Abstract
The aquatic γ-proteobacterium Shewanella oneidensis is able to form two types of biofilms: a floating biofilm at the air–liquid interface (pellicle) and a solid surface-associated biofilm (SSA-biofilm). S. oneidensis possesses the Bpf system, which is orthologous to the Lap system first described in Pseudomonas fluorescens. In the Lap systems, the retention of a large adhesin (LapA) at the cell surface is controlled by LapD, a c-di-GMP effector protein, and LapG, a periplasmic protease targeting LapA. Here, we showed that the Bpf system is mandatory for pellicle biogenesis, but not for SSA-biofilm formation, indicating that the role of Bpf is somewhat different from that of Lap. The BpfD protein was then proved to bind c-di-GMP via its degenerated EAL domain, thus acting as a c-di-GMP effector protein like its counterpart LapD. In accordance with its key role in pellicle formation, BpfD was found to interact with two diguanylate cyclases, PdgA and PdgB, previously identified as involved in pellicle formation. Finally, BpfD was shown to interact with CheY3, the response regulator controlling both chemotaxis and biofilm formation. Altogether, these results indicate that biofilm formation in S. oneidensis is under the control of a large c-di-GMP network.
Keywords: biofilm, pellicle, diguanylate cyclase, secondary messenger, c-di-GMP, regulatory network, Shewanella
1. Introduction
Bacteria can adopt two lifestyles. They live either as planktonic cells able to move independently in their environment or as sessile cells forming a community called a biofilm. In nature, bacteria are mainly found in biofilms [1]. Several forms of biofilms have been described. Bacteria can adhere to biotic or abiotic surfaces forming what is called a solid surface-associated biofilm (SSA-biofilm) [2,3]. They can also form a floating biofilm at the air–liquid interface, also named a pellicle [4,5]. A biofilm is a community of cells encased in a self-produced extracellular matrix. The composition of the matrix can vary from one bacterium to another and can also differ from one type of biofilm to another even for the same bacterium. Nevertheless, the main components of the matrix are exopolysaccharides, extracellular proteins, and DNA [6,7].
The transition from planktonic to sessile lifestyles is governed by several molecular actors. A key player is the cyclic di-guanosine monophosphate (c-di-GMP), a secondary messenger involved not only in the regulation of biofilm formation but also in the regulation of motility, virulence, and differentiation [8,9,10]. The concentration of c-di-GMP in the cells is fine-tuned by a whole array of enzymes. C-di-GMP is synthetized from GTP by diguanylate cyclases (DGCs) and hydrolyzed by phosphodiesterases (PDEs). The DGCs are characterized by GGDEF domains containing the consensual motif GG(D/E)EF crucial for catalytic activity. They function as dimers in which each subunit binds a GTP molecule. Most DGCs also contain an inhibitory site (RXXD) located five residues upstream of the catalytic site. The fixation of c-di-GMP to this inhibitory site allows negative feedback control of DGC activity. There are two types of PDEs: the ones containing an EAL domain and hydrolyzing c-di-GMP into pGpG, and the others harboring an HD-GYP domain and hydrolyzing c-di-GMP into GMP. Once c-di-GMP is synthetized, it can interact with a third type of actor, the effectors, to induce a cellular response. Effectors can be RNA or proteins [11]. The effector proteins are less characterized than DGCs or PDEs and can contain different types of domains (PilZ, YadQ, etc.). In some cases, effector proteins contain degenerated GGDEF or EAL domains, in which the catalytic sites are not conserved, giving rise to proteins unable to synthetize or hydrolyze c-di-GMP but still able to bind to it.
A well-characterized system involved in biofilm formation and controlled by c-di-GMP is the Lap system of Pseudomonas fluorescens [12]. The main components of this system are LapA, an adhesin secreted to the cell surface by a type-I secretion system (LapBCE) and involved in cell-surface adhesion; LapG, a periplasmic protease targeting LapA; and LapD, an effector protein containing degenerated GGDEF and EAL domains and binding to c-di-GMP. In biofilm conditions, LapD receives c-di-GMP from a specific DGC GcbC [13]. LapD bound to c-di-GMP becomes active and can sequester the LapG protease. This latter is then unable to cleave LapA, leading to LapA accumulation at the surface of the cells and promoting cell-surface interaction. This constitutes the first step of biofilm formation. The Lap systems have been described in several bacteria belonging to different species such as Pseudomonas, Legionella, Bordetella, Vibrio, and Shewanella [14,15,16,17,18].
Shewanella oneidensis is a motile aquatic γ-proteobacterium. It was shown to form two types of biofilms: an SSA-biofilm when cultivated in agitated/aerated minimal medium and a pellicle when cultivated in static rich medium [19,20,21,22]. A cluster of genes (so_4317 to so_4323) was previously shown to encode proteins sharing similarities to the Lap proteins of P. fluorescens [15]. The system of S. oneidensis was named Bpf (Biofilm-Promoting Factor) and contains BpfA (SO_4317), a homolog of the adhesin LapA; BpfD (SO_4323), a homolog of the effector LapD; BpfG (SO_4322), the unique homolog of LapG in S. oneidensis; and a type-I secretion system composed of AggC (SO_4318, homologous to LapB), AggB (SO_4319, homologous to LapC), and AggA (SO_4320, homologous to LapE). A mutant of bpfA and a mutant of either aggA, aggB, or aggC were reported to be totally impaired in biofilm formation, while a mutant of bpfG was greatly affected but not totally impaired and a bpfD mutant was only partially impaired [15,23]. It is noteworthy that the conditions used to perform these tests were not discriminant enough to distinguish the SSA-biofilm from the pellicle. It was hypothesized that the Bpf system of S. oneidensis is an ortholog of the Lap system of P. fluorescens since the BpfA adhesin was proved to interact with the BpfG protease, which itself interacts with the BpfD protein [15]. Even though c-di-GMP was shown to be required for biofilm formation in S. oneidensis like in other bacteria [22,24], it is not currently known whether BpfD is able to bind c-di-GMP or not and, if it is the case, whether BpfD receives c-di-GMP from a specific DGC or not. Several DGCs were identified to play a role in biofilm formation in S. oneidensis. Two DGCs, PdgA and PdgB, were found to restore pellicle formation in the cheY3 pellicle-deficient mutant [24]. A similar approach, used in the context of SSA-biofilm formation, led to the identification of two additional DGCs [25]. Interestingly, three out of these four DGCs interact with CheY3, the response regulator controlling both chemotaxis and biofilm formation in S. oneidensis. Moreover, the complex regulatory network governing biofilm formation and centered around CheY3 also contains an effector protein, MxdA. MxdA was shown to bind c-di-GMP and to interact with both CheY3 and PdgA, and was suspected to trigger exopolysaccharide synthesis via the Mxd machinery [24].
In this study, we first showed that the Bpf system is required for pellicle formation, but is dispensable for SSA-biofilm formation. We then proved that BpfD binds c-di-GMP via its degenerated EAL domain and is therefore an effector protein. Finally, we found that BpfD belongs to a complex regulatory network governing pellicle formation by interacting not only with CheY3 but also with two DGCs (PdgA and PdgB).
2. Results
2.1. The Bpf System Is Not Required for SSA-Biofilm Formation in S. oneidensis
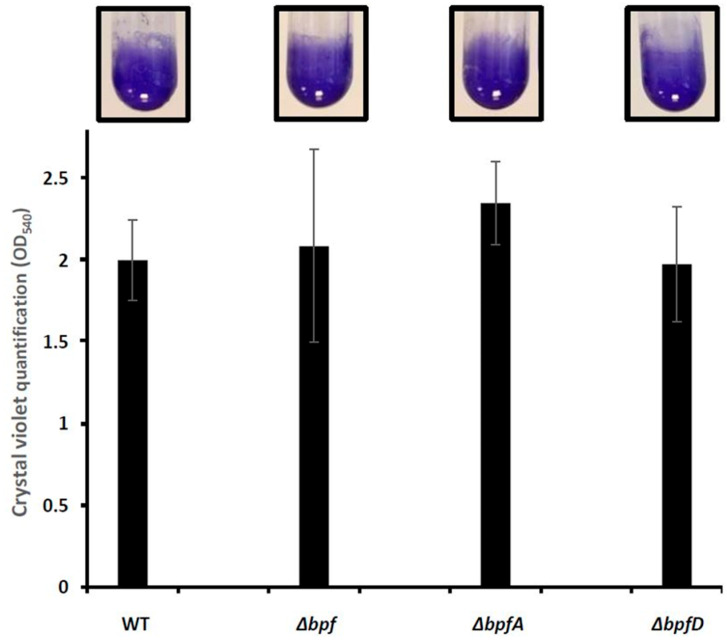
In order to test the involvement of the bpf gene cluster in biofilm formation, we constructed various deletion mutants starting from the S. oneidensis MR1-R strain (referred to as wild-type). The ΔbpfA and ΔbpfD strains are deleted of the so_4317 and so_4323 genes, respectively, while the Δbpf strain is deleted of the entire cluster, i.e., from so_4317 to so_4323. The three resulting strains were first tested for their ability to form an SSA-biofilm. The wild-type and mutant strains were grown in LM (Lactate Medium) under shaking conditions for 24 h. The cells adhered to the tube walls were stained with crystal violet (CV). As expected, the wild-type strain formed an SSA-biofilm in these conditions (Figure 1). Unexpectedly, the ΔbpfA, ΔbpfD, and Δbpf mutants were also able to form an SSA-biofilm (Figure 1). CV quantification indicated that the biomass of the bpf mutants is similar to that of the wild-type. These results show that, in these conditions, the Bpf system is not required for SSA-biofilm formation in S. oneidensis.
Figure 1.
The bpf mutants of Shewanella oneidensis are able to form an SSA-biofilm. The wild-type MR1-R (WT), ΔbpfA, ΔbpfD, and Δbpf strains were grown at 28 °C under agitation in LM. After 24 h of incubation, biofilm formation was evaluated by crystal violet staining, photographed, and quantified by OD540 measurements. The graphs represent the means and standard deviations from two independent experiments conducted in duplicate.
2.2. The Bpf System Is Mandatory for Pellicle Formation in S. oneidensis
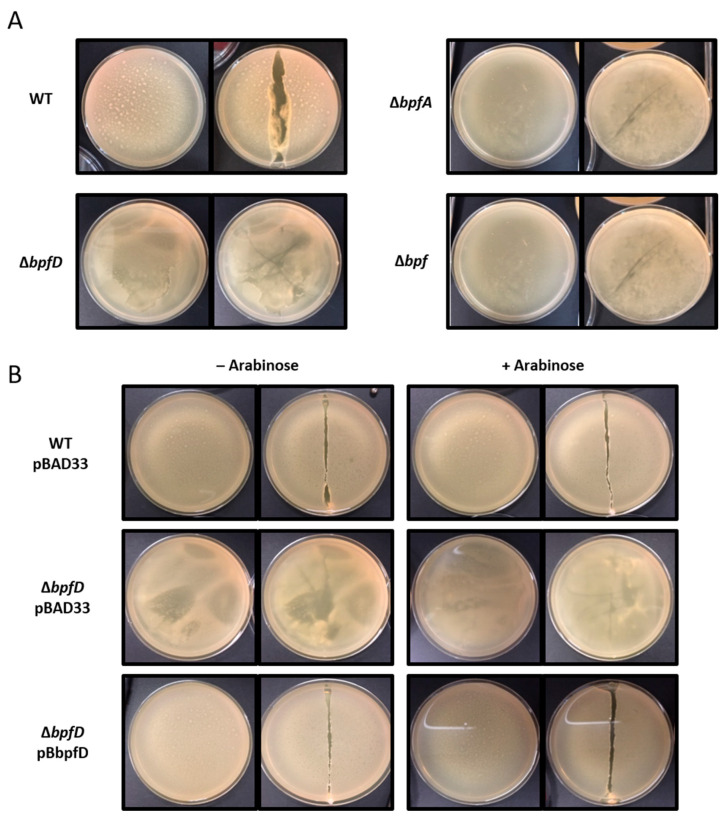
Since S. oneidensis is also able to form a pellicle, we wondered whether the Bpf system could be involved in this process. To test this, cells were grown in rich medium (LB) without shaking at 28 °C for 24 h. We then observed the formation of the pellicle and tested its integrity, thickness, robustness, and elasticity using a toothpick. As previously shown, the wild-type strain was able to form a mature and robust pellicle at the air–liquid interface (Figure 2A). On the contrary, the ΔbpfA, ΔbpfD, and Δbpf mutants were unable to form a mature pellicle, suggesting that the Bpf system is mandatory for pellicle formation (Figure 2A). Strikingly, the ΔbpfD mutant showed a phenotype which was slightly different from that of the ΔbpfA and Δbpf mutants. Indeed, pellicle fragments were observed on the edge of the plate, but the pellicle had not spread at the liquid surface (Figure 2A). It should be repeated that bpfD is the last gene of the bpf cluster, meaning that, in a ΔbpfD mutant, BpfA could be produced and exported and that only its cleavage is likely affected.
Figure 2.
The Bpf system is involved in Shewanella oneidensis pellicle formation. (A) The wild-type (WT), ΔbpfA, ΔbpfD, and Δbpf strains were grown at 28 °C in LB medium without agitation. (B) Strains containing either the pBAD33 vector or pBbpfD plasmid were grown at 28 °C in LB medium without agitation in the presence (+) or absence (−) of arabinose (0.2%). Pictures were taken after a 24 h incubation. For each strain, the pellicle phenotype was observed before (left panel) and after (right panel) disruption by a toothpick.
To ascertain that the defect of the ΔbpfD mutant is due to bpfD gene deletion and not to a polar effect, the bpfD gene was cloned under the control of an arabinose-inducible promoter in the pBAD33 vector, and the resulting pB-bpfD plasmid and the empty vector were then introduced into the ΔbpfD mutant. The strains were then cultured in the presence or absence of arabinose (Figure 2B). As expected, the presence of the pBAD33 vector did not modify the phenotypes of the wild-type and ΔbpfD strains. As shown in Figure 2B, the presence of the pB-bpfD plasmid restored pellicle formation in the ΔbpfD mutant, whether arabinose was added or not. This result confirms that the defect for pellicle formation observed in the ΔbpfD mutant was only due to the deletion of bpfD, meaning that BpfD is crucial for pellicle formation in S. oneidensis.
2.3. BpfD Acts as a c-di-GMP Effector
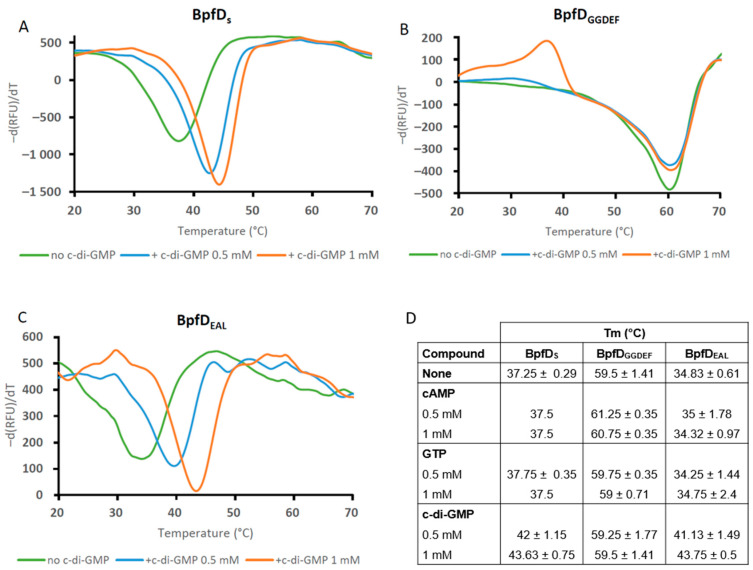
BpfD is homologous to LapD. As LapD, it is predicted to be anchored to the membrane by two transmembrane segments and to possess a cytoplasmic part containing an HAMP domain, a degenerated GGDEF domain (AAFEF), and a degenerated EAL domain (ELY). We therefore wondered whether BpfD could bind the secondary messenger, cyclic di-GMP (c-di-GMP), and act as an effector protein of the Bpf system. To test this, we overproduced and purified the cytoplasmic region of BpfD containing the degenerated GGDEF and EAL domains (called BpfDS). We then performed thermal shift assays (TSAs) using purified BpfDS alone or incubated with either c-di-GMP or other nucleotides (cAMP and GTP were used as control). The curve of the first derivative of fluorescence emission relative to temperature revealed one peak for BpfDS alone with a melting temperature (Tm) of 37.25 °C (Figure 3A,D). When the TSA was performed in the presence of 0.5 mM and 1 mM c-di-GMP, the Tm of BpfDS rose to 42 °C and 43.63 °C, respectively. The resulting ΔTm (Tmprotein + ligand − Tmprotein alone) was therefore about 4.75 and 6.38 °C, respectively. When the TSA was carried out in the presence of cAMP or GTP, the Tm of BpfDS remained unchanged compared to the condition without any ligand (Figure 3D). An additional control was performed using the CheY3 protein. The Tm of CheY3 was not changed by the addition of c-di-GMP (Figure S1). These results indicate that BpfDS directly and specifically binds c-di-GMP.
Figure 3.
BpfD binds c-di-GMP via its degenerated EAL domain. Thermal shift assays (TSAs) were performed using the cytoplasmic part of BpfD (BpfDS) (A), its GGDEF domain (B), or EAL (C) domain and c-di-GMP, cAMP and GTP. The various domains of BpfD (7.5 µM) were incubated in the presence of SYPRO Orange and various concentrations of the different compounds. The mix was then submitted to a temperature gradient from 20 to 70 °C. Graphs represent the first derivative of the fluorescence emission (-d(RFU)/dT, RFU: Raw Fluorescence Unit) as a function of temperature. The melting temperatures (Tm) of each protein are listed in the table (mean values with standard deviation, n = 2 to 6) (D). All graphs are representative of two independent experiments.
To determine whether BpfD binds c-di-GMP via its degenerated GGDEF or EAL domain, we produced and purified the two domains independently (called BpfDGGDEF and BpfDEAL, respectively). When BpfDGGDEF was incubated alone, a Tm of about 59.5 °C was observed (Figure 3B,D). In the presence of either c-di-GMP or GTP, the Tm of this protein was unchanged (Figure 3D). A slight increase in the Tm was only observed in the presence of cAMP, but the ΔTm was below 2 °C, suggesting that there was no binding (Figure 3D). These results indicate that the degenerated GGDEF domain of BpfD is not able to bind c-di-GMP. However, when we performed similar experiments with BpfDEAL, an increase in the Tm was specifically observed in the presence of c-di-GMP. The ΔTm was about 6.3 °C and 8.92 °C in the presence of 0.5 mM and 1 mM c-di-GMP, respectively (Figure 3C,D). Altogether, these results show that BpfD binds c-di-GMP via its EAL domain.
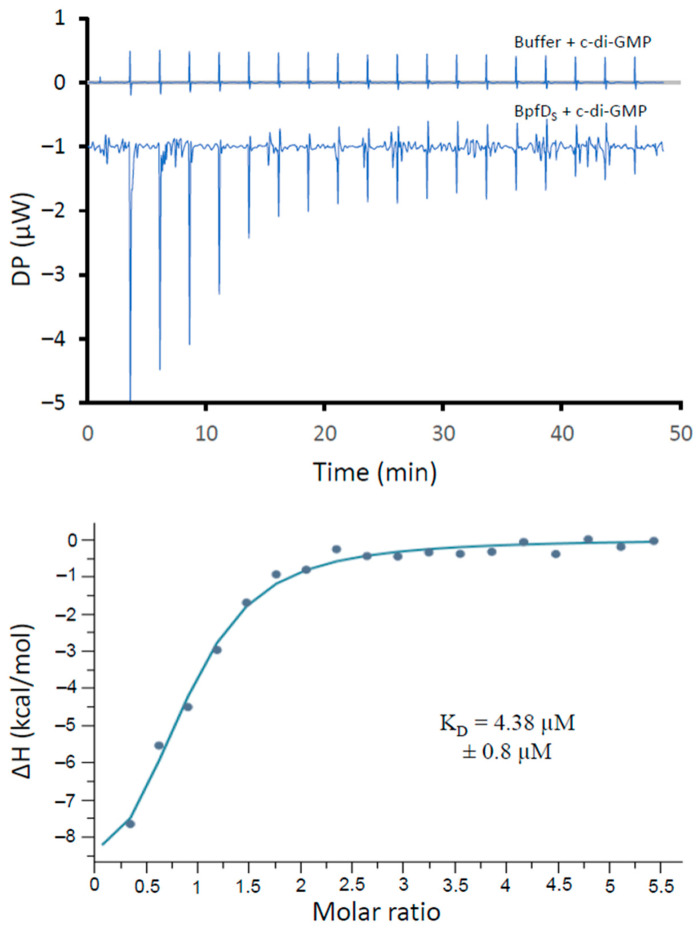
To further validate the direct binding of c-di-GMP to BpfD and to estimate the affinity of this interaction, we performed isothermal titration calorimetry (ITC) with the purified BpfDS protein. As shown in Figure 4, injection of c-di-GMP elicited an exothermic reaction, confirming that BpfDS binds c-di-GMP. Data were fitted using the “One Set of Sites” model, and the apparent dissociation constant (KD) was calculated. BpfDS binds c-di-GMP with a KD of 4.38 µM ± 0.8 µM, a value which is close to the estimated KD for the P. fluorescens LapD homolog (5.5 ± 2.8 µM) [26]. Altogether, TSA and ITC assays indicate that BpfDS directly binds c-di-GMP and can act as a c-di-GMP effector via its degenerated EAL domain.
Figure 4.
Interaction of BpfDS with c-di-GMP tested by isothermal titration calorimetry. BpfDS (20 µM) or dialysis buffer was submitted to several injections of c-di-GMP (575 µM). Top graphics show heat exchange upon ligand titration, either with dialysis buffer (control) or with BpfDS. The bottom graphic shows the integrated data after control subtraction with binding isotherms fitted according to a one-site binding model. The data shown are representative of two independent experiments.
2.4. BpfD Interacts with the PdgA and PdgB Diguanylate Cyclases as Well as with the CheY3 Regulator
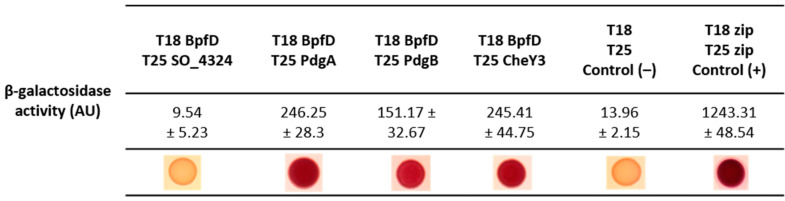
In the Lap system of P. fluorescens, the LapD protein was proved to physically interact with a specific diguanylate cyclase (GcbC) and proposed to receive c-di-GMP from GcbC [13]. We thus wondered whether this was also the case for BpfD. We therefore searched for a DGC partner of BpfD. First, we performed a Blast search in the genome of S. oneidensis using the sequence of GcbC as a query. This resulted in numerous diguanylate cyclases (44 hits), but did not pinpoint a close homolog with a similar domain organization. Second, we looked at the genetic context for the gcbC gene in P. fluorescens and found that it is close to the ftsZ, ftsA, and ftsQ genes. The homologs of these three genes in S. oneidensis are so_4215, so_4216, and so_4217, but no gene encoding a DGC was found close by. Interestingly, a gene encoding a putative diguanylate cyclase (so_4324) was found downstream of bpfD and separated from it by several tRNA genes. SO_4324 could be a good candidate as the DGC partner of BpfD. Moreover, as the Bpf system is mandatory for pellicle formation, we wondered whether the two DGCs (PdgA and PdgB) previously found to be involved in pellicle formation could be BpfD partners. We thus carried out bacterial two-hybrid assays using BpfD fused to the T18 domain of the adenylate cyclase, and the other proteins (PdgA, PdgB, SO_4324) fused to the T25 domain. After incubation on MacConkey–lactose plates, the cells producing T18-BpfD with either T25-PdgA or T25-PdgB turned red, while the cells producing T18-BpfD with T25-SO_4324 did not (Figure 5). Accordingly, β-galactosidase activities measured on the strains containing T18-BpfD and either T25-PdgA or T25-PdgB were significantly higher than those measured on the control strain containing the empty vectors or the strain containing T18-BpfD and T25-SO_4324 (Figure 5). These results suggest that PdgA and PdgB, but not SO_4324, interact with BpfD.
Figure 5.
BpfD interacts with two diguanylate cyclases and the response regulator CheY3. E. coli BTH101 containing pUT18-BpfD and pKT25-SO4324, CheY3, PdgA, or PdgB were tested in this experiment. As controls, pEB354 (T25) with pEB355 (T18) (negative) and pT18-zip with pT25-zip (positive) were used. β-galactosidase activity was measured at 420 nm using a TECAN™ spectrophotometer after the addition of ONPG (4 mg.mL−1). Measures are indicated as mean values in arbitrary units (AUs) and their standard deviations. Values are representative of three independent experiments. The same strains were also spotted on MacConkey plates containing lactose and photographed after 48 h of incubation. All pictures were taken from the same plate and are representative of at least three experiments.
Since pellicle formation is controlled by a complex regulatory network centered around the chemotaxis regulator CheY3, we wondered whether BpfD could interact with CheY3. We thus performed a bacterial two-hybrid assay using T18-BpfD and T25-CheY3. As shown in Figure 5, the cells containing T18-BpfD and T25-CheY3 turned red on MacConkey–lactose plates and the β-galactosidase activity was significantly higher than that of the control strain, suggesting that BpfD interacts with CheY3.
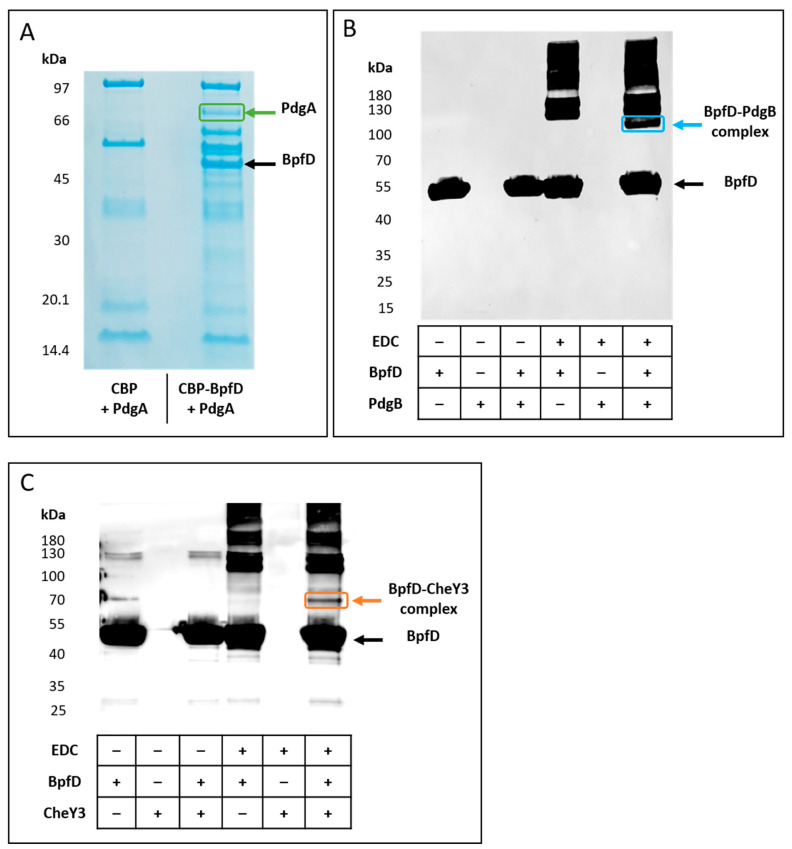
To confirm these results, we first performed pull-down assays. To do so, the bpfD gene was cloned into the pBAD24-CBP-linker plasmid in which the cbp gene, encoding the calmodulin-binding protein, was placed under the control of the arabinose-inducible promoter. The resulting construction (pBcbp-bpfD) allows the production of a CBP-BpfD chimeric protein. An E. coli strain was then co-transformed with pBcbp-bpfD (or pBAD24-CBP-linker, used as control) and either pBpdgA, pBpdgB, or pBcheY3. The cells were then grown in the presence of arabinose, allowing the co-production of CBP-BpfD (or CBP only) with either PdgA, PdgB, or CheY3. Cell extracts were subsequently incubated with calmodulin-coated beads, allowing CBP-BpfD (or CBP) purification. The elution fractions were then submitted to SDS-PAGE and their protein contents were analyzed. When PdgA was co-produced with BpfD, a band was observed at a position which is in agreement with the molecular mass of PdgA (80 kDa), while this band was absent when the experiment was performed with CBP only (Figure 6A). The presence of PdgA was confirmed by a mass spectrometry experiment performed after excision of the band from gel (Figure S2 and Table S1). This result confirms that BpfD interacts with the diguanylate cyclase PdgA.
Figure 6.
Interaction of BpfD with PdgA, PdgB, and CheY3. (A) Co-purification assay of BpfD and PdgA. PdgA was co-produced either with CBP-BpfD or CBP. CBP-BpfD (or CBP) was purified using CBP affinity resin, and bound proteins were submitted to SDS-PAGE. The band surrounded in green was excised and analyzed by mass spectrometry. The presence of PdgA was confirmed (coverage % = 60; PSM number = 315; unique peptides = 35). (B,C) Crosslinking experiments. Purified Strep-tagged BpfDS and His-tagged PdgB (B) or Strep-tagged CheY3 (C) was incubated in the presence or in the absence of the crosslinker EDC (− indicates the absence of the proteins and EDC, while + indicates their presence). All samples were submitted to SDS-PAGE. After blotting, the membranes were revealed using anti-StrepTag II antibodies. Due to its low size, the Strep-tagged CheY3 is not detected in these electrophoretic conditions. The band surrounded in blue was analyzed by mass spectrometry. The presence of BpfD (coverage % = 73; PSM number = 220; unique peptides = 27) and PdgB (coverage % = 62; PSM number = 58; unique peptides = 17) was confirmed. The band surrounded in orange was analyzed by mass spectrometry. The presence of BpfD (coverage % = 66; PSM number = 125; unique peptides = 23) and CheY3 (coverage % = 65; PSM number = 46; unique peptides = 6) was confirmed.
Unfortunately, pull-down assays performed using PdgB or CheY3 in combination with CBP-BpfD did not show co-purification of BpfD with either one of the two proteins. This could be due to a low level of detection or either transient or weak interaction. Therefore, in order to confirm the interaction of BpfD with PdgB and CheY3, we performed crosslinking experiments using Strep-tagged BpfDS, His-tagged PdgB, and Strep-tagged CheY3 purified proteins. As shown on Figure 6B,C, when BpfDS, is incubated alone in the presence of the crosslinker, complexes of higher molecular masses are observed, suggesting that BpfD is able to multimerize. When BpfDS and PdgB were incubated in the presence of the crosslinker (EDC), an additional band was observed below the multimeric forms of BpfD (Figure 6B). The molecular mass of this complex is between 100 kDa and 130 kDa and could correspond to a monomer of BpfD (50.5 kDa) interacting with a dimer of PdgB (2 × 35 kDa). The presence of both BpfD and PdgB in this complex was confirmed by mass spectrometry (Figure S3 and Table S1).
When BpfDS and CheY3 were incubated in the presence of the crosslinker (EDC), an additional band was observed between the monomeric and the multimeric forms of BpfD (Figure 6C). The molecular mass of this complex is between 70 kDa and 100 kDa and could correspond to a monomer of BpfD (50.5 kDa) interacting with a dimer of CheY3 (2 × 16 kDa). The presence of both BpfD and CheY3 in this complex was confirmed by mass spectrometry (Figure S4 and Table S1).
Altogether, these results indicate that BpfD not only interacts with two diguanylate cyclases previously shown to be involved in pellicle formation but also with CheY3, which is at the center of a complex regulatory network controlling biofilm formation.
3. Discussion
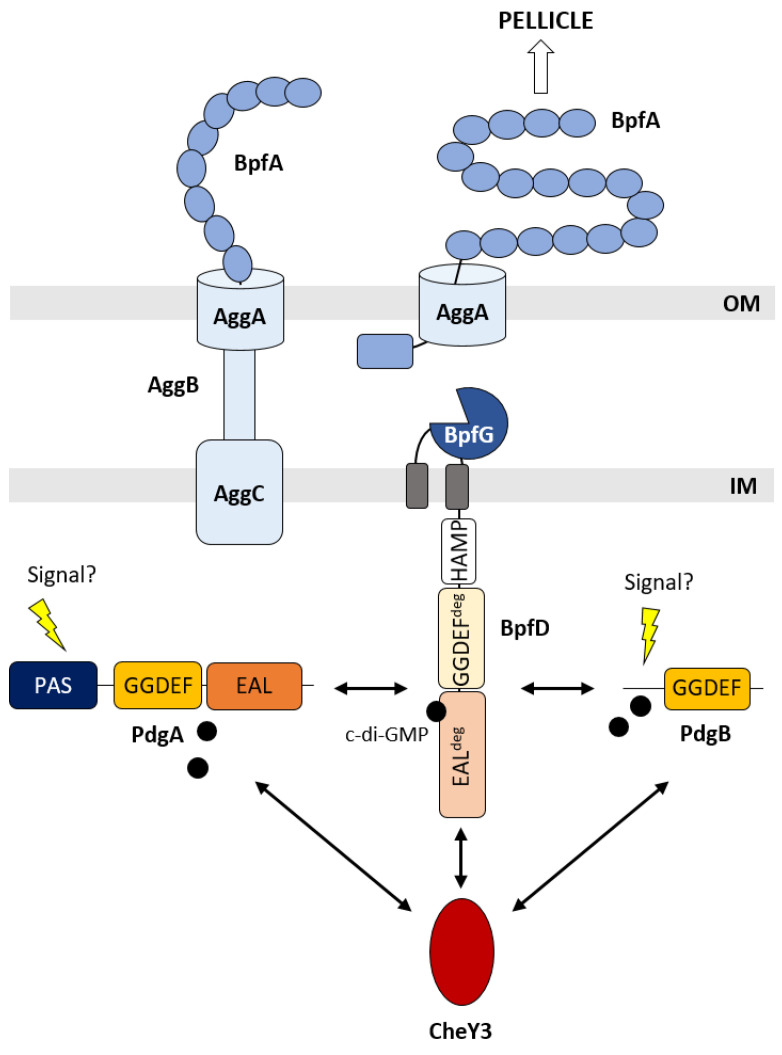
The Bpf system of S. oneidensis was proposed to be an ortholog of the Lap system of P. fluorescens. Indeed, in addition to sequence homologies and syntheny conservation, it was already shown that bpf mutants are either totally or partially impaired in biofilm formation and interactions between partners of the system are conserved [15,23]. However, several questions remained unanswered. (1) Is the Bpf system required for both pellicle and SSA-biofilm formation? (2) Is BpfD able to bind c-di-GMP, and if this is the case, is there a specific DGC delivering c-di-GMP directly to BpfD? (3) Is the Bpf system connected to the complex CheY3-centered regulatory network, which has been shown to control both pellicle and SSA-biofilm formation? Our study provides clues to answer these questions, as schematized in Figure 7.
Figure 7.
The Bpf system and its partners are involved in pellicle formation in S. oneidensis. BpfA is an adhesin secreted by a type-I secretion system composed of the AggA, AggB, and AggC proteins. BpfG is a periplasmic protease. BpfD is a membrane-anchored protein with an HAMP domain as well as degenerated GGDEF and EAL domains (GGDEFdeg and EALdeg). PdgA and PdgB are diguanylate cyclases and CheY3 is a response regulator proved to be mandatory for pellicle formation. Based on our results, we propose that BpfD is a c-di-GMP effector receiving the c-di-GMP secondary messenger from PdgA and PdgB, which could be activated by yet-unknown signals. BpfD also interacts with CheY3 and therefore belongs to the complex regulatory network controlling pellicle biogenesis. Fixation of c-di-GMP on the degenerated EAL domain of BpfD probably allows BpfD to sequester BpfG, leading to the accumulation of BpfA at the cell surface and pellicle formation. The protein–protein interactions, which were experimentally proven, are indicated by double arrows. IM: inner membrane; OM: outer membrane.
First, we showed that the bpf mutants are still able to form SSA-biofilm but are either totally (ΔbpfA and Δbpf) or partially (ΔbpfD) impaired in pellicle formation. Consistent with these results, a previous study reported that a mutant of aggA (so_4320), a BpfA type-I secretion system-encoding gene, is unable to form a pellicle [19]. This strongly suggests that the Bpf system is specific to pellicle formation. The difference in behavior between the ΔbpfA and ΔbpfD mutants is reminiscent of what was observed for the ΔlapA and ΔlapD mutants of P. fluorescens. Indeed, while the ΔlapA mutant is severely impaired for biofilm formation, the ΔlapD mutant is still able to form a biofilm somewhat different from the wild-type strain [27,28]. The authors proposed that the difference could be due to the fact that LapA protein is absolutely required for biofilm formation, while LapD only controls LapA secretion. This hypothesis could also apply to the Bpf system. Nevertheless, it should be mentioned that the role of BpfA is probably different from that of LapA. While LapA was shown to be involved in the interaction between the cells and the surfaces, BpfA does not seem to be necessary for adhesion to surfaces since the ΔbpfA mutant is still able to form an SSA-biofilm [29]. One hypothesis could be that BpfA is involved in cell–cell or cell–matrix interactions. Interestingly, BpfA and LapA, while both belonging to the RTX adhesion family, present different domain architectures, which could explain the difference in function. It is noteworthy that CdrA, an adhesin of P. aeruginosa different from LapA but also controlled by LapG, was shown to bind to the matrix Psl exopolysaccharide, leading to robust biofilms [30,31].
Second, we showed that the cytoplasmic domain of BpfD specifically binds c-di-GMP with an apparent KD value in the low micromolar range like its LapD counterpart. This interaction involves the degenerated EAL domain of BpfD, as also demonstrated for LapD [26]. BpfD is therefore acting as a c-di-GMP effector protein and probably controls the maintenance of BpfA at the cell surface by sequestering BpfG. An interaction between the periplasmic region of BpfD and the BpfG protein was indeed observed using bacterial two-hybrid experiments [15]. We then identified two DGCs physically interacting with BpfD, namely PdgA and PdgB, while no interaction was found with the SO_4324 DGC encoded by a gene close to the bpf operon. This makes sense, since PdgA and PdgB were identified to be involved in pellicle formation for which the Bpf system is mandatory [24].
Interaction between LapD and the DGC GcbC was shown to involve the α2 helix of the LapD EAL domain (α2-EAL:462GRFLPWLER470) and the α5 helix of the GcbC GGDEF domain (α5-GGDEF:477EQLLFAADK485) [13]. Interestingly, BpfD contains the sequence GQFMPYIEL at the position corresponding to the α2-EAL of LapD, while PdgA and PdgB contain GQLISLADT and EDTLKRADA, respectively, at the position corresponding to the α5-GGDEF of GcbC. It is therefore possible that these sequences are involved in the interaction between BpfD and its two partners PdgA and PdgB.
The fact that BpfD interacts with two DGCs is not so surprising. Indeed, a large-scale interaction study using bacterial two-hybrid experiments has shown that LapD of P. fluorescens could interact with 15 different partners, among which are 12 DGCs [32]. Each DGC could respond to a specific signal triggering its diguanylate cyclase activity. Actually, many DGC proteins encompass detecting modules such as PAS, Cache, GAF, CZB, etc. In the case of GcbC, a periplasmic Cache domain senses the presence of citrate [33]. Both PdgA and PdgB are predicted to be cytoplasmic proteins and have an N-terminal extension upstream of the GGDEF domain. While no known sensory domain is predicted in the N-terminal region of PdgB, a PAS domain is present in PdgA. Interestingly, CdgF from Bacillus cereus has a similar architecture to that of PdgA, i.e., PAS-GGDEF-EAL, and was shown to contain a flavin cofactor bound to the PAS domain. This bifunctional enzyme possesses a prominent diguanylate cyclase activity when the flavin cofactor is in the oxidized form, while the phosphodiesterase activity is upregulated when the PAS domain flavin cofactor is reduced [34]. We can imagine that PdgA could behave similarly and be active in oxygenated conditions, which is in good agreement with its role in biofilm formation at the air–liquid interface (pellicle).
Finally, we showed that BpfD also interacts with the CheY3 response regulator. The latter was previously demonstrated to be mandatory for both pellicle and SSA-biofilm formation, and proposed to be at the center of a complex regulatory network composed of several DGCs and a c-di-GMP effector protein (MxdA) [24,25]. It seems that this network is even more complex and includes another c-di-GMP effector protein, BpfD. This is reminiscent of the Hub-based model proposed for local c-di-GMP signaling [35]. Although interesting, many questions remain on how these Hub systems function, in particular whether or not they are modular depending on the environmental cues. In the case of S. oneidensis, we hypothesize that the network could be responsive to specific cues, since pellicle and SSA-biofilm-controlling networks seem to involve specific components but share a common knot, namely CheY3.
4. Materials and Methods
4.1. Strains and Growth Conditions
In this study, we used S. oneidensis and E. coli strains, which were routinely grown in lysogeny broth (LB) medium at 28 °C for S. oneidensis and at 37 °C for E. coli. Antibiotics were added when necessary: rifampicin (10 µg.mL−1), ampicillin (50 µg.mL−1), kanamycin (25 µg.mL−1), or chloramphenicol (25 µg.mL−1). All strains used in this work are listed in Table 1.
Table 1.
Strains used in the study.
| Strains | Relevant Characteristics | Sources |
|---|---|---|
| S. oneidensis strains | ||
| MR1-R | Rifampicin-resistant derivative of MR1 | [36] |
| ΔbpfA | MR1-R deleted of bpfA (so_4317) | This work |
| ΔbpfD | MR1-R deleted of bpfD (so_4323) | This work |
| Δbpf | MR1-R deleted from bpfA to bpfD (from so_4317 to so_4323) | This work |
| E. coli strains | ||
| BL21 (DE3) | F- ompT hsdSB (rB−mB−) dcm gal (DE3) | Novagen |
| BTH101 | F- cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | [37] |
| C600 | F- tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 λ− | [38] |
| CC118 λpir | Δ(ara-leu) araDE ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE (Am) recA1 λpir | [39] |
4.2. Construction of Deletion Mutants
The ΔbpfA, ΔbpfD, and Δbpf mutant strains were constructed as previously described [40]. Briefly, upstream and downstream regions flanking the gene (s) to be deleted were cloned into the suicide vector pKNG101. The ligation product was introduced into E. coli CC118 λpir. The resulting plasmid was introduced into the S. oneidensis MR1-R strain by conjugation using the E. coli helper strain 1047/pRK2013. The plasmid was integrated into the chromosome by a first recombination event and removed by a second recombination event in the presence of 6% sucrose. Deletions were confirmed by PCR (Figure S5).
4.3. Plasmid Constructions
All plasmids used in this work are listed in Table 2. To construct the plasmid pBbpfD, the entire coding sequence of bpfD (so_4323) was PCR-amplified using chromosomal S. oneidensis DNA as a template with primers containing the appropriate restriction sites and an optimized Shine Dalgarno. After digestion, the PCR product was inserted into the pBAD33 vector.
Table 2.
Plasmids used in the study.
| Plasmids | Relevant Characteristics | Sources |
|---|---|---|
| pBAD33 | Vector containing pBAD promoter with a p15A origin of replication (CmR) | [41] |
| pBbpfD | Sequence coding for BpfD (SO_4323) cloned into pBAD33 | This work |
| pBpdgA | Sequence coding for PdgA (SO_4552) cloned into pBAD33 | [24] |
| pBpdgB | Sequence coding for PdgB (SO_0796) cloned into pBAD33 | [24] |
| pBcheY3 | Sequence coding for CheY3 (SO_3209) cloned into pBAD33 | [20] |
| pET52b | Vector containing the T7 phage promoter and the coding sequence of StrepTagII (ApR) | Novagen |
| pETbpfD | Sequence coding for the cytoplasmic part (S225 to E639) of BpfD (SO_4323) cloned into pET52b | This work |
| pETbpfDGGDEF | Sequence coding for the GGDEF domain (S225 to T396) of BpfD (SO_4323) cloned into pET52b | This work |
| pETbpfDEAL | Sequence coding for the EAL domain (E397 to E639) of BpfD (SO_4323) cloned into pET52b | This work |
| pETcheY3 | Sequence coding for CheY3 (SO_3209) cloned into pET52b | [24] |
| pETpdgB | Sequence coding for PdgB (SO_0796) into pET21b | [24] |
| pEB355 | pUT18C derivative, coding for the T18 domain of the adenylate cyclase of Bordetella pertussis | [37] |
| pUT18-bpfD | Sequence coding for the cytoplasmic part (S225 to E639) of BpfD (SO_4323) cloned in frame at the 3’ extremity of the sequence coding for the T18 domain into pEB355 | This work |
| pUT18-zip | Sequence coding for a leucine zipper region cloned in-frame with the T18 domain (positive control) | [37] |
| pEB354 | pKT25 derivative, coding for the T25 domain of the adenylate cyclase of B. pertussis | [37] |
| pKT25-SO4324 | Sequence coding for SO_4324 cloned in frame at the 3’ end of the sequence coding for the T25 domain into pEB354 | This work |
| pKT25-cheY3 | Sequence coding for CheY3 (SO_3209) cloned in frame at the 3’ end of the sequence coding for the T25 domain into pEB354 | [24] |
| pKT25-pdgA | Sequence coding for PdgA (SO_4552) cloned in frame at the 3’ end of the sequence coding for the T25 domain into pEB354 | [24] |
| pKT25-pdgB | Sequence coding for PdgB (SO_0796) cloned in frame at the 3’ end of the sequence coding for the T25 domain into pEB354 | [24] |
| pKT25-zip | Sequence coding for a leucine zipper region cloned in-frame with the T25 domain (positive control) | [37] |
| pBAD24 | Vector containing pBAD promoter (ApR) | [41] |
| pBAD24-CBP-linker | Sequence coding for the calmodulin-binding protein (CBP) cloned into pBAD24 | [42] |
| pBcbp-bpfD | Sequence coding for the cytoplasmic part (S225 to E639) of BpfD (SO_4323) cloned in frame at the 3’ extremity of the sequence coding for the calmodulin-binding protein (CBP) into pBAD24-CBP-linker | This work |
| pKNG101 | R6K-derived suicide plasmid containing StrR and sacB | [39] |
| pRK2013 | RK2-Tra1 RK2-Mob1 KmR ori ColE1 | [43] |
To construct the plasmids pETbpfD, pETbpfDGGDEF, and pETbpfDEAL, the coding sequence of so_4323 (from position 225 to 639, from position 225 to 396, and from position 397 to 639, respectively) was PCR-amplified from S. oneidensis genomic DNA and cloned into pET52b with a sequence encoding a Strep-Tag upstream on the vector (Novagen).
For two-hybrid experiments, the so_4324 coding sequence from S. oneidensis was cloned in-frame at the 3’ end of the sequence coding for the T25 domain of adenylate cyclase into pEB354, leading to pKT25-SO4324. The bpfD (so_4323) coding sequence (from position 225 to 639) was cloned in-frame at the 3’ end of the sequence coding for the T18 domain of adenylate cyclase into pEB355, leading to pUT18-bpfD.
To construct pBcbp-bpfD, the bpfD (so_4323) coding sequence (from position 225 to 639) was cloned in-frame at the 3’ end of the sequence coding for the calmodulin-binding domain into pBAD24-CBP-linker.
All constructs were checked by DNA sequencing using appropriate primers.
4.4. SSA-Biofilm Formation Assay
For SSA-biofilm formation, S. oneidensis cells were cultivated in poor medium under agitation as previously established [24,25]. S. oneidensis cells were first grown overnight on LB plates and resuspended in 10 mL of LB medium. They were then diluted in LM (Lactate Medium) (0.2 g.L−1 yeast extract, 0.1 g.L−1 peptone, 10 mM HEPES (pH 7.4), 10 mM NaHCO3 and 20 mM lactate) at an optical density (OD) of 0.05 at 600 nm. For each tested condition, 2 mL of cells was put into borosilicate glass tubes. Incubation was performed at 28 °C with shaking for 24 h. Each tube was then emptied, filled with 3.5 mL of 0.2% crystal violet, and colored for 10 min. The tubes were then rinsed several times with water, in order to remove unbound crystal violet, and photographed. Spectrophotometric quantification was then performed: crystal violet was solubilized in 3 mL of 30% acetic acid and OD540 was measured. Strains containing plasmids were grown overnight in the presence of chloramphenicol.
4.5. Pellicle Formation Assay
For pellicle formation, S. oneidensis cells were cultivated in rich medium under static conditions as previously established [20,24]. S. oneidensis cells were first grown overnight on LB plates and resuspended in LB medium before being diluted in the same medium to reach an OD600 of 0.2. The suspensions were then transferred into Petri dishes and incubated for 24 h at 28 °C without agitation. Strains containing plasmids were grown overnight on plates containing antibiotics. When indicated, arabinose was added at 0.2%. Pictures were taken above the plates before and after the use of a toothpick on the pellicle surface.
4.6. Expression and Purification of Recombinant Proteins
Recombinant proteins Strep-BpfDS (cytoplasmic part of BpfD only), Strep-CheY3, and PdgB-His were produced from E. coli BL21 (DE3) strains containing pETbpfD, pETcheY3, and pETpdgB, respectively. The strains were grown aerobically to reach an OD600 of 0.8. Overproduction of the proteins was then allowed by adding isopropyl-β-D-thiogalactopyranoside (1 mM) and incubating for 1 h (pETbpfD) or 3 h (pETpdgB and pETcheY3) at 37 °C. The cells were then collected by centrifugation (10 min at 8000 rpm and 4 °C), resuspended in 100 mM Tris-HCl pH 7.5 and 150 mM NaCl (Strep-BpfD/CheY3) or 20 mM phosphate buffer pH 7.4 (PdgB-His), disrupted by a French press, and centrifuged at 11,000 rpm and 4 °C for 15 min. The supernatant was then centrifuged at 45,000 rpm for 1 h at 4 °C. For the Strep-tagged proteins, the resulting supernatant was loaded on a Strep-Tactin resin (IBA), while a HisTrapFF resin (GE Healthcare) was used for PdgB-His. The recombinant proteins were purified according to the manufacturer’s protocol. Protein concentrations were estimated by Bradford assays (Bio-Rad).
4.7. Bacterial Two-Hybrid Assays
Bacterial two-hybrid experiments were performed as described by Battesti and Bouveret [37] with some modifications. Two-hybrid plasmids were co-transformed into the reporter strain E. coli BTH101 lacking the adenylate cyclase gene, and the clones were selected on LB agar containing 50 µg.mL−1 of kanamycin and 100 µg.mL−1 of ampicillin. For positive controls, we used pUT18-zip and pKT25-zip, while pEB354 and pEB355 were used as negative controls. The plates were incubated for 4 days at 28 °C. Ten isolated clones were then inoculated overnight in fresh LB with kanamycin, ampicillin, and 0.5 mM IPTG. Then, 2 µL of the cultures was spotted on MacConkey plates containing lactose (Difco™ MacConkey agar), kanamycin, and ampicillin. MacConkey plates were scanned after 48 h incubation at 28 °C. For β-galactosidase assays, cells were lysed by adding PopCulture Reagent solution (Agilent) and lysozyme at 1 mg.mL−1 for 15 min prior to adding Z buffer (100 mM phosphate buffer pH 7, 1 mM MgSo4, 10 mM KCl and 50 mM β-mercaptoethanol). Finally, 2.2 mM of ortho-nitrophenyl-β-galactoside (ONPG) was added. Afterwards, β-galactosidase activity was measured using a modified Miller assay adapted for use in a Tecan Spark microplate reader according to Baaziz et al. [44].
4.8. Thermal Shift Assays
For buffer exchange, purified proteins were loaded onto an NAP-5 desalting column (GE Healthcare) and recovered in 100 mM Tris-HCl (pH 7.5) containing 150 mM NaCl. Thermal shift assays (TSAs) were performed using a BioRad CFX96 Touch Real-Time PCR instrument. Samples were prepared in a total volume of 20 µL as described previously [24,45]. BpfDS (7.5 µM), BpfDGGDEF (7.5 µM), or BpfDEAL (7.5 µM) were incubated in the presence of 10x SYPRO Orange (Sigma Life Science) with or without cAMP, GTP, or c-di-GMP (500 µM and 1 mM). Samples were then heated from 20 °C to 70 °C at a scan rate of 0.5 °C per 30 s. The protein unfolding curves were monitored by detecting changes in SYPRO Orange fluorescence. Melting temperatures were determined using the first derivative values of raw fluorescence data using Bio-Rad CFX Manager 3.1 software.
4.9. Isothermal Titration Calorimetry
The purified BpfDS was dialyzed at 4 °C three times (for 1 h each) against Tris-HCl 100 mM (pH 7.4) and NaCl 150 mM. Isothermal titration calorimetry (ITC) experiments were performed using the MicroCal PEAQ-ITC (Malvern Panalytical, Palaiseau, France) at 25 °C. Then, 575 µM of c-di-GMP (Sigma-Aldrich, Saint-Quentin Fallavier, France) was titrated using 19 injections of 2 µL against 20 µM of BpfDS in the sample cell of the ITC under a constant stirring speed of 750 rpm. C-di-GMP was also titrated against dialysis buffer and the resulting values were subtracted from the measured data with BpfDS. The PEAQ-ITC Analysis Software (version 1. 1. 0. 1262) was used to fit the collected data using a “One Set of Sites” model.
4.10. In Vivo Protein Interaction Assay
Pull-down experiments were adapted from Battesti et al. [46]. E. coli C600 cells containing pBcbp-bpfD and pBpdgA were grown at 37 °C. At an OD600 of 0.8, 0.2% arabinose was added and the incubation was prolonged for 1 h. Cells were then centrifuged at 10,000 rpm for 10 min at 4 °C and resuspended in CBP buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM Mg acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% Triton), disrupted by a French press, and pelleted at 8000 rpm for 10 min. The supernatant was incubated with 50 µL of calmodulin-binding peptide (CBP) affinity resin (Agilent) for 1 h on a stirring wheel. Then, the resin was centrifuged for 1 min at 2000 rpm, washed four times with 1 mL of the CBP buffer, resuspended in 40 µL loading buffer, and heat-denatured for 5 min at 95 °C. Proteins were then loaded on SDS-PAGE. A band excised from the gels was analyzed by mass spectrometry (LC-MS/MS).
4.11. In Vitro Protein Interaction Assay
Strep-tagged CheY3 (4.4 µM) or His-tagged PdgB (2.3 µM) were incubated with Strep-tagged BpfDS (4.4 µM or 5.4 µM, respectively). Then, 5 mM of crosslinker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was added for 1 h 15 min at room temperature before stopping the interactions with Tris-HCl (1M pH 8). Interactions were then analyzed by Western blotting after SDS-PAGE using StrepTag II Antibody HRP Conjugate (Novagen). In parallel, the reactions were also loaded on SDS-PAGE and bands excised from the gel were analyzed by mass spectrometry (LC-MS/MS).
Acknowledgments
Our special thanks go to Vincent Méjean for the fruitful discussions we had over the years and for his unfailing support. We thank Deborah Byrne from the Protein Expression facility (IMM) and Henri-Pierre Fierobe (LCB) for technical advice concerning ITC. We thank Florian Boullee for technical support during his internship. Rémy Puppo, Pascal Mansuelle, Christophe Verthuy, Maya Belghazi, and Régine Lebrun from the Proteomics Facility of the Mediterranean Institute of Microbiology, CNRS FR3479, “Marseille Protéomique” (labeled by IBiSA and Aix Marseille Univ), are acknowledged for mass spectrometry analyses. We would also like to thank Claudine Baraquet for critical reading of the manuscript. We are very grateful to the members of the Bip1 team for their helpful discussions.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms25179697/s1.
Author Contributions
Conceptualization, C.J.-C.; formal analysis, J.-P.P., A.B., A.L., A.V.-B., A.A.C., M.-T.G.-O., M.F. and C.J.-C.; investigation, J.-P.P., A.B., A.L., A.V.-B., A.A.C. and C.J.-C.; methodology, J.-P.P., A.B., A.L., A.V.-B. and A.A.C.; supervision, C.J.-C.; validation, C.J.-C.; writing—original draft, J.-P.P. and C.J.-C.; writing—review and editing, J.-P.P., A.B., M.-T.G.-O., M.F. and C.J.-C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
This research was funded by the Centre National de la Recherche Scientifique (www.cnrs.fr), Aix-Marseille Université (www.univ-amu.fr) and HTS-BIO (www.htsbio.com). A.B. was supported by an MESR fellowship and by an ATER position (Aix-Marseille Université). J.-P.P. was supported by a postdoctoral fellowship (Université de Corse Pasquale Paoli). A.V.-G. is supported by HTS-BIO.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Flemming H.-C., Wuertz S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 2.Stoodley P., Sauer K., Davies D.G., Costerton J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 3.Lappin-Scott H., Burton S., Stoodley P. Revealing a World of Biofilms—The Pioneering Research of Bill Costerton. Nat. Rev. Microbiol. 2014;12:781–787. doi: 10.1038/nrmicro3343. [DOI] [PubMed] [Google Scholar]
- 4.Armitano J., Méjean V., Jourlin-Castelli C. Gram-Negative Bacteria Can Also Form Pellicles. Environ. Microbiol. Rep. 2014;6:534–544. doi: 10.1111/1758-2229.12171. [DOI] [PubMed] [Google Scholar]
- 5.Kovács Á.T., Dragoš A. Evolved Biofilm: Review on the Experimental Evolution Studies of Bacillus subtilis Pellicles. J. Mol. Biol. 2019;431:4749–4759. doi: 10.1016/j.jmb.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Flemming H.-C., Wingender J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 7.Karygianni L., Ren Z., Koo H., Thurnheer T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Römling U., Galperin M.Y., Gomelsky M. Cyclic Di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. MMBR. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hengge R., Gründling A., Jenal U., Ryan R., Yildiz F. Bacterial Signal Transduction by Cyclic Di-GMP and Other Nucleotide Second Messengers. J. Bacteriol. 2016;198:15–26. doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenal U., Reinders A., Lori C. Cyclic Di-GMP: Second Messenger Extraordinaire. Nat. Rev. Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 11.Chou S.-H., Galperin M.Y. Diversity of Cyclic Di-GMP-Binding Proteins and Mechanisms. J. Bacteriol. 2016;198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins A.J., Smith T.J., Sondermann H., O’Toole G.A. From Input to Output: The Lap/c-Di-GMP Biofilm Regulatory Circuit. Annu. Rev. Microbiol. 2020;74:607–631. doi: 10.1146/annurev-micro-011520-094214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlstrom K.M., Giglio K.M., Collins A.J., Sondermann H., O’Toole G.A. Contribution of Physical Interactions to Signaling Specificity between a Diguanylate Cyclase and Its Effector. mBio. 2015;6:e01978-15. doi: 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee D., Boyd C.D., O’Toole G.A., Sondermann H. Structural Characterization of a Conserved, Calcium-Dependent Periplasmic Protease from Legionella pneumophila. J. Bacteriol. 2012;194:4415–4425. doi: 10.1128/JB.00640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G., Yuan J., Gao H. Regulation of Biofilm Formation by BpfA, BpfD, and BpfG in Shewanella oneidensis. Front. Microbiol. 2015;6:790. doi: 10.3389/fmicb.2015.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrosis N., Boyd C.D., O Toole G.A., Fernández J., Sisti F. Homologs of the LapD-LapG c-Di-GMP Effector System Control Biofilm Formation by Bordetella bronchiseptica. PLoS ONE. 2016;11:e0158752. doi: 10.1371/journal.pone.0158752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooley R.B., Smith T.J., Leung W., Tierney V., Borlee B.R., O’Toole G.A., Sondermann H. Cyclic Di-GMP-Regulated Periplasmic Proteolysis of a Pseudomonas aeruginosa Type Vb Secretion System Substrate. J. Bacteriol. 2016;198:66–76. doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitts G., Giglio K.M., Zamorano-Sánchez D., Park J.H., Townsley L., Cooley R.B., Wucher B.R., Klose K.E., Nadell C.D., Yildiz F.H., et al. A Conserved Regulatory Circuit Controls Large Adhesins in Vibrio cholerae. mBio. 2019;10:e02822-19. doi: 10.1128/mBio.02822-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Y., Gao H., Chen J., Dong Y., Wu L., He Z., Liu X., Qiu G., Zhou J. Pellicle Formation in Shewanella oneidensis. BMC Microbiol. 2010;10:291. doi: 10.1186/1471-2180-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armitano J., Méjean V., Jourlin-Castelli C. Aerotaxis Governs Floating Biofilm Formation in Shewanella oneidensis. Environ. Microbiol. 2013;15:3108–3118. doi: 10.1111/1462-2920.12158. [DOI] [PubMed] [Google Scholar]
- 21.Thormann K.M., Saville R.M., Shukla S., Pelletier D.A., Spormann A.M. Initial Phases of Biofilm Formation in Shewanella oneidensis MR-1. J. Bacteriol. 2004;186:8096–8104. doi: 10.1128/JB.186.23.8096-8104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thormann K.M., Duttler S., Saville R.M., Hyodo M., Shukla S., Hayakawa Y., Spormann A.M. Control of Formation and Cellular Detachment from Shewanella oneidensis MR-1 Biofilms by Cyclic Di-GMP. J. Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theunissen S., De Smet L., Dansercoer A., Motte B., Coenye T., Van Beeumen J.J., Devreese B., Savvides S.N., Vergauwen B. The 285 KDa Bap/RTX Hybrid Cell Surface Protein (SO4317) of Shewanella oneidensis MR-1 Is a Key Mediator of Biofilm Formation. Res. Microbiol. 2010;161:144–152. doi: 10.1016/j.resmic.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Gambari C., Boyeldieu A., Armitano J., Méjean V., Jourlin-Castelli C. Control of Pellicle Biogenesis Involves the Diguanylate Cyclases PdgA and PdgB, the c-Di-GMP Binding Protein MxdA and the Chemotaxis Response Regulator CheY3 in Shewanella oneidensis. Environ. Microbiol. 2019;21:81–97. doi: 10.1111/1462-2920.14424. [DOI] [PubMed] [Google Scholar]
- 25.Boyeldieu A., Ali Chaouche A., Ba M., Honoré F.A., Méjean V., Jourlin-Castelli C. The Phosphorylated Regulator of Chemotaxis Is Crucial throughout Biofilm Biogenesis in Shewanella oneidensis. Npj Biofilms Microbiomes. 2020;6:54. doi: 10.1038/s41522-020-00165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newell P.D., Monds R.D., O’Toole G.A. LapD Is a Bis-(3′,5′)-Cyclic Dimeric GMP-Binding Protein That Regulates Surface Attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinsa S.M., O’Toole G.A. Biofilm Formation by Pseudomonas fluorescens WCS365: A Role for LapD. Microbiol. Read. Engl. 2006;152:1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 28.Boyd C.D., Smith T.J., El-Kirat-Chatel S., Newell P.D., Dufrêne Y.F., O’Toole G.A. Structural Features of the Pseudomonas fluorescens Biofilm Adhesin LapA Required for LapG-Dependent Cleavage, Biofilm Formation, and Cell Surface Localization. J. Bacteriol. 2014;196:2775–2788. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinsa S.M., Espinosa-Urgel M., Ramos J.L., O’Toole G.A. Transition from Reversible to Irreversible Attachment during Biofilm Formation by Pseudomonas fluorescens WCS365 Requires an ABC Transporter and a Large Secreted Protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 30.Reichhardt C., Wong C., Passos da Silva D., Wozniak D.J., Parsek M.R. CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing. mBio. 2018;9:e01376-18. doi: 10.1128/mBio.01376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borlee B.R., Goldman A.D., Murakami K., Samudrala R., Wozniak D.J., Parsek M.R. Pseudomonas aeruginosa Uses a Cyclic-Di-GMP-Regulated Adhesin to Reinforce the Biofilm Extracellular Matrix. Mol. Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlstrom K.M., Collins A.J., Doing G., Taroni J.N., Gauvin T.J., Greene C.S., Hogan D.A., O’Toole G.A. A Multimodal Strategy Used by a Large C-Di-GMP Network. J. Bacteriol. 2018;200:e00703-17. doi: 10.1128/JB.00703-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giacalone D., Smith T.J., Collins A.J., Sondermann H., Koziol L.J., O’Toole G.A. Ligand-Mediated Biofilm Formation via Enhanced Physical Interaction between a Diguanylate Cyclase and Its Receptor. mBio. 2018;9:e01254-18. doi: 10.1128/mBio.01254-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fagerlund A., Smith V., Røhr Å.K., Lindbäck T., Parmer M.P., Andersson K.K., Reubsaet L., Økstad O.A. Cyclic Diguanylate Regulation of Bacillus cereus Group Biofilm Formation. Mol. Microbiol. 2016;101:471–494. doi: 10.1111/mmi.13405. [DOI] [PubMed] [Google Scholar]
- 35.Vasenina A., Fu Y., O’Toole G.A., Mucha P.J. Local Control: A Hub-Based Model for the c-Di-GMP Network. mSphere. 2024;9:e0017824. doi: 10.1128/msphere.00178-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordi C., Iobbi-Nivol C., Méjean V., Patte J.-C. Effects of ISSo2 Insertions in Structural and Regulatory Genes of the Trimethylamine Oxide Reductase of Shewanella oneidensis. J. Bacteriol. 2003;185:2042–2045. doi: 10.1128/JB.185.6.2042-2045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battesti A., Bouveret E. The Bacterial Two-Hybrid System Based on Adenylate Cyclase Reconstitution in Escherichia coli. Methods. 2012;58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Appleyard R.K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrero M., de Lorenzo V., Timmis K.N. Transposon Vectors Containing Non-Antibiotic Resistance Selection Markers for Cloning and Stable Chromosomal Insertion of Foreign Genes in Gram-Negative Bacteria. J. Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baraquet C., Théraulaz L., Iobbi-Nivol C., Méjean V., Jourlin-Castelli C. Unexpected Chemoreceptors Mediate Energy Taxis towards Electron Acceptors in Shewanella oneidensis. Mol. Microbiol. 2009;73:278–290. doi: 10.1111/j.1365-2958.2009.06770.x. [DOI] [PubMed] [Google Scholar]
- 41.Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose PBAD Promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battesti A., Bouveret E. Improvement of Bacterial Two-Hybrid Vectors for Detection of Fusion Proteins and Transfer to PBAD-Tandem Affinity Purification, Calmodulin Binding Peptide, or 6-Histidine Tag Vectors. Proteomics. 2008;8:4768–4771. doi: 10.1002/pmic.200800270. [DOI] [PubMed] [Google Scholar]
- 43.Figurski D.H., Helinski D.R. Replication of an Origin-Containing Derivative of Plasmid RK2 Dependent on a Plasmid Function Provided in trans. Proc. Natl. Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baaziz H., Gambari C., Boyeldieu A., Ali Chaouche A., Alatou R., Méjean V., Jourlin-Castelli C., Fons M. ChrASO, the Chromate Efflux Pump of Shewanella oneidensis, Improves Chromate Survival and Reduction. PLoS ONE. 2017;12:e0188516. doi: 10.1371/journal.pone.0188516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyeldieu A., Ali Chaouche A., Méjean V., Jourlin-Castelli C. Combining Two Optimized and Affordable Methods to Assign Chemoreceptors to a Specific Signal. Anal. Biochem. 2021;620:114139. doi: 10.1016/j.ab.2021.114139. [DOI] [PubMed] [Google Scholar]
- 46.Battesti A., Hoskins J.R., Tong S., Milanesio P., Mann J.M., Kravats A., Tsegaye Y.M., Bougdour A., Wickner S., Gottesman S. Anti-Adaptors Provide Multiple Modes for Regulation of the RssB Adaptor Protein. Genes Dev. 2013;27:2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.