Abstract
Among oncogenic adenoviruses, human adenovirus type 9 (Ad9) is unique in eliciting exclusively estrogen-dependent mammary tumors in rats and in not requiring viral E1 region transforming genes for tumorigenicity. Instead, studies with hybrid viruses generated between Ad9 and the closely related nontumorigenic virus Ad26 have roughly localized an Ad9 oncogenic determinant(s) to a segment of the viral E4 region containing open reading frame 1 (E4-ORF1), E4-ORF2, and part of E4-ORF3. Although subsequent findings have shown that E4-ORF1 codes for an oncoprotein essential for tumorigenesis by Ad9, it is not known whether other E4 region functions may similarly play a role in this process. We report here that new results with Ad9/Ad26 hybrid viruses demonstrated that the minimal essential Ad9 E4-region DNA sequences include portions of both E4-ORF1 and E4-ORF2. Investigations with Ad9 mutant viruses additionally showed that the E4-ORF1 protein and certain E4-ORF2 DNA sequences are necessary for Ad9-induced tumorigenesis, whereas the E4-ORF2 and E4-ORF3 proteins are not. In fact, the E4-ORF3 protein was found to antagonize this process. Also pertinent was that certain crucial nucleotide differences between Ad9 and Ad26 within E4-ORF1 and E4-ORF2 were found to be silent with respect to the amino acid sequences of the corresponding proteins. Furthermore, supporting a prominent role for the E4-ORF1 oncoprotein in Ad9-induced tumorigenesis, an E1 region-deficient Ad5 vector that expresses the Ad9 but not the Ad26 E4-ORF1 protein was tumorigenic in rats and, like Ad9, promoted solely mammary tumors. These findings argue that the E4-ORF1 oncoprotein is the major oncogenic determinant of Ad9 and that an undefined regulatory element(s) within the E4 region represents a previously unidentified second function likewise necessary for tumorigenesis by this virus.
Human adenoviruses, which are classified into six subgroups (A through F), cause a variety of human illnesses associated with infections of the respiratory and gastrointestinal tracts, as well as the eye (15). Although these viruses are not linked to human cancers, a subset of them, including all subgroup A and B viruses and two members of the subgroup D viruses, have the capacity to promote tumors in rodents. Following subcutaneous inoculation of animals, the subgroup A and B viruses induce undifferentiated sarcomas at the site of injection (13), whereas the subgroup D viruses Ad9 (adenovirus serotype 9) and Ad10 cause exclusively estrogen-dependent mammary tumors (1, 2, 17). Furthermore, it has been established that tumorigenesis by subgroup A and B viruses relies solely on their E1 region-encoded E1A and E1B oncoproteins (37). On the contrary, we have shown that tumorigenesis by subgroup D virus Ad9 lacks such a requirement for E1 region-encoded gene products and rather depends on the viral E4 region-encoded open reading frame 1 (E4-ORF1) oncoprotein (20, 41). Thus, two classes of oncogenic human adenoviruses can be distinguished based on the types of tumors they elicit in animals and the viral oncoproteins responsible for their tumorigenic potential.
One rationale for studying DNA tumor viruses such as adenovirus stems from the fact that such investigations have contributed greatly to our understanding of mechanisms responsible for the development of cancer (9). For example, the tumorigenic potentials of the nuclear adenovirus E1A and E1B oncoproteins, as well as the nuclear simian virus 40 (SV40) large T antigen, have been shown to depend in part on their abilities to complex with products of the pRb and p53 tumor suppressor genes (16, 31), two of the most commonly mutated genes in human cancers. These findings, together with succeeding studies of such interactions, have proven instrumental in defining functions for these two remarkably important tumor suppressor proteins.
While the mechanisms underlying the tumor-promoting capacity of the cytoplasmic Ad9 E4-ORF1 polypeptide have not been determined, our results suggest that transformation by this viral oncoprotein depends in part on its ability to complex with a select group of cellular PDZ domain-containing proteins, including DLG, MUPP1, and MAGI-1 (11, 23, 24, 44). These types of cellular factors generally act as scaffolding proteins in cell signaling (6, 8, 32), yet precise functions for the Ad9 E4-ORF1-associated PDZ proteins are not known. Nevertheless, DLG is a functional homologue of the Drosophila discs large (dlg) tumor suppressor protein (25, 28, 42) and, significantly, is likewise a cellular target for both the Tax oncoprotein of human T-cell leukemia virus type 1 and the E6 oncoproteins of high-risk but not low-risk human papillomaviruses (10, 21, 24, 40). These observations hint that important new mechanisms of oncogenesis may be uncovered through studies of the Ad9 tumor model system.
A prior study of hybrid viruses generated between Ad9 and the closely related nontumorigenic subgroup D virus Ad26 has shown that an essential determinant(s) for Ad9-induced tumorigenesis is encoded somewhere within the Ad9 E4 region DNA sequences encompassing E4-ORF1, E4-ORF2, and part of E4-ORF3 (18). The experiments reported here were undertaken to establish whether the genetic differences responsible for the disparate tumorigenic phenotypes of Ad9 and Ad26 map to one or more of these three E4 region functions. In light of the fact that the Ad9 E4-ORF1 oncoprotein, but not the E4-ORF2 or E4-ORF3 protein, has been found to represent a crucial determinant for Ad9-induced tumorigenesis (20), however, we anticipated that all pertinent genetic differences between Ad9 and Ad26 would be confined to E4-ORF1 DNA sequences. We report that new results with Ad9/Ad26 hybrid viruses unexpectedly demonstrated that such genetic differences actually localize within both the E4-ORF1 and E4-ORF2 coding regions. Additional findings presented in this study indicated that although the E4-ORF1 protein is the principal oncogenic determinant of Ad9, the essential E4-ORF1 and E4-ORF2 DNA sequences also likely define a previously unrecognized E4 region regulatory element(s) similarly necessary for tumorigenesis by Ad9.
MATERIALS AND METHODS
Cells.
Human 293 (ATCC CRL-1573) and A549 (ATCC CCL-185) cell lines were maintained in culture medium (Dulbecco's modified Eagle medium supplemented with gentamicin [20 μg/ml] and 10% fetal bovine serum) under a humidified 5% CO2 atmosphere at 37°C.
Construction of Ad5 recombinant vectors.
E1 region-deficient, replication-defective Ad5 recombinant vectors that express either Ad9 or Ad26 E4-ORF1 from a cytomegalovirus (CMV) promoter-driven cassette (dl327/9E4ORF1 or dl327/26E4ORF1, respectively) were constructed as described previously (35). Briefly, Ad9 or Ad26 E4-ORF1 coding sequences (nucleotides [nt] 471 to 855) were first introduced into the BamHI and EcoRI sites of the CMV expression cassette situated between the two correctly oriented Ad5 DNA fragments 0 to 1.3 and 9.3 to 17 map units (mu) within plasmid pACCMVpLpA (12). Ad5 recombinant vectors were generated by cotransfection of recombinant pACCMVpLpA plasmids and BstBI-digested dl327Bstβ-gal virion DNA into 293 cells. Resultant viral plaques failing to stain blue in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were isolated, and the cloned viruses were amplified and titrated on 293 cells. Virion DNAs were prepared from these viruses as described previously (41) and subjected to restriction enzyme analyses to verify proper genomic structures.
Construction of Ad9/Ad26 hybrid and Ad9 mutant viruses.
Two different two-step procedures were used to construct Ad9/Ad26 hybrid viruses. First, with the use of common restriction enzyme sites or by PCR methods, Ad9/Ad26 hybrid E4 regions were assembled either within plasmid p26XbaIA (18), containing the Ad26 DNA fragment XbaI-A (65 to 100 mu), or within plasmid p26EcoRI(B+C), containing correctly oriented Ad26 DNA fragments EcoRI-C (0 to 7.5 mu) and EcoRI-B (89.5 to 100 mu). Second, for isolation of hybrid viruses by overlap recombination (5), Ad9/Ad26 hybrid p26XbaIA plasmids were cotransfected into A549 cells with plasmid p26EcoRI(A+C) containing the Ad26 DNA fragment 0 to 89.5 mu (18). Alternatively, for isolation of hybrid viruses from whole virus genome plasmids (41), the Ad26 virion-derived DNA fragment EcoRI-A (7.5 to 89.5 mu) was introduced in the correct orientation into the unique EcoRI site of Ad9/Ad26 hybrid p26EcoRI(B+C) plasmids, and resultant infectious, whole virus genome p26EcoRI(A+B+C) plasmids were transfected into 293 cells.
For construction of Ad9 mutant viruses, E4 region-containing DNA fragments were mutated either by disruption of appropriate restriction enzyme sites or by PCR methods. With the use of convenient restriction enzyme sites, mutant E4 regions were subsequently introduced into plasmid p9EcoRI(B+C) (41), containing correctly oriented Ad9 DNA fragments EcoRI-B (0 to 7.5 mu) and EcoRI-C (95 to 100 mu). The virion-derived Ad9 DNA fragment EcoRI-A (7.5 to 95 mu) was subsequently introduced in the correct orientation into the unique EcoRI site of p9EcoRI(B+C) mutant plasmids. For isolation of mutant viruses, resultant infectious, whole virus genome p9EcoRI(A+B+C) plasmids were transfected into 293 cells.
Ad9/Ad26 hybrid and Ad9 mutant viruses were amplified and titrated in either 293 or A549 cells (19). A combination of limited sequence and restriction enzyme analyses was used to verify the genomic structures of all viruses.
Tumor assays.
One- or two-day-old male and female Wistar/Furth rats (Harlan Sprague-Dawley, Indianapolis, Ind.) were inoculated subcutaneously on both flanks with the indicated dose of virus and then monitored for tumors over an 8-month period as previously described (19). Portions of tumors were either fixed in 10% neutral-buffered formalin for histological examination or frozen at −80°C for DNA and protein analyses. Caring and handling of animals were in accordance with institutional guidelines.
Antisera, cell extracts, and immunoblot assays.
Ad9 E4-ORF2 coding sequences, PCR amplified with primers 5′AGC TGG ATC CAT GCT TCA GCG ACG CG3′ and 5′CGC GAA TTC TCA TAA TAG AAA CAG ATC C3′, were introduced between the BamHI and EcoRI sites of plasmid pGEX-2T (Pharmacia) in frame with the glutathione S-transferase (GST) gene. GST–Ad9 E4-ORF2 fusion protein was expressed in bacteria, purified, and used as an antigen to generate rabbit polyclonal antisera (39).
At 24 h postinfection, virus-infected A549 or 293 cells (10 PFU/cell) were washed in ice-cold phosphate-buffered saline and lysed in sample buffer (0.065 M Tris-HCl [pH 6.8], 2% [wt/vol] sodium dodecyl sulfate, 10% [vol/vol] glycerol, 1% [vol/vol] β-mercaptoethanol, 0.0015% [wt/vol] bromophenol blue). Resultant cell extracts were boiled and centrifuged (16,000 × g, 10 min). Protein concentrations of these cleared cell extracts were determined by the Bradford assay (4). For immunoblot analyses, proteins from cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes, which were sequentially incubated with blocking buffer (5% nonfat dry milk in TBST [50 mM Tris-HCl {pH 7.4}, 200 mM NaCl, 2% Tween 20]), with Ad9 E4-ORF1 antiserum (20) or Ad9 E4-ORF2 antiserum (1:5,000 in TBST), and then with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibodies (Southern Biotechnology Associates). Membranes were developed by enhanced chemiluminescence (Pierce).
PCR assays.
Virion, cellular, and tumor DNAs were isolated by standard methods (41). PCR amplifications were performed with Taq polymerase (Stratagene) as recommended by the manufacturer. Ad9 E4-ORF1 expression cassette and Ad5 E1 region DNA sequences were PCR amplified with primer pairs Ad9 E4-ORF1 [nt 471-494] (5′ATG GCT GAA TCT CTG TAT GCT TTC3′)/SV40 cassette (5′GCG GAA TTC TTC AGG GGG AGG TGT GGG AG3′) and Ad5 [nt 2504-2525] (5′GCA GCC AGG GGA TGA TTT TGA G3′)/Ad5 [nt 3053-3075] (5′CCT CGC AGT TGC CAC ATA CCA TG3′), respectively.
RESULTS
Tumorigenesis by Ad9 depends on DNA sequences located within both E4-ORF1 and E4-ORF2.
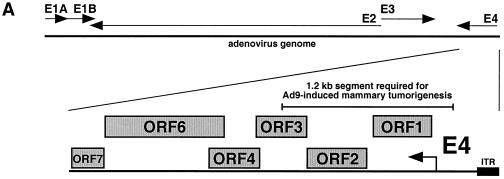
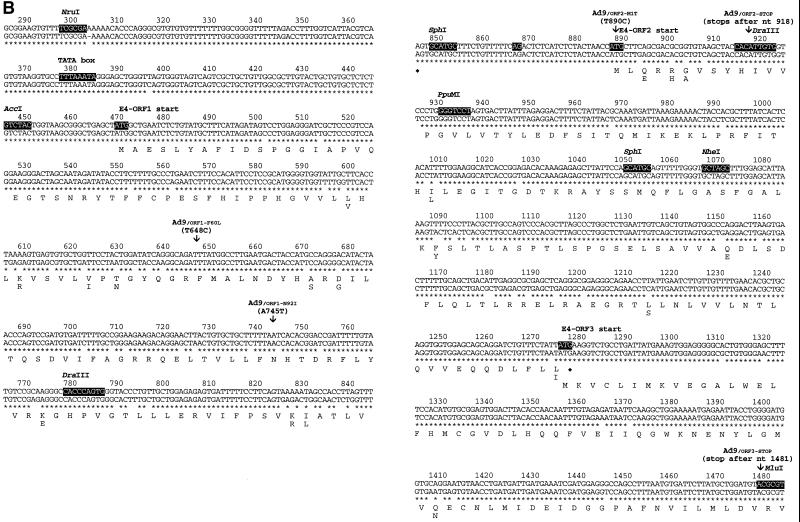
Our previous results with Ad9/Ad26 hybrid viruses indicate that an approximately 1.2-kb segment of the Ad9 E4 region encodes a determinant(s) for tumorigenesis by Ad9 (Fig. 1A) (18). With the extreme right end of the adenovirus genome defined as nt 1, this segment extends from the NruI site at nt 299 to the MluI site at nt 1481 (Fig. 1B). These sequences either partially or completely code for four separate functions, including the E4 promoter/5′ untranslated region, E4-ORF1, E4-ORF2, and E4-ORF3. Additionally, alignment of these Ad9 sequences with the corresponding Ad26 sequences reveals a total of 74 nucleotide differences distributed within all four functional elements contained in the defined segment (Fig. 1B). Although this particular finding fails to aid more precise localization of the Ad9 E4 region oncogenic determinant(s), we have previously shown that Ad9 E4-ORF1 codes for a transforming protein (20, 46) and that expression of this polypeptide is required for Ad9-induced tumorigenesis (20). From these observations, we postulated that specific nucleotide differences within the E4-ORF1 genes of Ad9 and Ad26 may be solely responsible for the divergent tumorigenic phenotypes of these viruses.
FIG. 1.
(A) Illustration of the Ad9 E4 region showing the location of the 1.2-kb segment required for Ad9-induced mammary tumorigenesis. ITR, inverted terminal repeat. (B) Alignment of E4 region DNA sequences of Ad9 (top) and Ad26 (bottom) extending from nt 299 to 1481. Within the Ad9 DNA sequences, common restriction enzyme sites used to construct Ad9/Ad26 hybrid viruses, the E4 promoter TATA box, the initiator methionine codons of E4 proteins, and a putative splice acceptor site at nt 868 used to generate E4-ORF2 mRNAs are highlighted. Shown below the DNA sequences are the amino acid sequences of Ad9 E4 proteins, beneath which are indicated Ad26 amino acid residues differing from those of Ad9. The mutations of viruses shown in Table 1 are also described. Asterisks denote nucleotide identity between the Ad9 and Ad26 DNA sequences.
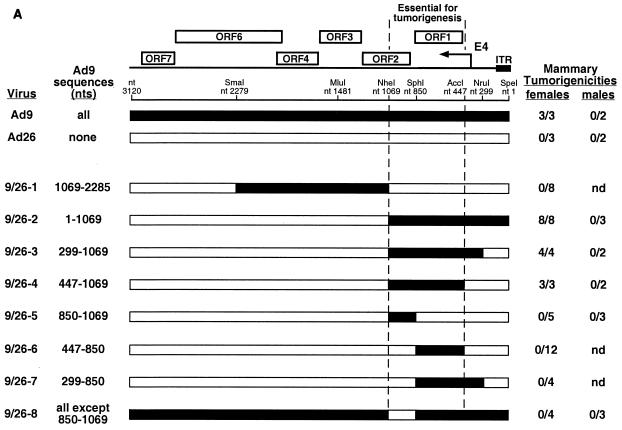
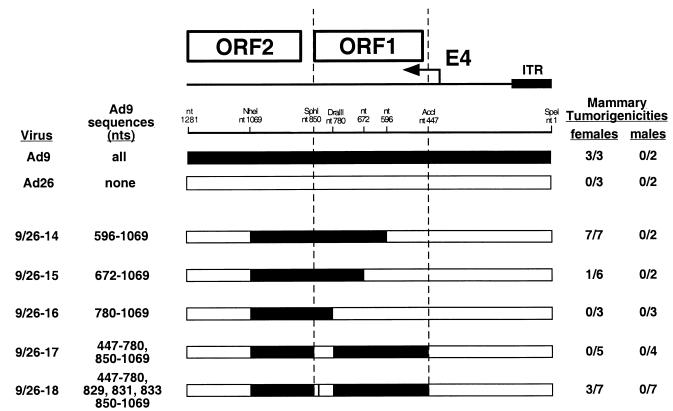
We initially tested this idea by constructing the eight different Ad9/Ad26 hybrid viruses (group 1 hybrid viruses) shown in Fig. 2A. Seven of these viruses, 9/26-1 to 9/26-7, have an Ad26 genome in which specific blocks of the E4 region were replaced by equivalent Ad9 sequences, whereas virus 9/26-8 has an Ad9 genome in which the 5′ half of E4-ORF2 was replaced by equivalent Ad26 sequences. To determine the tumorigenic potentials of these viruses, we inoculated newborn rats subcutaneously with 7 × 107 PFU of each virus and then monitored the animals for the development of tumors for 8 months. In these assays, the hybrid viruses behaved identically to either the tumorigenic parental virus Ad9, which generates solely mammary tumors in 100% of infected female rats, or the nontumorigenic parental virus Ad26 (18).
FIG. 2.
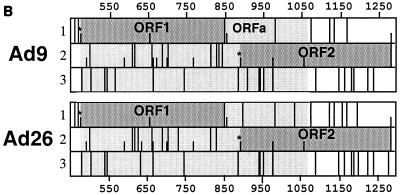
(A) Genomic structures and tumorigenic potentials of group 1 Ad9/Ad26 hybrid viruses. Newborn rats were inoculated subcutaneously on each flank with a total of 7 × 107 PFU of the indicated virus, and the animals were monitored for tumor formation over an 8-month period. Filled and open genomic regions depict Ad9 and Ad26 DNA sequences, respectively; restriction enzyme sites used to construct these hybrid viruses are indicated. The two vertical dashed lines delimit the segment of the Ad9 E4 region shown by results with these hybrid viruses to be essential for tumorigenesis by Ad9. nd, not determined; ITR, inverted terminal repeat. (B) ORF maps spanning the E4-ORF1 and E4-ORF2 DNA sequences of Ad9 and Ad26. The three reading frames (1, 2, and 3) of the E4 region sense strand are shown. Darkly shaded regions highlight E4-ORF1 and E4-ORF2 coding sequences, whereas the lightly shaded region covers the 622-bp essential E4 region segment extending from nt 447 to 1069. Vertical lines segmenting each reading frame indicate stop codons, whereas shorter vertical ticks denote methionine codons. Asterisks mark the initiator methionine-codons of E4-ORF1 and E4-ORF2.
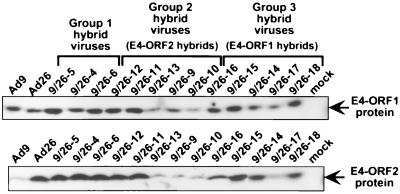
Among the wild-type tumorigenic hybrid viruses shown in Fig. 2A, virus 9/26-4 contained the least amount of Ad9 E4 region DNA sequences, 622 bp, extending from the AccI site at nt 447 to the NheI site at nt 1069 (Fig. 1B). This segment of the E4 region contains the entire E4-ORF1 gene, a 40-bp noncoding region between the E4-ORF1 and E4-ORF2 genes, and the 5′ half of the E4-ORF2 gene. For both Ad9 and Ad26, additional methionine codon-containing E4-ORFs having the capacity to code for peptides longer than 15 residues are absent in this segment, with the exception of nonconserved Ad9 E4-ORFa (nt 852 to 980, ORF1) which potentially expresses a 42-residue polypeptide (Fig. 2B). Whereas their 40-bp noncoding regions exhibit 100% sequence identity, Ad9 and Ad26 display 39 nucleotide differences in E4-ORF1 and 10 nucleotide differences in the defined portion of E4-ORF2, producing 9 amino acid differences in the E4-ORF1 protein and 4 amino acid differences in the E4-ORF2 protein, respectively (Fig. 1B). It was also noteworthy that hybrid viruses 9/26-6 and 9/26-5, containing only the corresponding Ad9 E4-ORF1 or Ad9 E4-ORF2 sequences of tumorigenic virus 9/26-4, respectively, were nontumorigenic in rats (Fig. 2A), as these results suggested that Ad9-induced tumorigenesis depends on two separate E4 region functions. Furthermore, viruses 9/26-4, 9/26-5, and 9/26-6 similarly expressed wild-type levels of the E4-ORF1 and E4-ORF2 proteins during lytic infections of human A549 cells (Fig. 3).
FIG. 3.
Expression of E4-ORF1 and E4-ORF2 proteins by Ad9/Ad26 hybrid viruses. Cell extracts were prepared from human A549 cells mock infected or lytically infected with the indicated virus (10 PFU/cell, 24 h postinfection) and then subjected to immunoblot analyses with antisera raised against either an Ad9 E4-ORF1 or Ad9 E4-ORF2 fusion protein. The E4-ORF1 and E4-ORF2 proteins have molecular masses of 14 and 14.5 kDa, respectively.
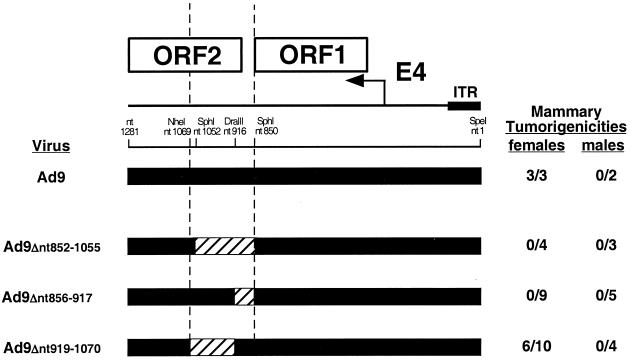
Because it was surprising to uncover a requirement for E4-ORF2 DNA sequences in tumorigenesis by Ad9, we performed a deletion mutation analysis of the 220-bp region immediately downstream of E4-ORF1, including the 40-bp noncoding region (nt 849 to 888) and the 5′ half of E4-ORF2 (nt 889 to 1069). For this purpose, we engineered three different Ad9 deletion mutant viruses (Ad9Δnt852-1055, Ad9Δnt856-917, and Ad9Δnt919-1070) (Fig. 4) which, consistent with their deletions, expressed approximately wild-type amounts of the E4-ORF1 protein in A549 cells yet did not express the E4-ORF2 protein (Fig. 5). The results of experiments examining the tumorigenic potentials of these mutant viruses showed that both Ad9Δnt852-1055 and Ad9Δnt856-917 failed to elicit tumors, whereas Ad9Δnt919-1070 exhibited a partially tumorigenic phenotype in that it generated mammary tumors in only 60% of infected females (Fig. 4). In contrast, wild-type Ad9 invariably promotes mammary tumors in 100% of infected females (Fig. 4) (17). Moreover, compared to wild-type Ad9-induced tumors, the tumors elicited by virus Ad9Δnt919-1070 showed an extended latency period (4 to 5 months versus 3 months) and were also typically smaller (data not shown). These results with mutant viruses provided further evidence indicating that a previously unrecognized oncogenic determinant for Ad9 is located within the 220-bp region immediately downstream of E4-ORF1.
FIG. 4.
Genomic structures and tumorigenic potentials of Ad9 deletion mutant viruses. Methods for determining the tumorigenicity of viruses are described in the legend to Fig. 2A. Filled and hatched genomic regions represent Ad9 and deleted DNA sequences, respectively; locations of restriction enzyme sites used to create these deletions are indicated. The two vertical dashed lines delimit the 220-bp region immediately downstream of E4-ORF1 implicated in Ad9-induced tumorigenesis by results with group 1 hybrid viruses (Fig. 2A). ITR, inverted terminal repeat.
FIG. 5.
Expression of E4-ORF1 and E4-ORF2 proteins by Ad9 mutant viruses. Immunoblot analyses used to detect E4-ORF1 and E4-ORF2 proteins in extracts of A549 cells mock infected or lytically infected with the indicated virus (10 PFU/cell, 24 h postinfection) were carried out as described in the legend to Fig. 3.
The Ad9 E4-ORF1 protein, but not the E4-ORF2 or E4-ORF3 protein, is a critical oncogenic determinant for Ad9.
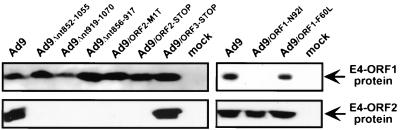
As the results described thus far demonstrated a requirement for certain E4-ORF2 DNA sequences in Ad9-induced tumorigenesis, we were prompted to reevaluate a possible role for the E4-ORF2 protein in this process. For this purpose, we constructed two different Ad9 mutant viruses specifically unable to express a functional E4-ORF2 polypeptide. In the virus Ad9/ORF2-MIT, the E4-ORF2 initiator methionine codon was changed to a threonine codon, whereas in virus Ad9/ORF2-STOP, stop codons in all three reading frames were inserted after E4-ORF2 histidine codon 10 (Fig. 1B). It is also notable that the putative 42-amino-acid residue product of Ad9 E4-ORFa (Fig. 2B) was truncated to 22 residues in virus Ad9/ORF2-STOP. As controls, we engineered three additional Ad9 mutant viruses, Ad9/ORF1-N92I, Ad9/ORF1-F60L, and Ad9/ORF3-STOP. Ad9/ORF1-N92I carries the N92I mutant E4-ORF1 gene, which is transformation defective due to unstable protein expression, whereas Ad9/ORF1-F60L carries the F60L mutant E4-ORF1 gene, which expresses a transformation-proficient protein (Fig. 1B) (43). In addition, Ad9/ORF3-STOP has a 4-bp deletion within the MluI site at nt 1481, thereby truncating the 117-residue E4-ORF3 polypeptide to a 68-residue amino-terminal peptide (Fig. 1B). This mutation is identical to that of mutant hybrid virus inMluI reported previously (20).
The results of experiments assessing the tumorigenic potentials of these Ad9 mutant viruses are presented in Table 1. We found that despite their inability to express the E4-ORF2 protein (Fig. 5), viruses Ad9/ORF2-M1T and Ad9/ORF2-STOP displayed wild-type tumorigenic phenotypes in rats. Sequencing of E4-ORF2 genes PCR amplified from tumor DNAs confirmed that the tumors were caused by these mutant viruses and that their mutations had not reverted (data not shown). The findings with E4-ORF2 mutant viruses differed from those obtained with virus Ad9/ORF1-N92I, in which a specific failure to express the E4-ORF1 protein (Fig. 5) led to a nontumorigenic phenotype. The latter result was specific because virus Ad9/ORF1-F60L, which expressed wild-type levels of its transformation-proficient E4-ORF1 protein (Fig. 5), displayed wild-type tumorigenicity in animals. Though Ad9/ORF3-STOP was likewise tumorigenic in rats, mammary tumors promoted in females by this virus arose with a shortened latency period compared to tumors induced by wild-type Ad9 (2 months versus 3 months). Similar results have been reported for virus inMluI (20). Interestingly, Ad9/ORF3-STOP also generated mammary tumors in all of the male rats with a 5-month latency period. Introducing an identical E4-ORF3 mutation into Ad26, however, did not confer it with a tumorigenic phenotype (data not shown). While exhibiting substantially enhanced tumorigenicity in rats, virus Ad9/ORF3-STOP expressed wild-type rather than elevated levels of the E4-ORF1 protein in A549 cells (Fig. 5). These findings with Ad9 mutant viruses corroborated and extended our previous results indicating a requirement for the E4-ORF1 protein, but not for the E4-ORF2 and E4-ORF3 proteins, in tumorigenesis by Ad9 (20).
TABLE 1.
The E4-ORF1 oncoprotein but not the E4-ORF2 or E4-ORF3 protein is required for mammary tumorigenesis by Ad9a
| Virusb | No. of rats that developed tumors/no. infected
|
|
|---|---|---|
| Females | Males | |
| Wild-type | ||
| Ad9 | 3/3 | 0/2 |
| E4-ORF1 mutants | ||
| Ad9/ORF1-N92I | 0/5 | 0/4 |
| Ad9/ORF1-F60L | 4/4 | 0/3 |
| E4-ORF2 mutants | ||
| Ad9/ORF2-M1T | 5/5 | 0/5 |
| Ad9/ORF2-STOP | 13/13 | 0/3 |
| E4-ORF3 mutant | ||
| Ad9/ORF3-STOP | 3/3 | 3/3 |
Evidence that an undefined regulatory element(s) represents a second E4 region oncogenic determinant for Ad9.
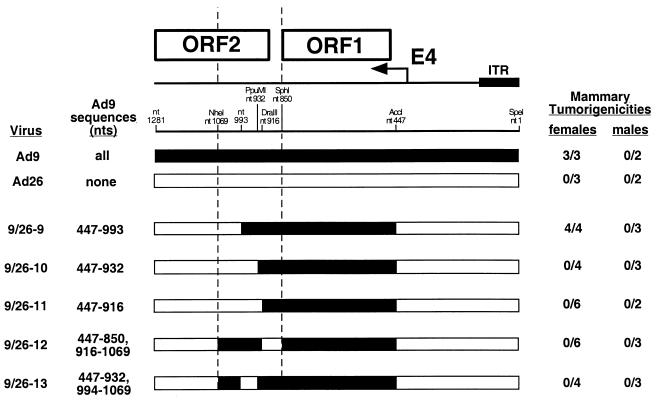
As results with hybrid viruses showed that tumorigenesis by Ad9 depends in part on DNA sequences within the 5′ half of E4-ORF2 (Fig. 2A), we were interested in identifying the crucial nucleotide differences between Ad9 and Ad26 in this region. For this purpose, we constructed five different Ad9/Ad26 hybrid viruses (group 2 hybrid viruses) having a wild-type tumorigenic virus 9/26-4 genome (Fig. 2A) in which specific segments of the Ad9 E4-ORF2 sequences were replaced by equivalent Ad26 sequences (viruses 9/26-9 to 9/26-13) (Fig. 6). In A549 cells, two of these hybrid viruses (9/26-11 and 9/26-12) expressed wild-type levels of the E4-ORF1 and E4-ORF2 proteins, but the remaining three hybrid viruses (9/26-9, 9/26-10, and 9/26-13) expressed lower levels of both polypeptides (Fig. 3). Despite expressing reduced amounts of the E4-ORF1 and E4-ORF2 proteins, however, virus 9/26-9 showed wild-type tumorigenicity following inoculation into rats, whereas the remaining four hybrid viruses were found to be nontumorigenic (Fig. 6).
FIG. 6.
Genomic structures and tumorigenic potentials of group 2 Ad9/Ad26 hybrid viruses. Methods for determining the tumorigenicity of viruses are described in the legend to Fig. 2A. Filled and open genomic regions represent Ad9 and Ad26 sequences, respectively; locations of restriction enzyme sites or PCR primers used to construct these hybrid viruses are indicated. The two vertical dashed lines delimit the 220-bp region immediately downstream of E4-ORF1 implicated in Ad9-induced tumorigenesis by results with group 1 hybrid viruses (Fig. 2A). ITR, inverted terminal repeat.
The results with these group 2 hybrid viruses implicated two separate E4-ORF2 segments, nt 850 to 916 and 932 to 993, in tumorigenesis by Ad9. An important role for the nt 850–916 segment was demonstrated by the fact that replacing these Ad9 sequences of tumorigenic virus 9/26-4 with equivalent Ad26 sequences resulted in nontumorigenic virus 9/26-12 (Fig. 6). In this 66-bp segment, Ad9 and Ad26 display four nucleotide and three amino acid differences at the amino terminus of the E4-ORF2 polypeptide (Fig. 1B). The nt 932–993 segment was likewise important because replacing these Ad9 sequences of tumorigenic viruses 9/26-4 and 9/26-9 with equivalent Ad26 sequences resulted in nontumorigenic viruses 9/26-13 and 9/26-10, respectively (Fig. 6). Significantly, in this 61-bp segment, Ad9 and Ad26 display two nucleotide differences (nt 969 and 993), both of which are silent with respect to the amino acid sequence of the E4-ORF2 polypeptide (Fig. 1B).
A similar strategy was used to identify crucial nucleotide differences between Ad9 and Ad26 within the essential E4-ORF1 sequences. In this case, we constructed five different Ad9/Ad26 hybrid viruses (group 3 hybrid viruses) having a wild-type tumorigenic virus 9/26-4 genome (Fig. 2A) in which specific segments of the Ad9 E4-ORF1 gene were replaced by equivalent Ad26 sequences (viruses 9/26-14 to 9/26-18) (Fig. 7). After inoculation into rats, these hybrid viruses manifested one of three distinct phenotypes, including wild-type tumorigenicity (virus 9/26-14), partial tumorigenicity (viruses 9/26-15 and 9/26-18), or nontumorigenicity (viruses 9/26-16 and 9/26-17) (Fig. 7). In addition, though possessing diminished tumorigenic potentials, the latter four viruses were found to express wild-type amounts of the E4-ORF1 protein in A549 cells (Fig. 3).
FIG. 7.
Genomic structures and tumorigenic potentials of group 3 Ad9/Ad26 hybrid viruses. Methods for determining the tumorigenicity of viruses are described in the legend to Fig. 2A. Filled and open genomic regions represent Ad9 and Ad26 sequences, respectively; locations of restriction enzyme sites or PCR primers used to construct these hybrid viruses are indicated. The two vertical dashed lines delimit the 403-bp E4-ORF1 region implicated in Ad9-induced tumorigenesis by results with group 1 hybrid viruses (Fig. 2A). ITR, inverted terminal repeat.
The results with these group 3 hybrid viruses revealed involvement of two separate E4-ORF1 segments, nt 596 to 780 and nt 780 to 850, in tumorigenesis by Ad9. The importance of the nt 596–780 segment was shown by the fact that replacing these Ad9 sequences of tumorigenic virus 9/26-14 with equivalent Ad26 sequences resulted in nontumorigenic virus 9/26-16 (Fig. 7). The partially tumorigenic phenotype of virus 9/26-15 further suggested that crucial nucleotide differences within E4-ORF1 are distributed both upstream and downstream of nt 672. In the defined 184-bp segment, Ad9 and Ad26 display 23 nucleotide and 7 amino acid differences in the middle of the E4-ORF1 polypeptide (Fig. 1B).
The nt 780–850 segment, on the other hand, was implicated by the fact that replacing these Ad9 sequences of tumorigenic virus 9/26-4 with equivalent Ad26 sequences resulted in nontumorigenic virus 9/26-17 (Fig. 7). In this 70-bp segment, Ad9 and Ad26 display 12 nucleotide differences, three of which produce two amino acid differences near the carboxyl terminus of the E4-ORF1 polypeptide (Fig. 1B). Virus 9/26-18 was constructed to distinguish whether the three amino acid-altering nucleotide differences (nt 829, 831, and 833) or the remaining nine silent nucleotide differences in this segment of E4-ORF1 were important. In this regard, virus 9/26-18 is identical to nontumorigenic virus 9/26-17 except that the three Ad26-derived amino acid-altering nucleotides of virus 9/26-17 are converted to the respective Ad9 nucleotides in virus 9/26-18. Conversely, virus 9/26-18 is likewise identical to wild-type tumorigenic virus 9/26-4 except that the nine Ad9-derived silent nucleotides of virus 9/26-4 are replaced with the respective Ad26 nucleotides in virus 9/26-18 (Fig. 1B and 7). Therefore, the fact that virus 9/26-18 exhibited a partially tumorigenic phenotype in rats (Fig. 7) suggested that both amino acid-altering and silent nucleotide differences in E4-ORF1 contribute to the divergent tumorigenic phenotypes of Ad9 and Ad26.
Collectively, our findings with group 2 and group 3 hybrid viruses revealed that amino acid-altering nucleotide differences in E4-ORF1, as well as silent nucleotide differences in both E4-ORF1 and E4-ORF2, are responsible for the divergent tumorigenic phenotypes of Ad9 and Ad26. The observed role for silent nucleotide differences, together with results demonstrating that the E4-ORF2 polypeptide is dispensable for tumorigenesis by Ad9 (Table 1), suggested that an undefined E4 region regulatory element(s), rather than a protein function, represents a second determinant for Ad9-induced tumorigenesis (see Discussion).
The E4-ORF1 oncoprotein is the major oncogenic determinant of Ad9.
Compared with other oncogenic adenoviruses, Ad9 is unique both in targeting tumorigenesis to the mammary glands of animals and in having the E4-ORF1 protein as an oncogenic determinant (17, 18, 20). Considering these observations, we hypothesized that the strict propensity of Ad9 to promote mammary tumors may be largely due to unique activities associated with its E4-ORF1 oncoprotein. This question was addressed by examining whether an otherwise nontumorigenic subgroup C human adenovirus (Ad5) that is engineered to express the Ad9 E4-ORF1 oncoprotein would become tumorigenic and, if so, whether this virus would also acquire the unique capacity to elicit exclusively mammary tumors in animals.
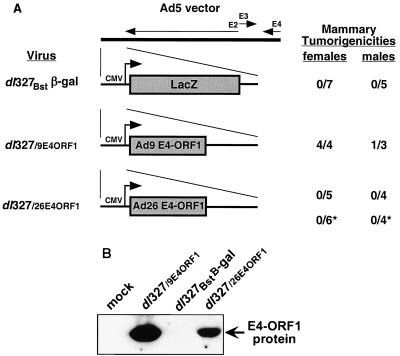
Because E1 region transforming functions of Ad9 are dispensable for its tumorigenic potential (41), we chose to utilize the Ad5 recombinant vector dl327Bstβ-gal, in which the Ad5 E1 region genes are deleted and replaced by a lacZ expression cassette (35). By replacing the lacZ cassette of this Ad5 vector with CMV promoter-driven Ad9 E4-ORF1 and Ad26 E4-ORF1 cassettes, we isolated viruses dl327/9E4ORF1 and dl327/26E4ORF1, respectively (Fig. 8A). Because our main interest was to investigate the E4-ORF1 protein determinant in these experiments, the expression cassettes contained only E4-ORF1 coding sequences and therefore lacked sequences downstream of this gene. Using our Ad9 E4-ORF1 antiserum, which reacts with subgroup D but not subgroup C adenovirus E4-ORF1 proteins (45), we detected heterologous E4-ORF1 protein expression by both dl327/9E4ORF1 and dl327/26E4ORF1 but not by dl327Bstβ-gal during lytic infections of human 293 cells (Fig. 8B), a cell line that complements the replication defects of such Ad5 vectors by stably expressing Ad5 E1 region gene products (14). Also noteworthy was that the E4-ORF1 protein levels observed for both dl327/9E4ORF1 and dl327/26E4ORF1 in these assays are substantially higher than those achieved by wild-type Ad9 after infection of 293 cells at the same multiplicity of infection (data not shown).
FIG. 8.
(A) Genomic structures and tumorigenic potentials of E1 region-deficient Ad5 recombinant virus vectors that express the LacZ (dl327Bstβ-gal), Ad9 E4-ORF1 (dl327/9E4ORF1), or Ad26 E4-ORF1 (dl327/26E4ORF1) protein. Methods for determining the tumorigenicity of viruses are described in the legend to Fig. 2A. Asterisks indicate results obtained after inoculating rats with 7 × 108 PFU of virus, as opposed to the 7 × 107-PFU inoculum used with other rats in this experiment. (B) E4-ORF1 protein expression by Ad5 vectors. Immunoblot analyses used to detect heterologous E4-ORF1 protein in extracts of 293 cells mock infected or lytically infected with the indicated virus (10 PFU/cell, 24 h postinfection) were carried out as described in the legend to Fig. 3.
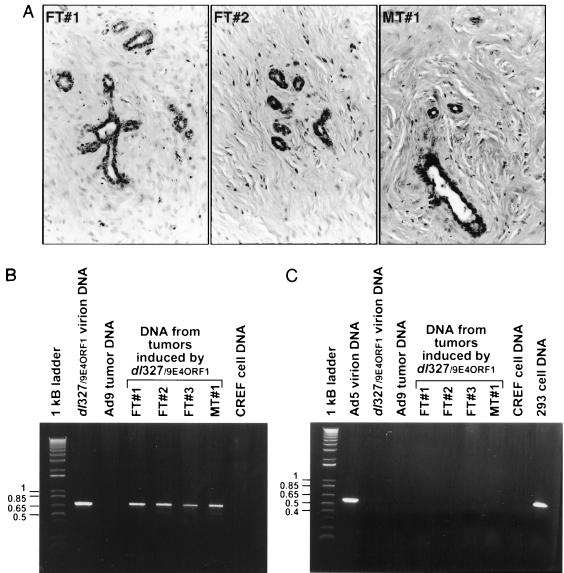
Following subcutaneous inoculation of newborn rats with 7 × 107 PFU of each Ad5 vector, we found that dl327Bstβ-gal and dl327/26E4ORF1 failed to induce tumors, whereas dl327/9E4ORF1 generated tumors in all of the females (3-month tumor latency) and in one male (5-month tumor latency) (Fig. 8A). As somewhat lower E4-ORF1 protein expression was observed for dl327/26E4ORF1 than for dl327/9E4ORF1 (Fig. 8B), we also demonstrated that rats inoculated with a 10-fold-higher dose of dl327/26E4ORF1 likewise failed to develop tumors of any kind (Fig. 8A). More important, histological analyses of the dl327/9E4ORF1-induced male and female tumors indicated that they were exclusively mammary fibroadenomas, identical to those generated by wild-type Ad9 (Fig. 9A) (17). Virus dl327/9E4ORF1 rather than a wild-type Ad9 contaminant promoted these tumors because, using primers specific for the Ad9 E4-ORF1 cassette of dl327/9E4ORF1, we succeeded in PCR amplifying the predicted 700-bp product from DNAs of dl327/9E4ORF1-induced tumors but not from DNA of an Ad9-induced tumor or CREF cells (Fig. 9B). Additional results also showed that dl327/9E4ORF1-induced tumors express the Ad9 E4-ORF1 protein at levels slightly above those seen in wild-type Ad9-induced tumors (data not shown). An inability to PCR amplify a 540-bp Ad5 E1 region product from these tumor DNAs also confirmed that the tumorigenic potential of dl327/9E4ORF1 is not dependent on Ad5 E1 region functions (Fig. 9C). These findings are significant in demonstrating that an otherwise nontumorigenic E1 region-deficient Ad5 vector that heterologously expresses the Ad9 E4-ORF1 oncoprotein not only becomes tumorigenic but also promotes solely mammary tumors like those induced by wild-type Ad9.
FIG. 9.
(A) dl327/9E4ORF1-induced tumors are histologically identical to mammary tumors generated by wild-type Ad9. Female tumor 1 (FT#1), female tumor 2 (FT#2), and male tumor 1 (MT#1) are fibroademomas. The stromal portion of MT#1, however, is more sclerotic, indicating higher collagen composition. (B) dl327/9E4ORF1-induced mammary tumors contain Ad9 E4-ORF1 cassette DNA sequences (C) but not Ad5 E1 region DNA sequences. Tumor DNAs (2 μg) or virion DNAs (1 ng) were subjected to PCR amplification using primer pairs specific for either the Ad9 E4-ORF1 cassette of virus dl327/9E4ORF1 or the Ad5 E1 region (see Materials and Methods). DNAs from three different dl327/9E4ORF1-induced mammary tumors from females (FT#1, FT#2, and FT#3), as well as one from a male (MT#1), were examined. Water or DNA from Ad5 virions, CREF cells, 293 cells, or an Ad9-induced tumor served as controls. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide.
DISCUSSION
The work presented in this paper was undertaken to precisely define the E4 region DNA sequences that determine mammary tumorigenesis by Ad9. Findings with Ad9/Ad26 hybrid viruses and Ad9 mutant viruses localized these essential DNA sequences to portions of both E4-ORF1 and E4-ORF2 (Fig. 2A and 4). We also showed that abrogating E4-ORF1 protein expression by introducing a single nucleotide substitution into the E4-ORF1 gene of Ad9 abolished its tumorigenic potential (virus Ad9/ORF1-N92I) whereas, conversely, maintaining transformation-competent E4-ORF1 protein expression by introducing a different nucleotide substitution into the E4-ORF1 gene of Ad9 preserved its wild-type tumorigenic potential (virus Ad9/ORF1-F60L) (Fig. 5; Table 1). These results demonstrate an absolute requirement for the Ad9 E4-ORF1 oncoprotein in tumorigenesis by Ad9. Results with Ad9/Ad26 hybrid viruses further suggested that certain amino acid differences between the E4-ORF1 polypeptides of Ad9 and Ad26 contribute to the dramatically divergent tumorigenic phenotypes of these viruses (Fig. 7). Because Ad9 and Ad26 E4-ORF1 expression plasmids display similar transforming potentials in CREF rat embryo fibroblasts in vitro (unpublished results) (18), we hypothesize that such amino acid differences cause the Ad26 E4-ORF1 protein to have stability or perhaps functional deficiencies specifically in cells of the rat mammary gland. Alternatively, similar to the Ad5 E1A protein (22), the Ad26 E4-ORF1 protein may provoke a strong inflammatory response, leading to clearance of infected cells. With respect to the essential Ad9 DNA sequences identified within E4-ORF2, however, results with mutant viruses indicated that the E4-ORF2 polypeptide is dispensable for Ad9-induced tumorigenesis (Table 1). Taken together, these findings argue that the tumorigenic potential of Ad9 depends on two separate E4 region determinants, only one of which represents a protein function.
Supporting the existence of a nonprotein oncogenic determinant in the Ad9 E4 region, results with Ad9/Ad26 hybrid viruses revealed that silent nucleotide differences with respect to the E4-ORF1 and E4-ORF2 polypeptides are also partly responsible for the divergent tumorigenic phenotypes of Ad9 and Ad26 (Fig. 6 and 7). With respect to other potentially important protein-encoding ORFs within the essential E4 region DNA sequences, Ad9 E4-ORFa is the only one, besides E4-ORF1 and E4-ORF2, capable of encoding a peptide larger than 15 residues (Fig. 2B). The fact that a stop codon interrupted Ad9 E4-ORFa in tumorigenic virus Ad9/ORF2-STOP, however, argues against a role for this ORF in Ad9-induced tumorigenesis. Therefore, we postulate that the essential Ad9 sequences define a novel E4 region regulatory element(s) which, like the Ad9 E4-ORF1 oncoprotein, is also necessary for Ad9-induced tumorigenesis.
For tumorigenic virus 9/26-9, it was shown that only two silent substitutions, at nt 969 and 993, in E4-ORF2 are required to produce nontumorigenic virus 9/26-10 (Fig. 6). This finding indicates that these two nucleotides are critical for the function of the proposed E4 region regulatory element. Consequently, it was surprising that virus Ad9Δnt919-1070, in which the segment extending from nt 919 to 1070 is deleted, displayed a partial tumorigenic rather than nontumorigenic phenotype (Fig. 4). The reason for this disparity is not known, but a possible explanation could be that for virus Ad9Δnt919-1070, DNA sequences immediately downstream of the deleted region are able to partially complement the missing component of the regulatory element in a position-dependent manner, thereby endowing this mutant virus with weak but measurable tumorigenic potential.
A function for the proposed regulatory element(s) has not been established, but we presume that it would act at the level of transcription, mRNA stability, or splicing within the Ad9 E4 region transcription unit. It may be relevant, however, that the crucial, silent nucleotide differences within E4-ORF1 and E4-ORF2 flank both sides of a conserved 40-nt noncoding region containing the putative splice acceptor site at nt 868 used to generate E4-ORF2 mRNAs (Fig. 1B). Additionally, the regions affected by these silent nucleotide differences have sequences and locations reminiscent of splicing branch sites and exonic splicing enhancers, respectively (3). Recruitment of splicing factors to such elements in pre-mRNAs facilitates splicing through the formation of protein networks across introns and exons. Further considering that alternative splicing plays a central role in regulating gene expression by the adenovirus E4 region (36), we favor the idea that the proposed element(s) functions to modulate splicing of certain E4 region mRNAs. Perhaps related to this possibility, future studies will examine whether reduced E4-ORF2 splice acceptor site selection during production of E4 region mRNAs is responsible for the decreased E4-ORF2 protein expression observed for Ad9 compared to Ad26 in lytically infected A549 cells (Fig. 3).
While the proposed regulatory element(s) is expected to directly control the abundance of specific E4 region mRNAs in cells, this activity likely ultimately alters expression of one or more E4 proteins. Two different models in which the element either increases or decreases protein expression can be envisioned. In our first model, the abundance of E4 proteins that enhance tumorigenesis by Ad9 is increased. For example, an increase in E4-ORF1 protein levels might result if the element were to block conversion of the primary E4 region transcript, which expresses the E4-ORF1 protein (7), into spliced mRNA species coding for other E4 proteins. Although this idea is appealing because the element and E4-ORF1 share overlapping sequences, evidence for such an activity was not obtained in experiments examining E4-ORF1 protein levels in A549 cells infected with hybrid or mutant viruses (Fig. 3). It must be considered, however, that this postulated function for the element may be restricted to specific cell types, such as those of the rat mammary gland. Additionally, besides possibly increasing accumulation of the E4-ORF1 oncoprotein in cells, the proposed element could likewise potentially augment levels of other adenovirus E4 proteins known to possess transforming potential, including E4-ORF3 (30), E4-ORF6 (27, 29), and E4-ORF6/7 (47). In our second model, we imagine that the abundance of E4 proteins that suppress tumorigenesis by Ad9 is decreased by the proposed element. The E4-ORF4 protein, which triggers programmed cell death (26, 38), and the Ad9 E4-ORF3 protein, which antagonizes tumorigenesis by Ad9 (see below), represent plausible candidates for this scenario.
Although the Ad5 E4-ORF3 protein has been reported to possess transforming potential (30), our results with virus Ad9/ORF3-STOP indicate that the Ad9 E4-ORF3 gene product is dispensable for Ad9-induced tumorigenesis (Table 1). In fact, compared to wild-type Ad9, Ad9/ORF3-STOP displayed enhanced tumorigenicity in rats, as revealed by the shortened tumor latency period in females and by the occurrence of tumors in males. These results show that the Ad9 E4-ORF3 protein actually inhibits Ad9-induced tumorigenesis. Furthermore, the fact that Ad9/ORF3-STOP elicited mammary tumors in males with 100% frequency is particularly noteworthy because tumors induced by wild-type Ad9 are known to be absolutely dependent on estrogen for growth and maintenance (1, 17). Thus, our findings with Ad9/ORF3-STOP are significant in arguing that the Ad9 E4-ORF3 protein is responsible, in part, for the strict estrogen dependence of Ad9-induced mammary tumors. In this regard, one interesting possibility may be that the Ad9 E4-ORF3 protein possesses an activity that attenuates the response of estrogen receptor to its hormone ligand in mammary cells.
It is remarkable that the E1 region-deficient Ad5 vector dl327/9E4ORF1, which heterologously expresses the Ad9 E4-ORF1 protein, not only was tumorigenic in rats but also generated exclusively mammary tumors identical to those induced by wild-type Ad9 (Fig. 8A and 9A). Expression of the Ad9 E4-ORF1 oncoprotein is specifically required for this effect because the Ad5 vector dl327Bstβ-gal or dl327/26E4ORF1, which instead heterologously expresses the Ad26 E4-ORF1 protein, was nontumorigenic in rats. Additional work is needed to determine whether, in the context of the Ad5 vector, both silent and amino acid-altering nucleotide differences are responsible for the inability of Ad26 E4-ORF1 to promote mammary tumors. Considering that the E1 region codes for the major transforming functions of Ad5 (37), it is also notable that Ad5 E1 region genes were found to be dispensable for tumorigenesis by virus dl327/9E4ORF1 (Fig. 9C). This observation is in agreement with our previous findings showing that the Ad9 E1 region is likewise unnecessary for mammary tumorigenesis by Ad9 (41). Nonetheless, because the Ad5 vector used in these studies possesses an intact E4 region, it is feasible that Ad5 E4 proteins contribute to the tumorigenic potential of dl327/9E4ORF1. Regardless of this possibility, however, the fact that virus dl327/9E4ORF1 and Ad9 display nearly identical oncogenic properties in rats provides a compelling argument that the major oncogenic determinant of Ad9 is its E4-ORF1 oncoprotein.
The reason that Ad9 generates only mammary tumors in rats has not been established. Its select tropism is unlikely due to restricted infection of mammary cells in animals because Ad9 binds to the same widely expressed cellular receptor used by Ad5 (33, 34). Additional findings also suggest that transcription from the Ad9 E4 region is neither enhanced in nor limited to estrogen receptor-expressing cells stimulated by hormone (unpublished results; 19). Thus, the finding that the Ad5 vector dl327/9E4ORF1, like Ad9, caused exclusively mammary tumors in rats is significant because it indicates that certain activities associated with the Ad9 E4-ORF1 oncoprotein serve to promote tumorigenesis by Ad9 selectively in cells of the rat mammary gland. These observations lead us to hypothesize that in rats inoculated with Ad9, a wide variety of cell types become infected, yet only within certain mammary cells are the novel activities of the Ad9 E4-ORF1 oncoprotein sufficient to induce oncogenic transformation. With respect to this possibility, it may be found that cellular PDZ protein targets of the Ad9 E4-ORF1 oncoprotein are particularly important regulators of growth and proliferation in cells of the mammary gland in vivo.
In this study, we identified several different E4 region functions that represent key factors in determining the unique tumorigenic properties of Ad9, including the propensity to elicit only mammary tumors and the strict estrogen dependence of these neoplasms. Our results also suggest that Ad9-induced tumorigenesis is governed by a complex interplay between these E4 region functions, which act both positively and negatively to influence this process. This new information is expected to lead to a more complete understanding of the mechanisms responsible for tumorigenesis by Ad9.
ACKNOWLEDGMENTS
We thank Stephen Hoang and Ileana Silva for assistance in constructing plasmids and viruses. We also thank Siu Sylvia Lee, Britt Glaunsinger, Isabel Latorre, and Kris Frese for many helpful discussions.
D.L.T. was supported by National Research Service Award CA09197 from the National Cancer Institute and by the Federal Work-Study program of the U.S. Department of Education. This work was funded by grants from the National Cancer Institute (R01 CA/AI58541), American Cancer Society (RP6-97-068-01-VM), and Department of the Army (DAMD17-97-1-7082) to R.J. and from the National Institutes of Health (RO1 HL58344) to J.S.
REFERENCES
- 1.Ankerst J, Jonsson N. Adenovirus type 9-induced tumorigenesis in the rat mammary gland related to sex hormonal state. J Natl Cancer Inst. 1989;81:294–298. doi: 10.1093/jnci/81.4.294. [DOI] [PubMed] [Google Scholar]
- 2.Ankerst J, Jonsson N, Kjellen L, Norrby E, Sjogren H O. Induction of mammary fibroadenomas in rats by adenovirus type 9. Int J Cancer. 1974;13:286–290. doi: 10.1002/ijc.2910130303. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe B J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chinnadurai G, Chinnadurai S, Brusca J. Physical mapping of a large-plaque mutation of adenovirus type 2. J Virol. 1979;32:623–628. doi: 10.1128/jvi.32.2.623-628.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven S E, Bredt D S. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 7.Dix I, Leppard K N. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J Virol. 1993;67:3226–3231. doi: 10.1128/jvi.67.6.3226-3231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning A S, Anderson J M. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Investig. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint S J, Enquist L W, Krug R M, Racaniello V R, Skalka A M. Principles of virology: molecular biology, pathogenesis, and control. Washington, D.C.: ASM Press; 2000. pp. 552–593. [Google Scholar]
- 10.Gardiol D, Kuhne C, Glaunsinger B, Lee S S, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 11.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene, in press. [DOI] [PMC free article] [PubMed]
- 12.Gomez-Foix A M, Coats W S, Baque S, Alam T, Gerard R D. Adenovirus-mediated transfer of the muscle glycogen phosphorylase gene into hepatocytes confers altered regulation of glycogen metabolism. J Biol Chem. 1992;267:25129–25134. [PubMed] [Google Scholar]
- 13.Graham F. Transformation by and oncogenicity of human adenoviruses. In: Ginsberg H, editor. The adenoviruses. New York, N.Y: Plenum Press; 1984. pp. 339–398. [Google Scholar]
- 14.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2149–2171. [Google Scholar]
- 16.Imreh S, Szekely L, Wiman K G, Klein G. Oncogenes and signal transduction. In: Peters G, Vousden K H, editors. Oncogenes and tumour suppressors. New York, N.Y: Oxford University Press; 1997. pp. 3–32. [Google Scholar]
- 17.Javier R, Raska K, Jr, Macdonald G J, Shenk T. Human adenovirus type 9-induced rat mammary tumors. J Virol. 1991;65:3192–3202. doi: 10.1128/jvi.65.6.3192-3202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javier R, Raska K, Jr, Shenk T. Requirement for the adenovirus type 9 E4 region in production of mammary tumors. Science. 1992;257:1267–1271. doi: 10.1126/science.1519063. [DOI] [PubMed] [Google Scholar]
- 19.Javier R, Shenk T. Mammary tumors induced by human adenovirus type 9: a role for the viral early region 4 gene. Breast Cancer Res Treat. 1996;39:57–67. doi: 10.1007/BF01806078. [DOI] [PubMed] [Google Scholar]
- 20.Javier R T. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krantz C K, Routes B A, Quinlan M P, Cook J L. E1A second exon requirements for induction of target cell susceptibility to lysis by natural killer cells: implications for the mechanism of action. Virology. 1996;217:23–32. doi: 10.1006/viro.1996.0089. [DOI] [PubMed] [Google Scholar]
- 23.Lee S S, Glaunsinger B, Mantovani F, Banks L, Javier R T. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J Virol. 2000;74:9680–9693. doi: 10.1128/jvi.74.20.9680-9693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lue R A, Marfatia S M, Branton D, Chishti A H. Cloning and characterization of hdlg: the human homologue of the Drosophila discs large tumor suppressor binds to protein 4.1. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcellus R C, Lavoie J N, Boivin D, Shore G C, Ketner G, Branton P E. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J Virol. 1998;72:7144–7153. doi: 10.1128/jvi.72.9.7144-7153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller B M, Kistner U, Veh R W, Cases-Langhoff C, Becker B, Gundelfinger E D, Garner C C. Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs-large tumor suppressor protein. J Neurosci. 1995;15:2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevels M, Tauber B, Kremmer E, Spruss T, Wolf H, Dobner T. Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol. 1999;73:1591–1600. doi: 10.1128/jvi.73.2.1591-1600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevins J R, Vogt P K. Cell transformation by viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 301–343. [Google Scholar]
- 32.Ranganathan R, Ross E M. PDZ domain proteins: scaffolds for signaling complexes. Curr Biol. 1997;7:R770–R773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 33.Roelvink P W, Kovesdi I, Wickham T J. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaack J, Langer S, Guo X. Efficient selection of recombinant adenoviruses by vectors that express β-galactosidase. J Virol. 1995;69:3920–3923. doi: 10.1128/jvi.69.6.3920-3923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp P A. Adenovirus transcription. In: Ginsberg H, editor. The adenoviruses. New York, N.Y: Plenum Press; 1984. pp. 173–204. [Google Scholar]
- 37.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 38.Shtrichman R, Kleinberger T. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol. 1998;72:2975–2982. doi: 10.1128/jvi.72.4.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith D B, Corcoran L M. Expression and purification of glutathione-S-transferase fusion proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1994. pp. 16.7.1–16.7.7. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T, Ohsugi Y, Uchida-Toita M, Akiyama T, Yoshida M. Tax oncoprotein of HTLV-1 binds to the human homologue of Drosophila discs large tumor suppressor protein, hDLG, and perturbs its function in cell growth control. Oncogene. 1999;18:5967–5972. doi: 10.1038/sj.onc.1203008. [DOI] [PubMed] [Google Scholar]
- 41.Thomas D L, Shin S, Jiang B H, Vogel H, Ross M A, Kaplitt M, Shenk T E, Javier R T. Early region 1 transforming functions are dispensable for mammary tumorigenesis by human adenovirus type 9. J Virol. 1999;73:3071–3079. doi: 10.1128/jvi.73.4.3071-3079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas U, Phannavong B, Muller B, Garner C C, Gundelfinger E D. Functional expression of rat synapse-associated proteins SAP97 and SAP102 in Drosophila dlg-1 mutants: effects on tumor suppression and synaptic bouton structure. Mech Dev. 1997;62:161–174. doi: 10.1016/s0925-4773(97)00658-8. [DOI] [PubMed] [Google Scholar]
- 43.Weiss R S, Gold M O, Vogel H, Javier R T. Mutant adenovirus type 9 E4 ORF1 genes define three protein regions required for transformation of CREF cells. J Virol. 1997;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss R S, Javier R T. A carboxy-terminal region required by the adenovirus type 9 E4 ORF1 oncoprotein for transformation mediates direct binding to cellular polypeptides. J Virol. 1997;71:7873–7880. doi: 10.1128/jvi.71.10.7873-7880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss R S, Lee S S, Prasad B V, Javier R T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss R S, McArthur M J, Javier R T. Human adenovirus type 9 E4 open reading frame 1 encodes a cytoplasmic transforming protein capable of increasing the oncogenicity of CREF cells. J Virol. 1996;70:862–872. doi: 10.1128/jvi.70.2.862-872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamano S, Tokino T, Yasuda M, Kaneuchi M, Takahashi M, Niitsu Y, Fujinaga K, Yamashita T. Induction of transformation and p53-dependent apoptosis by adenovirus type 5 E4orf6/7 cDNA. J Virol. 1999;73:10095–10103. doi: 10.1128/jvi.73.12.10095-10103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]