Abstract
N-terminal modifications of the chemokine RANTES bind to C-C chemokine receptor 5 (CCR5) and block human immunodeficiency virus type 1 (HIV-1) infection with greater efficacy than native RANTES. Modified RANTES compounds induce rapid CCR5 internalization and much slower receptor reexpression than native RANTES, suggesting that receptor sequestration is one mode of anti-HIV activity. The rates of CCR5 internalization and reexpression were compared using the potent n-nonanoyl (NNY)-RANTES derivative and CD4+ T cells derived from donors with different CCR5 gene polymorphisms. NNY-RANTES caused even more rapid receptor internalization and slower reexpression than aminooxypentane (AOP)-RANTES. Polymorphisms in the promoter and coding regions of CCR5 significantly affected the receptor reexpression rate after exposure of cells to NNY-RANTES. These observations may be relevant for understanding the protective effects of different CCR5 genotypes against HIV-1 disease progression.
C-C chemokine receptor 5 (CCR5) binds the chemokines MIP-1α, MIP-1β, RANTES, MCP-2, MCP-3, MCP-4, MCP-1, and eotaxin (5, 53) and also serves as the primary coreceptor for human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus entry into target cells (1, 3, 4, 10, 12, 16, 19). Although multiple other coreceptors are permissive for fusion of virus and transfected target cells (11, 14, 21–23, 26, 30, 32, 44, 48, 49, 51, 52, 54, 56, 57), only CCR5 and CXCR4 seem to be important for virus infection in human lymphoid cells (9, 38, 59, 63, 67). RANTES is a weak inhibitor of HIV-1 infection of T cells (13), but N-terminal modifications of RANTES such as aminooxypentane (AOP)- or n-nonanoyl (NNY)-RANTES are more potent inhibitors of both T-cell and macrophage infection (39, 50, 58). This inhibition of virus infection could be explained by occupancy of CCR5 and blocking of interaction with the CD4-gp120 complex, but it is more likely to be due to receptor sequestration following internalization and diminished CCR5 recycling or transport to the cell surface (33, 43, 64). In this study, we compared the rates of CCR5 internalization and reexpression triggered by exposure to AOP- or NNY-RANTES in primary CD4+ T lymphocytes isolated from donors who differed in CCR5 genotype. NNY-RANTES induced more rapid internalization of CCR5 and slower reexpression than AOP-RANTES, a finding that correlates with the more potent inhibition of virus infection by NNY-RANTES (39). Two unlinked polymorphisms, V64I in CCR2 (27, 29) and Δ32 in exon 4 of CCR5 (25), were found to affect the rate of CCR5 receptor reexpression. The rate of receptor reexpression was largely independent of the baseline surface expression of CCR5, and the internalization rate was not correlated with the reexpression rate. The impact of polymorphisms at the CCR5 locus on receptor reexpression after prolonged internalization and degradation may be an independent predictor of disease progression following HIV-1 infection.
MATERIALS AND METHODS
Donor genotyping.
Anonymous DNA samples from adult donors participating in the volunteer blood donor pool of The Scripps Research Institute General Clinical Research Center were amplified with PCR primers flanking CCR5 promoter polymorphisms or the 32-bp deletion in exon 4. The V64I allelic variant in CCR2 was detected by restriction fragment length polymorphism by a modification of the procedure described previously (37). The 3′ primer for amplification of the polymorphic region of CCR2 was replaced with the antisense primer 5′ ATGAGAGGGTAACACCCGAG 3′. The 129-bp product was subsequently digested as described elsewhere (37). Single nucleotide polymorphisms were detected by automated sequencing of the PCR products (5 to 10 clones per donor). A subset of donors were typed for the G/A polymorphism at position −2459 (59029) by David McDermott at the National Institutes of Health as previously described (36).
Cell isolation and separation.
Peripheral blood mononuclear cells were separated from whole blood from coded, genotyped donors by Ficoll-Hypaque density sedimentation. Purified CD4+ T cells were separated from other mononuclear cells by depletion of CD8+, CD14+, CD16+, and CD19+ cells by antibody treatment and magnetic bead separation. The purified CD4+ T cells were activated by 3 days of exposure to phytohemagglutinin (PHA-P; 2 μg/ml; Boehringer Mannheim) followed by the addition of 20 U of recombinant human interleukin-2. Activated CD4+ T cells were used for CCR5 modulation experiments after 10 to 14 days of activation, a time chosen to maximize baseline CCR5 expression (6, 66).
Analysis of CCR5 expression.
Surface-exposed CCR5 was identified with the anti-CCR5 antibody PA12 (42), kindly provided by Progenics Pharmaceuticals, Inc. (Tarrytown, N.Y.). Staining of CCR5 by PA12 is not affected by binding of RANTES (42). Preliminary experiments confirmed that PA12 staining was not inhibited by prior incubation of CD4+ T cells with NNY-RANTES at 4°C, but that staining with the anti-CCR5 antibody 2D7 (66) was inhibited. All staining of cells was performed at 4°C in the presence of 0.02% sodium azide to prevent further internalization of CCR5. Binding of PA12 antibody was detected with phycoerythrin-conjugated donkey anti-mouse immunoglobulin G (Jackson ImmunoResearch). The number of stained cells and the intensity of staining were evaluated by flow cytometry using a FACScalibur instrument and CellQuest software (Becton Dickinson Immunocytometry Systems, Mountain View, Calif.). The collected data (usually 10,000 events) were gated by forward and right angle light scatter to distinguish small cells from lymphoblasts. Under the culture conditions used, 85 to 90% of the CD4+ T cells were small and 5 to 10% were lymphoblasts. Unless otherwise indicated, results are presented for the predominant small cell population.
CCR5 recycling rates.
The rate of CCR5 internalization following addition of ligands was calculated from plots of an index of CCR5 expression versus time (minutes) after ligand addition (see the legend to Fig. 1). The CCR5 expression index was calculated by multiplying the geometric mean channel number of CCR5 staining by the percentage of positive cells and dividing the resulting product by 100. The decline in CCR5 expression after ligand binding was exponential, and a curve fitting program was used to calculate the power exponent of the slope. The same program was used to calculate the exponential slope of CCR5 reexpression after removal of ligands. Replicate experiments (two to three) showed that the rates of CCR5 internalization and reexpression were highly consistent for cells from a single donor.
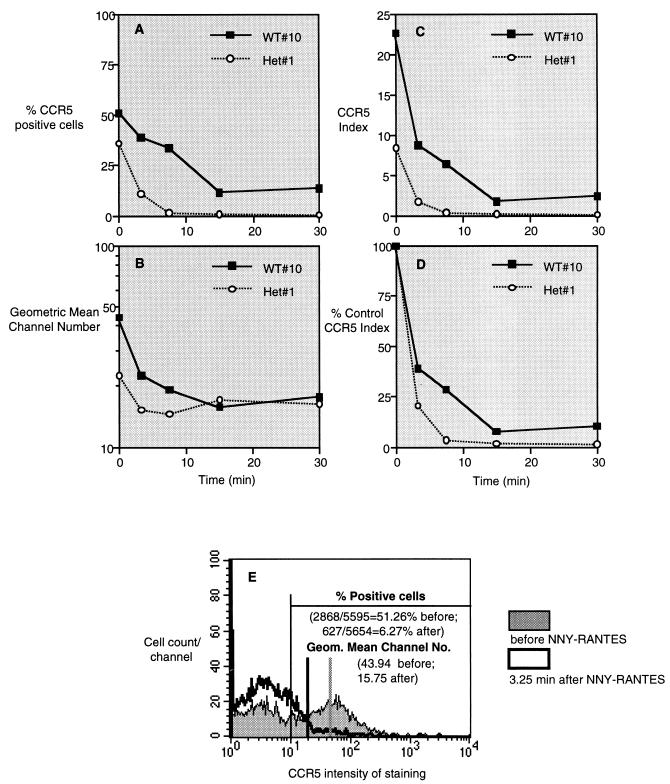
FIG. 1.
Down regulation of CCR5 expression by exposure to NNY-RANTES. NNY-RANTES was added to purified human CD4+ lymphocytes from a Δ32 wild-type (WT; +/+) or heterozygous (Het; Δ32/+) donor at time zero, and CCR5 expression was determined by staining with the anti-CCR5 antibody PA12 at the indicated time points. Data are expressed as the percentage of CCR5-positive cells (A), the geometric mean staining intensity (B), the CCR5 index (percent positive cells × mean intensity/100) (C), and the percent control CCR5 index (D). (E) Representative flow cytometry data used for these calculations. Cells were classified as CCR5 positive if the intensity of staining was greater than channel 10, a cutoff established by staining with an isotype control antibody. These results thus reflect virtually complete internalization of CCR5 and do not reflect minor decreases in CCR5 intensity. Antibody PA12 recognizes the N-terminal extracellular domain of CCR5, and its binding site is not blocked by chemokine binding (42).
AOP- and NNY-RANTES.
N-terminal modifications of RANTES were prepared by total chemical synthesis as described previously (39, 65). The purity of each batch of compounds was tested by mass spectrometry. Their ability to block HIV-1 infection was also confirmed prior to the CCR5 modulation experiments.
Cell fusion assays.
CCR5-tropic viral envelope-mediated cell fusion assays were carried out essentially as described elsewhere (18), using the cell lines HeLa-P5L and HeLa-Env-ADA (58), both of which were provided by the laboratory of M. Alizon (Paris, France). HeLa-P5L cells were seeded in 96-well plates (104 cells per well in 100 μl). Twenty-four hours later, medium was removed and medium containing 104 HeLa-Env-ADA cells per well plus chemokines was added to a final volume of 200 μl. After a further 24 h, cells were washed once in phosphate-buffered saline and lysed in 50 μl of phosphate-buffered saline–0.5% NP-40 for 15 min at room temperature. Lysates were assayed for for β-galactosidase activity by the addition of 50 μl 2× CPRG (chlorophenol red-β-d-galactopyranoside) substrate (16 mM CPRG 120 mM Na2HPO4, 80 mM NaH2PO4, 20 mM KCl, 20 mM MgSO4, 10 mM β-mercaptoethanol) followed by incubation for 1 to 2 h in the dark at room temperature. The A575 was then read on a Labsystems microplate reader. The reaction was stopped when the A575 for the positive control wells (no chemokine) reached 0.5 to 1, and results are expressed as 100 × (mean absorbance [treated] − mean absorbance [no envelope cells])/(mean absorbance [no chemokine] − mean absorbance [no envelope cells]). Experiments were performed in triplicate, and dose-inhibition curves were fitted using Prism software (GraphPad).
RESULTS
CCR5 internalization with different RANTES analogues.
Native RANTES, AOP-RANTES, or NNY-RANTES was added to purified human CD4+ T lymphocytes previously activated for 10 to 14 days to optimize CCR5 baseline expression (66). Surface expression of CCR5 was determined by staining with anti-CCR5 antibody PA12 (42) followed by flow cytometric analysis at 3.25, 7.5, 15, and 30 min after addition of each ligand at 100 ng/ml (∼12.7 nM). The methods for analyzing CCR5 expression are illustrated in Fig. 1, and the results of a representative experiment are shown in Fig. 2A. Addition of NNY-RANTES to CD4+ T cells resulted in a decrease in both the percentage of CCR5+ cells (Fig. 1A and E) and the mean intensity of CCR5 staining (Fig. 1B and E). The genotype of the CD4+ T-cell donor (in this case, CCR5 Δ32/+ versus +/+) influenced both of these parameters. The total expression of surface-exposed CCR5 was calculated as the CCR5 index (see Materials and Methods), as illustrated in Fig. 1C. The resulting data were then normalized by expressing the CCR5 index as a percentage of the time zero value prior to addition of CCR5 ligands (Fig. 1D). Subsequent figures use the data presentation format shown in Fig. 1D to facilitate comparison between different ligands and cell donors.
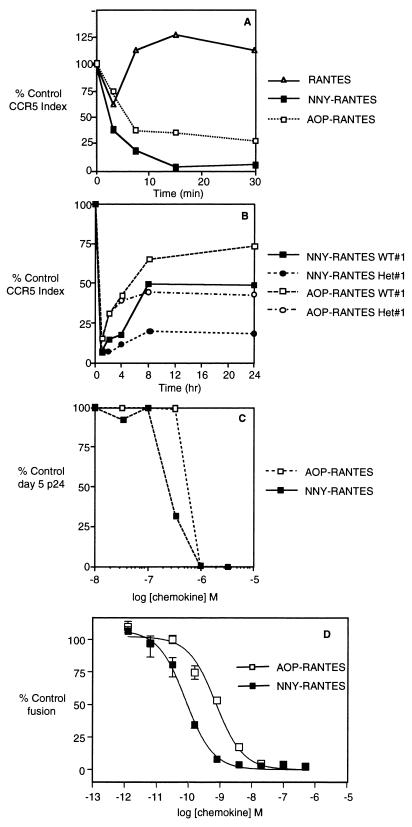
FIG. 2.
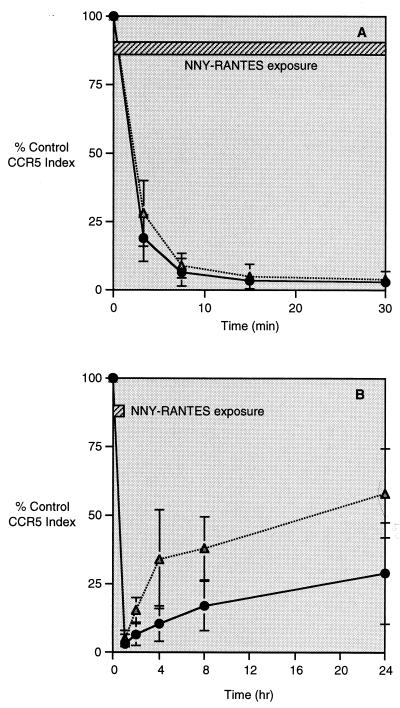
Modulation of CCR5 following binding of AOP-RANTES, NNY-RANTES, or native RANTES. Data are presented as in Fig. 1D. (A) CCR5 internalization following exposure of CD4+ T cells to native RANTES, NNY-RANTES, or AOP-RANTES. (B) CCR5 internalization and reexpression following 1 h of exposure to either AOP- or NNY-RANTES. WT or Het refers to the Δ32 genotype of the cell donor as defined in the legend to Fig. 1. (C) Inhibition of SF162 R5 HIV-1 infection by addition of AOP- or NNY-RANTES to cultures of purified human CD4+ T lymphocytes. (D) Inhibition of R5-tropic viral envelope-mediated fusion between HeLa-P5L cells and HeLa-Env-ADA cells. Fusion was measured in the presence of increasing concentrations of AOP-RANTES and NNY-RANTES (mean, maxima, and minima), as described in Material and Methods. In this representative experiment, the calculated IC50s are 730 pM for AOP-RANTES and 82 pM for NNY-RANTES. We have noted that the sensitivity of the assay, and hence the absolute IC50 for a given inhibitor, is subject to systematic variation, probably according to the passage number of both the envelope cell line (HeLa-Env-ADA) and the target cell line (HeLa-P5L). Over a series of tens of experiments, for example, the IC50s for AOP-RANTES lay in a range between 0.3 and 3 nM, and those of NNY-RANTES were between 40 and 300 pM. In a given experiment, the activity of one inhibitor relative to another varies to a much lesser extent, however, with NNY-RANTES consistently more active than AOP-RANTES by approximately an order of magnitude.
As shown in Fig. 2A, NNY-RANTES addition resulted in more rapid internalization of CCR5 than exposure to AOP-RANTES. Addition of native RANTES to CD4+ T cells resulted in only a transient and minor decrease in CCR5 intensity (Fig. 2A), followed by an apparent increase in CCR5 expression to values higher than baseline. In further analysis of these data, CD4+ lymphocytes were segregated into small cells and lymphoblasts were segregated on the basis of forward and side light scatter. Baseline CCR5 expression was higher on lymphoblasts, which accounted for ∼5% of total cells, and the extent of CCR5 internalization was less than on smaller cells (data not shown). Nonetheless, the initial rate of CCR5 internalization was similar or higher for lymphoblasts than for small cells. These results indicate that the rate and extent of CCR5 internalization depends on both the ligand and the state of cell activation. Subsequent experiments excluded lymphoblasts from analysis of CCR5 expression, since they represented a small proportion of the cultured CD4+ T lymphocytes and their inclusion would increase the heterogeneity of CCR5 modulation following ligand binding.
NNY-RANTES exposure causes prolonged receptor internalization.
Exposure of lymphocytes to AOP-RANTES causes prolonged internalization of CCR5 (33). The ability of NNY-RANTES to inhibit CCR5 receptor recycling was compared to that of AOP-RANTES. Purified CD4+ T lymphocytes were exposed to 100 ng of AOP- or NNY-RANTES per ml for 1 h to allow receptor internalization, and reexpression of CCR5 was measured as shown in Fig. 1 at 2, 4, 8, and 24 h. Representative results are plotted in Fig. 2B. Exposure to NNY-RANTES resulted in more prolonged internalization of CCR5 than exposure to AOP-RANTES, and this effect was more pronounced when the CD4+ T cells were derived from a donor of the Δ32/+ genotype than when they were derived from a +/+ genotype. NNY-RANTES is thus more potent than AOP-RANTES in accelerating CCR5 internalization and preventing CCR5 reexpression.
NNY-RANTES is more potent than AOP-RANTES in virus inhibition assays.
To confirm a correlation between CCR5 internalization and potency of N-terminal modifications of RANTES in virus inhibition assays, AOP- or NNY-RANTES was added to purified CD4+ T cells 5 days after activation. The cells were infected with the R5 HIV-1 isolate SF162, which is highly susceptible to inhibition by AOP-RANTES (58). NNY-RANTES was more potent at inhibiting virus infection than AOP-RANTES (Fig. 2C) in this and subsequent experiments. The ability of AOP- and NNY-RANTES to inhibit envelope-mediated cell fusion was also assessed. As shown in Fig. 2D, NNY-RANTES was about 1 log more potent (calculated 50% inhibitory concentrations [IC50s] are 730 pM for AOP-RANTES and 82 pM for NNY-RANTES) than AOP-RANTES in inhibiting cell fusion. NNY-RANTES is thus more potent than AOP-RANTES in modulating CCR5 expression and in inhibiting virus interaction with target cells.
Impact of CCR5 genotype on receptor internalization and reexpression.
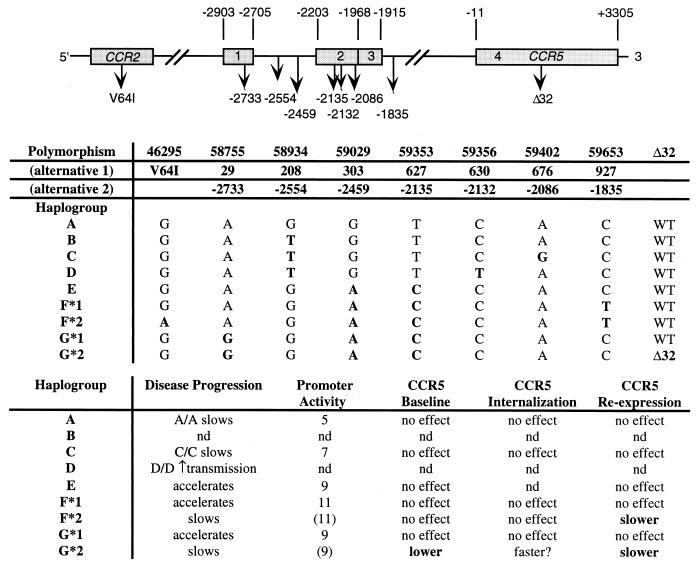
A number of polymorphisms in the 5′ untranslated region of the CCR5 locus and a coding polymorphism in the closely linked CCR2 gene have been described elsewhere (7, 28, 29, 35, 36, 40). The provisional nomenclature (alternative 2 7) for the most important of these polymorphisms as well as previous designations are summarized in Fig. 3. The affects of these polymorphisms and the 32-bp pair deletion in the CCR5 coding region (25) on disease progression or transmission are also summarized in Fig. 3. We have used the designation of CCR5 alleles based on their evolutionary history (40, 41) rather than the order of discovery (7) to simplify the data presentation and allow some grouping of related alleles, although the significance of the grouping may be open to question.
FIG. 3.
Definition of CCR5 genotypes and linkage to disease progression. Promoter activity represents relative enhancement of luciferase expression (data from Mummidi et al. 41). WT, wild type; nd, not determined.
In preliminary experiments, human donors were segregated based on the G/A polymorphism at position −2459 (59029). As reported by McDermott et al. (36), typing categorized approximately half of the donor pool as G/A heterozygotes, one quarter as G/G homozygotes (haplogroups A to D) and one quarter as A/A homozygotes (haplogroups E to G) (Fig. 3). Donors were also typed for the Δ32 mutation as previously described (47). We examined the internalization rate of CCR5 on purified CD4+ T cells from donors of each genotype following exposure to 100 ng of AOP-RANTES per ml. CCR5 internalization was most rapid with cells from A/A, Δ32/+ donors and slowest with cells from G/G, +/+ donors (data not shown). These initial findings suggested a linkage between donor genotype and CCR5 receptor modulation by N-terminal modifications of RANTES and prompted a more detailed investigation of this relationship.
A more extensive genotyping involving most of the polymorphisms shown in Fig. 3 was performed on a subset of the donor pool. In particular, all Δ32/+ donors were genotyped since the impact of one wild-type allele in the cis regulatory region could be measured with the other allele held constant (haplogroup G*2 [Fig. 3]). Since the protein product of the Δ32 locus is not expressed at the cell surface (31), surface expression of CCR5 on CD4+ T cells from Δ32/+ donors is derived solely from the other CCR5 allele (i.e., they are functionally homozygous).
CCR5 internalization and reexpression rate.
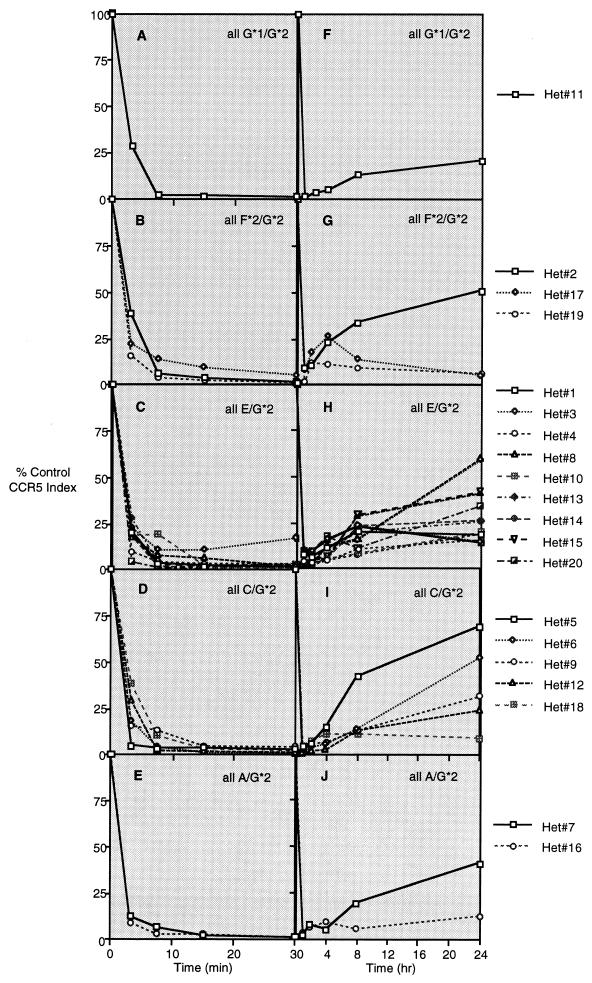
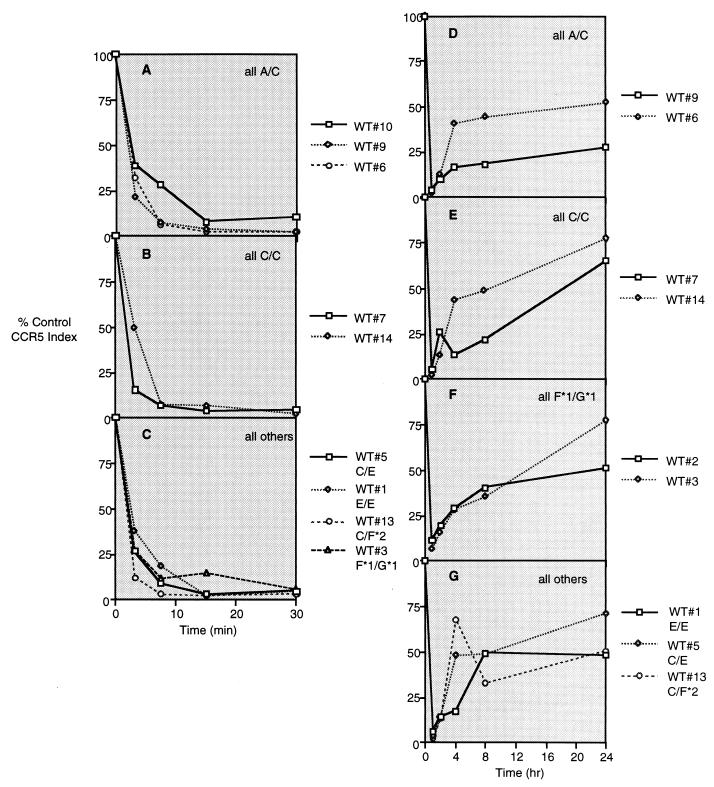
The internalization of CCR5 following receptor ligation and the rate of CCR5 reexpression after removal of ligands was determined for CD4+ T cells derived from 20 donors with the Δ32 genotype (G*2 [Fig. 3]) at one allele (Fig. 4) and 9 donors without the Δ32 mutation (Fig. 5). Fig. 4A to E presents histograms showing the CCR5 staining index on CD4+ T lymphocytes during 30 min of exposure to 100 ng of NNY-RANTES per ml, and Fig. 4F to J presents data for CCR5 reexpression at 1 (just after removal of ligand), 2, 4, 8, and 24 h. Results are grouped by genotype at the expressed CCR5 allele. The intensity of CCR5 expression and the number of positive cells are reduced by NNY-RANTES exposure, and both slowly recover over the next 24 h. There was no consistent pattern of CCR5 internalization or reexpression associated with a given haplotype, although two of three donors with the F*2/G*2 (V64I) mutation showed very limited reexpression of CCR5 (Fig. 4G). Individuals without the Δ32 mutation express the CCR5 protein product of both alleles. CCR5 internalization was similar in CD4+ T cells from these wild-type donors regardless of CCR5 haplotype. CCR5 reexpression was more rapid than in cells from Δ32/+ donors, and cells from one donor with the F*2 (V64I) mutation had an unusual pattern of CCR5 reexpression (Fig. 5G). The mean rates of CCR5 internalization and reexpression for cells from all 20 Δ32/+ donors and 9 +/+ donors are shown in Fig. 6. CCR5 internalization following NNY-RANTES binding is marginally faster in cells from Δ32/+ donors, but CCR5 reexpression is substantially and significantly (P = 0.004, two-tailed t test) slower in Δ32/+ cells (Fig. 6B).
FIG. 4.
Internalization and reexpression of CCR5 after incubation with NNY-RANTES followed by its removal. Twenty different Δ32/+ (Het) donors were examined, since cell surface expression of CCR5 protein is restricted to the product of the unmutated allele. The CCR5 haplotype (using the nomenclature presented in Fig. 3) of each donor is given in the corresponding panel. (A to E) Data for CCR5 internalization; (F to J) data for CCR5 reexpression after removal of NNY-RANTES.
FIG. 5.
Internalization and reexpression of CCR5 after exposure to NNY-RANTES. Data are plotted as in Fig. 4 except that the nine donors are wild type (WT; +/+). CCR5 genotypes are indicated in the panels and keys to symbols.
FIG. 6.
Mean rates of CCR5 internalization (A) or reexpression (B) for CD4+ T lymphocytes from all Δ32/+ (circles) or +/+ (triangles) donors. Error bars indicate the standard deviation from the mean.
Calculation of internalization and reexpression rates.
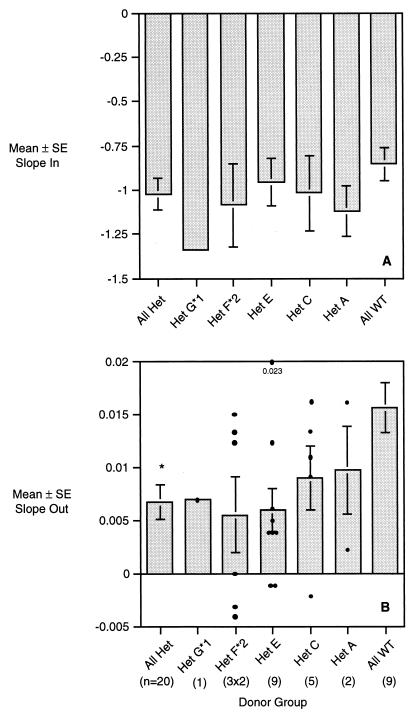
The rates of CCR5 internalization and reexpression were both exponential functions, with early rapid phases and later slower phases. The exponential function describes the rate of internalization or the rate of reexpression. The many observation points for each donor illustrated in Fig. 4 and 5 were reduced to two numbers, the internalization rate (a negative slope) and a reexpression rate (a positive slope for all but five donors). The mean (± standard error) internalization slope of CCR5 following ligation with NNY-RANTES for CD4+ T cells from 9 +/+ donors and 20 Δ32/+ donors grouped by CCR5 haplotype is shown in Fig. 7A, and the mean reexpression slope for the same groups is shown in Fig. 7B. The mean CCR5 reexpression rate is more than twice as fast for cells from +/+ donors (Fig. 7B), also a highly significant difference (P = 0.005). Five Δ32/+ donors had negative slopes of CCR5 reexpression; i.e., the CCR5 index continued to decline even after removal of NNY-RANTES. Two of these donors shared the F*2(V64I)/G*2 haplogroup. The mean CCR5 reexpression rate was slightly higher for Δ32/+ cells from CCR5 haplogroups A and C (Fig. 7B), but this difference was not statistically significant.
FIG. 7.
Mean CCR5 internalization and reexpression rates for donor groups segregated by CCR5 genotype (Het, Δ32/+; WT, wild type). (A) The internalization slope (slope in) is the power function of the exponential curve best fitting the data shown in Fig. 4 and 5. The larger the negative value, the faster the rate of CCR5 internalization. (B) CCR5 reexpression rates for the same donor groups. The slope is the exponential function of the curve best fitting the data shown in Fig. 4 and 5 for time points from 1 to 24 h. The larger the positive value of the slope, the faster the rate of CCR5 reexpression. The number of donors in each group is shown in parentheses. The data for CCR5 reexpression for F*2 heterozygotes represent duplicate assays on the three donors.
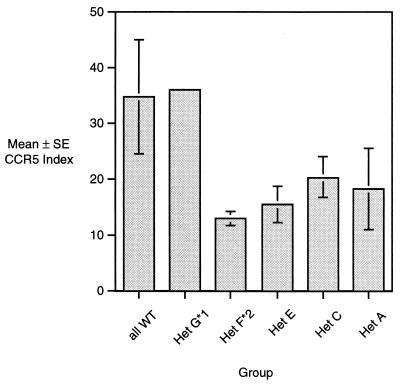
Baseline CCR5 expression might affect rates of receptor internalization or reexpression. Baseline expression (prior to addition of NNY-RANTES) of CCR5 is shown in Fig. 8. Baseline expression was lower in T cells from Δ32/+ donors, as previously observed (46). The lowest expression was seen in cells from donors who had the F*2/G*2 CCR5 haplotype. We analyzed the correlation between baseline CCR5 intensity versus internalization or reexpression rates for all 20 Δ32/+ donors. Both rates showed a weak correlation with baseline CCR5 expression (higher baseline CCR5 tended to accelerate both internalization and reexpression), but neither correlation was statistically significant. Baseline CCR5 expression thus appears to be a poor predictor of either internalization or reexpression rates.
FIG. 8.
Baseline CCR5 expression in the same donor groups shown in Fig. 7. CCR5 expression is calculated as given in the legend to Fig. 1C.
DISCUSSION
These experiments demonstrate that modulation of CCR5 expression following binding of modified RANTES ligands is dependent on both the ligand and the CCR5 genotype of the lymphocyte donor. The NNY-RANTES modification induced more rapid internalization of CCR5 following its addition and slower reexpression of CCR5 following its removal than AOP-RANTES. The observed kinetics of CCR5 modulation correlate with the antiviral activity of NNY-RANTES versus AOP-RANTES (reference 39 and Fig. 2), which suggests but does not prove that coreceptor sequestration is responsible for preventing HIV-1 infection. The mechanisms by which AOP- and NNY-RANTES slow receptor reexpression remain to be determined, although slowing the normal process of receptor recycling and/or enhancing receptor degradation are obvious possibilities. Native RANTES causes extremely rapid CCR5 internalization, and reexpression occurs even in the presence of RANTES (Fig. 2), which may explain why RANTES is not very effective at inhibiting HIV-1 infection (13, 58). Prolonged receptor internalization following binding of modified RANTES compounds may lead to degradation and enhance the role of new protein synthesis in receptor reexpression.
The second finding presented here is that the CCR5 genotype of lymphocyte donors affected the rate of CCR5 reexpression after binding and removal of NNY-RANTES. CD4+ T cells from donors who were heterozygous for the Δ32 deletion in exon 4 of CCR5 showed slower reexpression of CCR5 than cells from donors with other genotypes. As previously reported (66), cells from Δ32/+ donors had lower baseline expression of CCR5 than cells from +/+ donors (Fig. 8). However, baseline CCR5 expression was a poor predictor of either CCR5 internalization or reexpression rates.
A second polymorphism, V64I in CCR5 (haplogroup F*2 [Fig. 3]), may affect CCR5 reexpression after ligand-induced internalization. Figure 4 shows slower CCR5 reexpression in CD4+ T cells from two of three donors who shared the F*2/G*2 genotype. A third donor with this genotype did not show slower CCR5 re-expression, however; thus, more observations are necessary to confirm these preliminary results. As noted above, cells from these donors express only the protein product of the V64I allele on their surface, and so any biological deficits may be attributed to this protein or its turnover rate. However, the nonsecreted, truncated protein product of the Δ32 allele may have a dominant negative effect by dimerization with full-length CCR5 protein (2), and this effect might accentuate minor defects associated with the V64I F*2 allele. In infected patients, the protective effect of the V64I mutation is independent of the Δ32 allele (60).
We initially observed delayed CCR5 internalization in donors who were G/G at position −2459 (59029, haplogroups A to D [Fig. 3]), but examination of additional donors did not confirm a dominant effect of this allele. A larger sample of donors will be necessary to confirm any impact of the −2459 polymorphism on CCR5 receptor reexpression.
The observations presented here were made with nonnative ligands that cause rapid internalization and very slow reexpression of CCR5. CCR5 reexpression is much faster with native RANTES (Fig. 2), and it would be difficult to calculate rates of CCR5 receptor recycling if it were used as the CCR5 ligand. Slowing receptor recycling with NNY-RANTES may exaggerate subtle differences in receptor recycling linked to genetic polymorphisms either by extending the time frame of recycling or by altering the underlying mechanisms. CCR5 is internalized after association with arrestins and dynamin followed by endocytosis via clathrin-coated pits (64). Reexpression may be due to recycling from late endosomal compartments, transport from presynthesized protein stores, or new protein synthesis. Preliminary experiments indicate that receptor recycling or transport is more important than protein synthesis (C. Pastore et al., unpublished data).
Polymorphisms at the CCR5 locus have been noted to affect the rate of disease progression or transmission (7, 8, 15, 20, 27, 28, 31, 35–37, 40, 55, 60, 62). These effects are summarized in Fig. 3, as are the results obtained in this study for CCR5 internalization and reexpression. Rare polymorphisms in the coding region of CCR5 affect both chemokine responses and HIV-1 infection (7, 24), but because of their rarity and the absence of homozygotes, no effect on disease progression can be evaluated. Several reports suggest a correlation between higher CCR5 expression (in terms of both positive cells and intensity of CCR5 staining) and more rapid disease progression (17, 45). These effects can be most easily understood as increasing the available target cell pool for the predominant R5 virus population and allowing more efficient spreading of virus infection. CCR5 polymorphisms that increase total expression should increase disease progression, and those that decrease CCR5 expression should be protective. It is clear that the Δ32 mutation decreases surface expression of CCR5, an observation that is consistent with this explanation. However, there is no evidence that the protective CCR2 V64I (F*2) or 59029 G/G (A to D) alleles decrease CCR5 expression (34, 36), and the mechanism of their protective effect is unclear.
The results presented here suggest a modification of the target cell hypothesis that adds a dynamic function. The available target cell pool for R5 virus spread may be a product of the number of CCR5 positive cells times the relative intensity of CCR5 expression times the duration of CCR5 surface expression. CCR5 receptor internalization could be triggered by any of the native chemokine ligands or by contact with CD4-bound or activated-gp120 on adjacent infected cells (61). Polymorphisms in the cis-regulatory region of CCR5 could affect protein synthesis rates, which in turn could affect rates of CCR5 recovery. However, steady-state levels of CCR5 expression may be largely independent of CCR5 reexpression rates. We therefore propose that CCR5 receptor reexpression rate may be an independent variable that affects the rate of HIV-1 disease progression.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI43645 and M01-RR00833 (TSRI GCRC) and by the Swiss National Science Foundation AIDS Commission (project 3339-62032-00). C. Pastore was supported by a fellowship from the Italian Istituto Superiore di Sanita.
We thank David McDermott for genotyping donors, and we thank Michael Neal and Gabriel Kuenzi for technical assistance.
Footnotes
Publication 13345-IMM from The Scripps Research Institute.
REFERENCES
- 1.Alkhatib G, Locati M, Kennedy P E, Murphy P M, Berger E A. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 2.Benkirane M, Jin D Y, Chun R F, Koup R A, Jeang K T. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5-Δ32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 3.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanpain C, Migeotte I, Lee B, Vakili J, Doranz B J, Govaerts C, Vassart G, Doms R W, Parmentier M. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
- 6.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CXCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington M, Dean M, Martin M P, O'Brien S J. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8:1939–1945. doi: 10.1093/hmg/8.10.1939. [DOI] [PubMed] [Google Scholar]
- 8.Carrington M, Kissner T, Gerrard B, Ivanov S, O'Brien S J, Dean M. Novel alleles of the chemokine-receptor gene CCR5. Am J Hum Genet. 1997;61:1261–1267. doi: 10.1086/301645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Gettie A, Ho D D, Marx P A. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology. 1998;246:113–124. doi: 10.1006/viro.1998.9174. [DOI] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Combadiere C, Salzwedel K, Smith E D, Tiffany H L, Berger E A, Murphy P M. Identification of CX3CR1. A chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J Biol Chem. 1998;273:23799–23804. doi: 10.1074/jbc.273.37.23799. [DOI] [PubMed] [Google Scholar]
- 15.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.de Roda Husman A M, Blaak H, Brouwer M, Schuitemaker H. CC chemokine receptor 5 cell-surface expression in relation to CC chemokine receptor 5 genotype and the clinical course of HIV-1 infection. J Immunol. 1999;163:4597–4603. [PubMed] [Google Scholar]
- 18.Dragic T, Hazan U, Alizon M. Methods in molecular genetics. San Diego, Calif: Academic Press, Inc.; 1995. pp. 218–236. [Google Scholar]
- 19.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 20.Easterbrook P J, Rostron T, Ives N, Troop M, Gazzard B G, Rowland-Jones S L. Chemokine receptor polymorphisms and human immunodeficiency virus disease progression. J Infect Dis. 1999;180:1096–1105. doi: 10.1086/314997. [DOI] [PubMed] [Google Scholar]
- 21.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efremov R, Truong M J, Darcissac E C, Zeng J, Grau O, Vergoten G, Debard C, Capron A, Bahr G M. Human chemokine receptors CCR5, CCR3 and CCR2B share common polarity motif in the first extracellular loop with other human G-protein coupled receptors implications for HIV-1 coreceptor function. Eur J Biochem. 1999;263:746–756. doi: 10.1046/j.1432-1327.1999.00553.x. [DOI] [PubMed] [Google Scholar]
- 23.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard O M, Shirakawa A K, Turpin J A, Maynard A, Tobin G J, Carrington M, Oppenheim J J, Dean M. Naturally occurring CCR5 extracellular and transmembrane domain variants affect HIV-1 co-receptor and ligand binding function. J Biol Chem. 1999;274:16228–16234. doi: 10.1074/jbc.274.23.16228. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 26.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 27.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 28.Kostrikis L G, Neumann A U, Thomson B, Korber B T, McHardy P, Karanicolas R, Deutsch L, Huang Y, Lew J F, McIntosh K, Pollack H, Borkowsky W, Spiegel H M, Palumbo P, Oleske J, Bardeguez A, Luzuriaga K, Sullivan J, Wolinsky S M, Koup R A, Ho D D, Moore J P. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–10271. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B, Doranz B J, Rana S, Yi Y, Mellado M, Frade J M, Martinez A C, O'Brien S J, Dean M, Collman R G, Doms R W. Influence of the CCR2–V64I polymorphism on human immunodeficiency virus type 1 coreceptor activity and on chemokine receptor function of CCR2b, CCR3, CCR5, and CXCR4. J Virol. 1998;72:7450–7458. doi: 10.1128/jvi.72.9.7450-7458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 32.Loetscher M, Amara A, Oberlin E, Brass N, Legler D, Loetscher P, D'Apuzzo M, Meese E, Rousset D, Virelizier J L, Baggiolini M, Arenzana-Seisdedos F, Moser B. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Curr Biol. 1997;7:652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- 33.Mack M, Luckow B, Nelson P J, Cihak J, Simmons G, Clapham P R, Signoret N, Marsh M, Stangassinger M, Borlat F, Wells T N, Schlondorff D, Proudfoot A E. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariani R, Wong S, Mulder L C, Wilkinson D A, Reinhart A L, LaRosa G, Nibbs R, O'Brien T R, Michael N L, Connor R I, Macdonald M, Busch M, Koup R A, Landau N R. CCR2–64I polymorphism is not associated with altered CCR5 expression or coreceptor function. J Virol. 1999;73:2450–2459. doi: 10.1128/jvi.73.3.2450-2459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O'Brien T R, Hilgartner M W, Vlahov D, O'Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 36.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 37.Michael N L, Louie L G, Rohrbaugh A L, Schultz K A, Dayhoff D E, Wang C E, Sheppard H W. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat Med. 1997;3:1160–1162. doi: 10.1038/nm1097-1160. [DOI] [PubMed] [Google Scholar]
- 38.Michael N L, Nelson J A, KewalRamani V N, Chang G, O'Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O'Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosier D E, Picchio G R, Gulizia R J, Sabbe R, Poignard P, Picard L, Offord R E, Thompson D A, Wilken J. Highly Potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O'Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 41.Mummidi S, Bamshad M, Ahuja S S, Gonzalez E, Feuillet P M, Begum K, Galvis M C, Kostecki V, Valente A J, Murthy K K, Haro L, Dolan M J, Allan J S, Ahuja S K. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorpic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18949–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 42.Olson W C, Rabut G E, Nagashima K A, Tran D N, Anselma D J, Monard S P, Segal J P, Thompson D A, Kajumo F, Guo Y, Moore J P, Maddon P J, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oppermann M, Mack M, Proudfoot A E, Olbrich H. Differential effects of CC chemokines on CC chemokine receptor 5 (CCR5) phosphorylation and identification of phosphorylation sites on the CCR5 carboxyl terminus. J Biol Chem. 1999;274:8875–8885. doi: 10.1074/jbc.274.13.8875. [DOI] [PubMed] [Google Scholar]
- 44.Owman C, Garzino-Demo A, Cocchi F, Popovic M, Sabirsh A, Gallo R C. The leukotriene B4 receptor functions as a novel type of coreceptor mediating entry of primary HIV-1 isolates into CD4-positive cells. Proc Natl Acad Sci USA. 1998;95:9530–9534. doi: 10.1073/pnas.95.16.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paxton W A, Kang S, Koup R A. The HIV type 1 coreceptor CCR5 and its role in viral transmission and disease progression. AIDS Res Hum Retroviruses. 1998;14(Suppl. 1):S89–S92. [PubMed] [Google Scholar]
- 46.Paxton W A, Liu R, Kang S, Wu L, Gingeras T R, Landau N R, Mackay C R, Koup R A. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244:66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 47.Picchio G R, Gulizia R J, Mosier D E. Chemokine receptor CCR5 genotype influences the kinetics of human immunodeficiency virus type 1 infection in human PBL-SCID mice. J Virol. 1997;71:7124–7127. doi: 10.1128/jvi.71.9.7124-7127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pleskoff O, Treboute C, Alizon M. The cytomegalovirus-encoded chemokine receptor US28 can enhance cell-cell fusion mediated by different viral proteins. J Virol. 1998;72:6389–6397. doi: 10.1128/jvi.72.8.6389-6397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pohlmann S, Krumbiegel M, Kirchhoff F. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J Gen Virol. 1999;80:1241–1251. doi: 10.1099/0022-1317-80-5-1241. [DOI] [PubMed] [Google Scholar]
- 50.Proudfoot A E, Power C A, Hoogewerf A J, Montjovent M O, Borlat F, Offord R E, Wells T N. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 51.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubbert A, Combadiere C, Ostrowski M, Arthos J, Dybul M, Machado E, Cohn M A, Hoxie J A, Murphy P M, Fauci A S, Weissman D. Dendritic cells express multiple chemokine receptors used as coreceptors for HIV entry. J Immunol. 1998;160:3933–3941. [PubMed] [Google Scholar]
- 53.Ruffing N, Sullivan N, Sharmeen L, Sodroski J, Wu L. CCR5 has an expanded ligand-binding repertoire and is the primary receptor used by MCP-2 on activated T cells. Cell Immunol. 1998;189:160–168. doi: 10.1006/cimm.1998.1379. [DOI] [PubMed] [Google Scholar]
- 54.Samson M, Edinger A L, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms R W, Parmentier M. ChemR23, a putative chemoattractant receptor, is expresed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol. 1998;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 55.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu N, Soda Y, Kanbe K, Liu H Y, Jinno A, Kitamura T, Hoshino H. An orphan G protein-coupled receptor, GPR1, acts as a coreceptor to allow replication of human immunodeficiency virus types 1 and 2 in brain-derived cells. J Virol. 1999;73:5231–5239. doi: 10.1128/jvi.73.6.5231-5239.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu N, Soda Y, Kanbe K, Liu H Y, Mukai R, Kitamura T, Hoshino H. A putative G protein-coupled receptor, RDC1, is a novel coreceptor for human and simian immunodeficiency viruses. J Virol. 2000;74:619–626. doi: 10.1128/jvi.74.2.619-626.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 59.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O'Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 61.Speck R F, Esser U, Penn M L, Eckstein D A, Pulliam L, Chan S Y, Goldsmith M A. A trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr Biol. 1999;9:547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- 62.van Rij R P, de Roda Husman A M, Brouwer M, Goudsmit J, Coutinho R A, Schuitemaker H. Role of CCR2 genotype in the clinical course of syncytium-inducing (SI) or non-SI human immunodeficiency virus type 1 infection and in the time to conversion to SI virus variants. J Infect Dis. 1998;178:1806–1811. doi: 10.1086/314522. [DOI] [PubMed] [Google Scholar]
- 63.Verani A, Pesenti E, Polo S, Tresoldi E, Scarlatti G, Lusso P, Siccardi A G, Vercelli D. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998;161:2084–2088. [PubMed] [Google Scholar]
- 64.Vila-Coro A J, Mellado M, Martin de Ana A, Martinez A C, Rodriguez-Frade J M. Characterization of RANTES- and aminooxypentane-RANTES-triggered desensitization signals reveals differences in recruitment of the G protein-coupled receptor complex. J Immunol. 1999;163:3037–3044. [PubMed] [Google Scholar]
- 65.Wilken J, Hoover D, Thompson D A, Barlow P N, McSparron H, Picard L, Wlodawer A, Lubkowski J, Kent S B. Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES. Chem Biol. 1999;6:43–51. doi: 10.1016/S1074-5521(99)80019-2. [DOI] [PubMed] [Google Scholar]
- 66.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, He T, Talal A, Wang G, Frankel S S, Ho D D. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]