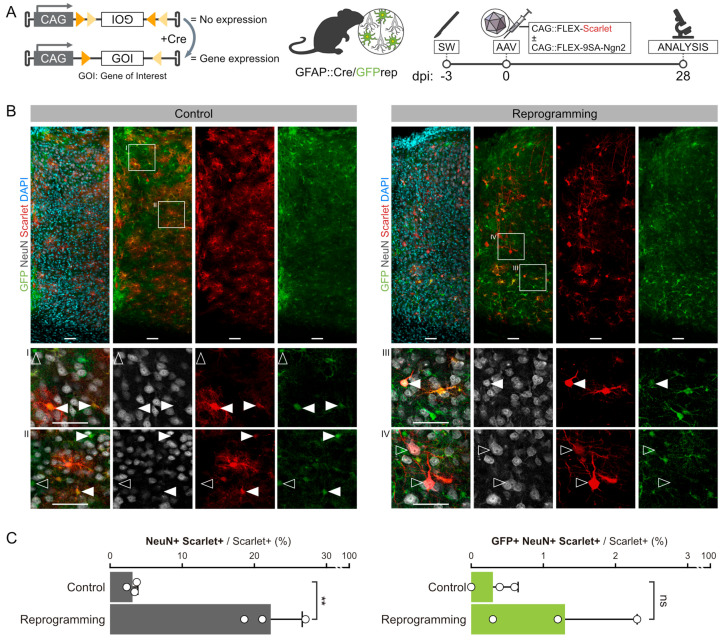
Figure 1.
Genetic lineage tracing of endogenous astrocytes during FLEX-AAV transduction. (A) Schematic of the Cre-dependent FLEX-switch (left) and surgical procedures used to lineage trace endogenous astrocytes in SW-injured GFAP::Cre/GFPrep mice after the injection of FLEX-AAVs (right). (B) Confocal pictures of a 15 μm stack of the 28 dpi SW-injured motor cortex of GFAP::Cre/GFPrep mice injected with control (left, light grey) or 9SA-Ngn2 (right, dark grey) viral mixes. Single optical plane magnifications of a representative group of cells are reported in inserts I-IV. AAV-transduced cells (red) were tested for the expression of the lineage tracing reporter GFP (green) and neuronal marker NeuN (white). For control experiments, GFP+ astrocytes targeted by the FLEX-AAVs are pointed out by full arrowheads, while non-infected astrocytes are pointed out by empty arrowheads (inserts I and II). For reprogramming experiments, one of the rare astrocyte-derived GFP+/NeuN+/mScarlet+ cells is pointed out by a full arrowhead in insert III, while GFP-/NeuN+/mScarlet+ endogenous neurons are pointed out by empty arrowheads in inserts III and IV. Scalebars: 50 μm. (C) Quantification and lineage tracing analysis of the NeuN+/mScarlet+ cells belonging to the control or 9SA-Ngn2 experimental groups. mScarlet+ cells in all cortical layers that were less than 340 μm distant from the SW were included in the quantification. Every dot represents the values from one animal, for which three sections have been averaged. Statistical differences in the numbers of NeuN+/mScarlet+ or GFP+/NeuN+/mScarlet+ cells between control and SA-Ngn2 mice were calculated by an unpaired t-test. **: p-value < 0.01; ns: no statistically relevant difference.