Abstract
We have characterized the properties of the maedi-visna virus (MVV) glycoprotein, which has a long cytoplasmic C-terminal domain, and of a panel of C-terminally truncated and C-terminally chimeric MVV-Env constructs. Cells expressing wild-type MVV glycoprotein form syncytia with target cells from many different species and tissues, demonstrating that the MVV-Env cellular receptor is widely distributed. Similar to the situation with other lentiviral glycoproteins, truncation of the C-terminal domain of MVV-Env significantly increases its membrane fusion capacity. However, despite their presence in a fusogenic form at the cell surface, neither the wild-type nor any of the C-terminally modified MVV-Env constructs, these latter lacking sterically inhibitory C termini, were able to successfully pseudotype murine leukemia virus- or human immunodeficiency virus-derived vector particles.
The molecular processes which play a role in determining whether a particular surface glycoprotein can be incorporated into the membrane of retroviral particles and mediate virus infectivity are not yet entirely understood. Incorporation is not restricted to the homologous glycoprotein, and pseudotyping of retroviral vector particles can be achieved with a number of heterologous glycoproteins (e.g., the glycoproteins of amphotropic murine leukemia virus [MuLV-Env], vesicular stomatitis virus [VSV-G], and gibbon ape leukemia virus give rise to high vector titers on the order of 105 to 106 IU/ml). Further examples of pseudotyping include the use of the glycoproteins of Ebola virus (45), lymphocytic choriomeningitis virus (27), and fowl plague virus (hemagglutinin) (8, 13). Additionally, incorporation of several cellular glycoproteins into retroviral and rhabdoviral particles has been demonstrated directly elsewhere (1, 10, 14, 15, 25, 36, 47). In several instances in which the incorporated proteins represent cellular receptors for virus uptake, the pseudotyped virus particles were infectious for cells expressing the respective viral glycoprotein (1, 10, 25, 36). All of these results demonstrate that foreign C-terminal regions on incorporated viral or cellular glycoproteins can be compatible with retroviral particle infectivity. However, it is of note that the naturally occurring C termini on these incorporable viral and cellular glycoproteins are usually relatively short (in the range of 15 to 35 amino acids [aa]). In contrast to most viral glycoproteins, which have only short C-terminal domains, most lentiviral glycoproteins have long cytoplasmic C termini (150 to 200 aa). The functions of these conserved regions during wild-type virus infection have not been completely elucidated but may, in part, involve binding to calmodulin (26, 40). Bulky heterologous C termini have been implicated to be inhibitory to incorporation and pseudotyping in many (15, 24, 37, 44) but apparently not all (18) cases. Thus, wild-type human immunodeficiency virus (HIV) glycoprotein, with its naturally occurring long C terminus (151 aa), was not able to pseudotype murine retroviral vector particles, but a C-terminally truncated mutant (7 aa) (43) was able to do so (24, 37).
Visna virus, referred to in this paper as maedi-visna virus (MVV), is the prototype virus of the family of Lentiviridae and causes chronic pneumonia and/or a progressive demyelinating disease in sheep. In common with most other lentiviral glycoproteins, the MVV glycoprotein has a very long cytoplasmic domain (126 aa). We were thus interested in establishing whether the MVV-Env C terminus would prevent incorporation of MVV glycoprotein into and pseudotyping of heterologous retroviral (HIV-like) particles by MVV glycoprotein and, if so, whether this situation could be reversed by removing this region. For this purpose, we have generated a panel of C-terminally truncated and C-terminally chimeric MVV glycoproteins and analyzed their properties with respect to fusion function toward different target cells and pseudotyping capability.
Expression of functional MVV-Env and distribution of its cellular receptor.
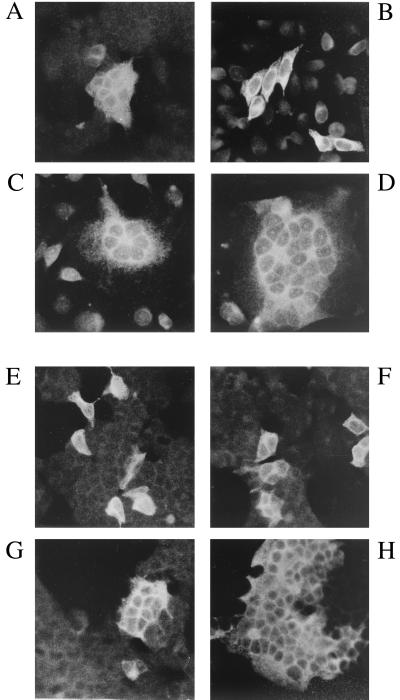
Plasmid pLV1-1KS1 (accession no. M60609 and M37977) (41) carries the entire proviral sequence of the replication-competent MVV strain 1514. A fragment (nucleotides 5401 to 9221), which contains the entire open reading frames (ORFs) of the MVV env and rev genes, was inserted into the eucaryotic expression vector pKEx (34) to yield pKEx-MVVenvWt (Fig. 1A). Expression, in transfected 293T cells, of MVV-EnvWt was analyzed by indirect immunofluorescence employing sera from sheep infected with MVV. Already at early time points posttransfection (p.t.) (24 h), transfected cells showed bright fluorescent staining and, as occurs after infection of susceptible sheep choroid plexus cells with MVV (12), large syncytia (Fig. 2A) were seen. This shows that the expressed MVV glycoprotein is functional and that human 293T cells (30) express the cellular receptor for MVV-Env at the cell surface. This was an indication that the cellular receptor for MVV-Env may be widely distributed, and in fact, transfection experiments in a limited number of different cell lines confirmed that large syncytia were formed in most instances. HeLa cells were, however, an exception, and transfected HeLa cells expressing MVV-Env remained predominantly as single cells (Fig. 2B). This allowed us to examine the distribution of the MVV cellular receptor in a coculture assay in which HeLa cells, transfected with pKEx-MVVenvWt, were trypsinized from the dish at 24 h p.t. and replated on glass coverslips with an excess (approximately three times more) of potential target cells. Twenty-four hours later, cocultured cells were subjected to indirect immunofluorescence with sheep anti-MVV serum. Figure 2C shows an example of cocultivation of MVVEnvWt-expressing HeLa cells with COS-7 cells. The presence of multinucleated syncytia showed that COS-7 cells (derived from monkey kidney) express the MVV-Env cellular receptor. In fact, in addition to 293T cells, human ECV (cardiovascular endothelium) and A204 (muscle, ATCC HTB-82) cells, in addition to COS-7 cells, monkey Vero cells (kidney, ATCC CCL-81), sheep SCP cells (choroid plexus, ATCC CRL-1700), equine ED cells (epidermal, ATCC CCL-57), hamster BHK cells (kidney, ATCC CRL-10), murine NIH 3T3 cells (fibroblast, ATCC CRL-1658), and chicken embryo fibroblast cells, formed multinucleated syncytia on cocultivation with MVV-Env-expressing HeLa cells. A further example of a cell line apparently lacking the MVV cellular receptor (in addition to HeLa cells, which show only weak membrane fusion) was HaCat cells. These are spontaneously transformed human keratinocytes (2). With these exceptions, MVV-Env appears to have a wide host range and mediates membrane fusion with cells from many species and tissues. Thus, the host range restriction of MVV infection is not likely to be due to a block at the level of receptor recognition and membrane fusion. In fact, it was previously shown that a large input of infectious MVV particles can induce syncytium formation in BHK cells, which are nonpermissive for MVV replication (12). Previous studies have pointed to the putative involvement of ovine major histocompatibility complex class II antigen as a component of the MVV-Env receptor (6), a possibility that was not further examined here. However, the results obtained demonstrate that if the major histocompatibility complex class II molecule is involved, its species origin would not appear to be important for functionality as a cellular receptor.
FIG. 1.
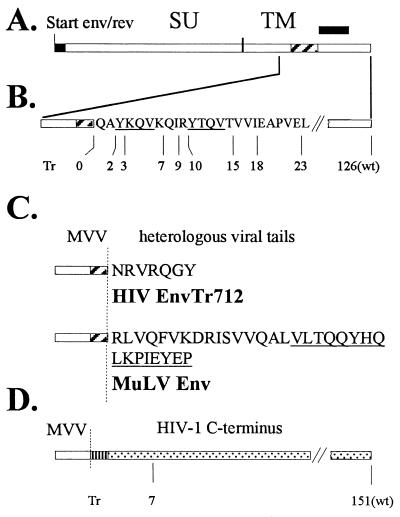
MVV constructs. (A) Genomic organization of MVV env and rev. MVV env and MVV rev begin in frame at the same initiation codon. The two rev exons are filled in black; the TMD region of the env gene is hatched. The border between the Env surface (SU) and transmembrane (TM) subunit is shown. (B) Relative positions of the C termini of the truncated MVV glycoproteins within the amino acid sequence of the expanded C terminus are as indicated. The two tyrosine-based motifs representing potential endocytosis signals within the C terminus of MVV-Env are underlined. (C) Amino acid sequences of the C termini of chimeric MVV glycoproteins with the TMD of MVV-Env. The C termini have been derived from HIV-EnvTr712 and MuLV-Env as indicated. The sequence of the MuLV R peptide is underlined. The vertical dotted line demarcates the border between MVV and heterologous sequences. (D) MVV-HIV chimeric constructs with the TMD of HIV-Env. The vertical dotted line demarcates the border between MVV and HIV sequences. The TMD of HIV-Env is vertically striped. The position of the C-terminal truncation in MVV-HIV-1TMDEnvTr712, with 7 remaining C-terminal amino acids, is indicated.
FIG. 2.
Membrane fusion capacity of wild-type and C-terminally truncated MVV glycoprotein. The figure shows indirect immunofluorescence (with anti-MVV serum) of 293T cells (A) or HeLa cells (B), transfected with pKEx-MVVenvWt; cocultivation of HeLa cells transfected with pKEx-MVVenvWt (C) or pKEx-MVVenvTr10 (D) with an excess of untransfected COS-7 cells; and indirect immunofluorescence of 293T cells expressing with MVV-HIV-1TMDEnvTr712 (plus MVV-Rev) (E), MVV-EnvTr2 (F), MVVTMD-MuLVEnv (plus MVV-Rev) (G), and MVV-EnvTr15 (H).
C-terminally truncated and C-terminally chimeric MVV-Env constructs.
Based on its hydrophobicity profile, the putative transmembranal domain (TMD) of the MVV glycoprotein has been proposed to extend from aa 832 to 863 (taking the initiation methionine as aa 1), i.e., it is encoded by the sequence from nucleotides 8450 to 8545 (3). This means that the cytoplasmic C-terminal domain contains 126 aa. An extensive panel of MVV-Env molecules with different truncations at the C terminus (schematically illustrated in Fig. 1B) was generated by introduction of in-frame stop codons downstream of the TMD sequence in pKEx-MVVenvWt by fusion PCR technology (16, 17). Additionally, in order to more easily identify mutated plasmids, new restriction sites were introduced downstream of the respective stop codons. The resultant plasmids were designated pKEx-MVVenvTr0 pKEx-MVVenvTr2, pKEx-MVVenvTr3, etc. (Tr for truncated), indicating the presence of 0, 2, or 3 aa, etc., remaining at the cytoplasmic C terminus of the respective mutant glycoproteins. The MVV env gene product carries two tyrosine-based motifs (Y-X-X-φ with X being any amino acid and φ being L, I, F, V, or M) within the cytoplasmic tail in close proximity to the lipid membrane (illustrated in Fig. 1B). In other systems, such tyrosine motifs have been implicated to be involved in endocytosis of the respective surface protein (5, 29). It is important to note that the different C-terminal truncations within the MVV-Env C terminus result in gene products encoding no (MVV-Env Tr0, Tr2, and Tr3), one (MVV-Env Tr7, Tr9, and Tr10), or both (MVV-Env Tr15, Tr18, and Tr23) of these tyrosine-based motifs. Additionally, chimeric MVV-Env proteins carrying heterologous cytoplasmic domains were generated again by fusion PCR technology (16, 17). The C-terminal regions were chosen to be those which are compatible with vector transduction by HIV- or MuLV-derived vector particles. In pKEx-MVVTMD-HIV-1envTr712, the C terminus of the HIV-Env truncation mutant Tr712 (43) was fused to the TMD of MVV-Env (Fig. 1C). This was achieved by inserting the appropriate sequence, encoding 7 aa downstream of the MVV TMD sequence, and by changing the following MVV-env codon to a stop codon. In pKEx-MVVTMD-MuLVenv, the entire cytoplasmic C terminus of MuLV-Env, including the R peptide, has been fused to the TMD of MVV and replaces downstream MVV-Env sequences (Fig. 1C). This was chosen since MuLV-Env itself with this C terminus has been shown elsewhere to efficiently pseudotype HIV-derived vector particles (20, 28). Additionally, two further MVV-HIV chimeras were generated in which both the TMD and the C terminus of wild-type HIV-Env (in pKEx-MVV-HIV-1TMDenvWt) or of HIV-EnvTr712 (in pKEx-MVV-HIV-1TMDenvTr712) were fused to the extracellular domain of MVV-Env, again replacing downstream MVV-Env sequences (Fig. 1D). In all cases, the entire sequences of the cloned PCR fragments were determined in order to confirm that only the intended mutations were present. Finally, an expression vector encoding MVV-Rev in the absence of Env, designated pKEx-MVVrev, was generated by introducing a large deletion (nucleotide 7326 to 8106) into pKEx-MVVenvWt.
Expression of truncated and chimeric MVV-Env glycoproteins.
In the plasmids encoding truncated MVV glycoproteins (Fig. 1B), the introduction of the premature stop codons, and the diagnostic restriction sites downstream of these, resulted in up to three amino acid changes in the overlapping rev ORF (nucleotide 8549 to 8910 [7]) (Rev changes summarized in Table 1). In the plasmids encoding the C-terminally chimeric MVV glycoproteins, except in the case of pKEx-MVVTMD-HIV-1envTr712 (see below), the second rev exon has been deleted. Thus, the expression of the different mutated MVV-env constructs was examined in the presence and absence of additional MVV-Rev coexpression from pKEx-MVVrev. Figure 3A, upper and middle panels, shows wild-type and C-terminally truncated MVV glycoproteins (the constructs depicted in Fig. 1B), immunoprecipitated with sheep anti-MVV serum, from lysates and culture supernatants of transfected cells, metabolically labeled for 5 h at 48 h p.t., as described previously (31). Additional Rev protein has been provided from pKEx-MVVrev. In all the cell lysates, predominant bands, migrating at the position of the glycoprotein precursor (approximately 159kDa) (Pr-MVV-EnvWt) or slightly faster (in the cases of the truncated constructs, consistent with the absence of 103 to 126 cytoplasmic aa in these cases), were observed. In most experiments, no distinct radioactive species, migrating at the position of the MVV surface glycoprotein (MVV-Env-SU), could be observed in the cell lysates. In contrast, in all the culture supernatants, only single bands migrating at the position of MVV-Env-SU (approximately 124kDa) were immunoprecipitated (Fig. 3A, middle panel). This indicates that proteolytic processing of Pr-MVV-Env (wild type and truncated) has occurred and that most of the generated SU has been shed into the culture supernatant. Quantitation of radioactivity in the respective bands indicates that the ratio of MVV-Env-SU to Pr-MVV-Env is similar for the wild type and most of the C-terminally truncated mutants (SU represents about 25% of the total MVV-Env present in cells plus culture supernatant). As shown in Fig. 3A, bottom panel, expression of all the C-terminally truncated MVV glycoproteins occurred in the absence of additional MVV-Rev expression. Quantitation of radioactivity in the respective bands indicates that, in all cases, the amounts of expressed MVV glycoproteins were not significantly altered whether MVV-Rev was coexpressed or not. Some of the changes in the domain affected by the mutations (Rev aa 50 to 72 [Table 1]) are nonconservative and also involve residues which are conserved between HIV-Rev and MVV-Rev (42). Thus, this region, which lies N-terminal to the functionally important basic domain, may not be critical for MVV-Rev function (at any rate as measured by expression of MVV-Env protein) and may thus be able to tolerate amino acid changes. In all experiments, there was a reproducible trend that MVV-EnvTr0 and MVV-EnvTr2 were expressed to higher levels than was MVVenvWt.
TABLE 1.
Properties of the different C-terminally truncated and chimeric MVV glycoproteins
| MVV glycoprotein | MVV-Rev amino acid changesa | Revc | Syncytium formationd | Pseudotypingf |
|---|---|---|---|---|
| MVV-EnvWt | None | − | + | − |
| MVV-EnvTr0 | (L49L) | − | −/+ | − |
| MVV-EnvTr2 | Q50D, S52C | − | −/+ | − |
| MVV-EnvTr3 | Q50L, (A51A), (S52S) | − | + | − |
| MVV-EnvTr7 | A54V, Q56L | − | ++ | − |
| MVV-EnvTr9 | Q56L, I57R, T59P | − | ++ | − |
| MVV-EnvTr10 | Y58N, N61T | − | ++ | − |
| MVV-EnvTr15 | S61I, (G63G), D64F | − | ++ | − |
| MVV-EnvTr18 | R65I, T67F | − | ++ | − |
| MVV-EnvTr23 | (I70I), G72S | − | + | − |
| MVVTMD-HIV-1-EnvTr712 | Noneb | + | + | − |
| MVVTMD-MuLV-Env | Deleted | + | + | − |
| MVV-HIV-1TMD-EnvWt | Deleted | + | − | − |
| MVV-HIV-1TMD-EnvTr712 | Deleted | + | − | − |
| VSV-G | − | ++ (low pH)e | 105–106 IU/ml |
Amino acid changes in Rev resulting from the mutations in the env ORF. Parentheses indicate the presence of mutations which do not result in an amino acid change.
No Rev amino acid changes but changes in the vicinity of the rev splice acceptor site (see text).
Requirement (+) or no requirement (−) for Rev expression from pKEx-MVVrev.
−, only single MVV-expressing cells; −/+, predominantly single MVV-expressing cells and a few small syncytia; +, syncytia with an average of 15 nuclei; ++, large syncytia containing >15 nuclei, often involving the whole culture.
Syncytia observed after treatment of transfected cells with low pH.
Pseudotyping was measured as the ability to mediate transduction of MuLV- or HIV-derived vectors carrying the β-galactosidase reporter gene. e, no. titer.
FIG. 3.
Analysis of wild-type and modified MVV glycoproteins. (A) Gel electrophoresis and autoradiography of immunoprecipitates from cell lysates (top and bottom panels) and culture supernatants (middle panel) of 293T cells transfected with plasmids encoding wild-type and C-terminally truncated MVV-Env in the presence (top and middle panels) and absence (bottom panel) of cotransfection with pKEx-MVV-rev. Lane 1, wild-type MVV-Env; lanes 2 to 10, truncated MVV-Env proteins Tr0, Tr2, Tr3, Tr7, Tr9, Tr10, Tr15, Tr18, and Tr23, respectively. The exposure time of the top panel is shorter than that of the middle and bottom panels. No sample has been applied in lane 4, bottom panel, but it was confirmed in further experiments that MVV-EnvTr3 is also equally expressed in the absence of additional MVV-Rev. (B) (Top panels) Gel electrophoresis of immunoprecipitates from cell lysates (left) and culture supernatants (right) of 293T cells transfected with plasmids encoding C-terminally chimeric MVV-Env in the presence of pKEx-MVV-rev. Lanes 1 and 6, MVV-HIV-1TMDEnvTr712; lanes 2 and 7, MVV-HIV-1TMDEnvWt; lanes 3 and 8, MVVTMD-MuLVEnv; lanes 4 and 9, MVVTMD-HIV-1EnvTr712; lanes 5 and 10, mock transfection. The right panel has been exposed longer to film than has the left panel. (Bottom panel) Gel electrophoresis of immunoprecipitates from cell lysates of 293T cells transfected with (+) and without (−) pKEx-MVV-rev and pKEx-MVV-HIV-1TMDEnvTr712 (lanes 1), pKEx-MVV-HIV-1TMDEnvWt (lanes 2), pKEx-MVVTMD-MuLVEnv (lanes 3), and pKEx-MVVTMD-HIV-1EnvTr712 (lanes 4). The positions of the respective MVV-Env components are given on the left, and those of molecular weight markers (in thousands) are given on the right.
As was to be expected, expression of the chimeric MVV glycoproteins with heterologous C termini (these are schematically depicted in Fig. 1C and D) is strictly dependent on coexpression of additional MVV-Rev (Fig. 3B). Thus, only on cotransfection of pKEx-MVVrev could predominant bands, migrating at the positions of the respective glycoprotein precursors, be observed in the cell lysates and cleaved MVV-Env-SU be observed in the culture supernatants. In the case of pKEx-MVV-HIV-1TMDenvWt, a weaker band of precursor glycoprotein could also be observed in the culture supernatant. The two constructs with the TMD of HIV-Env (encoded by pKEx-MVV-HIV-1TMDenvWt and pKEx-MVV-HIV-1TMDenvTr712) were reproducibly expressed to higher levels than were the other C-terminally chimeric MVV-Env proteins. As mentioned above, in the case of pKEx-MVVTMD-HIV-1envTr712, the second rev exon has not been deleted and, in fact, the amino acid sequence of the Rev ORF has actually not been changed. However, seven heterologous codons and a point mutation have been introduced 3 and 2 nucleotides upstream of the rev exon 2 splice acceptor site, respectively, which perhaps interferes with splicing required for production of mRNA for Rev.
Increased membrane fusion capacity of C-terminally truncated MVV-Env glycoproteins.
Indirect immunofluorescence of 293T cells transfected with the vectors encoding the truncated and chimeric MVV-Env proteins demonstrated that these resulted in different levels of membrane fusion (Fig. 2E to H). pKEx-MVV-HIV-1TMDenvWt and pKEx-MVV-HIV-1TMDenvTr712, with the TMD of HIV type 1 (HIV-1)-Env, did not result in syncytium formation at all (Fig. 2E), and cells expressing MVV-EnvTr0 and MVV-EnvTr2, with 0 and 2 C-terminal aa, respectively, also remained predominantly as single cells with some few, very small, syncytia (Fig. 2F). This is not simply the result of gross malfolding and retention in the endoplasmic reticulum, since all of these proteins were proteolytically cleaved (Fig. 3), a process which occurs in a late compartment of the Golgi complex. Lack of fusion in the cases of pKEx-MVV-HIV-1TMDenvWt and pKEx-MVV-HIV-1TMDenvTr712 could indicate that the TMD of MVV-Env is required during the processes leading to membrane fusion either directly or by influencing the conformation of the extracellular domain. On the other hand, it is conceivable that the deduced dimensions of the MVV-Env TMD (MVV-Env aa 832 to 863), which were determined only by analysis of the hydrophobicity profile of MVV-Env (3), are too large. In this case, the replacement of the deduced MVV-Env TMD region with the TMD of HIV-1 would lead to the deletion of a few amino acids in the extracellular domain of MVV-Env which may be required for membrane fusion function. In the case of the truncation mutants, MVV-EnvTr0 and MVV-EnvTr2, it is possible that the lack of at least a certain minimal size of cytoplasmic C terminus is in some way detrimental to functional glycoprotein folding at some stage in the fusion process. All of the other MVV-Env glycoproteins resulted in the formation of large syncytia. MVV-EnvTr3, MVV-EnvTr23, and the two chimeric constructs, MVVTMD-HIV-1EnvTr712 and MVVTMD-MuLVEnv, formed syncytia with sizes approximately the same as those formed by MVVEnvWt (Fig. 2G). It is of note that the MuLV-Env cytoplasmic domain present in MVVTMD-MuLV-Env contains the MuLV R-peptide sequence (Fig. 1C). This region has been shown previously to prevent membrane fusion both in the homologous context (i.e., MuLV-Env) and in a chimeric simian immunodeficiency virus glycoprotein containing the cytoplasmic C terminus of MuLV-Env (46). Nevertheless, 293T cells expressing MVVTMD-MuLV-Env were still able to form multinucleated syncytia, perhaps indicating that this region is not functional in the context of MVVTMD-MuLV-Env or that the strong fusion potential of truncated MVV-Env is partially overcoming the inhibitory effect of the R peptide. The remaining C-terminally truncated MVV-Env proteins (Tr7, Tr9, Tr10, Tr15, and Tr18) all formed larger syncytia than did MVV-EnvWt (Fig. 2H). In an experiment parallel to that shown in Fig. 2C for MVV-EnvWt, Fig. 2D shows syncytium formation between HeLa cells expressing one of these mutants (MVV-EnvTr10) and COS-7 cells. On average, three times as many nuclei are present per syncytium. This is analogous to the situation with HIV-Env (43) and simian immunodeficiency virus Env (33), in which cases truncation of the respective C termini also increased Env-mediated membrane fusion capacity. The properties of the different MVV-Env constructs are summarized in Table 1. It is worth noting that, in the metabolic labeling experiment described above, those proteins, which do not result in efficient syncytium formation, were more strongly labeled, indicating expression to higher levels.
As indicated above, the different truncations within the MVV-Env C terminus result in the removal of one or both of the tyrosine-based motifs within this region. However, we could not discern any correlation between the properties of the different constructs and the presence or absence of these motifs.
Pseudotyping retroviral vector particles with truncated and chimeric MVV-Env glycoproteins.
Since several of the C-terminally truncated and C-terminally chimeric MVV-Env constructs were functionally present at the cell surface and carried C-terminal domains which should not be sterically inhibitory, it was of interest to examine whether they could pseudotype retroviral vector particles. For the generation of murine retroviral particles, the packaging construct pSV-Ψ-MLV-env- (21) and the MuLV vector pBAG (32) encoding β-galactosidase were employed. For the generation of HIV-1-derived lentiviral particles, the packaging construct pCMV D8.91 and the HIV-derived vector pHR-CMVlacZ SIN18 (49) were employed. The respective constructs were transiently transfected into 293T cells and plasmids, encoding the individual glycoprotein under analysis, supplied by cotransfection. Culture supernatants, containing potentially pseudotyped vector particles (in the case of HIV-derived particles normalized by enzyme-linked immunosorbent assay for HIV-CA), were applied to fresh 293T cells as targets. Both VSV-G (49) and MuLV-Env (38), employed as positive controls, gave rise to high vector titers (in the range of 105 to 106 IU/ml). In our initial pseudotyping experiments with murine retroviral vectors, we reconfirmed, as described previously (24, 37), that pseudotyping with C-terminally truncated HIV-EnvTr712, with 7 instead of 151 aa at its cytoplasmic C terminus, but not with wild-type HIV-Env, resulted in transduction of appropriate CD4-positive target cells (data not shown). In the cases of pseudotyping with the different MVV-Env constructs, only very small numbers of cells expressing the marker gene (β-galactosidase), reflecting a titer of maximally 100 to 300 IU/ml, could be observed. However, these β-galactosidase-positive cells lost marker gene expression on passage, showing that the β-galactosidase gene had not been stably integrated in the cell genome, as is the case after genuine retrovirus-mediated transduction. It is possible that the low numbers of cells, showing transient expression of β-galactosidase, are the result of gene transfer mediated by membrane vesicles, as has been reported elsewhere to occur with VSV-G (35). It is of note that, in the cases of MVVTMD-MuLV-Env and MVVTMD-HIV-1EnvTr712, the respective C termini have proven, in the context of the respective homologous glycoprotein, to be compatible with both MuLV- and HIV-derived vector particle systems (23, 28, 37). In fact, a similar strategy, i.e., transfer of the C-terminal domain of MuLV-Env to a heterologous glycoprotein, has been shown elsewhere to improve pseudotyping of MuLV-derived vector particles with the glycoprotein of human foamy virus by a factor of 10 to 30 (22). The MuLV R peptide does not interfere with infectivity in the HIV system, since it can be proteolytically processed by the HIV-1 protease (20). However, despite these potentially advantageous properties, the results obtained show that there has been no genuine transduction at all by vector particles pseudotyped by any of the MVV-Env constructs.
Incorporation of MVV glycoproteins into HIV-like particles.
In order to gain insight as to why the MVV-Env constructs have failed to pseudotype retroviral vector particles, incorporation analysis of wild-type MVV-Env, MVV-EnvTr10, and MVVTMD-MuLV-Env into HIV-like particles was performed. All of these glycoproteins are membrane fusion competent, and the C termini of MVV-EnvTr10 and MVVTMD-MuLV-Env should potentially be sterically compatible with incorporation. pKEx-Tr-EGFR, expressing C-terminally truncated human epithelial growth factor receptor (Tr-EGFR), which is efficiently incorporated into HIV-like particles (15), was additionally employed as a positive control. HIV-like particle expression was achieved by employing pKEx-HIVΔEnv3, which encodes all of the HIV-1 genes except nef and env (14). After metabolic labeling of cells, transfected with expression vectors for the respective glycoproteins with or without pKEx-HIVΔEnv3, for 16 h at 32 to 48 h p.t., culture supernatants were clarified by filtering through a 45-μm-pore-size filter and centrifuged through a cushion of 32% sucrose for 3 h at 200,000 × g. Figure 4A shows electrophoresis of immunoprecipitated glycoproteins and of HIV-CA, employing rabbit antiserum directed against the HIV-1 capsid protein (CA), p24, from cell lysates and confirms that the expression levels of the respective glycoproteins were comparable. Figure 4B shows direct analysis, without immunoprecipitation, of equal amounts, as determined by enzyme-linked immunosorbent assay detecting HIV-CA (Innogenetics, Ghent, Belgium), of centrifuged viruslike particles. Radioactive CA can be observed in all cases in which pKEx-HIVΔenv3 has been expressed and Tr-EGFR can be seen to have been efficiently incorporated. However, no specific bands representing MVV-EnvWt, MVV-EnvTr10, or MVVTMD-MuLV-Env can be observed at all. This is also not the case after specific immunoprecipitation of the viruslike particles with anti-MVV serum and long exposure of the gel, indicating that incorporation of these components, if it occurs at all, is below the detection level of the methodology used here. As illustrated in Fig. 3, in common with many other retroviral glycoproteins, there is shedding of the MVV-SU protein into the medium. In fact, MVV-SU shedding is very pronounced, since MVV-SU could not be clearly seen in the cell lysates at all. Shedding could thus be the reason for the failure to detect MVV-Env-SU in particles. It is, however, clear that enough SU subunit remains attached to the TM subunit on the surface of MVV-Env-expressing cells (and this would presumably also be the case for released virions), since these form large syncytia. The fact that no radioactively labeled MVV-Env-TM moieties (neither wild type nor truncated) could be directly detected in virus particles is in line with the fact that, in our hands, it is also very difficult to directly detect metabolically labeled HIV-Env-TM (gp41) in infectious HIV particle preparations (data not shown). This may indicate that lentiviral glycoproteins are, in fact, incorporated only in low, but, in the case of HIV, obviously sufficient, amounts into particles. As described above, MVV-Env results in the induction of syncytia in transfected cells, and we considered the possibility that this potentially cytotoxic situation may have a general negative influence on glycoprotein incorporation into virus particles. To examine this, we analyzed incorporation of Tr-EGFR into particles released from cells additionally expressing either MVV-EnvTr10 or MVVTMD-MuLV-Env. In both cases, specific incorporation of Tr-EGFR could be detected. In the case of coexpression of MVV-EnvTr10, incorporation of Tr-EGFR was reduced by a factor of 5 to 10, pointing, indeed, to a negative influence of the large syncytia generated by this construct. However, in the case of coexpression of MVVTMD-MuLV-Env, which results in the formation of smaller syncytia, similar to those induced by MVV-EnvWt, the amount of incorporated Tr-EGFR was not significantly affected (data not shown). The reasons which can account for a failure of incorporation of surface glycoproteins lacking sterically inhibitory C termini are the subject of speculation and present investigation. Interactions of such glycoproteins with other cellular components may prevent incorporation either sterically or by directing the localization of the respective glycoprotein to a cell surface location distinct from the virus particle assembly site (14). In fact, Johnson et al. (19) have demonstrated that HIV glycoprotein constructs, which fail to be incorporated into VSV particles, do not localize with budding VSV.
FIG. 4.
Analysis of the incorporation of MVV glycoprotein into HIV-like particles. (A) Gel electrophoresis of immunoprecipitates from lysates of cells transfected with pKEx-MVV-rev and pKEx-MVVenvWt (lanes 1), pKEx-MVVenvTr15 (lanes 2), pKEx-EGFR-Tr (lanes 3), and pKEx-MVVTMD-MuLVEnv (lanes 4) with (+) or without (−) pKEx-HIVΔenv3 encoding HIV-1 Gag. (Upper panel) Immunoprecipitation of glycoprotein (anti-MVV-Env, lanes 1, 2, and 4; anti-EGFR, lanes 3); (lower panel) immunoprecipitation with anti-CA. (B) Electrophoresis (no immunoprecipitation) of the pellets obtained on centrifugation of culture supernatants of cells transfected as in panel A. The positions of the respective protein components are given on the left, and those of molecular weight markers (in thousands) are given on the right.
It is, however, of note that there have been reports of high titers of pseudotyped retroviral particles (spleen necrosis virus vector particles) in the absence of detectable particle-associated glycoprotein (4). Thus, we have to also consider the possibility that low, but potentially sufficient, amounts of the MVV-Env constructs have been incorporated into particles but that these are not detectable with our methods. In this case, the lack of pseudotyping potential would demonstrate that the respective glycoprotein cannot mediate all the steps required for virus uptake into the target cell. In this context, it is of note that, apart from HIV-EnvTr712, which is functional in mediating HIV-1 infectivity at least in certain cell lines, numerous further C-terminally truncated HIV-Env constructs are unable, or only very poorly able, to do so (9, 11, 44, 48). This is so although many of the described C-terminally truncated HIV glycoproteins are able to induce cell-cell fusion and are incorporated into virions. This means that, in addition to a putative requirement for short length for pseudotyping, the exact nature of the cytoplasmic region may be important in determining if a particular glycoprotein can mediate infectivity in the homologous (i.e., same virus) as well as in the heterologous (pseudotyping) context. The simplest explanation for this lack of function would be that, in the context of the viral membrane, in which the artificial C terminus is located in close proximity to the viral matrix layer, there are detrimental effects on the conformation of the extracellular domain at some stage in the fusion process. Several studies report on the effects of cytoplasmic C termini on the conformation of the extracellular domains of surface glycoproteins (e.g., see reference 39). On the other hand, although less easy to envisage, it is possible that membrane fusion is mediated normally by the viral glycoproteins with artificially truncated C termini but that infection is blocked at a subsequent step. Our present studies are aimed at distinguishing between these possibilities.
Acknowledgments
We thank K. Staskus for providing the MVV proviral plasmid, pLV1-1KS1; J. Clements and V. Andresdottir for MVV antisera; E. Pfaff for sheep choroid plexus and equine dermal cells; and R. Zufferey and D. Trono for plasmids pCMV D8.91, pHR′-CMVlacZ SIN18, and pMD-G. The following reagent was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pSV-Ψ-MLV-env- from Nathaniel Landau. We thank E. Pfaff for help and M. Pawlita and T. Wilk for discussion.
This work was supported, in part, by grant 01-KI-9412 from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie.
REFERENCES
- 1.Balliet J W, Bates P. Efficient infection mediated by viral receptors incorporated into retroviral particles. J Virol. 1998;72:671–676. doi: 10.1128/jvi.72.1.671-676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boukamp P, Petrusevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun M J, Clements J E, Gonda M A. The visna virus genome: evidence for a hypervariable site in the env gene and sequence homology among lentivirus envelope proteins. J Virol. 1987;61:4046–4054. doi: 10.1128/jvi.61.12.4046-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu T-H T, Dornburg R. Toward highly efficient cell-type-specific gene transfer with retroviral vectors displaying single-chain antibodies. J Virol. 1997;71:720–725. doi: 10.1128/jvi.71.1.720-725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collawn J F, Lai A, Domingo D, Fitch M, Hatton S, Trowbridge I S. Transferrin receptor internalisation sequence YTRF implicates a tight turn as the structural recognition motif for endocytosis. J Biol Chem. 1993;268:21686–21692. [PubMed] [Google Scholar]
- 6.Dalziel R G, Hopkins J, Watt N J, Dutia B M, Clark H A K, McConnell I. Identification of a putative cellular receptor for the lentivirus visna virus. J Gen Virol. 1991;72:1905–1911. doi: 10.1099/0022-1317-72-8-1905. [DOI] [PubMed] [Google Scholar]
- 7.Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong J, Roth M G, Hunter E. A chimeric avian retrovirus containing the influenza virus hemagglutinin gene has an expanded host range. J Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endres M J, Jaffer S, Haggerty B, Turner J D, Doranz B J, O'Brian P J, Kolson D L, Hoxie J A. Targeting of HIV- and SIV-infected cells by CD4-chemokine receptor pseudotypes. Science. 1997;278:1462–1464. doi: 10.1126/science.278.5342.1462. [DOI] [PubMed] [Google Scholar]
- 11.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harter D H, Choppin P W. Cell-fusing activity of Visna virus particles. Virology. 1967;31:270–288. doi: 10.1016/0042-6822(67)90172-9. [DOI] [PubMed] [Google Scholar]
- 13.Hatziioannou T, Valsesia-Wittmann S, Russell S J, Cosset F-L. Incorporation of fowl plague virus hemagglutinin into murine leukemia virus particles and analysis of the infectivity of the pseudotyped retroviruses. J Virol. 1998;72:5313–5317. doi: 10.1128/jvi.72.6.5313-5317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriksson P, Bosch V. Inhibition of cellular glycoprotein incorporation into human immunodeficiency virus-like particles by coexpression of additional cellular interaction partner. Virology. 1998;251:16–21. doi: 10.1006/viro.1998.9403. [DOI] [PubMed] [Google Scholar]
- 15.Henriksson P, Pfeiffer T, Zentgraf H, Alke A, Bosch V. Incorporation of wild-type and C-terminally truncated human epidermal growth factor receptor into human immunodeficiency virus-like particles: insight into the processes governing glycoprotein incorporation into retroviral particles. J Virol. 1999;73:9294–9302. doi: 10.1128/jvi.73.11.9294-9302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease R L. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Horton R M, Hunt H D, Ho S N, Pullen J K, Paese L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 18.Indraccolo S, Minuzzo S, Feroli F, Mammano F, Calderazzo F, Chieco-Bianchi L, Amadori A. Pseudotyping of Moloney leukemia virus-based retroviral vectors with simian immunodeficiency virus envelope leads to targeted infection of human CD4+ lymphoid cells. Gene Ther. 1998;5:209–217. doi: 10.1038/sj.gt.3300603. [DOI] [PubMed] [Google Scholar]
- 19.Johnson J E, Rodgers W, Rose J K. A plasma membrane localisation signal in the HIV-1 envelope cytoplasmic domain prevents localisation at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology. 1998;251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- 20.Kiernan R E, Freed E. Cleavage of the murine leukemia virus transmembrane env protein by human immunodeficiency virus type 1 protease: transdominant inhibition by matrix mutations. J Virol. 1998;72:9621–9627. doi: 10.1128/jvi.72.12.9621-9627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusso P, di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco S E, Kalyanaraman V S, Gallo R C. Expanded HIV-1 cellular tropism by phenotypic mixing with endogenous retroviruses. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 24.Mammano F, Salvatori F, Indraccolo S, De Rossi A, Chieco-Bianchi L, Göttlinger H. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol. 1997;71:3341–3345. doi: 10.1128/jvi.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mebatsion T, Finke S, Weiland F, Conzelmann K-K. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 26.Micoli K J, Pan G, Wu Y, Williams J P, Cook W J, McDonald J M. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J Biol Chem. 2000;275:1233–1240. doi: 10.1074/jbc.275.2.1233. [DOI] [PubMed] [Google Scholar]
- 27.Miletic H, Bruns M, Tsiakas K, Vogt B, Rezai R, Baum C, Kuhlke K, Cosset F L, Ostertag W, Lother H, von Laer D. Retroviral vectors pseudotyped with lymphocytic choriomeningitis virus. J Virol. 1999;73:6114–6116. doi: 10.1128/jvi.73.7.6114-6116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of non-dividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 29.Ochsenbauer C, Dubay S R, Hunter E. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol Cell Biol. 2000;20:249–260. doi: 10.1128/mcb.20.1.249-260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 32.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritter G D, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 34.Rittner C, Stöppler H, Pawlita M, Sczakiel G. Versatile eucaryotic vectors for strong and constitutive transient and stable gene expression. Methods Mol Cell Biol. 1991;2:176–181. [Google Scholar]
- 35.Rolls M M, Webster P, Balba N H, Rose J K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 36.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1 infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 37.Schnierle B S, Stitz J, Bosch V, Nocken F, Merget-Millitzer H, Engelstädter M, Kurth R, Groner B, Cichutek K. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc Natl Acad Sci USA. 1997;94:8640–8645. doi: 10.1073/pnas.94.16.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spies C P, Ritter G D, Jr, Mulligan M M, Compans R W. Truncation of the cytoplasmic domain of the immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivas S K, Srinivas R V, Anantharamaiah G M, Compans R W, Segrest J P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 41.Staskus K A, Retzel E F, Lewis E D, Silsby J L, St. Cyr S, Rank J M, Wietgrefe S W, Haase A T, Cook R, Fast D, Geiser P T, Harty J T, Kong S H, Lahti C J, Neufeld T P, Porter T E, Shoop E, Zachow K R. Isolation of replication competent molecular clones of Visna virus. Virology. 1991;181:228–240. doi: 10.1016/0042-6822(91)90488-w. [DOI] [PubMed] [Google Scholar]
- 42.Tiley L S, Malim M H, Cullen B R. Conserved functional organization of the human immunodeficiency virus type 1 and visna virus Rev proteins. J Virol. 1991;65:3877–3881. doi: 10.1128/jvi.65.7.3877-3881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 44.Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- 45.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young J A T, Bates P, Willert K, Varmus H E. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science. 1990;250:1421–1423. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]