Abstract
Rabies virus nucleoprotein (N) was produced in insect cells, in which it forms nucleoprotein-RNA (N-RNA) complexes that are biochemically and biophysically indistinguishable from rabies virus N-RNA. We selected recombinant N-RNA complexes that were bound to short insect cellular RNAs which formed small rings containing 9 to 11 N monomers. We also produced recombinant N-RNA rings and viral N-RNA that were treated with trypsin and that had lost the C-terminal quarter of the nucleoprotein. Trypsin-treated N-RNA no longer bound to recombinant rabies virus phosphoprotein (the viral polymerase cofactor), so the presence of the C-terminal part of N is needed for binding of the phosphoprotein. Both intact and trypsin-treated recombinant N-RNA rings were analyzed with cryoelectron microscopy, and three-dimensional models were calculated from single-particle image analysis combined with back projection. Nucleoprotein has a bilobed shape, and each monomer has two sites of interaction with each neighbor. Trypsin treatment cuts off part of one of the lobes without shortening the protein or changing other structural parameters. Using negative-stain electron microscopy, we visualized phosphoprotein bound to the tips of the N-RNA rings, most likely at the site that can be removed by trypsin. Based on the shape of N determined here and on structural parameters derived from electron microscopy on free rabies virus N-RNA and from nucleocapsid in virus, we propose a low-resolution model for rabies virus N-RNA in the virus.
Negative-strand RNA viruses such as rabies virus are enveloped viruses with an RNA genome in the opposite sense from the mRNA. The first viral activity after entry into the cell is transcription of the viral RNA (vRNA) into mRNA. The vRNA is complexed to the viral nucleoprotein (N) to form an N-RNA complex in which the nucleoprotein binds to the phosphate-sugar backbone and exposes the nucleotide bases (11, 18, 21, 22). This N-RNA complex (and not the naked vRNA) serves as the actual template for the vRNA-dependent RNA polymerase. For the nonsegmented negative-strand RNA viruses, this polymerase consists of a large protein (L), which is the actual enzyme (31), and a polymerase cofactor, the phosphoprotein (P) (11).
The binding of L to the N-RNA is mediated by P (24). As shown for the paramyxovirus Sendai virus, the carboxy-terminal part of N is involved in the binding of P or the L-P complex to the N-RNA (2, 8). The carboxy-terminal part of paramyxovirus N is often cleaved off in vivo, resulting in a protease-resistant fragment and a reduction in size of N from about 60 kDa to 41 to 45 kDa (15, 25, 26). This C-terminal part of N seems to be located on the outside of the nucleocapsid (3), and removal of this peptide from the paramyxovirus nucleocapsid does not lead to gross changes in appearance in electron microscopy (EM) (16, 25, 26). Trypsin cleavage of rabies virus N also leads to removal of the C-terminal part of N (23), but, as with the paramyxovirus nucleocapsids, this has no major effect on the appearance of the rabies virus nucleocapsid as observed by EM after negative staining: same length of the N subunit and same N-to-N spacing along the nucleocapsid but perhaps a smaller depth at one end of the N subunit (17) (Table 1).
TABLE 1.
Dimensions of rabies virus N protein in N-RNA from cryo-EM and negative stain EM
| N dimensions (Å)
|
|||
|---|---|---|---|
| Length | Height | N-N spacing | |
| Negative staining with sodium silicotungstate | |||
| Intact | 84 ± 5 | 53 ± 3 | 34 ± 1 |
| After trypsin treatment | 84 ± 3 | 48 ± 4 | 34 ± 2 |
| Negative staining with uranyl acetate | |||
| Intact | 90 | 50 | 33 ± 1 |
| Cryo-EM | |||
| Intact | 94 | 52a | 35b |
| After trypsin treatment | 94 | 43a | 35b |
Height in projection.
At mid-height.
The N-RNA complexes of the various negative-strand RNA viruses are all helical structures with different overall morphologies, depending on the viral families, but all rather flexible, which precludes high-resolution image analysis of electron micrographs (10). Here we have studied recombinant rabies virus N protein produced in insect cells that forms helical structures that are indistinguishable from those of regular viral nucleocapsids when bound to long insect cellular RNAs but which forms small rings when it is bound to short cellular RNAs. The longer recombinant helical structures and the small rings are bound to cellular RNAs with the same protein-RNA stoichiometry as viral nucleocapsids (9 nucleotides per N monomer), and they have the same reactivities towards proteases and RNases and the same density in a CsCl gradient as viral nucleocapsids (17). However, whereas the recombinant and the viral helical structures have variable helical parameters and are difficult to analyze with EM and image analysis, the small rings are more regular and can be analyzed by single-particle analysis. A similar analysis was recently performed on small circular, recombinant influenza virus RNPs that were also much more regular than the full-length viral RNPs (30).
We have performed cryo-EM on the recombinant rabies virus N-RNA rings and calculated a three-dimensional (3D) structure for the rabies virus nucleoprotein by back projection. We have also shown that trypsin-treated rabies virus nucleocapsids have lost the binding site for rabies virus P. Comparison of the intact recombinant rabies virus N-RNA rings with an independent three-dimensional reconstruction of trypsin-treated rings indicates the binding site for P at one end of the two-lobed nucleoprotein. The morphological information on N has been combined with EM observations on nucleocapsids isolated by CsCl gradient centrifugation and on nucleocapsid/matrix protein skeletons in order to produce a model for the packing of N in free nucleocapsids and in virus.
MATERIALS AND METHODS
Binding of phosphoprotein to intact and trypsin-treated rabies virus nucleocapsids.
Viral nucleocapsids were isolated from virus-infected cells and digested with trypsin as described (17). After digestion the trypsin was removed by a second CsCl gradient step. Recombinant His-tagged P protein was produced in Escherichia coli as described by Gigant et al. (14). The intact and digested nucleocapsids were incubated with 30 μg of P per ml in 20 mM Tris-HCl (pH 7.5)–150 mM NaCl for 1.5 h at room temperature. This mixture was deposited on a 20% sucrose cushion, which was spun for 2.5 h at 25,000 rpm and 4°C in an SW55 rotor with an adapter for 500 μl. The pellet and the supernatant were then analyzed by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (SDS-PAGE).
Recombinant N-RNA rings and trypsin digestion.
Production of rabies virus N-RNA rings in insect cells by the baculovirus system and their purification were done as described (17, 32). Since this preparation contains rings with various diameters and with 8 to 13 N monomers per ring (even though most rings had 10 monomers [17]), this sample was further purified over an additional 7.5 to 15% (vol/vol) continuous glycerol gradient (SW41 rotor); 300-μl fractions were taken from the bottom of the tube. The peak fractions were dialyzed against 20 mM Tris-HCl (pH 7.5)–150 mM NaCl and contained rings with 9, 10, and 11 monomers. However, the image analysis programs were powerful enough to distinguish the different species in the sample (see below) (35).
Trypsin treatment of the N-RNA rings was also done as described by Iseni et al. (17) at a final trypsin concentration of 100 μg/ml for 16 h at room temperature in 150 mM NaCl–10 mM Tris-HCl (pH 7.5). The digested rings were then once more purified by centrifugation on a 7.5 to 15% (vol/vol) glycerol gradient (SW41 rotor) and dialyzed against 20 mM Tris-HCl (pH 7.5)–150 mM NaCl.
Production of N-RNA rings with bound P.
Rabies virus N and P were coexpressed in insect cells by coinfection with two baculoviruses as described by (32) at a multiplicity of infection of 5 for both baculoviruses. During this double infection, most of N does not bind to RNA but to P, forming soluble N°-P complexes that do not contain RNA (32; Iseni et al., unpublished results). However, we found that a small proportion of P forms P oligomers and also that a small proportion of N does bind to RNA and forms the same kind of rings as when N is expressed alone. These N-RNA rings bind to free P oligomers, and these N-RNA-P complexes can be separated from the N°-P complexes and from the free P oligomers on the basis of their size on a Superdex 200 gel filtration column. A UV absorption spectrum showed the presence of RNA through an absorbance maximum at 260 nm.
EM. (i) Negative staining.
Protein samples at 0.1 mg/ml in 50 mM NaCl–20 mM Tris-HCl (pH 7.5) were applied to the clean side of carbon on mica (carbon-mica interface) and negatively stained with 2% ammonium molybdate (pH 7.4) or 1% sodium silicotungstate (pH 7.0). Micrographs were taken under low-dose conditions with a JEOL 1200 EX II microscope at 100 kV and a nominal magnification of ×40,000.
(ii) Cryo-EM.
The method of Dubochet et al. (9) was used to vitrify the sample. Native or digested N-RNA rings (3 μl at 1 mg/ml) were applied to a thin holey carbon film on a 400-mesh copper grid without glow discharge. The excess of sample was blotted with filter paper for 1 to 2 s and rapidly plunged into liquid ethane. The vitrified grids were observed with a Gatan 626 cryotransfer stage on a Philips CM20 LaB6 microscope at ×38,000 magnification. Images were recorded under low-dose conditions (<10 e−/Å2) on Kodak SO163 films, with a defocus chosen so that the first zero of the contrast transfer function was beyond (25 Å)−1.
Image processing and 3D reconstruction.
The quality of the best images was checked on an optical bench and then digitized on a Zeiss scanner with a pixel size of 21 μm (native N-RNA rings) or on an Optronics microdensitometer with a pixel size of 25 μm (digested N-RNA rings). At the level of the sample, the pixel size corresponds to 5.5 Å for intact N-RNA rings and 6.5 Å for the digested rings. There were many fewer side views of the complex than top views. We selected approximately the same number of top views as side views and also all intermediate views for the two types of N-RNAs. In total, 6,200 particles were selected from 26 films for the intact rings and 4,500 particles from 40 films for the digested rings with Ximdisp (7). The images included rings with 9, 10, and 11 subunits per ring. All particles were band pass filtered between 200 and 25 Å without contrast transfer function correction and then normalized to the same mean and standard deviation. For each set of N-RNA rings (intact and digested), three starting models were created with 9-, 10-, and 11-fold symmetry. These models were generated by back-projection of one top view and truncation of the 3D model to the thickness of a side view (95 Å) (35). All images were aligned against all the reprojections of the three starting models. The best match for each image determined its assignment to one of the three structures. All class averages were used to generate a new model by back-projection. After 20 iterative cycles of this projection-matching procedure with SPIDER (12, 34), the orientations of the raw images were stable. For both N-RNA preparations, the proportion of views matching the three models were equivalent: more than 55% of the views matched the rings made of 10 subunits, 25% matched the rings with 9 subunits, and 20% matched the 11-fold rings. The quality of the reconstructions of the rings with 9 and 11 subunits was quite poor but good enough to segregate the images with 9 and 11 subunits from those with 10 subunits (not shown). Here we only present the 3D structure of the rings with 10 subunits. The reconstructions were calculated by back-projecting 60 class averages distributed equally in the asymmetric unit. The resolution was determined by Fourier shell correlation (FSC; 0.5 level) to be 28 and 29 Å for the intact and digested N-RNA ring, respectively. The threshold of the two reconstructions was chosen to include the right molecular mass (550 and 380 kDa), assuming an average protein density of 0.84 Da · Å−3.
Image analysis of N-RNA rings with bound P.
Recombinant N-RNA-P complexes were isolated and prepared for EM by negative staining. EM negatives were digitized on the Optronics densitometer. The sample contained rings with 10 and 11 subunits of N, but the spots inside the rings were better visible for the rings with 11 subunits. Therefore, 100 rings with 11 subunits and with attached P were selected by eye. We selected one image with a particle inside the ring and one image with a particle attached to the outside of the ring and centered the rings. When we aligned the 100 images against the two selected images, 75 particles aligned with the image that had a particle inside the ring and 25 aligned with the one with a particle outside. We averaged the 10 best-matching pictures of each of the two classes and went through the same operation five times. The numbers of images matching each class were stable at 75 and 25. The final averages of the 10 best-matching pictures are shown in Fig. 5. The analysis on negatively stained N-RNA rings without P was performed on a preparation from cells that had only been infected with the baculovirus that expresses N. The same procedure was followed as for the negatively stained N-RNA-P complexes, and selection was done for rings with 11 subunits.
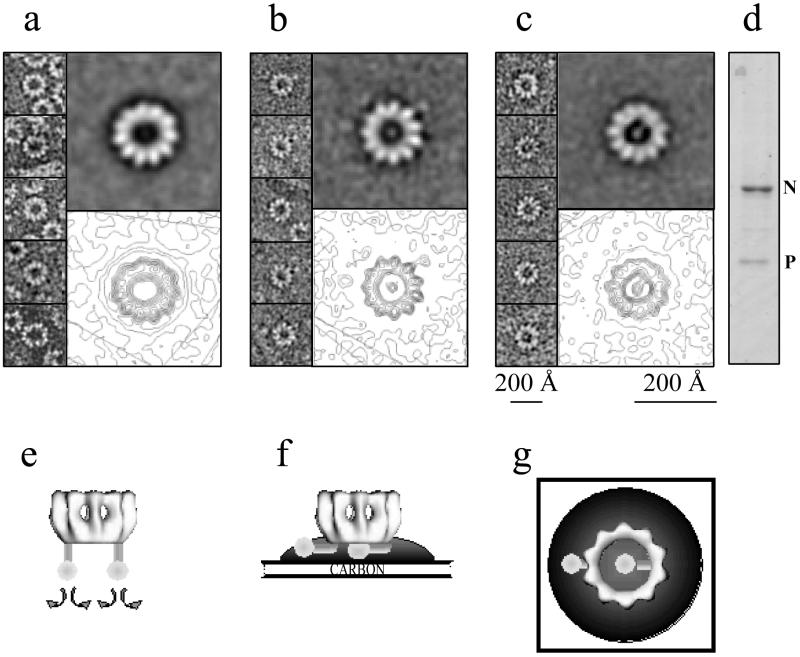
FIG. 5.
Visualization of P bound to N-RNA rings by negative staining with sodium silicotungstate. (a to c) Galleries in the left parts of selected top views of N-RNA rings with 11 monomers of N and in the right parts at the top the averages of 10 raw images. The lower right parts show the same averages but with contour levels in order to outline the connection between P and the N-RNA rings. (a) N-RNA rings without P. (b) N-RNA rings with P sticking out from the rings (but also with some density inside). (c) Rings with one P oligomer inside the ring. Calibration bars indicate the sizes of the raw images (smaller bar) and of the averaged images (larger bar). (d) SDS-PAGE (stained with Coomassie blue) of the complexes, showing that they contain more N than P. Panels e to g represent cartoons of our interpretation of the images in panels a to c. (e) N-RNA-P complex in solution (shown upside down). (f) Side view of the N-RNA-P complex bound to carbon film and embedded in a stain drop. (g) Top view of the situation in panel f.
RESULTS AND DISCUSSION
Binding of P to intact and trypsin-digested rabies virus nucleocapsids.
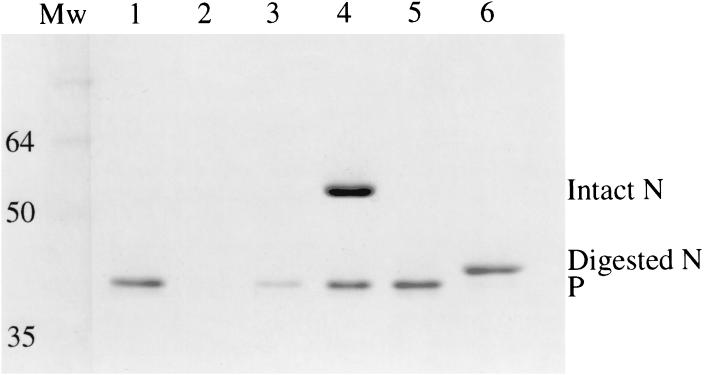
When rabies virus nucleocapsids or recombinant rabies virus N-RNA rings are digested with trypsin, a 17-kDa fragment is removed from N by cleavage at lysine-376 (17, 23). We first tested binding of recombinant rabies virus P protein to intact and trypsin-treated nucleocapsids by incubating P with the nucleocapsids and then sedimentation through a 20% (wt/vol) sucrose cushion, which allows the nucleocapsid-bound P to pellet with the nucleocapsid and unbound P to remain in the supernatant. Figure 1 shows that almost all of P stays in the supernatant in the absence of nucleocapsids and that most of P cosediments with intact nucleocapsids. However, there was little or no binding of P to trypsin-treated nucleocapsids. This suggests that, as for the paramyxoviruses, the C-terminal part of rabies virus N is necessary for the binding of P to the N-RNA complex.
FIG. 1.
Rabies virus P binds to intact but not to trypsin-treated nucleocapsids. The SDS–12% polyacrylamide gel shows supernatant (lanes 1, 3, and 5) and pellet fractions (lanes 2, 4, and 6) from centrifugation through sucrose cushions of P protein alone (lanes 1 and 2), P mixed with intact nucleocapsids (lanes 3 and 4), and P mixed with trypsin-treated nucleocapsids (lanes 5 and 6). Lane Mw, size markers (in kilodaltons). The gel was stained with Coomassie blue.
Image reconstruction of intact and trypsin-digested N-RNA rings.
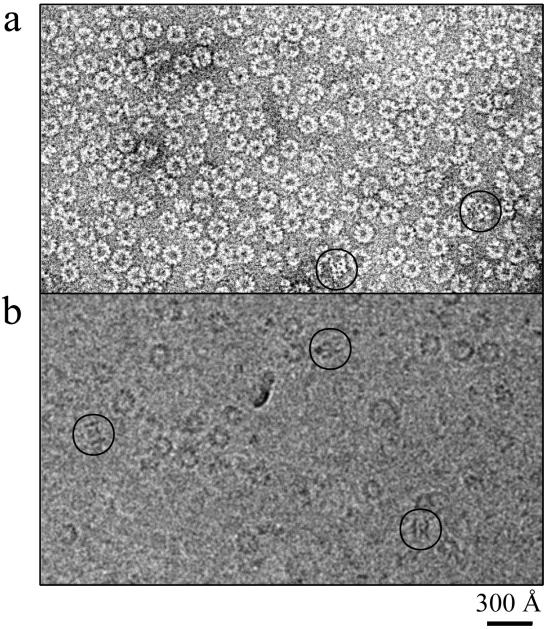
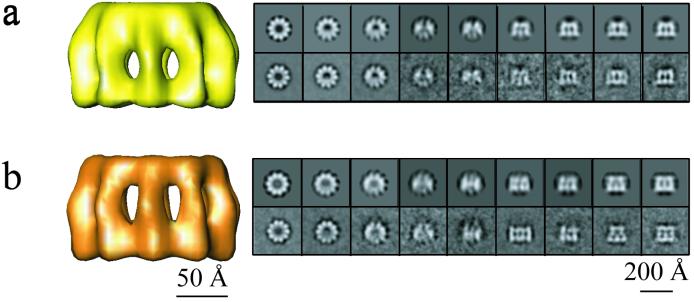
Both intact and trypsin-digested recombinant N-RNA rings were then studied by negative stain and cryo-EM (Fig. 2). In both types of EM there are many more top views than side views. Some of the side views that can be distinguished in Fig. 2 are circled. The negative stain images are of much higher contrast and easier to interpret than the cryo-images. However, since the cryo-images best represent the structure in solution and because the side views in the cryo-images are not flattened by the staining and air-drying treatment, we performed image analysis on the cryo-pictures. The Materials and Methods section describes extensively how the images were analyzed and how the 3D models of the intact and digested rings containing 10 N monomers were calculated. Figure 3 shows both models, the intact rings in yellow and the digested rings in orange, together with tilt series of averages of the actual data and tilt series of reprojections of the calculated models. The two data series correspond closely to the model series, indicating that the 3D reconstructed models agree very well with the EM images. Here, we only show the reconstructions of the rings with 10 subunits of N because the reconstructions of the rings with 9 and 11 subunits were of lower quality than would be expected from the number of images that were used. This is perhaps because the rings with 10 subunits are intrinsically more stable or less flexible or because the encapsidated insect cellular RNA leads to more stable rings with 10 subunits (too short for 11 or too long for 9; see also below). It is also possible that the rings with 9 or 11 monomers are less symmetrical than the rings with 10 monomers.
FIG. 2.
Comparison of negative staining with ammonium molybdate (a) and cryo-EM (b) images of recombinant rabies virus N-RNA rings. The heterogeneity of the ring size in the top views of the rings can be observed, with rings of 9 to 11 subunits of N. The circles show side views or tilted views of rings. Bar, 300 Å.
FIG. 3.
Side views of the 3D models of intact and digested N-RNA rings containing 10 subunits of N: (a) intact in yellow and (b) digested in orange. Next to the reconstructions are the averaged views (lower panel) and corresponding reprojections of each model (upper panel). The top view orientations are shown on the left, with the views progressively tilted, and ending with the side views on the right. There is excellent agreement between averaged views (actual data) and reprojections of the 3D structures (model). The protein density is in white.
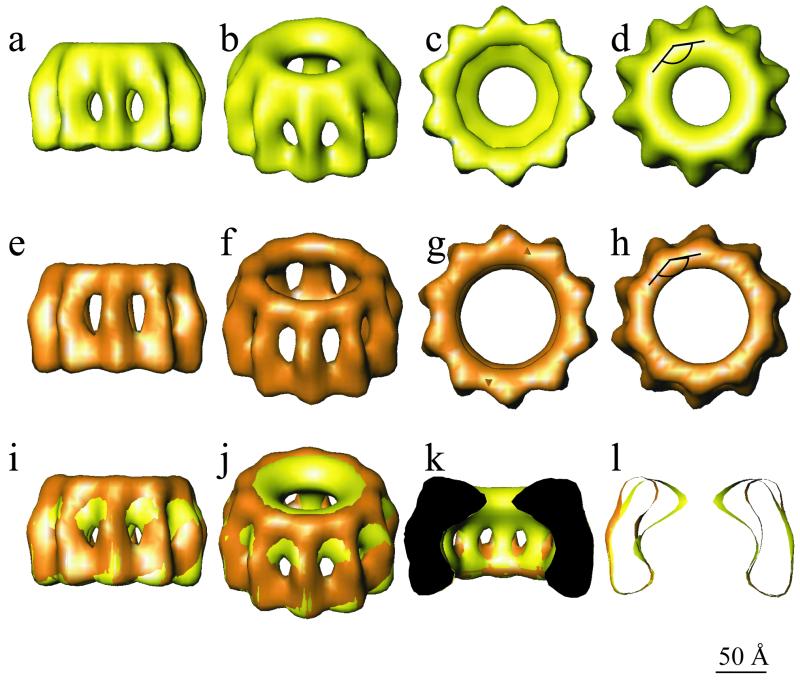
Figure 4 shows various tilted views of both models as well as a superposition of the two models in which it can be seen that trypsin treatment removes the inner part of the ring at its top end. Since P does not bind to trypsinized nucleocapsids, it is inferred that this inner part of the ring constitutes the binding site for P. The dimensions of the N subunits in the rings are given in Table 1, where they are compared with previously published dimensions derived from negatively stained nucleocapsids and rings. The values for N-N spacing and the axial width and height of N agree well among all methods, but the value for the length of N is higher by cryo-EM than by negative stain EM. This is related to the fact that in cryo-EM the density of the protein is observed directly, whereas in negative staining EM the stain-excluding imprint of the molecule is seen, which may be shorter than the actual molecule itself. However, the principal conclusions on the effect of tryptic digestion of N in N-RNA derived from negative-staining EM are confirmed by the model derived from cryo-EM: the length does not change but the N molecules have become thinner at one end (17) (Table 1). Another difference between the intact and digested rings is that the top part of digested N moves to the outside of the ring by 15 Å.
FIG. 4.
3D reconstructions of native (yellow) and digested (orange) N-RNA rings. The maps are shown in side views (a and e), tilted views (b and f), and end views from the bottom (c and g) and from the top (d and h). The lower panel shows a superposition of the native and digested rings in side view (i), tilted view (j), with the front half cut off (k), and in axial slice (l). The two structures are very similar except at the top end, where the inner part of the circle is missing for the digested rings. The angle between N-N-N shown in panels d and h measures 144°.
Each N monomer has two sites of interaction with each of its neighbor N molecules in both the intact and digested rings. This has consequences for designing site-directed mutagenesis experiments on N with the purpose of obtaining a recombinant molecule that is soluble for crystallization experiments. It also has consequences for ideas on how N° is kept soluble by P, since both N-N interaction sites need to be protected by P. This could be one of the reasons why rabies virus P has two binding sites for N° (6, 13, 37). Furthermore, paramyxovirus P binds to two sites on N° (1). It is also possible that there is a conformational difference between N° bound to P and mature N bound to RNA, as suggested by Kawai et al. (20). However, our results indicate that the C-terminal part of N is the only binding site for P on N-RNA.
Visualization of P bound to N-RNA rings.
If P binds to the tip of the N-RNA rings, then P may be visible in negatively stained N-RNA-P complexes when the rings lie with their top end on the carbon support film. We coinfected insect cells with two baculoviruses containing the coding sequences for N or for P. Préhaud et al. (33) have shown that in these coinfections, soluble N°-P complexes are formed. We reproduced these results but also found a small percentage of N-RNA rings complexed with P (see Materials and Methods). Figure 5 shows a comparison of negatively stained N-RNA rings alone with N-RNA rings that have P attached and also a gel showing the bands for both proteins, there being more N than P in these complexes (Fig. 5d). Figure 5a shows an averaged image of negatively stained N-RNA rings without P, and Fig. 5b and c show averaged N-RNA rings that have P bound. In the averaged image in Fig. 5b, there is a light spot inside the ring and also a spot outside the ring that is attached to the ring of N by a thin connection. This suggests that this class average represents N-RNA rings that have two P oligomers bound. In the actual micrographs, thin connections between the spot inside these rings and N can be seen, but since the images were aligned on the spot outside the ring, these inside connections were averaged out. In the average image in Fig. 5c, there is only a spot in the inside of the ring that also shows a connection to the ring.
The staining agent that was used for these images outlines only those parts of microscopic objects that are in direct contact with the carbon support film (for example, helical filaments are only stained on the carbon-near side, as judged from optical diffraction of micrographs [DiCapua and Ruigrok, unpublished results]). The cartoons in Fig. 5e to g indicate how we interpret the images in Fig. 5a to c, respectively. P is probably bound to the top of the rings (shown upside down in Fig. 5e) in a flexible manner, as indicated by the curved arrows. When this N-RNA-P complex binds with its tip to the carbon support film, the flexibly linked P molecules have to bend away so that the rings can touch the carbon film. P can bend either towards the outside of the rings or towards the inside, as shown in Fig. 5f and g. This implies that we assume that the P molecules seen inside the rings are attached to the same site on N as the P molecules seen outside of the rings, but are bent in another direction. The length of P measured from these images is 85 Å for P inside the rings (going from the end of P to the middle of N) and 75 Å for P on the outside. This probably means that P binds closer to the inside of the rings, in agreement with the difference between the models for intact and trypsin-digested N. The diameter of the blob is 40 Å.
The P protein of Sendai virus is a tetramer (38), and that of rabies virus is a trimer or a tetramer (14). The density for P in the averaged images in Fig. 5b and c would seem too low for an oligomeric form of P. However, if P is flexibly linked to N, then the images represent an average of P in different positions and thus with a lower occupation than the N molecules in the rings. A similar case of averaged-out density was reported for the polymerase complex on the ellipsoid RNP complexes described by Ortega et al. (30). These authors have explained the absence of density for the polymerase complex in averaged ellipsoids by the fact that the weight of the N monomers in the ellipsoids was higher than that of the polymerase complex that was attached at various positions compared to the ellipsoid axes.
The N-RNA rings that were used for the averages in Fig. 5 had 11 monomers of N rather than the 10 in the rings that gave the best reconstructions of intact and digested rings. We did observe rings with 10 monomers that had a light spot inside, but there the connection between P and N was not so clear. This is possibly due to a slightly smaller inside of the rings with 10 subunits, leading to differences in stain accumulation. Note that these rings with 11 monomers in the averaged image (Fig. 5a) are less regular than those with 10 monomers (averaged image in Fig. 3) and that one side of the rings is better defined than the opposite side. This absence of symmetry may be why the reconstruction of the rings with 11 monomers was poor (see Materials and Methods).
Model of the rabies virus nucleocapsid.
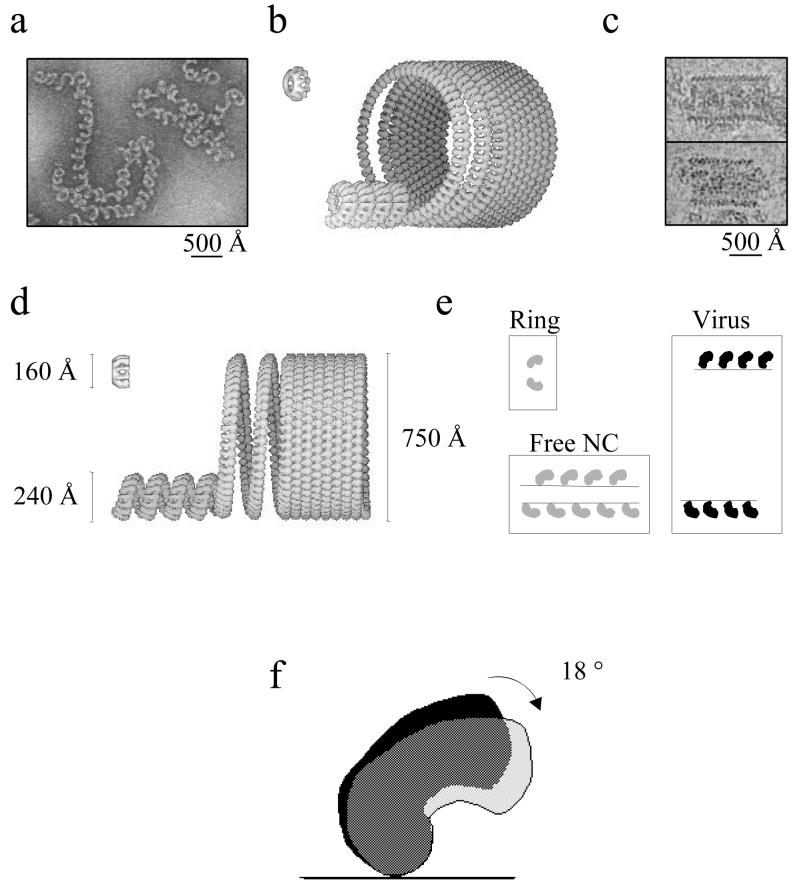
Inside the rabies virus particle, the nucleocapsid forms a tightly wound helix with a small pitch but a large diameter. When the nucleocapsid is released, the helical parameters change drastically, and the nucleocapsid can be found as a loosely coiled helix with a variable pitch and a smaller diameter (17). This difference in pitch and diameter between free and virus-incorporated nucleocapsids has been more extensively studied for vesicular stomatitis virus (VSV). The condensation of the free VSV nucleocapsid into the tightly coiled structure inside the virus is caused by the interaction of the M protein with the nucleocapsid (27, 28, 29). In order to show the relationship between the structure of the recombinant N-RNA rings and the morphologies of the viral nucleocapsids, we have modeled the free nucleocapsid and the coil inside the virus using the structure of the recombinant N-RNA rings, the morphology of negatively stained, CsCl-isolated viral N-RNA, and cryo-EM images of rabies virus (Fig. 6). Although the details in this new model are different, the principles are the same as those of the model described for VSV by Nakai and Howatson (27).
FIG. 6.
Models of free nucleocapsid and nucleocapsid inside virus. Negatively stained images of viral nucleocapsids isolated with CsCl gradient centrifugation (a) and cryo-EM images of rabies virus skeletons that were present in an untreated virus preparation (c) were used to determine the diameter of the two helical structures shown in panels b and d. (b) Tilted view of the recombinant rings, the free nucleocapsid, and nucleocapsid inside virus; (d) side views with an indication of the diameters. (e) Tilt angles between N and the symmetry axis of the rings or the helical axes in the two types of nucleocapsids. The difference of 18° between the tilt angles of N in rings or free nucleocapsids and N in virus is also shown (f).
In the recombinant rings with 10 monomers, the angle between the lines connecting three N monomers is 144° (Fig. 4). The numbers of monomers in each helical turn of both viral structures were determined by the diameters of the helices and by the size of the N monomer determined in the model for the recombinant rings of intact N-RNA. In order to go from the ring to the free nucleocapsid (Fig. 6a), the diameter had to increase from 160 to 240 Å (Fig. 6d), leading to an increase from 10 to 15 monomers per turn, corresponding to an increase in the angle between two N monomers from 144° to 156°. We also included a translation of two N heights per turn, as was inferred from the negative-stain images, in order to turn the rings into a helix. The tilt angles between N and the symmetry axis in the rings or the helical axis in the nucleocapsid were the same, 32°, as indicated (Fig. 6e). The external diameter of the helical coil in the virus was measured from the cryomicrographs (Fig. 6c) to be about 750 Å (Fig. 6d), leading to about 53.5 N monomers per turn. The angle between two N monomers was increased to 173°. The tilt angle of N inside the virus is 50° compared to the helix axis, a value that was measured from the cryomicrographs of the virus skeletons as shown (Fig. 6c). This introduces a tilt increase of 18° compared to N in the N-RNA rings (Fig. 6e and f). However, apart from the tilt angle, the contacts between the N monomers did not have to be modified. If this model were true, then the major effect of the interaction of the M protein with the nucleocapsid, compacting it from the free nucleocapsid to the tight coil inside the virus, would be an increase in this tilt angle. A consequence of this model would be that the part of N that is removed by trypsin and that probably constitutes the binding site for P would be positioned at the inside of the coil inside the virus. As mentioned above, the changes needed to go from the rings through the free nucleocapsid to the nucleocapsid coil inside the virus are only minimal in this model. The changes that would be needed in order to position the binding site for P to the outside of the coil in the virus would be much more drastic.
The geometry of the contacts between the N subunits in the rings and in the helical free nucleocapsids cannot be identical. The same holds for the contacts between the N subunits in the free and virion nucleocapsids. Along the same lines, the N-N contacts in the various helical states of the Sendai virus nucleocapsid cannot be identical, as discussed by Egelman et al. (10). For icosahedral viruses, the term quasi-equivalence is used to describe the differences between subunit contacts at fivefold vertices and those at sixfold symmetry axes (5). The same term could be applied for the interactions between the N subunits in negative-strand RNA virus nucleocapsids. Recently, Canady et al. (4) and Zhang et al. (40) have described quite extensive conformational changes in coat protein subunits and in the angles between the subunits when the morphology of icosahedral procapsids was compared with that of the mature capsids. These procapsids were stabilized by scaffolding proteins. It would be an interesting parallel to compare the effect of the rhabdovirus matrix protein on the helical parameters of the nucleocapsid and tilting angles of N with the effect of a scaffolding protein on an icosahedral capsid.
Position of the RNA in the N-RNA rings.
At the resolution obtained, 28 Å for the intact rings and 29 Å for the digested rings, the RNA in the rings is not visible. Although N binds to the RNA backbone and exposes the nucleotide bases (in the case of VSV [18]), we do not know the path of the single-stranded RNA in N-RNA. Rhabdoviral N-RNA contains 9 nucleotides per monomer of N (17, 39). The inside perimeter of the N-RNA rings at mid-height in the groove between the two lobes measures 315 Å, and the outside perimeter of the rings is 525 Å long (following the outside surface). A stretched-out nucleotide is about 5.5 Å long, so a stretched-out oligonucleotide of 90 residues, bound to 10 N monomers, would be about 495 Å long, which seems to exclude positioning at the outside of the rings. Another localization of the RNA could be at the top of the rings, close to the proposed binding site of P. This position would make sense, since P is supposed to place L onto the nucleotide sequence of the vRNA. The top of the digested rings forms a circle with a perimeter of 325 Å. In the nucleocapsid inside the virus, this position would also place the vRNA at the interior side of the coil, like the RNA in Tobacco Mosaic Virus (19, 36).
ACKNOWLEDGMENTS
The cryo-EM and scanning of the negatives were performed at the Institut de Biologie Structurale in Grenoble. The image shown in Fig. 6c was taken by Elizabeth Hewat of the Institut de Biologie Structurale.
We thank N. Ranson and H. Saibil (Birkbeck College, London) for providing the latest versions of the single-particle analysis procedure running on SPIDER and Yves Gaudin for critical comments on the text.
REFERENCES
- 1.Bankamp B, Horikami S M, Thompson P D, Huber M, Billeter M, Moyer S A. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology. 1996;216:272–277. doi: 10.1006/viro.1996.0060. [DOI] [PubMed] [Google Scholar]
- 2.Buchholz C J, Retzler C, Homann H E, Neubert W J. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology. 1994;204:770–776. doi: 10.1006/viro.1994.1592. [DOI] [PubMed] [Google Scholar]
- 3.Buckland R, Giraudon P, Wild F. Expression of measles virus nucleoprotein in Escherichia coli: use of deletion mutants to locate the antigenic sites. J Gen Virol. 1989;70:435–441. doi: 10.1099/0022-1317-70-2-435. [DOI] [PubMed] [Google Scholar]
- 4.Canady M A, Tihova M, Hanzlik T N, Johnson J E, Yeager M. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV) J Mol Biol. 2000;299:573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- 5.Caspar D L D, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–12. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 6.Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 7.Crowther R A, Henderson R, Smith J M. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 8.Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J Virol. 1993;67:4358–4364. doi: 10.1128/jvi.67.7.4358-4364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubochet J, Adrian M, Chang J J, Homo J C, Lepault J, McDowall A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 10.Egelman E H, Wu S-S, Amrien M, Portner A, Murti G K. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson S U. Transcription of vesicular stomatitis virus. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum; 1987. pp. 245–269. [Google Scholar]
- 12.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualisation of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 13.Fu Z F, Zheng Y, Wunner W H, Koprowski H, Dietzschold B. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology. 1994;200:590–597. doi: 10.1006/viro.1994.1222. [DOI] [PubMed] [Google Scholar]
- 14.Gigant B, Iseni F, Gaudin Y, Knossow M, Blondel D. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J Gen Virol. 2000;81:1757–1761. doi: 10.1099/0022-1317-81-7-1757. [DOI] [PubMed] [Google Scholar]
- 15.Graves M C. Measles virus polypeptides in infected cells studied by immune precipitation and one-dimensional peptide mapping. J Virol. 1981;38:224–230. doi: 10.1128/jvi.38.1.224-230.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heggeness M H, Scheid A, Choppin P W. The relationship of conformational changes in the Sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology. 1981;114:555–562. doi: 10.1016/0042-6822(81)90235-x. [DOI] [PubMed] [Google Scholar]
- 17.Iseni F, Barge A, Baudin F, Blondel D, Ruigrok R W H. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol. 1998;79:2909–2919. doi: 10.1099/0022-1317-79-12-2909. [DOI] [PubMed] [Google Scholar]
- 18.Iseni F, Baudin F, Blondel D, Ruigrok R W H. Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA. 2000;6:270–281. doi: 10.1017/s135583820099109x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeng T W, Crowther R A, Stubbs G, Chiu W. Visualization of alpha-helices in tobacco mosaic virus by cryo-electron microscopy. J Mol Biol. 1989;205:251–257. doi: 10.1016/0022-2836(89)90379-3. [DOI] [PubMed] [Google Scholar]
- 20.Kawai A, Toriumi H, Tochikura T S, Takahashi T, Honda Y, Morimoto K. Nucleocapsid formation and/or subsequent conformational change of rabies virus nucleoprotein is a prerequisite step for acquiring the phosphatase-sensitive epitope of monoclonal antibody 5-2-26. Virology. 1999;263:395–407. doi: 10.1006/viro.1999.9962. [DOI] [PubMed] [Google Scholar]
- 21.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klumpp K, Ruigrok R W H, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouznetzoff A, Buckle M, Tordo N. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J Gen Virol. 1998;79:1005–1013. doi: 10.1099/0022-1317-79-5-1005. [DOI] [PubMed] [Google Scholar]
- 24.Mellon M G, Emerson S U. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27:560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountcastle W E, Compans R W, Caliguiri L A, Choppin P W. Nucleocapsid protein subunits of simian virus 5, newcastle disease virus, and Sendai virus. J Virol. 1970;6:677–684. doi: 10.1128/jvi.6.5.677-684.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mountcastle W E, Compans R W, Lackland H, Choppin P W. Proteolytic cleavage of subunits of the nucleocapsid of the paramyxovirus simian virus 5. J Virol. 1974;14:1253–1261. doi: 10.1128/jvi.14.5.1253-1261.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakai T, Howatson A F. The fine structure of vesicular stomatitis virus. Virology. 1968;35:268–281. doi: 10.1016/0042-6822(68)90267-5. [DOI] [PubMed] [Google Scholar]
- 28.Newcomb W W, Brown J C. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J Virol. 1981;39:295–299. doi: 10.1128/jvi.39.1.295-299.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb W W, Tobin G J, McGowan J J, Brown J C. In vitro reassembly of vesicular stomatitis virus skeletons. J Virol. 1982;41:1055–1062. doi: 10.1128/jvi.41.3.1055-1062.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega J, Martín-Benito J, Zürcher T, Valpuesta J M, Carrascosa J L, Ortín J. Ultrastructural and functional analysis of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;12:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Préhaud C, Nel K, Bishop D H. Baculovirus-expressed rabies virus M1 protein is not phosphorylated: it forms multiple complexes with expressed rabies N protein. Virology. 1992;189:766–770. doi: 10.1016/0042-6822(92)90602-l. [DOI] [PubMed] [Google Scholar]
- 33.Préhaud C, Harris R D, Fulop V, Koh C L, Wong J, Flamand A, Bishop D H. Expression, characterization, and purification of a phosphorylated rabies nucleoprotein synthesized in insect cells by baculovirus vectors. Virology. 1990;178:486–497. doi: 10.1016/0042-6822(90)90346-s. [DOI] [PubMed] [Google Scholar]
- 34.Roseman A M, Chen S, White H, Braig K, Saibil H R. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 35.Schoehn G, Quaite-Randall E, Jiménez J L, Joachimiak A, Saibil H. Three conformations of an archeal chaperonin, TF55 from Sulfolobus shibatae. J Mol Biol. 2000;296:813–819. doi: 10.1006/jmbi.2000.3505. [DOI] [PubMed] [Google Scholar]
- 36.Stubbs G, Warren S, Holmes K. Structure of RNA and RNA binding site in tobacco mosaic virus from 4-A map calculated from X-ray fibre diagrams. Nature. 1977;267:216–221. doi: 10.1038/267216a0. [DOI] [PubMed] [Google Scholar]
- 37.Tacacs A M, Das T, Banerjee A K. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarbouriech N, Curran J, Ebel C, Ruigrok R W H, Burmeister W P. On the domain structure and the polymerization state of the Sendai virus P protein. Virology. 2000;266:99–109. doi: 10.1006/viro.1999.0066. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Greene B, Thuman-Commike P A, Jakana J, Prevelige P E, Jr, King J, Chiu W. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J Mol Biol. 2000;297:615–626. doi: 10.1006/jmbi.2000.3601. [DOI] [PubMed] [Google Scholar]