Abstract
The objective of this study was to determine prevalence and perform genomic analysis of Salmonella spp. and Campylobacter spp. isolated from different stages of an integrated NAE broiler complex. Environmental samples were screened with 3M-Molecular Detection System (MDS) and MDS positive samples were further processed for confirmation of results and identification. Core genome-based phylogenies were built for both bacteria isolated from this study along with selected NCBI genomes. The odds ratios and 95% confidence limits were compared among stages and sample types (α < 0.05) using multivariable model. Based on MDS results, 4% and 18% of total samples were positive for Salmonella spp. and Campylobacter spp. respectively. The odds of Salmonella detection in hatchery samples were 2.58 times as likely as compared to its detection in production farms’ samples (P = 0.151) while the odds of Campylobacter detection in production farms’ samples were 32.19 times as likely as its detection in hatchery (P = 0.0015). Similarly, the odds of Campylobacter detection in boot swabs, soil, water, and miscellaneous samples were statistically significant (P < 0.05) as compared with fly paper as reference group. The serovars identified for Salmonella were Typhimurium, Barranquilla, Liverpool, Kentucky, Enteritidis, Luciana, and Rough_O:r:1,5. For Campylobacter, the species identified were Campylobacter jejuni and Campylobacter coli. Phylogeny results show close genetic relatedness among bacterial strains isolated from different locations within the same stage and between different stages. The results show possibility of multiple entry points of such bacteria entering broiler complex and can potentially contaminate the final raw product in the processing plant. It suggests the need for a comprehensive control strategy with strict biosecurity measures and best management practices to minimize or eliminate such pathogens from the poultry food chain.
Key words: Salmonella, Campylobacter, serovar, phylogeny, environmental sample
INTRODUCTION
Salmonella and Campylobacter continue to be the leading bacterial foodborne pathogens frequently associated with consumption or handling of raw poultry meat and meat products, a serious food safety concern (Scallan et al., 2011; Silva et al., 2011; Antunes et al., 2016; Skarp et al., 2016). About 17.3% of Salmonella illnesses were attributed to chicken meat in 2020 (IFSAC report, 2022) while 65% of Campylobacter illness were attributed to chicken meat in 2019 in the United States (IFSAC report, 2021). However, the pathogens are not only limited to poultry meat and meat products alone. In most of the cases, these foodborne infections in humans are usually self-limiting and subside within a week without any medications, but sometimes, the infections can be fatal, especially in vulnerable population groups such as young, immunocompromised, and elderly people (Skarp et al., 2016; Turgeon et al., 2018). The Centers for Disease Control and Prevention (CDC) estimates approximately 1.35 million infections, 26,500 hospitalizations, and 420 deaths annually in the United States due to non-typhoidal Salmonella infections (CDC, 2023a). Similarly, Campylobacter is considered as one of the 4 major global causes of diarrheal diseases worldwide (WHO, 2020). Moreover, the annual economic burden of foodborne illness due to Salmonella spp. in chicken and Campylobacter in poultry account for $2.8 and $6.9 billion respectively every year (Scharff, 2020).
These pathogens are typically present in the gastrointestinal tracts of animal and bird as reservoirs (Corry and Atabay, 2001; Newell and Fearnley, 2003; Eng et al., 2015). The ingestion of food or drinking water contaminated with feces usually causes foodborne illness in humans which is characterized by gastroenteritis, abdominal cramps and headache (Eng et al., 2015; Skarp et al., 2016; CDC, 2023b). In rare cases, Campylobacter jejuni (C. jejuni) may lead to post-infectious Guillain–Barré syndrome, a severe demyelinating neuropathy (Nachamkin et al., 1998). In birds, the Campylobacter spp. infection is usually asymptomatic (Newell and Fearnley, 2003). However, C. hepaticus and C. bilis was reported to decrease egg production in layer chickens and cause spotty liver disease (Moore et al., 2019; Van et al., 2023).
Despite the significant improvements made over past few years to minimize these pathogens along poultry food chain, the major challenges is to reduce the risk of introduction/spread of such bacteria in flocks, and to achieve performance standards at processing plants. Salmonella can survive desiccation and can persist in dry environments and foods for years (Estrada et al., 2023) while birds’ ceca provide a suitable condition for Campylobacter to multiply and spread rapidly across the flock (Corry and Atabay, 2001; Munoz et al., 2023). Moreover, epidemiological studies that could explain the introduction of these pathogens along the chain and their transmissions resulting in the contamination of raw poultry products in the processing plant are lacking (Obe et al., 2020). Previously, Pulsed Field Gel Electrophoresis (PFGE) fingerprinting was often used in most of the previous longitudinal studies to track pathogens along food chain (Peters, 2009). However, at present, whole genome sequencing (WGS) and Single Nucleotide Polymorphism (SNP) based analysis allow in-depth understanding of the epidemiology and population dynamics of these pathogens along the food chain (Octavia et al., 2015; Medina-Santana et al., 2022).
In the United States, most commercial broiler companies are vertically integrated that comprises of pullet rearing farms, rooster rearing farms, breeder farms, hatcheries, broiler grow-out farms, feed mills, transport facilities and processing plants (NCC, 2023). Almost 50% of total broiler production in the United States in 2020 were accounted from NAE-raised birds (Poultry Health Today, 2021). This shift to NAE from conventional antibiotic-fed birds was partially the result of the Food and Drug Administration's (FDA) antibiotic withdrawal policy in 2017. However, the impact of this shift to NAE has been rarely studied on the prevalence and tracking of Salmonella spp. and Campylobacter spp. on the farms and facilities. “NAE” label refers to no use of any sort of antibiotics either classified as important in human medicine or others during raising of broiler chicken; “organic” label refers to no use of any sort of antibiotics as well as chemical coccidiostats in the life of broiler chickens while conventional refers to use of antibiotics or growth promoters as standard feed additive via feed or drinking water (Smith, 2019). However, “antibiotic free” or “chemical free” labels are considered misleading since claims cannot be conclusively proven (Smith, 2019). It is important to consider that if a sick broiler flock needs to be treated with antibiotics, meat from that flock cannot be labeled as NAE or organic and must be separated and diverted to and labeled as a conventional product.
The objective of this study was to determine the prevalence status and phylogenetic analysis of Salmonella spp. and Campylobacter spp. isolated via environmental sampling of various farms/ facilities of an integrated broiler complex. To our knowledge, this is the first longitudinal study from pullets through final raw product at the processing plant to determine critical entry points and transmission pattern of such bacteria for improved control strategies. We hoped to determine the possible entry points along different stages of an integrated broiler complex and movement pattern of pathogens along the poultry food chain.
MATERIALS AND METHODS
Study Design
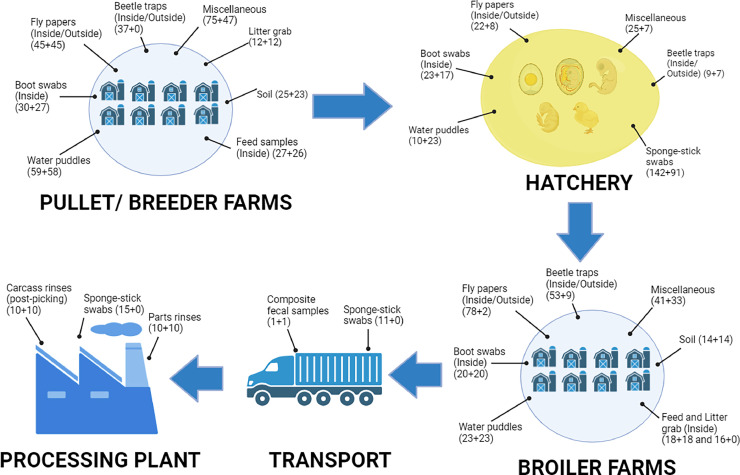
This study was conducted in the south-east region of the United States from December 2021 to November 2022 in an integrated broiler complex that consisted of approximately 8 pullet farms, 14 breeder farms, 74 broiler farms, 1 hatchery and 1 processing plant in total. The complex has been in operation for more than 5 y. With an aim to include at least 10% of total farms/ facilities, a total of 856 and 531 environmental samples were collected from 2 pullet farms at 16 wk-of-age, 4 breeder farms at 27, 36, 37 and 42 wk-of-age, a hatchery, 9 broiler farms at age 20, 21, 23, 28 × 2, 37, 41, 44 and 45 d-of-age, 2 transport trucks with transport cages, and 1 processing plant. The farms sampled were selected by the company personnel for the convenience of their routine schedule. The sample types and numbers collected in this study are presented in Figure 1 and Table 1.
Figure 1.
Environmental samples from farms and facilities considered in this study. (This figure displays the various stages: pullet farms, breeder farms, hatchery, broiler farms, transport, and processing plant of an integrated broiler complex along with the possible farm/ facilities’ environmental samples that were considered in this study for the isolation of Salmonella and Campylobacter. The numbers inside each bracket represent number of samples collected for isolation of Salmonella and Campylobacter respectively.) (Figure created with BioRender.com)
Table 1.
Total number of samples based on stages of production and sample types.
| Samples for Salmonella isolation |
Samples for Campylobacter isolation |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. no. | Sample types | Pullet | Breeder | Hatchery | Broiler | Transport | Processing | Total | Pullet | Breeder | Hatchery | Broiler | Transport | Processing | Total |
| 1 | Boot swabs | 12 | 18 | 23 | 20 | 0 | 0 | 73 | 12 | 15 | 17 | 20 | 0 | 0 | 64 |
| 2 | Sponge- Stick swabs | 0 | 0 | 142 | 0 | 11 | 15 | 168 | 0 | 0 | 91 | 0 | 0 | 0 | 91 |
| 3 | Beetle traps | 13 | 24 | 9 | 53 | 0 | 0 | 99 | 0 | 0 | 7 | 9 | 0 | 0 | 16 |
| 4 | Fly papers | 18 | 27 | 22 | 78 | 0 | 3 | 148 | 18 | 27 | 8 | 2 | 0 | 0 | 55 |
| 5 | Litter grab | 0 | 12 | 0 | 16 | 1 | 0 | 29 | 0 | 12 | 0 | 0 | 1 | 0 | 13 |
| 6 | Feed | 12 | 15 | 0 | 18 | 0 | 0 | 45 | 12 | 14 | 0 | 18 | 0 | 0 | 44 |
| 7 | Soil samples | 8 | 17 | 0 | 14 | 0 | 0 | 39 | 8 | 15 | 0 | 14 | 0 | 0 | 37 |
| 8 | Water puddles/ drainage | 9 | 50 | 10 | 23 | 0 | 0 | 92 | 9 | 49 | 23 | 23 | 0 | 0 | 104 |
| 9 | Carcass rinses | 0 | 0 | 0 | 0 | 0 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 20 | 20 |
| 10 | Feather fluff | 0 | 0 | 12 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | Miscellaneous | 35 | 40 | 13 | 41 | 0 | 2 | 131 | 10 | 37 | 7 | 33 | 0 | 0 | 87 |
| Total | 107 | 203 | 231 | 263 | 12 | 40 | 856 | 69 | 169 | 153 | 119 | 1 | 20 | 531 | |
The miscellaneous samples include cow feces, mouse intestine, unknown feces, wild bird feather, goat feces, dog feces and others that could potentially transmit these bacteria into the house/ facilities.)
Pullet, Breeder, and Broiler Farm Samples
Typically, broiler farms consisted of 4 houses per farm while the number of houses varied from 4 to 8 houses per farm in case of pullet and breeder farms. In total, 12 houses from 2 pullet farms, 14 houses from 4 breeder farms and a total of 18 houses from 9 broiler farms were sampled for this study. From inside the houses, 2 boot swabs (one for Salmonella and Campylobacter), 2 flypapers, 2 beetle traps, 1 litter grab and 1 feed sample were collected. Sterile boot swabs (Envirobootie, Hardy Diagnostics, CA) were placed over the boot covers just prior to entry into the house and the person wearing the boots walked along the feed and drinker lines from one end to the other end of the house, with an emphasis to step on fresh feces (McCrea et al., 2008). The boot swabs were then collected aseptically into sterile bags. The flypapers and beetle traps were placed near the entrance of houses a week before and were collected on the sampling day. The fly papers were dual-sided yellow sticky traps 6 × 8 inches in size (Yellow Sticky Traps, Gilead, ME) to collect flies and dust particles. Similarly, for collecting beetles/crawling insects, the beetle traps were prepared as described by Hess et al. (2008). A polyvinyl chloride (PVC) pipe of 23 cm long and 3.8 cm diameter with a small hole in each end (to hold the pipe in place on the litter) was prepared and a rolled piece of cardboard (20 × 30 cm) was inserted inside the pipe.
From outside the houses, 2 flypapers, 2 beetle traps and 1 soil sample were collected near the entrance. In addition, water puddles/ drainage and miscellaneous samples were collected from around the house. The miscellaneous samples consisted of any samples that can potentially transmit pathogens from the outside environment to the birds inside, such as fecal samples from cows, wild birds, dogs, goats, deer, or unknown origin. When available, samples from dead rodents’ intestines, wild bird feathers and nests, ants, spiders, and dead wild birds’ intestines were also collected.
Hatchery Samples
Samples were collected from 2 different visits to a single-stage hatchery. Like the farms, fly papers and beetle traps were collected from inside and outside the facility. Incubators, hatchers, egg buggies, chick trays, faucet and door handle of restroom were swabbed with sterile 3M sponge-stick swabs premoistened with 10 mL buffered peptone water (Schroeder et al., 2014). In addition, boot swabs, feather fluff from hatchers and composite broken eggs were collected. Moreover, swabs were collected from waters and floor of hatchers after sanitation and disinfection.
Transport and Processing Plant Samples
There was a total of 9 different sampling visits to pullet, breeder, hatchery, and broiler farms. On the tenth visit, we followed the broiler flocks to transport and finally to processing plant. We collected boot swabs, fly papers and beetle traps from broiler farms. Similarly, we swabbed the transport cages’ floor with sponge-stick swabs and collected composite fecal samples from the floor of transport trailer and transport cages. From the processing plant, sponge stick swab samples were collected from live birds unloading areas, conveyor belts, floor near scalding and evisceration areas, and 10 carcass rinses after scalding and picking, and 10 tenders’ parts rinses post-chilling and deboning.
Pathogen Isolation
To isolate the pathogen, samples were pre-enriched with buffered peptone water (DifcoTM Buffered Peptone Water, Becton, Dickinson and Company, Sparks, MD) for Salmonella isolation and with 3MTM Campy enrichment broth CE250 for Campylobacter spp. isolation and incubated for 18 to 24 h at 37°C and 42°C respectively. The enrichments were screened with 3M Molecular Detection System (MDS), a PCR-based system which works on the principle of loop-mediated isothermal amplification of DNA (Notomi, 2000; Pooja et al., 2014). The MDS positive samples were further processed to obtain pure isolates. Salmonella spp. and Campylobacter spp. were isolated using USDA FSIS microbiology laboratory guidelines (United States Department of Agriculture – Food Safety and Inspection Service (USDA-FSIS). MLG4.13 2023, United States Department of Agriculture – Food Safety and Inspection Service (USDA-FSIS). MLG41.05 2021 respectively). For the isolation of Salmonella spp., the suspected positive samples were further selectively enriched with tetrathionate broth and Rappaport-Vassiliadis broth for 24 h at 41°C after which they were streaked on Xylose Lysine Tergitol (XLT4) agar and then incubated for 24 h at 37°C. After that, 4 typical colonies were further streaked on XLT4 and chrome agar along with biochemical confirmation by Triple Sugar Iron (TSI), Lysine Iron Agar (LIA) and Urease agar slants. The pure isolates were then sero-grouped based on Poly O antiserum and O grouping antiserum. Finally, the pure isolates were sent for serotyping to National Veterinary Service Laboratory (NVSL), Ames, IA. Similarly, for Campylobacter isolation, the suspect positive samples were directly streaked onto Campy Cefex agar (HiMedia Laboratories Pvt. Ltd., Mumbai, India) then incubated at 42°C for 48 h under microaerobic conditions (5% O2 and 10% CO2, 85% N2). The pure isolates were identified based on typical translucent or mucoid, glistening, and pink color colonies. All isolates were stored in cryotubes containing Brucella broth with 20% glycerol with beads at -80°C for further analysis.
DNA Extraction and Sequencing
DNA were extracted using E. Z. N. A. Bacterial DNA Kit (Omega Bio-tek, Inc., GA). Concentrations and quality parameters were measured using a Qubit Fluorometer (Invitrogen/ Thermo Fisher Scientific, Waltham, MA) and NanoDrop (Thermo Fisher Scientific Waltham, MA) to determine 260/280 and 260/230 ratios. All 16 Salmonella isolates and 8 Campylobacter isolates were whole genome sequenced using MiniSeq (Illumina, San Diego, CA) in our laboratory. However, remaining 10Campylobacter isolates were sent to SEQCENTER (Pittsburgh, PA) for Illumina WGS. Briefly, the DNA library prep was performed based on the protocol described in Illumina DNA Prep Reference Guide that involves the workflow steps: tagment genomic DNA, post tagmentation cleanup, amplify tagmented DNA, clean up libraries and pool libraries (Illumina, 2020). The kits used for DNA library prep were IDT® for Illumina DNA/RNA UD Indexes Set A and Miniseq High Output Reagent Cartridge (300 cycles) with FC-420-1003. Final denaturation and dilution of pool libraries and PhiX (PhiX control v3®) to loading concentration were performed based on Denature and Dilute Libraries Guide Protocol A: Standard Normalization Method and Denature and Dilute PhiX Control protocol respectively (Illumina, 2019). Finally, paired end fastq files were obtained following demultiplexing.
Bioinformatics Analyses
Quality check of raw reads was performed using FastQC (Andrews, 2010). All raw reads with median quality score above 34 were then subjected for adapter trimming and quality trimming with BBDuk tools (Bushnell, 2014). The de novo assembly was performed using SPADEs (Prjibelski et al., 2020) with a custom k-mer values of 25, 33, 55, 77, 95 and 127 kmer lengths. The genome assembly statistics of each genome were obtained using Quast (Quast et al., 2012). The Salmonella serovars were identified using both SeqSero 1.2 service of CGE and classical serotyping using polyvalent and single factor antisera to determine the O and H antigens (National Veterinary Services Laboratories, USDA, Ames, IA). For the identification of Campylobacter spp., KmerFinder 3.2 (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018) and SpeciesFinder 2.0 (Camacho et al., 2009; Quast et al., 2012; Clausen et al., 2018) of Center for Genomic Epidemiology (CGE) were used.
Genomic Analyses
In addition to 16 Salmonella genomes from this study, 316 assembled genomes of Salmonella enterica from NCBI were also considered to build a phylogenetic tree. For serovars: Liverpool, Barranquilla, and Luciana, all the available assembled genomes were downloaded from NCBI. However, for serovars: Kentucky, Typhimurium, Enteritidis and Rough O: r:1,5, KmerFinder 3.2 service was used to find the closely related strain of specific serovar, and SNP clusters were identified via SNP Tree Viewer. The SNP clusters considered for serovars: Kentucky, Typhimurium, Enteritidis and Infantis were PDG000000002.2908/PDS000032535.11, PDG00000000 2.2908/PDS000100251.130, PDG000000002.2908/PDS000083226.595 and PDG000000002.2908/PDS000171305.18 respectively. Based on specific node in SNP tree, only 195 Liverpool, 67 Barranquilla, 12 Luciana, 20 Kentucky, 16 Typhimurium, 2 Enteritidis and 5 Infantis genomes were downloaded from NCBI for further phylogenetic analyses. Similarly, in addition to 18 Campylobacter genomes from this study, 7 assembled genomes of C. jejuni and 8 genomes of C. coli were downloaded from NCBI. KmerFinder 3.2 service of CGE was used to find the closely related strain of species and SNP clusters were identified via SNP Tree Viewer in NCBI. The SNP clusters considered for C. jejuni were PDG000000003.2136/PDS000022040.148, and for C. coli were PDG000000003.2136/PDS000043406.2 and PDG000000003.2136/ PDS000010524.4.
The assembled genomes were subjected to SNP-based core genome phylogeny using ParSNP (Treangen et al., 2014) and the phylogenetic trees were obtained using iTOL (Letunic and Bork, 2019). Furthermore, CSI Phylogeny 1.4 service of CGE (Kaas et al., 2014) was used to determine the SNP distance matrices among strains within the Salmonella serovars or Campylobacter species. The genetic relatedness among the Salmonella isolates were analyzed as mentioned in Octavia et al. (2015) which used the cutoff mutation of 4 SNPs within 30 d and 9 SNPs for 120 d time intervals for possibility to consider as same strains.
Statistical Analyses
Data were analyzed with R version 4.3.1 (Team, 2015) using Generalized Linear Modeling for binomial distribution. The odds ratio and 95% confidence limits were calculated for stages of productions and different sample types using multivariable model. Statistical significance was set to P -values < 0.05. The model assessed the effects of stages and sample types on the presence/ absence of Salmonella. The pairwise comparison of variables within a group were estimated by Tukey method to separate means among different variables using “emmeans” package. The statistical analyses and interpretations are based on MDS results since it provides more reliable information about the presence/absence of pathogens’ DNA even if the pathogen is not culturable.
RESULTS
Based on MDS results, 4% (38/856) of samples were positive for Salmonella while 18% (97/531) of samples were positive for Campylobacter. Out of 15 different farms, 6 farms (40%) were found to be contaminated with Salmonella and 12 farms (80%) were found to be contaminated with Campylobacter. However, based on culture results, 2% (16/856) of samples were positive for Salmonella while 3% (18/531) of samples were positive for Campylobacter. Detailed information about MDS and culture results among various stages for Salmonella spp. and Campylobacter spp. are presented in Table 2A, Table 2B respectively.
Table 2A.
Prevalence of Salmonella based on MDS and culture results.
| Stages | MDS positive | Culture positive | Serovars |
|---|---|---|---|
| Pullet | 5/107 (Boot swab, soil, and beetle trap) | 1/107 (Boot swab) | Rough_O: r:1,5 |
| Breeder | 0/203 | 0/203 | - |
| Hatchery | 17/231 (water sample, sponge-stick swabs, boot swabs, miscellaneous) | 5/231 (water sample, Swab from incubator, hatcher, chick box, and egg buggy) | Enteritidis (1) and Typhimurium (4) |
| Broiler | 13/263 (water sample, fly papers, miscellaneous, beetle trap) | 7/247 (Water near doors- 2, Unknown feces, Fly papers inside/outside-4) | Liverpool (1), Kentucky (1), Luciana (1) and Barranquilla (4) |
| Transport | 1/12 (swabs) | 1/12 (swabs) | Kentucky (1) |
| Processing | 2/40 (fly papers) | 2/40 (fly paper outside-2) | Liverpool (2) |
| Total | 38/856 | 16/856 |
Table 2B.
Prevalence of Campylobacter based on MDS and culture results.
| Stages | MDS positive | Culture positive | Species |
|---|---|---|---|
| Pullet farms | 12/69 (boot swabs, soil samples, miscellaneous) | 1/69 (Mouse intestine) | C. jejuni |
| Breeder farm | 58/169 (boot swabs, soil samples, fly papers, water samples, miscellaneous) | 5/169 (Mouse intestine, cow feces-3 and fly paper inside) | C. coli |
| Hatchery | 2/152 (fly paper, sponge-stick swab) | 0/152 | - |
| Broiler farms | 15/119 (water sample, soil sample, boot swabs, miscellaneous) | 3/119 (Cow feces and boot swabs) | C. coli and C. jejuni |
| Transport | 0/2 | 0/2 | - |
| Processing | 10/20 (rinses) | 9/20 (rinses) | C. coli |
| Total | 97/531 | 18/531 |
Salmonella Prevalence Among Different Stages and Sample types
For Salmonella spp., none of the samples from breeder farms (0/203) were MDS positive. However, the pathogen tested positive for MDS in the other stages and 7 different sample types. The prevalence percentages in production farms, hatchery, transport, and processing plant were 3.14% (18/573), 7.34% (17/231), 8.33% (1/12), and 5% (2/40) respectively. The prevalence percentages in boot swabs, sponge-stick swabs, fly papers, miscellaneous samples, soil, water, and beetle traps were 9.59% (7/73), 6.55% (11/168), 6.08% (9/148), 3.05% (4/131), 2.56% (1/39), 4.35% (4/92), and 2.02% (2/99) respectively. The odds of Salmonella spp. detection in hatchery samples were 2.58 times (1.03–6.11; 95% CLs) as likely as its detection in production farms’ samples (P = 0.151). Similarly, the odds of Salmonella detection in boot swabs samples were 4 times (0.9–27.93; 95% CLs) as likely as its detection in beetle traps samples (P = 0.637). However, it was not statistically significant (Table 3). Of 38 positive samples after screening with MDS, only 16 pure isolates were recovered in culture.
Table 3.
Generalized linear modeling for MDS positive Salmonella spp. results with multivariable analysis.
| STAGES | Positive/Total | Odds ratio | Lower CL | Upper CL | P value (Tukey) |
|---|---|---|---|---|---|
| Production farms | 18/500 | Reference group | |||
| Hatchery | 17/219 | 2.582 | 1.03 | 6.11 | 0.151 |
| Transport | 1/11 | 3.967 | 0.53 | 17.64 | 0.651 |
| Processing plant | 2/20 | 3.844 | 0.19 | 30 | 0.397 |
| SAMPLE TYPES | |||||
| Beetle trap | 2/99 | Reference group | |||
| Boot swab | 7/73 | 4.00 | 0.90 | 27.93 | 0.637 |
| Fly paper | 9/148 | 2.81 | 0.69 | 18.83 | 0.853 |
| Miscellaneous | 4/131 | 1.46 | 0.28 | 10.73 | 0.999 |
| Soil sample | 1/39 | 1.46 | 0.06 | 15.74 | 0.999 |
| Water sample | 4/92 | 2.16 | 0.41 | 15.90 | 0.976 |
| Sponge-stick swab | 11/168 | 1.39 | 0.29 | 10.31 | 0.999 |
(The table displays the odds ratio results for both stages and sample types. We observed that the odds of Salmonella spp. detection in hatchery samples were 2.582 times (1.03–6.11; 95% CLs) as likely as its detection in production farms’ samples (P = 0.151). Similarly, the odds of Salmonella spp. detection in boot swabs samples were 4 times (0.9–27.93; 95% CLs) as likely as its detection in beetle trap samples (P =0.637). However, none of the stages and sample types were statistically significant for likelihood of occurrence of Salmonella spp.)
Campylobacter Prevalence Among Different Stages and Sample types
For Campylobacter spp., samples from all the stages were positive with MDS. The prevalence percentages in production farms, hatchery, and processing plant were 23.81% (85/357), 1.32% (2/152), and 45.45% (10/22) respectively. The prevalence percentages in soil, boot swabs, water, miscellaneous samples, fly papers, sponge-stick swabs and carcass rinses were 43.24% (16/37), 42.19% (27/64), 22.16% (23/104), 18.39% (16/87), 7.27% (4/55), 1.10% (1/91) and 50% (10/20) respectively. The odds of Campylobacter spp. detection in production farms’ samples were 32.19 times (6.57–583.62; 95% CLs) as likely as its detection in hatchery samples (P = 0.0008). Among the production farms, the odds of Campylobacter spp. detection in breeder farms’ samples were 7.724 times (3.67–17.65; 95% CLs) as likely as its detection in broiler farms’ samples (P < 0.001). Similarly, the odds of Campylobacter spp. detection in boot swabs were 48.85 times (13.52 – 241.41; 95% CLs) as likely as its detection in fly paper samples (P < 0.0001) (Table 4). Of 97 positive samples after screening with MDS, only 18 pure isolates were recovered in culture.
Table 4.
Generalized linear modeling for MDS positive Campylobacter spp. results with multivariable analysis.
| STAGES | Positive/Total | Odds ratio | Lower CL | Upper CL | P value (Tukey) |
|---|---|---|---|---|---|
| Hatchery | 2/145B | Reference group | |||
| Production farms | 85/292A | 32.19 | 6.57 | 583.62 | 0.0008 |
| Broiler | 15/92B | Reference group | |||
| Pullet | 12/57B | 1.815 | 0.699 | 4.675 | 0.4302 |
| Breeder | 58/143A | 7.724 | 3.666 | 17.647 | <0.0001 |
| SAMPLE TYPES | |||||
| Fly paper | 3/47D | Reference group | |||
| Boot swab | 27/47A | 48.85 | 13.52 | 241.41 | <0.0001 |
| Soil sample | 16/37AB | 20.75 | 5.65 | 102.46 | 0.0002 |
| Water sample | 23/81BC | 6.57 | 2.04 | 29.48 | 0.0348 |
| Miscellaneous | 16/80CD | 5.08 | 1.52 | 23.29 | 0.1135 |
(The table displays the odds ratio results for both stages and sample types. We observed that the odds of Campylobacter spp. detection in production farms samples were 32.19 times (6.57–583.62; 95% CLs) as likely as its detection in hatchery samples (P = 0.0008). Among the production farms, the odds of Campylobacter spp. detection in breeder farms samples were 7.724 times (3.67–17.65; 95% CLs) as likely as its detection in broiler farms samples (P < 0.0001). Similarly, the odds of Campylobacter spp. detection in boot swabs were 48.85 times (13.52–241.41; 95% CLs) as likely as its detection in fly papers (P < 0.0001). The alphabets A, B, C, and D in superscript represent statistically different groups when level of significance was set to 0.05.)
Salmonella Serovars and Campylobacter Species
The Salmonella serovars identified in this study were Typhimurium (25%), Barranquilla (25%), Liverpool (18.75%), Kentucky (12.5%), Enteritidis (6.25%), Luciana (6.25%), and Rough O: r:1,5 (6.25%) as presented in Table 2A. Interestingly, the serovars of foodborne importance: S. Enteritidis and S. Typhimurium were isolated from various locations inside and outside the hatchery. Similarly, the Campylobacter species identified were C. jejuni (11.11%) and C. coli (88.89%) as presented in Table 2B.
Phylogeny Results
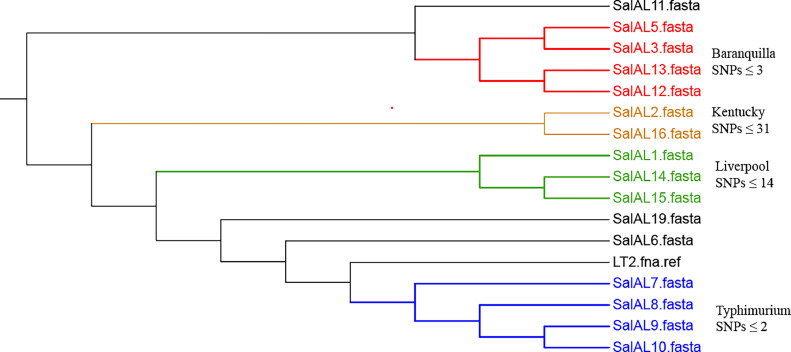
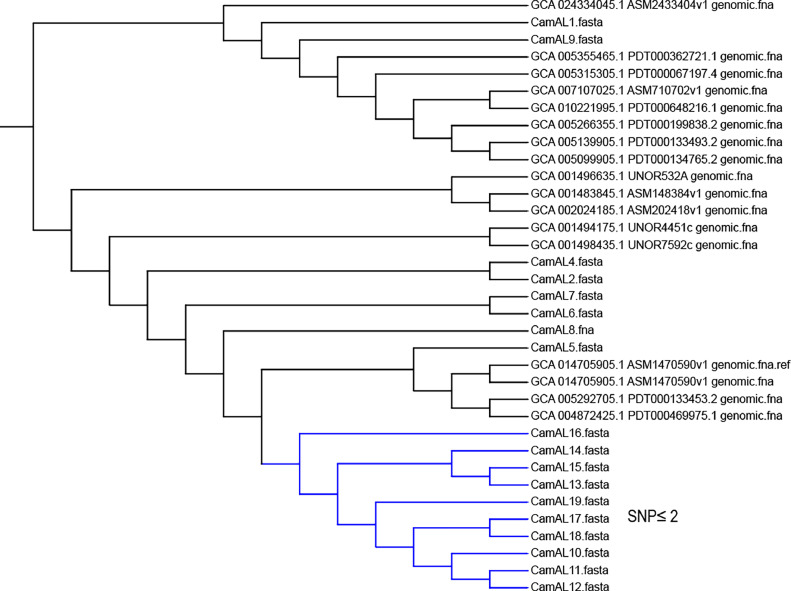
From core genome-based phylogeny of Salmonella spp. (Figure 2), we observed the clusters of S. Typhimurium, S. Barranquilla, S. Liverpool, and S. Kentucky strains isolated from this study. Furthermore, the CSI phylogeny showed S. Typhimurium strains (SalAL7, SalAL8, SalAL9 and SalAL10) recovered from hatchery surroundings with SNPs≤ 2 which suggests the possibility of transmission of same Typhimurium strain along these locations. Similarly, the S. Barranquilla strains (SalAL3, SalAL5 and (SalAL12 and SalAL13) recovered from fly papers placed outside and inside the broiler houses had SNPs≤ 3 which also suggests the possibility of transmission of same Barranquilla strain across outside and inside house environment via insects, flies, dust particles or any other mechanical vectors. Interestingly, the S. Liverpool strain (SalAL1) isolated from a water sample near the entry door of a broiler house and from fly papers outside the processing plant near unloading dock area (SalAL14 and SalAL15) had SNPs≤14 which suggest the possibility of transmission of same Liverpool strain or different strains but closely genetically related strains. Similarly, the SNPs between S. Kentucky strains (SalAL2 and SalAL16) recovered from a water sample near the entry door of a broiler house and from swabs of transport truck and cages’ floor was 31 which suggests possibility of close genetic relations between isolates, but not necessarily the same strains. Similarly, from the core genome-based phylogeny of Campylobacter spp. (Figure 3), we observed the clusters of C. jejuni and C. coli strains from this study in the phylogenetic tree. Moreover, the CSI phylogeny results showed that C. coli strains from boot swab in broiler farms (CamAL10) and carcass rinses from processing plant (CamAL11 to CamAL19) had SNPs≤2 which suggests the possibility of same C. coli strain being transmitted from broiler farms to carcass rinses in the processing plant.
Figure 2.
Core genome-based phylogeny of 16 Salmonella strains isolated from this study with reference S. enterica strain LT2. (The figure displays the different serovars’ clusters represented by different colors: Baranquilla (red), Kentucky (brown), Liverpool (green) and Typhimurium (blue). Based on Octavia et al. (2015), SNPs ≤ 9 were considered as cutoff reference to be same Salmonella strains. The strains of serovars: Barranquilla and Typhimurium had SNPs (≤ 3) which suggests the possibility of same strain transmitting along different locations within the hatchery and broiler farms. In the case of serovar Liverpool, the SNPs (≤14) suggest the possibility of same strains or closely related strains transmitted from broiler houses to processing plant while for serovar Kentucky, the SNPs (= 31) and time interval of sample collection was 4 months which suggests the possibility of transmission of strains with common origin and may had some mutations over time.)
Figure 3.
Core genome-based phylogeny of 18 Campylobacter spp. isolated from this study, and 8 C. jejuni and 9 C. coli strains downloaded from NCBI. (The figure displays single cluster with SNPs (≤ 2) highlighted with blue color which consists of C. coli strains (CamAL11 to CamAL19) recovered from carcass rinses in processing plant and strain isolated from boot swab in the broiler house (CamAL10) which suggest the possibility of transmission of same Campylobacter strain from broiler house to the processing plant.)
In case of Salmonella, the phylogenetic trees of each serovars were constructed with strains from this study and closely related NCBI strains present in specific nodes of SNP tree viewer (Supplementary Figure 1: A to G). However, based on CSI phylogeny results, almost all strains from this study had distant genetic relationships (SNPs>30) with NCBI strains except 2 S. Typhimurium strains isolated from broiler environment and stool clinical sources in Canada had SNPs≤ 19 and 2 S. Liverpool strains isolated from comminuted turkey and cattle feces in USA had SNPs≤ 25. However, none of the Campylobacter strains from this study were found to be genetically related with NCBI strains since SNPs> 500 between bacterial strains.
Multiple Entry Points and Transmission Pattern
In this study, both bacteria were recovered from multiple environmental samples and different stages of poultry food chain. It suggests multiple entry points of such bacteria along the chain. In addition, all Salmonella serovars and/ or strains that were present in the former stages were not present in the latter stages of broiler production chain and vice-versa. Similarly, all Campylobacter strains that were recovered from surroundings of pullet and breeder farms did not make up to transport and processing plant. Alternatively, the phylogeny results show distantly related C. coli strains in the surrounding of broiler farms as compared to strains from breeder farms.
DISCUSSION
The MDS as well as the culture results showed that both pathogens were present on most of the sectors of an integrated NAE broiler complex. Based on MDS results, about 40 and 80% of production farms such as pullet, breeder, and broiler farms were positive for Salmonella spp. and Campylobacter spp. respectively. In addition, both pathogens were present in hatchery, transport truck/cages and processing plant. Similar to the findings of our study, previous studies (Jones et al., 1991; Bailey et al., 2001, 2002; Heyndrickx et al., 2002; Liljebjelke et al., 2005; Volkova et al., 2010; Thakur et al., 2013; Berghaus et al., 2013; Trimble et al., 2013; Velasquez et al., 2018; Crabb et al., 2018; Cargnel et al., 2023) reported the prevalence of Salmonella spp. in production farms, hatcheries, transport and processing plant. Similarly, other studies (Thakur et al., 2013; Trimble et al., 2013; Schroeder et al., 2014; Schets et al., 2017; Sahin et al., 2024) reported the prevalence of Campylobacter spp. in production farms, and other facilities. However, the prevalence rates were reported comparatively higher than the results of this study. The reason may be because the previous studies either considered cross-sectional study design or were considered direct sampling from birds in addition to other environmental samples. However, only non-invasive environmental sampling was considered in this study. Other factors might be seasonal variation, differences in locations, biosecurity measures and farm managemental practices across the farms and farm types. Moreover, the NAE broiler complex we sampled was proactive for controlling Salmonella spp. via vaccination and approved biosecurity measures.
This study showed that the odds of Salmonella spp. detection were most likely in hatchery samples as compared to its detection in production farms which suggests significant deficiencies in standard hygiene practices and biosecurity measures in the surroundings of hatchery. In addition, Salmonella spp. can survive desiccation and can persist in dry environments and foods for several years (Estrada et al., 2023). As expected, the odds of Campylobacter spp. detection in production farms’ samples were significantly higher compared to its detection in hatchery samples. It is because the broiler birds generally become infected with bacteria with high cecal colonization level at 2 to 3 wk of age. Though only environmental samples were collected rather than samples from live birds in this study, most of the sample type that were positive for Campylobacter were boot swabs samples. In addition, the organism is thermotolerant and microaerophilic which cannot survive or persist for long periods under environmental conditions; however, birds’ ceca provide a suitable condition for this organism to multiply (Corry and Atabay, 2001). Similarly, the odds of Campylobacter spp. detection in breeder farms’ surroundings were significantly higher as compared to broiler farms’ surrounding in this study. However, similar prevalence of Campylobacter spp. was reported in both breeder and broiler flocks by Cox et al. (2002). The difference in results might be because of differences in sample types and management practices in the farms considered during the study.
In this study, Salmonella spp. was isolated from 6 fly papers, 2 water samples and each of boot swab, unknown feces, swab from transport trailer/cages’ floor, incubators, hatchers, chick trays and egg buggies. Similar to the findings of this study, higher recovery of Salmonella spp. was reported from fly papers, boot swabs and outside dirt near the entry doors by Bailey et al. (2001) during a multistate epidemiological investigation with an integrated poultry operations while Trimble et al. (2013) reported Salmonella spp. contamination in soil, compost, and wastewater from a small-scale pasture-raised broiler farms. Aerosolized Salmonella spp. can be present in the dust particles (Liljebjelke et al., 2005; Marin et al., 2011; Pal et al., 2021) and the swabbing of dust particles from poultry houses and facilities may also have a high chance of detecting pathogens. Fly paper sampling is an economical and simple method to screen houses and facilities for contamination with pathogens. Also, it suggests that the flies or dust attached to the papers might be mechanical vectors or fomites for the transmission of pathogen. Similarly, the recovery of pathogen from water puddles near entrances suggests the need of efficient foot baths at the entrance of houses to avoid the cross-contamination of pathogen during the movement of personnel among farmhouses. The recovery of pathogens in unknown feces suggests that the wild animals and birds outside the houses can also be potential sources for pathogen spread. Rodents, wild birds, insects, and snakes are also considered to play an important role in the transmission of Salmonella among birds and farms (Heyndrickx et al., 2002; Wales et al., 2007; Marin et al., 2011). The Salmonella spp. recovered from the transport truck and cages suggests that bird transport is another potential source for further pathogen spread (Heyndrickx et al., 2002).
Similarly, boot swabs, soil, water drainage, miscellaneous samples and carcass rinses were found to be useful for Campylobacter detection. In consistent to the findings of this study, previous studies (Ross and Donnison, 2003; Leatherbarrow et al., 2004; Trimble et al., 2013; Mughini-Gras et al., 2021) also reported the prevalence of C. jejuni and C. coli from cattle feces, water drainage and soil samples. Boot swabs were reported to be an effective tool for sampling most of the enteric bacteria or pathogens inside the poultry houses (McCrea et al., 2008; Marin et al., 2011; Berghaus et al., 2013). Schets et al. (2017) reported the detection of Campylobacter spp. in environmental samples like soil and surface water, not in dust or flies. The higher number of C. coli was usually recovered from carcass rinses followed by cow feces collected outside the houses. Cattle farms were found close to some of the production houses which suggests that cattle or other ruminants could be potential reservoirs for Campylobacter spp. contamination in the birds. Previous studies (Grau, 1988; Møller Nielsen et al., 1997; Stanley and Jones, 2003; Mughini-Gras et al., 2021) reported livestock such as cattle, pigs, and sheep as a reservoir for the Campylobacter spp. In contrast to the findings of most previous studies, higher percentage of C. coli was identified in this study as compared to C. jejuni. The reason might be because of high prevalence of C. coli in the broiler flock which was followed to transport and then to the processing plant. In addition, the percentage is high because almost all post pick whole carcass rinses (9/10) were positive for C. coli in the processing plant. A high prevalence of C. coli has also been reported by other researchers (Schets et al., 2017; Kemp et al., 2005). Similarly, only 4% C. coli and 10% C. jejuni were detected from 1,973 samples in Poland from children, domestic animals, poultry meat and surface water (Szczepanska et al., 2017). However, C. jejuni infection is more common in poultry and can cause foodborne illness in humans via consumption of uncooked contaminated chicken meat and meat products. Another important thing to consider in this study was that most of Campylobacter MDS positive samples were not recovered in culture. The reason for this may be because of the presence of viable but not culturable (VBNC) form, typically observed with Campylobacter (Moore, 2001; Pokhrel et al., 2022). Another possibility is the isolation procedure adopted in this study was based on USDA-FSIS MLG41.05 and utilized the MDS. These methodologies were validated for poultry rinses, sponges and raw poultry samples and not for some of the samples collected. This could be the limitation for lower recovery rate of bacteria. These results support the need to develop novel protocols or an accepted standard method for the isolation and identification of Campylobacter, particularly from environmental samples.
The phylogeny results in this study show strong genetic relatedness among bacterial strains isolated from same stages or among different stages. The very closely related or same strains of S. Typhimurium isolated from various locations of hatchery suggest hatchery’ environment as reservoir for the pathogen and can be easily transmitted from one place to another via cross-contamination (Bailey et al. 2002). However, Liljebjelke et al. (2005) reported breeders to have significant role in transmission and persistence of Salmonella spp. within an integrated broiler production system. The CSI phylogeny results of S. Barranquilla, S. Kentucky and S. Liverpool strains and the degree of their genetic relatedness within each serovar suggest strong possibility of transmission of same Salmonella strain between outdoor and indoor farm environments as well as between the surroundings of broiler farms, transport and processing plant. Similarly, the phylogeny results of Campylobacter spp. suggest possibility of transmission of same C. coli strain from broiler farm to carcass rinses in the processing plant (SNPs ≤ 2). Cox et al. (2002) reported that the broiler breeder flocks may serve as the source for Campylobacter contamination in the respective broiler flocks. However, the results reported here are similar to the finding of Prachantasena et al. (2016), in which Campylobacter strains isolated from breeder farms were not genetically related to strains recovered from broiler flocks and carcass rinses. In addition, Francesca Menna et al. (2005) reported the contamination of breeder flocks with C. jejuni and C. coli and suggested poultry breeder farms as a reservoir of Campylobacter. However, the vertical transmission of Campylobacter from parent flocks to their progenies is debatable since no strong evidence was reported to support its vertical transmission (Zhang and Sahin, 2020).
Furthermore, this study suggests multiple entry points and complex diversity of such bacteria along the different stages of broiler production chain. From the results, it was observed that all Salmonella or Campylobacter strains were not necessarily transmitted from former to latter stages along the chain. Alternatively, those strains present in the latter stages were not present in the upstream stages. Similar pattern of occurrence of Salmonella serovars were reported by Cason et al. (2024) in turkey flocks in the United States. It is important to consider that the current study might have some limitations since it was conducted in a single broiler complex. The results that was observed in this study may vary if high number of farms and facilities within a broiler complex or multiple complexes were included in the study. However, more than 10% of total production farms and facilities were included in this study to represent the single broiler complex. Another limitation can be the analysis of a single colony instead of multiple colonies isolated from the same sample during bacterial isolation procedure as mentioned in Siceloff et al. (2022). Moreover, multiple follow-up visits from broiler farms to processing plants could help to further validate the results of this study.
CONCLUSIONS
From this study, we can conclude that Salmonella spp. and Campylobacter spp. were present along various stages of an integrated NAE broiler complex which possess potential food safety risks. The odd of detecting Salmonella is more likely in the hatchery as compared to production farms suggest significant deficiencies in biosecurity measures in the hatchery. Similarly, the odds of Campylobacter detection is more likely on production farms as compared to hatchery since the organism is fastidious, microaerophilic, and thermotolerant in nature, that can better survive in the intestinal tracts, particularly in the ceca of birds. The Campylobacter spp. isolation method used in this study that was validated for samples from processing plants, was also able to detect bacteria in environmental samples. However, the efficiency of such protocol is unknown. Despite these facts, boot swabs, soil, water and miscellaneous samples could be considered as useful sample types for the detection of Campylobacter spp. during environmental sampling from farmhouses and facilities’ surroundings. The recovery of bacteria from different stages and sample types shows multiple entry points of introducing such bacteria into the complex. It suggests the need for a comprehensive control strategy that involves well-established biosecurity measures, best management practices and health programs as basic procedures in all stages of vertically integrated broiler complex. In addition, competitive exclusion with use of pre/ probiotics, acidified water, and vaccination of breeder hens could aid to minimize or eliminate these pathogens from the poultry food chain. The phylogeny results show close genetic relatedness among bacterial strains isolated from this study which suggest the possibility of transmission of same bacterial strain along different locations within the same stage and between different stages of same broiler chain. Moreover, the contaminated chicken meat and meat products with Salmonella spp., Campylobacter spp. and other zoonotic pathogens can potentially cause exposure to consumers and cause foodborne illness.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was funded by United States Department of Agriculture- Agricultural Research Service (USDA-ARS) Project Number: 6040-32000-085-002-S.
We would like to acknowledge USDA-ARS for providing the fund, Alabama Super Computer, Department of Poultry Science, Auburn University, company personnel, and contract growers for their help during the study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.104212.
Appendix. Supplementary materials
Supplementary Tables 1 and 2 consist of the list of 16 Salmonella spp. and 18 Campylobacter isolates respectively that were recovered from the NAE broiler complex along with their sources. The genome assembly statistics of assembled genomes for Salmonella spp. and Campylobacter spp. is listed in supplementary Tables 3 and 4 respectively. All assembled genomes from NCBI that are considered for building phylogenetic trees are listed with GenBank assembly number, isolation source, types, and locations in supplementary Tables 5 and 6 for Salmonella spp. and Campylobacter spp. respectively.
REFERENCES
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. Accessed Dec. 2023. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Antunes P., Mourão J., Campos J., Peixe L. Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 2016;22:110–121. doi: 10.1016/j.cmi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Bailey J.S., Cox N.A., Craven S.E., Cosby D.E. Serotype tracking of Salmonella through integrated broiler chicken operations. J. Food Prot. 2002;65:742–745. doi: 10.4315/0362-028x-65.5.742. [DOI] [PubMed] [Google Scholar]
- Bailey J.S., Stern N.J., Fedorka-Cray P., Craven S.E., Cox N.A., Cosby D.E., Ladely S., Musgrove M.T. Sources and movement of Salmonella through integrated poultry operations: a multistate epidemiological investigation. J. Food Prot. 2001;64:1690–1697. doi: 10.4315/0362-028x-64.11.1690. [DOI] [PubMed] [Google Scholar]
- Berghaus R.D., Thayer S.G., Law B.F., Mild R.M., Hofacre C.L., Singer R.S. Enumeration of Salmonella and Campylobacter spp. in environmental farm samples and processing plant carcass rinses from commercial broiler chicken flocks. Appl. Environ. Microbiol. 2013;79:4106–4114. doi: 10.1128/AEM.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, B. 2014. BBMap: a fast, accurate, splice-aware aligner. Accessed Dec. 2023. https://escholarship.org/uc/item/1h3515gn.
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnel M., Filippitzi M., Van Cauteren D., Mattheus W., Botteldoorn N., Cambier L., Welby S. Assessing evidence of a potential Salmonella transmission across the poultry food chain. Zoonoses Public Health. 2023;70:22–45. doi: 10.1111/zph.12998. [DOI] [PubMed] [Google Scholar]
- Cason E.E., Carlson A.V., Siemens A.L., Shariat N.W. High-resolution serotyping reveals Salmonella surveillance challenges in the turkey industry. J. Food Prot. 2024;100319 doi: 10.1016/j.jfp.2024.100319. [DOI] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention (CDC). 2023a. Salmonella. Accessed Dec. 2023. https://www.cdc.gov/Salmonella/faq.html.
- Center for Disease Control and Prevention (CDC). 2023b. Campylobacter (Campylobacteriosis). Accessed Dec. 2023. https://www.cdc.gov/Campylobacter/faq.html.
- Clausen P.T.L.C., Aarestrup F.M., Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018;19:307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry J., Atabay H. Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 2001;90:96S–114S. doi: 10.1046/j.1365-2672.2001.01358.x. [DOI] [PubMed] [Google Scholar]
- Cox N.A., Stern N.J., Hiett K.L., Berrang M.E. Identification of a new source of campylobacter contamination in poultry: transmission from breeder hens to broiler chickens. Avian Dis. 2002;46:535–541. doi: 10.1637/0005-2086(2002)046[0535:IOANSO]2.0.CO;2. [0535: IOANSO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Crabb H.K., Allen J.L., Devlin J.M., Firestone S.M., Wilks C.R., Gilkerson J.R. Salmonella spp. transmission in a vertically integrated poultry operation: Clustering and diversity analysis using phenotyping (serotyping, phage typing) and genotyping (MLVA) (ADS Sant'Ana, Ed.) PLOS ONE. 2018;13 doi: 10.1371/journal.pone.0201031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng S.-K., Pusparajah P., Mutalib N.-S.A.b, Ser H.-L., Chan K.-G., Lee L.-H. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015;8:284–293. [Google Scholar]
- Estrada E.M., Moyne A., Harris L.J. Characterizing the genetic diversity of Salmonella isolated from US Raw Inshell Pistachios using whole genome sequencing. J. Food Prot. 2023;86 doi: 10.1016/j.jfp.2023.100143. [DOI] [PubMed] [Google Scholar]
- Francesca Menna L., Matteoli G., Fontanella M., Cuomo A., De Paola A., Pepe T., Di Marco I., Dipineto L. Prevalence of Campylobacter jejuni in poultry breeder flocks. Ital. J. Anim. Sci. 2005;4:269–271. [Google Scholar]
- Grau F.H. Campylobacter Jejuni and Campylobacter Hyointestinalis in the intestinal tract and on the carcasses of calves and cattle. J. Food Prot. 1988;51:857–861. doi: 10.4315/0362-028X-51.11.857. [DOI] [PubMed] [Google Scholar]
- Hasman H., Saputra D., Sicheritz-Ponten T., Lund O., Svendsen C.A., Frimodt-Møller N., Aarestrup F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples (Y-W Tang, Ed.) J. Clin. Microbiol. 2014;52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Organization (WHO). 2020. Campylobacter. Accessed Dec. 2023. https://www.who.int/news-room/fact-sheets/detail/Campylobacter.
- Hess J., Macklin K., McCrea B. Litter beetle (Alphitobius diaperinus Panzer) counts in broiler houses treated with a range of insecticides. J. Appl. Anim. Res. 2008;33:127–131. [Google Scholar]
- Heyndrickx M., Vandekerchove D., Herman L., Rollier I., Grijspeerdt K., De Zutter L. Routes for salmonella contamination of poultry meat: epidemiological study from hatchery to slaughterhouse. Epidemiol. Infect. 2002;129:253–265. doi: 10.1017/s0950268802007380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina, I. 2019. MiSeq System denature and dilute libraries guide. Accessed Dec. 2023. https://support.illumina.com/content/dam/illumina-support/documents/documentation/system_documentation/miseq/miseq-denature-dilute-libraries-guide-15039740-10.pdf.
- Illumina. 2020. Illumina DNA Prep reference guide (1000000025416). Accessed Dec. 2023. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/illumina-dna-prep-reference-guide-1000000025416-09.pdf.
- Interagency Food Safety Analytics Collaboration (IFSAC). 2021. Foodborne illness source attribution estimates for 2015 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. Accessed Dec. 2023. https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2019-report-TriAgency-508.pdf.
- Interagency Food Safety Analytics Collaboration (IFSAC). 2022. Foodborne illness source attribution estimates for 2015 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. Accessed Dec. 2023. https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2020-report-TriAgency-508.pdf.
- Jones F., Axtell R., Rives D., Scheideler S., Tarver F., Walker R., Wineland m. A survey of salmonella contamination in modern broiler production. J. Food Prot. 1991;54:502–507. doi: 10.4315/0362-028X-54.7.502. [DOI] [PubMed] [Google Scholar]
- Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PloS One. 2014;9 doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp R., Leatherbarrow A., Williams N., Hart C., Clough H., Turner J., Wright E., French N. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 2005;71:1876–1882. doi: 10.1128/AEM.71.4.1876-1882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.V., Cosentino S., Lukjancenko O., Saputra D., Rasmussen S., Hasman H., Sicheritz-Pontén T., Aarestrup F.M., Ussery D.W., Lund O. Benchmarking of methods for genomic taxonomy (GA Land, Ed.) J. Clin. Microbiol. 2014;52:1529–1539. doi: 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow A.J.H., Hart C.A., Kemp R., Williams N.J., Ridley A., Sharma M., Diggle P.J., Wright E.J., Sutherst J., French N.P. Genotypic and antibiotic susceptibility characteristics of a Campylobacter coli population isolated from dairy farmland in the United Kingdom. Appl. Environ. Microbiol. 2004;70:822–830. doi: 10.1128/AEM.70.2.822-830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljebjelke K.A., Hofacre C.L., Liu T., White D.G., Ayers S., Young S., Maurer J.J. Vertical and horizontal transmission of salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2005;2:90–102. doi: 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- Marin C., Balasch S., Vega S., Lainez M. Sources of Salmonella contamination during broiler production in Eastern Spain. Prev. Vet. Med. 2011;98:39–45. doi: 10.1016/j.prevetmed.2010.09.006. [DOI] [PubMed] [Google Scholar]
- McCrea B., Macklin K., Norton R., Hess J., Bilgili S. Recovery and genetic diversity of Escherichia coli isolates from deep litter, shallow litter, and surgical shoe covers. J. Appl. Poult. Res. 2008;17:237–242. [Google Scholar]
- Medina-Santana J.L., Ortega-Paredes D., De Janon S., Burnett E., Ishida M., Sauders B., Stevens M., Vinueza-Burgos C. Investigating the dynamics of Salmonella contamination in integrated poultry companies using a whole genome sequencing approach. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2021.101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller Nielsen E., Engberg J., Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol. Med. Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- Moore J.E. Bacterial dormancy in Campylobacter: abstract theory or cause for concern? Int. J. Food Sci. Technol. 2001;36:593–600. [Google Scholar]
- Moore R.J., Scott P.C., Van T.H. Spotlight on avian pathology: Campylobacter hepaticus, the cause of spotty liver disease in layers. Avian Pathol. 2019;48:285–287. doi: 10.1080/03079457.2019.1602247. [DOI] [PubMed] [Google Scholar]
- Mughini-Gras L., Pijnacker R., Coipan C., Mulder A.C., Fernandes Veludo A., De Rijk S., Van Hoek A.H.A.M., Buij R., Muskens G., Koene M., Veldman K., Duim B., Van Der Graaf-van Bloois L., Van Der Weijden C., Kuiling S., Verbruggen A., Van Der Giessen J., Opsteegh M., Van Der Voort M., Castelijn G.A.A., Schets F.M., Blaak H., Wagenaar J.A., Zomer A.L., Franz E. Sources and transmission routes of campylobacteriosis: a combined analysis of genome and exposure data. J. Infect. 2021;82:216–226. doi: 10.1016/j.jinf.2020.09.039. [DOI] [PubMed] [Google Scholar]
- Munoz L.R., Krehling J.T., Bailey M.A., Bourassa D.V., Pacheco W.J., Chaves-Cordoba B., Escobar C., Orellana-Galindo L., Adhikari Y., Macklin K.S. The role of dietary supplementation of yeast cell walls in response to a Campylobacter jejuni inoculation in broiler chickens. Avian Dis. 2023;67:245–253. doi: 10.1637/aviandiseases-D-23-00003. [DOI] [PubMed] [Google Scholar]
- Nachamkin I., Allos B.M., Ho T. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Chicken Council (NCC). 2023. Broiler chicken industry key facts. Accessed Dec. 2023. https://www.nationalchickencouncil.org/statistic/broiler-industry-key-facts/.
- Newell D., Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 2003;69:4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obe T., Nannapaneni R., Schilling W., Zhang L., McDaniel C., Kiess A. Prevalence of Salmonella enterica on poultry processing equipment after completion of sanitization procedures. Poult. Sci. 2020;99:4539–4548. doi: 10.1016/j.psj.2020.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octavia S., Wang Q., Tanaka M.M., Kaur S., Sintchenko V., Lan R. Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J. Clin. Microbiol. 2015;53:1063–1071. doi: 10.1128/JCM.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A., Bailey M.A., Talorico A.A., Krehling J.T., Macklin K.S., Price S.B., Buhr R.J., Bourassa D.V. Impact of poultry litter Salmonella levels and moisture on transfer of Salmonella through associated in vitro generated dust. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, T. M. 2009. Pulsed-Field gel electrophoresis for molecular epidemiology of food pathogens. In: D. Caugant (eds), Molecular epidemiology of microorganisms. Methods in Molecular Biology™, vol 551. Humana Press, Totowa, NJ. 10.1007/978-1-60327-999-4_6. [DOI] [PubMed]
- Pokhrel D., Thames H.T., Zhang L., Dinh T.T.N., Schilling W., White S.B., Ramachandran R., Theradiyil Sukumaran A. Roles of aerotolerance, biofilm formation, and viable but non-culturable state in the survival of Campylobacter jejuni in poultry processing environments. Microorganisms. 2022;10:2165. doi: 10.3390/microorganisms10112165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooja S., Sudesh D., Poonam K., Joginder S.D., Suresh K.G. Loop-mediated isothermal amplification (LAMP) based detection of bacteria: a review. Afr. J. Biotechnol. 2014;13:1920–1928. [Google Scholar]
- Poultry Health Today No-antibiotics-ever production slips, but US producers remain committed to reducing antibiotic use. The Poultry Site. 2021 https://www.thepoultrysite.com/articles/no-antibiotics-ever-production-slips-but-us-producers-remain-committed-to-reducing-antibiotic-use-2 Accessed Dec. 2023. [Google Scholar]
- Prachantasena S., Charununtakorn P., Muangnoicharoen S., Hankla L., Techawal N., Chaveerach P., Tuitemwong P., Chokesajjawatee N., Williams N., Humphrey T., Luangtongkum T. Distribution and genetic profiles of campylobacter in commercial broiler production from breeder to slaughter in Thailand (P Fratamico, Ed.) PLOS ONE. 2016;11 doi: 10.1371/journal.pone.0149585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes de novo assembler. Curr. Protoc. Bioinforma. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C., Donnison A. Campylobacter and farm dairy effluent irrigation. N. Z. J. Agric. Res. 2003;46:255–262. [Google Scholar]
- Sahin O., Pang J., Pavlovic N., Tang Y., Adiguzel M.C., Wang C., Zhang Q. A longitudinal study on campylobacter in conventionally reared commercial broiler flocks in the United States: prevalence and genetic diversity. Avian Dis. 2024;67:317–325. doi: 10.1637/aviandiseases-D-23-00004. [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.-A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff R.L. Food attribution and economic cost estimates for meat- and poultry-related illnesses. J. Food Prot. 2020;83:959–967. doi: 10.4315/JFP-19-548. [DOI] [PubMed] [Google Scholar]
- Schets F.M., Jacobs-Reitsma W.F., Van Der Plaats R.Q.J., Heer L.K.-D., Van Hoek A.H.A.M., Hamidjaja R.A., De Roda Husman A.M., Blaak H. Prevalence and types of Campylobacter on poultry farms and in their direct environment. J. Water Health. 2017;15:849–862. doi: 10.2166/wh.2017.119. [DOI] [PubMed] [Google Scholar]
- Schroeder M.W., Eifert J.D., Ponder M.A., Schmale D.G. Association of Campylobacter spp. levels between chicken grow-out environmental samples and processed carcasses. Poult. Sci. 2014;93:734–741. doi: 10.3382/ps.2013-03646. [DOI] [PubMed] [Google Scholar]
- Siceloff A.T., Waltman D., Shariat N.W. Regional Salmonella differences in United States broiler production from 2016 to 2020 and the contribution of multiserovar populations to Salmonella surveillance. Appl. Environ. Microb. 2022;88:e00204–e00222. doi: 10.1128/aem.00204-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Leite D., Fernandes M., Mena C., Gibbs P.A., Teixeira P. Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarp C.P.A., Hänninen M.-L., Rautelin H.I.K. Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 2016;22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Smith J.A. Broiler production without antibiotics: United States field perspectives. Animal Feed Sci Technol. 2019;250:93–98. [Google Scholar]
- Stanley K., Jones K. Cattle and sheep farms as reservoirs of Campylobacter: campylobacter on cattle and sheep farms. J. Appl. Microbiol. 2003;94:104–113. doi: 10.1046/j.1365-2672.94.s1.12.x. [DOI] [PubMed] [Google Scholar]
- Szczepanska B., Andrzejewska M., Spica D., Klawe J.J. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017;17:80. doi: 10.1186/s12866-017-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.s. 2015. RStudio: integrated development for R. RStudio. Inc Boston MA 700:879.
- Thakur S., Brake J., Keelara S., Zou M., Susick E. Farm and environmental distribution of Campylobacter and Salmonella in broiler flocks. Res. Vet. Sci. 2013;94:33–42. doi: 10.1016/j.rvsc.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:1–15. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble L.M., Alali W.Q., Gibson K.E., Ricke S.C., Crandall P., Jaroni D., Berrang M., Habteselassie M.Y. Prevalence and concentration of Salmonella and Campylobacter in the processing environment of small-scale pastured broiler farms. Poult. Sci. 2013;92:3060–3066. doi: 10.3382/ps.2013-03114. [DOI] [PubMed] [Google Scholar]
- Turgeon P., Ng V., Murray R., Nesbitt A. Forecasting the incidence of salmonellosis in seniors in Canada: a trend analysis and the potential impact of the demographic shift (M Kirk, Ed.) PLOS ONE. 2018;13 doi: 10.1371/journal.pone.0208124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture – Food Safety and Inspection Service (USDA-FSIS). MLG4.13. 2023. Isolation and Identification of Salmonella from meat, poultry, pasteurized egg, carcass, and environmental sponges. Accessed Dec. 2023. https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-4.13.pdf
- United States Department of Agriculture – Food Safety and Inspection Service (USDA-FSIS). MLG41.05. 2021. Isolation and identification of Campylobacter jejuni/coli/lari from poultry rinse, sponge and raw product samples. Accessed Dec. 2023. https://www.fsis.usda.gov/sites/default/files/media_file/2021-06/MLG-41.pdf
- Van T.T.H., Phung C., Anwar A., Wilson T.B., Scott P.C., Moore R.J. Campylobacter bilis, the second novel Campylobacter species isolated from chickens with spotty liver disease, can cause the disease. Vet. Microbiol. 2023;276 doi: 10.1016/j.vetmic.2022.109603. [DOI] [PubMed] [Google Scholar]
- Velasquez C.G., Macklin K.S., Kumar S., Bailey M., Ebner P.E., Oliver H.F., Martin-Gonzalez F.S., Singh M. Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in southeastern United States. Poult. Sci. 2018;97:2144–2152. doi: 10.3382/ps/pex449. [DOI] [PubMed] [Google Scholar]
- Volkova V.V., Bailey R.H., Rybolt M.L., Dazo-Galarneau K., Hubbard S.A., Magee D., Byrd J.A., Wills R.W. Inter-relationships of Salmonella status of flock and grow-out environment at sequential segments in broiler production and processing. Zoonoses Public Health. 2010;57:463–475. doi: 10.1111/j.1863-2378.2009.01263.x. [DOI] [PubMed] [Google Scholar]
- Wales A., Breslin M., Carter B., Sayers R., Davies R. A longitudinal study of environmental salmonella contamination in caged and free-range layer flocks. Avian Pathol. 2007;36:187–197. doi: 10.1080/03079450701338755. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., and O. Sahin. 2020. Campylobacteriosis. In Diseases of Poultry (eds D.E. Swayne, M. Boulianne, C.M. Logue, L.R. McDougald, V. Nair, D.L. Suarez, S. Wit, T. Grimes, D. Johnson, M. Kromm, T.Y. Prajitno, I. Rubinoff and G. Zavala), 754–769. 10.1002/9781119371199.ch17. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1 and 2 consist of the list of 16 Salmonella spp. and 18 Campylobacter isolates respectively that were recovered from the NAE broiler complex along with their sources. The genome assembly statistics of assembled genomes for Salmonella spp. and Campylobacter spp. is listed in supplementary Tables 3 and 4 respectively. All assembled genomes from NCBI that are considered for building phylogenetic trees are listed with GenBank assembly number, isolation source, types, and locations in supplementary Tables 5 and 6 for Salmonella spp. and Campylobacter spp. respectively.