymidine synthase (Wyatt & Wilson, 2009). However, 5‐FU incorporation also affects RNA metabolism leading to inhibition of processing of multiple classes of RNA and effects on ribosomes causing altered translational efficiency and recoding of specific mRNAs (Chalabi‐Dchar et al., 2021; Ge et al., 2017; Mojardin et al., 2013; Therizols et al., 2022). We find that treatment of cells with 5‐FU leads to altered cellular and sEV RNA profiles, including miRNAs, snRDs, and snoRDs. The data support that these effects are driven by 5‐FU inhibition of pseudouridine modification because we detected significantly decreased levels of pseudouridine in purified sEV RNA, and because we observed enriched levels of uridine in RNAs that are differentially affected when comparing cells treated with 5‐FU to untreated cells. Surprisingly, we did not observe significant differential effects on rDRs or tDRs, despite the fact that these RNAs are known to contain abundant levels of pseudouridine (Borchardt et al., 2020). Thus, the overall effects of 5‐FU are pleiotropic and RNA‐subclass dependent. Focusing on miRNAs, significant decreased detection in sEVs may provide a potential biomarker for monitoring the effectiveness of 5‐FU treatment during cancer chemotherapy, consistent with a previous report that showed decreased levels of five miRNAs in plasma from patients treated with 5‐FU (Badr et al., 2023). We used RNAseq for an unbiased analysis of miRNA expression and trafficking and chose to examine relatively crude sEV preparations that encompass a heterogenous population of sEVs so as to not limit our analysis and also so that we could directly compare our data with previous small RNAseq data from DLD‐1 cells (Cha et al., 2015). For potential biomarker development, it may be preferable to analyze pseudouridine levels from a mix of sEVs, but future work may define specific sEV subclasses that are more affected by 5‐FU treatment. It also remains to be determined if the effects of 5‐FU are cell‐context dependent, similar to miRNA export being KRAS‐dependent, and because the sensitivity of colorectal cancer lines to 5‐FU is highly variable (Bracht et al., 2010; Cha et al., 2015). We are in the process of testing additional CRC lines and generating 5‐FU resistant cells.
Based on the abundance of pseudouridine modification, we expected to observe the greatest effect of 5‐FU treatment on rRNA and tRNA. To our surprise, the effects of 5‐FU exposure in DLD‐1 cells were most dramatic for miRNAs, snDRs, and snoRDs. For miRNAs, 5‐FU treatment led to a mix of both up‐ and down‐regulated cellular expression levels of miRNAs. In contrast, there was a dramatic change in sEV RNA profiles with significant inhibition of miRNA detection in sEVs after 5‐FU treatment. Remarkably, only six miRNAs were enriched in sEVs after treatment with 5‐FU, but four of these four may not be bona fide miRNAs, as curated by MirGeneDB (Fromm et al., 2015).
For spliceosomal snRNAs, we also observed a decrease in sEV snDR enrichment after 5‐FU treatment. However, in contrast to cellular miRNA expression changes which were both up‐ and down‐regulated, we observed only up‐regulation of cellular snRNAs after 5‐FU treatment. Based on that, one might predict increased detection of snDRs in sEVs, but that is not what we observed. We found that most snDRs were down‐regulated in sEVs, suggesting a strong trafficking defect after 5‐FU treatment.
For snoRNAs, we observed a third pattern with up‐regulation of both cellular and sEV expression of snoDRs after treatment with 5‐FU. snoRNAs function to target rRNA for 2′‐O‐methylation and pseudouridine modification through RNA:RNA base pairing and the catalytic action of the DKC pseudouridine synthase and fibrillarin, a 2′‐O‐methylase (Huang et al., 2022; Shubina et al., 2018). Given that we did not observe significant alteration in rDRs or tDRs in cells or sEVs after 5‐FU treatment, it seems likely that the increased expression and retention of snoDRs might be a cellular response to maintain pseudouridine levels in rRNA despite the presence of 5‐FU.
The dramatic decrease in miRNA reads that we observed in sEVs from cells treated with 5‐FU raises several possible mechanisms of action for miRNA‐mediated effects of 5‐FU. The first is that pseudouridine modification may be an essential mark for sEV export, perhaps in concert with specific‐sequence motifs. This model predicts the action of one or more RNA binding proteins that recognize pseudouridine residues (readers) to mediate miRNA export. To our knowledge, only 1 reader of pseudouridine has been proposed, a methionine aminoacyl tRNA synthetase (MetRS) that regulates translation (Levi & Arava, 2021). Perhaps additional readers will be identified since pseudouridine is such a prominent modification. If not, an alternative role for pseudouridine modification in sEV export could be related to RNA processing and/or RNA stability. It is known that pseudouridine modification can affect the structure, stability, and immunogenicity of RNA (Borchardt et al., 2020) and indeed, was critical to the success of the COVID‐19 mRNA vaccines (Morais et al., 2021). Previous reports have shown that pseudouridine formation can regulate miRNA processing and also that incorporation of 5‐FU can inhibit U2 snRNA modification and assembly (Kurimoto et al., 2020; Patton, 1991; Song et al., 2020). Thus, incorporation of 5‐FU could block pseudouridine modification of miRNAs and snRNAs leading to decreased processing and intracellular accumulation of precursor RNAs. For miRNAs, incomplete processing might lead to accumulation of more stable double stranded RNAs which could then also lead to intracellular accumulation.
One explanation for why we did not observe down‐regulation of rDRs and tDRs could be due to differential inhibition of pseudouridine synthases by 5‐FU. There are 13 pseudouridine synthases in human cells (Jin et al., 2022) and not all of them are inhibited by 5‐FU incorporation, indicating that these synthases are not functionally redundant (Spedaliere & Mueller, 2004). The synthases that act on rRNA or tRNA might not overlap with those that act on miRNAs and snRNAs. Also, it is important to recognize that there will be differences in expression and localization depending on the concentration and duration of 5‐FU exposure. We subjected DLD‐1 cells to a relatively moderate concentration of 5‐FU (Ge et al., 2017) for a limited time to observe initial changes in small RNA expression and localization. Future experiments are planned to compare transient and long‐term exposure to 5‐FU. Comparing the dose of 5‐FU used in patients to the levels we used for exposure to tissue culture cells is not entirely straightforward because reported human concentrations are highly variable due to estimates of body surface area, weight, and infusion rates (Abe et al., 2019). Although plasma concentrations can also vary due to sampling times and drug metabolism, our use of 10µM is similar to calculations derived from patients receiving infusions of 5‐FU over 24 h (Blaschke et al., 2012).
Among CRC cells, DLD‐1 cells are relatively resistant to 5‐FU (Bracht et al., 2010) so it will be interesting to expand to more sensitive cells and determine small RNA trafficking and PtxMs in the presence of differing concentrations of 5‐FU. Since 5‐FU can affect DNA metabolism, it will also be important to examine gene expression changes among DNA repair genes and their effects on small RNA trafficking in the presence and absence of 5‐FU.
Consistent with differential activity among pseudouridine synthases, chemotherapeutic resistance to 5‐FU is often associated with increased expression of one of more pseudouridine synthases (Jin et al., 2022). In hepatocellular carcinoma and colorectal cancer, higher expression of pseudouridine synthases (PUS1, PUS7, PusL1, RPUSD3, DKC1, PUS7L, PUS10, and RPUSD1) correlates with poor prognosis (Jin et al., 2022; Liang et al., 2022; Zhang, Zhu et al., 2023). Likewise, overexpression of the Dyskerin pseudouridine synthase that acts on rRNA (DKC1) predicts poor prognosis in breast cancer, while knockdown of DKC1 and Mek1/2 restricts colorectal cancer cell growth (Elsharawy et al., 2020; Kan et al., 2021). These studies are also consistent with work from S. cerevisiae where 5‐FU toxicity requires Cbf5p, a pseudouridine synthase (Hoskins & Butler, 2008). Data mining of published RNAseq data sets from 5‐FU resistant cells showed that six pseudouridine synthases were up‐regulated (PUS3, PUS7, PUS7L, Trub1, RPUSD3, and RPUSD1) with the remaining seven being either unaffected or down‐regulated (Chauvin et al., 2022). Analysis of a 5‐FU resistant colorectal cancer line (HCT116) found a negative correlation between 5‐FU sensitivity and PUS1 levels and stabilization of PUS1, PUS7, PUS10, and TRUB1 in mouse xenografts (Liang et al., 2022). In our RNAseq data after short term exposure of DLD‐1 to 10µM 5‐FU, we found that PUS7, TRUB1, TRUB2, RPUSD1, PUS10 and DKC1 were up‐regulated with the other pseudouridine synthases either unaffected (PUS1, PUS 7L, PUSL1) or down‐regulated (PUS3, RPUSD3, RPUSD4, and RPUSD2) (Table S1). Together, the data indicate that not all pseudouridine synthases are equivalent and raise the possibility that upon exposure to 5‐FU, cells adapt expression levels among the 13 members of the family to survive. This could be consistent with analysis of plasma miRNA levels in patients treated with 5‐FU which found that initial miRNA decreases gave way to a rebound effect restoring normal levels with extended treatment (Badr et al., 2023), presumably due to restoration of pseudouridine modification.
In addition to studying the effect of specific pseudouridine synthases, it will be important in the future to precisely map the location of pseudouridine residues within small RNAs and mutate specific residues to determine their effect on trafficking. With currently published technologies, mapping the precise location of pseudouridines within small RNA is a challenge, especially if there are multiple adjacent U residues (Dai et al., 2023; Gilbert, 2024; Zhang, Jiang et al., 2023). A recent pre‐print suggests that precise mapping may be possible, but additional work will be need to determine if mapping of residues within miRNA is possible (Xu et al., 2024).
PTxMs on cellular and extracellular small RNAs provide another layer of gene regulation. 5‐FU treatment significantly decreased the levels of pseudouridine in both cells and sEVs, as quantified using LC‐MS/MS. While we focused on pseudouridine because of inhibition by 5‐FU, we also detected changes in base modifications of adenosine, guanosine, cytosine, and inosine with significant increases in post‐transcriptional methylation (m1A, m1I, m3C, m5C, m1G, m2G, m7G, and m2 2G) on cellular, but not sEV, small RNAs. Mechanisms for how or why inhibition of pseudouridine formation affects other nucleoside modifications remain unknown but may suggest that overall RNA function, trafficking and/or stability could require coordination between different PTxMs. The development of mass spectrometry to analyze PTxMs will be an important powerful tool for future studies on small RNA trafficking and localization. Nevertheless, for this paper, the data support a role for pseudouridine in small RNA export to sEVs.
SUPPLEMENTARY DATA
Spreadsheets detailing DESeq2 analyses of raw sequencing data are included in Tables S1–S3. Quality control analyses of RNA sequencing data are included in Figure S1. Figure S2 includes differential expression analysis of tDRs, rDRs, and YDrs. Figure S3 includes mass spectrometry of post‐transcriptional modifications.
AUTHOR CONTRIBUTIONS
Shimian Qu: Data curation (equal); formal analysis (equal); methodology (equal); writing—review and editing (equal). Hannah M. Nelson: Data curation (equal); formal analysis (equal); methodology (equal); writing—original draft (equal); writing—review and editing (equal). Xiao Liu: Formal analysis (lead); methodology (equal). Yu Wang: Data curation (equal); formal analysis (equal). Elizabeth M. Semler: Data curation (equal); formal analysis (equal); methodology (equal). Danielle L. Michell: Data curation (equal); methodology (equal). Clark Massick: Data curation (equal); methodology (equal). Jeffrey L. Franklin: Data curation (equal); formal analysis (equal); funding acquisition (equal); investigation (equal); methodology (equal); writing—original draft (equal); writing—review and editing (equal). John Karijolich: Data curation (equal); writing—original draft (equal). Alissa M. Weaver: Funding acquisition (equal); investigation (equal); writing—original draft (equal). Robert J. Coffey: Funding acquisition (equal); investigation (equal); writing—original draft (equal). Qi Liu: Data curation (equal); formal analysis (lead); methodology (equal). Kasey C. Vickers: Data curation (equal); formal analysis (equal); investigation (equal); methodology (lead); writing—original draft (equal); writing—review and editing (equal). James G. Patton: Conceptualization (lead); data curation (equal); formal analysis (equal); funding acquisition (lead); investigation (equal); project administration (lead); writing—original draft (lead); writing—review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Figure S1. Quality control of small RNAseq. RNAseq was performed in triplicate on RNA (<200 nt) isolated from DLD‐1 cells and sEVs treated or not with 5‐FU. (A) Read numbers and percentage of reads mapping to the various categories are as indicated. (B) Read size distribution from the biological replicates in A with the indicated RNA subclasses including miRNA, tRNA‐derived fragments (tDRs), rRNA‐derived fragments (rDRs), Y RNA‐derived fragments (yDRs), spliceosomal snRNA‐derived fragments (snDRs), snoRNA‐derived fragments (snoDRs), and other assorted miscellaneous small RNA fragments (osDRs).
Figure S2. Differentially expressed tDRs and rDRs comparing DLD‐1 cells and sEVs grown in the presence or absence of 5‐FU. Volcano plots showing up‐ and down‐regulated tDRs (A), rDRs (B), and yDRs (C) in cells and sEVs treated or not with 5‐FU. Individual data points represent transcripts with significant differential gene expression changes, (padj < 0.05 and fold‐change > 1.5 or <0.67).
Figure S3. Effects of 5‐FU on RNA base modification in cells and sEVs. LC‐MS/MS approaches were used to quantify RNA base modifications in RNA from cells and sEVs. (A–E) Quantitation of the levels of the indicated base modification in cells and sEVs treated or not with 5‐FU. Base modifications include methylated uridine (m3U, m5U), methylated adenosine (m6A), Inosine, and dimethylated guanosine (m2 7G).
Table S1. Long RNAseq data from DLD‐1 cells exposed to 5‐FU. Sequencing data for libraries generated from RNA > 200 nt isolated from cells exposed to 5‐FU or not. Significant differentially expressed genes with padj < 0.05 and fold‐change > 1.5 or <0.67 were determined using DESeq2 (v1.30.1) (Love et al., 2014).
Table S2. Small RNAseq data from DLD‐1 cells and sEVs exposed to 5‐FU. Sequencing data for libraries generated from RNA <200 nt isolated from cells and sEVs exposed to 5‐FU or not. Significant differentially expressed genes with padj < 0.05 and fold‐change > 1.5 or <0.67 were determined using DESeq2 (v1.30.1) (Love et al., 2014). Tabs show sequencing data for miRNAs, rDRs, tDRs, snDRs, snoDRs and yDRs from both cells and sEVs.
Table S3. Interaction analysis: Comparison of miRNA Expression Patterns in Cells and sEVs. After 5‐FU treatment, the majority (278) of miRNAs showed significant changes in expression in sEVs (blue) with no significant change in cellular expression (red). 13 miRNAs showed significant changes in cellular expression (red) but not in sEVs (blue). 15 miRNAs showed significant changes in both cellular (red) and sEV (blue) expression after 5‐FU treatment.
ACKNOWLEDGEMENTS
The authors would like to thank all members of the Coffey, Weaver, and Patton labs for advice and suggestions. This work was supported by PO1 CA229123 to R.J.C., A.M.W., K.C.V., Q.L., and J.G.P.
Qu, S. , Nelson, H. M. , Liu, X. , Wang, Y. , Semler, E. M. , Michell, D. L. , Massick, C. , Franklin, J. L. , Karijolich, J. , Weaver, A. M. , Coffey, R. J. , Liu, Q. , Vickers, K. C. , & Patton, J. G. (2024). 5‐Fluorouracil treatment represses pseudouridine‐containing miRNA Export into extracellular vesicles. Journal of Extracellular Biology, 3, e70010. 10.1002/jex2.70010
Shimian Qu, Hannah M. Nelson and Xiao Liu contributed equally to this work.
Contributor Information
Kasey C. Vickers, Email: kasey.c.vickers@vumc.org.
James G. Patton, Email: james.g.patton@vanderbilt.edu.
DATA AVAILABILITY STATEMENT
Raw sequencing data have been deposited in GEO (Accession numbers GSE255875 (5FU RNA‐seq) and GSE255876 (5FU smallRNA‐seq). Processing of the data was performed as described in the Materials and Methods.
REFERENCES
- Abe, Y. , Sakuyama, N. , Sato, T. , Kishine, K. , Nagayasu, K. , Nakatani, A. , Kitajima, M. , Watanabe, T. , Nishimura, K. , Ochiai, T. , & Nagaoka, I. (2019). Evaluation of the 5‐fluorouracil plasma level in patients with colorectal cancer undergoing continuous infusion chemotherapy. Molecular and Clinical Oncology, 11, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abels, E. R. , & Breakefield, X. O. (2016). Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cellular and Molecular Neurobiology, 36, 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abner, J. J. , Franklin, J. L. , Clement, M. A. , Hinger, S. A. , Allen, R. M. , Liu, X. , Kellner, S. , Wu, J. , Karijolich, J. , Liu, Q. , Vickers, K. C. , Dedon, P. , Weaver, A. M. , Coffey, R. J. , & Patton, J. G. (2021). Depletion of METTL3 alters cellular and extracellular levels of miRNAs containing m(6)A consensus sequences. Heliyon, 7, e08519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, R. M. , Michell, D. L. , Cavnar, A. B. , Zhu, W. , Makhijani, N. , Contreras, D. M. , Raby, C. A. , Semler, E. M. , DeJulius, C. , Castleberry, M. , Zhang, Y. , Ramirez‐Solano, M. , Zhao, S. , Duvall, C. , Doran, A. C. , Sheng, Q. , Linton, M. F. , & Vickers, K. C. (2022). LDL delivery of microbial small RNAs drives atherosclerosis through macrophage TLR8. Nature Cell Biology, 24, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, R. M. , Zhao, S. , Ramirez Solano, M. A. , Zhu, W. , Michell, D. L. , Wang, Y. , Shyr, Y. , Sethupathy, P. , Linton, M. F. , Graf, G. A. , Sheng, Q. , & Vickers, K. C. (2018). Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. Journal of Extracellular Vesicles, 7, 1506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr, D. , Fouad, M. A. , Hussein, M. , Salem, S. , Zekri, A. , & Shouman, S. (2023). Rebound increase in microRNA levels at the end of 5‐FU‐based therapy in colorectal cancer patients. Scientific Reports, 13, 14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke, M. , Cameron, S. , Blumberg, J. , Wegner, U. , & Ramadori, G. (2012). Measurements of 5‐FU levels in plasma of patients with gastrointestinal cancer. Journal of Clinical Oncology, 30. [Google Scholar]
- Bolukbasi, M. F. , Mizrak, A. , Ozdener, G. B. , Madlener, S. , Ströbel, T. , Erkan, E. P. , Fan, J.‐B. , Breakefield, X. O. , & Saydam, O. (2012). miR‐1289 and “Zipcode”‐like Sequence Enrich mRNAs in Microvesicles. Molecular Therapy—Nucleic Acids, 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt, E. K. , Martinez, N. M. , & Gilbert, W. V. (2020). Regulation and function of RNA pseudouridylation in human cells. Annual Review of Genetics, 54, 309–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht, K. , Nicholls, A. M. , Liu, Y. , & Bodmer, W. F. (2010). 5‐Fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency. British Journal of Cancer, 103, 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas, E. I. (2023). The roles of extracellular vesicles in the immune system. Nature Reviews, 23, 236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, D. J. , Franklin, J. L. , Dou, Y. , Liu, Q. , Higginbotham, J. N. , Demory Beckler, M. , Weaver, A. M. , Vickers, K. , Prasad, N. , Levy, S. , Zhang, B. , Coffey, R. J. , & Patton, J. G. (2015). KRAS‐dependent sorting of miRNA to exosomes. Elife, 4, e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalabi‐Dchar, M. , Fenouil, T. , Machon, C. , Vincent, A. , Catez, F. , Marcel, V. , Mertani, H. C. , Saurin, J. C. , Bouvet, P. , Guitton, J. , Venezia, N. D. , & Diaz, J. J. (2021). A novel view on an old drug, 5‐fluorouracil: An unexpected RNA modifier with intriguing impact on cancer cell fate. NAR Cancer, 3, zcab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin, A. , Bergeron, D. , Vencic, J. , Levesque, D. , Paquette, B. , Scott, M. S. , & Boisvert, F. M. (2022). Downregulation of KRAB zinc finger proteins in 5‐fluorouracil resistant colorectal cancer cells. BMC Cancer, 22, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Thery, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Couch, Y. , Buzàs, E. I. , Di Vizio, D. , Gho, Y. S. , Harrison, P. , Hill, A. F. , Lötvall, J. , Raposo, G. , Stahl, P. D. , Théry, C. , Witwer, K. W. , & Carter, D. R. F. (2021). A brief history of nearly EV‐erything—The rise and rise of extracellular vesicles. Journal of Extracellular Vesicles, 10, e12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Q. , Zhang, L. S. , Sun, H. L. , Pajdzik, K. , Yang, L. , Ye, C. , Ju, C. W. , Liu, S. , Wang, Y. , Zheng, Z. , Zhang, L. , Harada, B. T. , Dou, X. , Irkliyenko, I. , Feng, X. , Zhang, W. , Pan, T. , & He, C. (2023). Quantitative sequencing using BID‐seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nature Biotechnology, 41, 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellar, E. R. , Hill, C. , Melling, G. E. , Carter, D. R. F. , & Baena‐Lopez, L. A. (2022). Unpacking extracellular vesicles: RNA cargo loading and function. Journal of Extracellular Biology, 1, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixson, A. C. , Dawson, T. R. , Di Vizio, D. , & Weaver, A. M. (2023). Context‐specific regulation of extracellular vesicle biogenesis and cargo selection. Nature Reviews Molecular Cell Biology. [DOI] [PMC free article] [PubMed]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: Ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharawy, K. A. , Mohammed, O. J. , Aleskandarany, M. A. , Hyder, A. , El‐Gammal, H. L. , Abou‐Dobara, M. I. , Green, A. R. , Dalton, L. W. , & Rakha, E. A. (2020). The nucleolar‐related protein Dyskerin pseudouridine synthase 1 (DKC1) predicts poor prognosis in breast cancer. British Journal of Cancer, 123, 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, J. V. , Heintz‐Buschart, A. , Ghosal, A. , Wampach, L. , Etheridge, A. , Galas, D. , & Wilmes, P. (2016). Sources and functions of extracellular small RNAs in human circulation. Annual Review of Nutrition, 36, 301–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm, B. , Billipp, T. , Peck, L. E. , Johansen, M. , Tarver, J. E. , King, B. L. , Newcomb, J. M. , Sempere, L. F. , Flatmark, K. , Hovig, E. , & Peterson, K. J. (2015). A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annual Review of Genetics, 49, 213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Martin, R. , Wang, G. , Brandao, B. B. , Zanotto, T. M. , Shah, S. , Kumar Patel, S. , Schilling, B. , & Kahn, C. R. (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature, 601, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, J. , Karijolich, J. , Zhai, Y. , Zheng, J. , & Yu, Y. T. (2017). 5‐Fluorouracil treatment alters the efficiency of translational recoding. Genes (Basel), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. V. (2024). Recent developments, opportunities, and challenges in the study of mRNA pseudouridylation. Rna, 30, 530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. , Liu, Y. , & Santi, D. V. (1999). The mechanism of pseudouridine synthase I as deduced from its interaction with 5‐fluorouracil‐tRNA. PNAS, 96, 14270–14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger, S. A. , Abner, J. J. , Franklin, J. L. , Jeppesen, D. K. , Coffey, R. J. , & Patton, J. G. (2020). Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Scientific Reports, 10, 15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, J. , & Butler, J. S. (2008). RNA‐based 5‐fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics, 179, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z. H. , Du, Y. P. , Wen, J. T. , Lu, B. F. , & Zhao, Y. (2022). snoRNAs: Functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discovery, 8, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Zhang, Q. , Franklin, J. L. , & Coffey, R. J. (2023). Extracellular vesicles and nanoparticles: Emerging complexities. Trends in Cell Biology, 33, P667–P681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. , Song, M. , Wang, J. , Zhu, W. , Sun, D. , Liu, H. , & Shi, G. (2022). Integrative multiomics evaluation reveals the importance of pseudouridine synthases in hepatocellular carcinoma. Frontiers in Genetics, 13, 944681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, G. , Wang, Z. , Sheng, C. , Chen, G. , Yao, C. , Mao, Y. , & Chen, S. (2021). Dual inhibition of DKC1 and MEK1/2 synergistically restrains the growth of colorectal cancer cells. Advanced Science (Weinh), 8, 2004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich, J. , Kantartzis, A. , & Yu, Y. T. (2010). RNA modifications: A mechanism that modulates gene expression. Methods in Molecular Biology, 629, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufe, D. W. , & Major, P. P. (1981). 5‐Fluorouracil incorporation into human breast carcinoma RNA correlates with cytotoxicity. Journal of Biological Chemistry, 256, 9802–9805. [PubMed] [Google Scholar]
- Kurimoto, R. , Chiba, T. , Ito, Y. , Matsushima, T. , Yano, Y. , Miyata, K. , Yashiro, Y. , Suzuki, T. , Tomita, K. , & Asahara, H. (2020). The tRNA pseudouridine synthase TruB1 regulates the maturation of let‐7 miRNA. Embo Journal, 39, e104708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi, O. , & Arava, Y. S. (2021). Pseudouridine‐mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Research, 49, 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. Y. , Bacanu, S. , Sreekumar, L. , Ramos, A. D. , Dai, L. , Michaelis, M. , Cinatl, J. , Seki, T. , Cao, Y. , Coffill, C. R. , Lane, D. P. , Prabhu, N. , & Nordlund, P. (2022). CETSA interaction proteomics define specific RNA‐modification pathways as key components of fluorouracil‐based cancer drug cytotoxicity. Cell Chemical Biology, 29, 572–585 e578. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Longley, D. B. , Harkin, D. P. , & Johnston, P. G. (2003). 5‐fluorouracil: Mechanisms of action and clinical strategies. Nature Reviews Cancer, 3, 330–338. [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, S. L. N. , Breakefield, X. O. , & Weaver, A. M. (2017). Extracellular vesicles: Unique intercellular delivery vehicles. Trends in Cell Biology, 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapator sequences from high‐throughput sequencing reads. EMBnetjournal, 17, 10–12. [Google Scholar]
- Mateescu, B. , Kowal, E. J. , van Balkom, B. W. , Bartel, S. , Bhattacharyya, S. N. , Buzás, E. I. , Buck, A. H. , de Candia, P. , Chow, F. W. , Das, S. , Driedonks, T. A. , Fernández‐Messina, L. , Haderk, F. , Hill, A. F. , Jones, J. C. , Van Keuren‐Jensen, K. R. , Lai, C. P. , Lässer, C. , Liegro, I. D. , … Nolte‐’t Hoen, E. N (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. Journal of Extracellular Vesicles, 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, A. J. , Hoshino, D. , Cha, D. J. , Franklin, J. L. , Coffey, R. J. , Patton, J. G. , & Weaver, A. M. (2016). KRAS‐MEK signaling controls Ago2 and miRNA sorting into exosomes. Cell Reports, 15, 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojardin, L. , Botet, J. , Quintales, L. , Moreno, S. , & Salas, M. (2013). New insights into the RNA‐based mechanism of action of the anticancer drug 5'‐fluorouracil in eukaryotic cells. PLoS One, 8, e78172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais, P. , Adachi, H. , & Yu, Y. T. (2021). The critical contribution of pseudouridine to mRNA COVID‐19 vaccines. Frontiers in Cell and Developmental Biology, 9, 789427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte‐’t Hoen, E. N. , Buermans, H. P. , Waasdorp, M. , Stoorvogel, W. , Wauben, M. H. , & t Hoen, P. A. (2012). Deep sequencing of RNA from immune cell‐derived vesicles uncovers the selective incorporation of small non‐coding RNA biotypes with potential regulatory functions. Nucleic Acids Research, 40, 9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordhuis, P. , Holwerda, U. , Van der Wilt, C. L. , Van Groeningen, C. J. , Smid, K. , Meijer, S. , Pinedo, H. M. , & Peters, G. J. (2004). 5‐Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Annals of Oncology, 15, 1025–1032. [DOI] [PubMed] [Google Scholar]
- Patton, J. R. (1991). Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Molecular and Cellular Biology, 11, 5998–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , Nijman, H. W. , Stoorvogel, W. , Liejendekker, R. , Harding, C. V. , Melief, C. J. , & Geuze, H. J. (1996). B lymphocytes secrete antigen‐presenting vesicles. The Journal of Experimental Medicine, 183, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, J. , Miekus, K. , Kucia, M. , Zhang, J. , Reca, R. , Dvorak, P. , & Ratajczak, M. Z. (2006). Embryonic stem cell‐derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia: Official Journal of the Leukemia Society of America, Leukemia Research Fund, UK, 20, 847–856. [DOI] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson, T. (1991). Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Research, 19, 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, L. , Giurato, G. , Cicchini, C. , Montaldo, C. , Mancone, C. , Tarallo, R. , Battistelli, C. , Alonzi, T. , Weisz, A. , & Tripodi, M. (2016). The RNA‐binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Reports, 17, 799–808. [DOI] [PubMed] [Google Scholar]
- Schaefer, M. , Kapoor, U. , & Jantsch, M. F. (2017). Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome.’. Open Biology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa, S. , Furuse, M. , Yokoyama, N. , & Sasazuki, T. (1993). Altered growth of human colon cancer cell lines disrupted at activated Ki‐ras. Science, 260, 85–88. [DOI] [PubMed] [Google Scholar]
- Shubina, M. Y. , Musinova, Y. R. , & Sheval, E. V. (2018). Proliferation, cancer, and aging‐novel functions of the nucleolar methyltransferase fibrillarin? Cell Biology International, 42, 1463–1466. [DOI] [PubMed] [Google Scholar]
- Shurtleff, M. J. , Temoche‐Diaz, M. M. , Karfilis, K. V. , Ri, S. , & Schekman, R. (2016). Y‐box protein 1 is required to sort microRNAs into exosomes in cells and in a cell‐free reaction. Elife, 5, e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff, M. J. , Yao, J. , Qin, Y. , Nottingham, R. M. , Temoche‐Diaz, M. M. , Schekman, R. , & Lambowitz, A. M. (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. PNAS, 114, E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , Sv, R. , Meijer, D. H. , Gainche, L. , Jr, W. T. C. , Carter, B. S. , Krichevsky, A. M. , & Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Zhuang, Y. , Zhu, C. , Meng, H. , Lu, B. , Xie, B. , Peng, J. , Li, M. , & Yi, C. (2020). Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nature Chemical Biology, 16, 160–169. [DOI] [PubMed] [Google Scholar]
- Spedaliere, C. J. , & Mueller, E. G. (2004). Not all pseudouridine synthases are potently inhibited by RNA containing 5‐fluorouridine. Rna, 10, 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito, M. L. , Baer, C. , Burdet, F. , Maderna, C. , Gilfillan, G. D. , Lyle, R. , Ibberson, M. , & De Palma, M. (2014). Endogenous RNAs modulate MicroRNA sorting to exosomes and transfer to acceptor cells. Cell Reports, 8, 1432–1446. [DOI] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V. K. , Mukherjee, S. , Ebert, B. L. , Gillette, M. A. , Paulovich, A. , Pomeroy, S. L. , Golub, T. R. , Lander, E. S. , & Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge‐based approach for interpreting genome‐wide expression profiles. PNAS, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka, M. , Nobe, Y. , Yamaki, Y. , Sato, K. , Ishikawa, H. , Izumikawa, K. , Yamauchi, Y. , Hirota, K. , Nakayama, H. , Takahashi, N. , & Isobe, T. (2018). Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Research, 46, 9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temoche‐Diaz, M. M. , Shurtleff, M. J. , Nottingham, R. M. , Yao, J. , Fadadu, R. P. , Lambowitz, A. M. , & Schekman, R. (2019). Distinct mechanisms of microRNA sorting into cancer cell‐derived extracellular vesicle subtypes. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols, G. , Bash‐Imam, Z. , Panthu, B. , Machon, C. , Vincent, A. , Ripoll, J. , Nait‐Slimane, S. , Chalabi‐Dchar, M. , Gaucherot, A. , Garcia, M. , Laforêts, F. , Marcel, V. , Boubaker‐Vitre, J. , Monet, M. A. , Bouclier, C. , Vanbelle, C. , Souahlia, G. , Berthel, E. , … & Diaz, J. J. (2022). Alteration of ribosome function upon 5‐fluorouracil treatment favors cancer cell drug‐tolerance. Nature Communications, 13, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery, C. , Duban, L. , Segura, E. , Veron, P. , Lantz, O. , & Amigorena, S. (2002). Indirect activation of naive CD4+ T cells by dendritic cell‐derived exosomes. Nature Immunology, 3, 1156–1162. [DOI] [PubMed] [Google Scholar]
- Thery, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … & Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts, L. M. , Chu, M. Y. , Hager, J. C. , Dexter, D. L. , & Calabresi, P. (1977). Chemotherapy of cell‐line‐derived human colon carcinomas in mice immunosuppressed with antithymocyte serum. Cancer, 40, 2651–2659. [DOI] [PubMed] [Google Scholar]
- Tkach, M. , & Thery, C. (2016). Communication by extracellular vesicles: Where we are and where we need to go. Cell, 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Turchinovich, A. , Drapkina, O. , & Tonevitsky, A. (2019). Transcriptome of extracellular vesicles: State‐of‐the‐art. Frontiers in Immunology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J. J. , & Lotvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Veziroglu, E. M. , & Mias, G. I. (2020). Characterizing extracellular vesicles and their diverse RNA contents. Frontiers in Genetics, 11, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya‐Beltri, C. , Gutierrez‐Vazquez, C. , Sanchez‐Cabo, F. , Perez‐Hernandez, D. , Vazquez, J. , Martin‐Cofreces, N. , Martinez‐Herrera, D. J. , Pascual‐Montano, A. , Mittelbrunn, M. , & Sanchez‐Madrid, F. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications, 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Vasaikar, S. , Shi, Z. , Greer, M. , & Zhang, B. (2017). WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research, 45, W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel, I. , Dror, S. , Kenific, C. M. , & Lyden, D. (2019). Exosome‐mediated metastasis: Communication from a distance. Developmental Cell, 49, 347–360. [DOI] [PubMed] [Google Scholar]
- Wyatt, M. D. , & Wilson, D. M., 3rd. (2009). Participation of DNA repair in the response to 5‐fluorouracil. Cellular and Molecular Life Sciences, 66, 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Kong, L. , Cheng, J. , Moussawi, K. A. , Chen, X. , Iqbal, A. , Wing, P. A. C. , Harris, J. M. , Tsukuda, S. , Embarc‐Buh, A. , Wei, G. , Castello, A. , Kriaucionis, S. , McKeating, J. A. , Lu, X. , & Song, C.‐X. (2024). Absolute quantitative and base‐resolution sequencing reveals comprehensive landscape of pseudouridine across the human transcriptome. BioRxiv, 2024.2001.2008.574649.
- Zaccara, S. , Ries, R. J. , & Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nature Reviews Molecular Cell Biology, 20, 608–624. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Zhu, Y. , Tan, Y. , Chen, B. , Shan, S. , Zhang, G. , & Lu, J. (2023). Higher expression of pseudouridine synthase 7 promotes non‐small cell lung cancer progression and suggests a poor prognosis. Journal of Cardiothoracic Surgery, 18, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Freitas, D. , Kim, H. S. , Fabijanic, K. , Li, Z. , Chen, H. , Mark, M. T. , Molina, H. , Martin, A. B. , Bojmar, L. , Fang, J. , Rampersaud, S. , Hoshino, A. , Matei, I. , Kenific, C. M. , Nakajima, M. , Mutvei, A. P. , Sansone, P. , Buehring, W. , … Lyden, D. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nature Cell Biology, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Jiang, Z. , Ma, Y. , Liu, W. , Zhuang, Y. , Lu, B. , Li, K. , Peng, J. , & Yi, C. (2023). Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nature Chemical Biology, 19, 1185–1195. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Higginbotham, J. N. , Jeppesen, D. K. , Yang, Y. P. , Li, W. , McKinley, E. T. , Graves‐Deal, R. , Ping, J. , Britain, C. M. , Dorsett, K. A. , Hartman, C. L. , Ford, D. A. , Allen, R. M. , Vickers, K. C. , Liu, Q. , Franklin, J. L. , Bellis, S. L. , & Coffey, R. J. (2019). Transfer of functional cargo in exomeres. Cell Reports, 27, 940–954 e946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Jeppesen, D. K. , Higginbotham, J. N. , Graves‐Deal, R. , Trinh, V. Q. , Ramirez, M. A. , Sohn, Y. , Neininger, A. C. , Taneja, N. , McKinley, E. T. , Niitsu, H. , Cao, Z. , Evans, R. , Glass, S. E. , Ray, K. C. , Fissell, W. H. , Hill, S. , Rose, K. L. , Huh, W. J. , … Coffey, R. J. (2021). Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nature Cell Biology, 23, 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. , Guo, Y. , Sheng, Q. , & Shyr, Y. (2014). Advanced heat map and clustering analysis using heatmap3. BioMed Research International, 2014, 986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , & Yu, Y. T. (2007). Incorporation of 5‐fluorouracil into U2 snRNA blocks pseudouridylation and pre‐mRNA splicing in vivo. Nucleic Acids Research, 35, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quality control of small RNAseq. RNAseq was performed in triplicate on RNA (<200 nt) isolated from DLD‐1 cells and sEVs treated or not with 5‐FU. (A) Read numbers and percentage of reads mapping to the various categories are as indicated. (B) Read size distribution from the biological replicates in A with the indicated RNA subclasses including miRNA, tRNA‐derived fragments (tDRs), rRNA‐derived fragments (rDRs), Y RNA‐derived fragments (yDRs), spliceosomal snRNA‐derived fragments (snDRs), snoRNA‐derived fragments (snoDRs), and other assorted miscellaneous small RNA fragments (osDRs).
Figure S2. Differentially expressed tDRs and rDRs comparing DLD‐1 cells and sEVs grown in the presence or absence of 5‐FU. Volcano plots showing up‐ and down‐regulated tDRs (A), rDRs (B), and yDRs (C) in cells and sEVs treated or not with 5‐FU. Individual data points represent transcripts with significant differential gene expression changes, (padj < 0.05 and fold‐change > 1.5 or <0.67).
Figure S3. Effects of 5‐FU on RNA base modification in cells and sEVs. LC‐MS/MS approaches were used to quantify RNA base modifications in RNA from cells and sEVs. (A–E) Quantitation of the levels of the indicated base modification in cells and sEVs treated or not with 5‐FU. Base modifications include methylated uridine (m3U, m5U), methylated adenosine (m6A), Inosine, and dimethylated guanosine (m2 7G).
Table S1. Long RNAseq data from DLD‐1 cells exposed to 5‐FU. Sequencing data for libraries generated from RNA > 200 nt isolated from cells exposed to 5‐FU or not. Significant differentially expressed genes with padj < 0.05 and fold‐change > 1.5 or <0.67 were determined using DESeq2 (v1.30.1) (Love et al., 2014).
Table S2. Small RNAseq data from DLD‐1 cells and sEVs exposed to 5‐FU. Sequencing data for libraries generated from RNA <200 nt isolated from cells and sEVs exposed to 5‐FU or not. Significant differentially expressed genes with padj < 0.05 and fold‐change > 1.5 or <0.67 were determined using DESeq2 (v1.30.1) (Love et al., 2014). Tabs show sequencing data for miRNAs, rDRs, tDRs, snDRs, snoDRs and yDRs from both cells and sEVs.
Table S3. Interaction analysis: Comparison of miRNA Expression Patterns in Cells and sEVs. After 5‐FU treatment, the majority (278) of miRNAs showed significant changes in expression in sEVs (blue) with no significant change in cellular expression (red). 13 miRNAs showed significant changes in cellular expression (red) but not in sEVs (blue). 15 miRNAs showed significant changes in both cellular (red) and sEV (blue) expression after 5‐FU treatment.
Data Availability Statement
Raw sequencing data have been deposited in GEO (Accession numbers GSE255875 (5FU RNA‐seq) and GSE255876 (5FU smallRNA‐seq). Processing of the data was performed as described in the Materials and Methods.


