Abstract
Objectives
To determine the prevalence, trends, and potential nosocomial transmission events of the hidden reservoir of rectal carriage of extended-spectrum beta-lactamase-producing Enterobacterales (ESBL-E).
Methods
From 2013 to 2022, yearly point prevalence surveys were conducted in a large Dutch teaching hospital. On the day of the survey, all admitted patients were screened for ESBL-E rectal carriage using peri-anal swabs and a consistent and sensitive selective culturing method. All Enterobacterales phenotypically suspected of ESBL production were analysed using whole genome sequencing for ESBL gene detection and clonal relatedness analysis.
Results
On average, the ESBL-E prevalence was 4.6% (188/4,119 patients), ranging from 2.1 to 6.6% per year. The ESBL-prevalence decreased on average 5.5% per year. After time trend correction, the prevalence in 2016 and 2020 was lower compared to the other year. Among the ESBL-E, Escherichia coli (80%) and CTX-M genes (85%) predominated. Potential nosocomial transmission events could be found in 5.9% (11/188) of the ESBL-E carriers.
Conclusions
The ESBL-E rectal carriage prevalence among hospitalized patients was 4.6% with a downward trend from 2013 to 2022. The decrease in ESBL-E prevalence in 2020 could have been due to the COVID-19 pandemic and subsequent countrywide measures as no nosocomial transmission events were detected in 2020. However, the persistently low ESBL-E prevalences in 2021 and 2022 suggest that the decline in ESBL-E prevalence goes beyond the COVID-19 pandemic, indicating that overall ESBL-E carriage rates are declining over time. Continuous monitoring of ESBL-E prevalence and transmission rates can aid infection control policy to keep antibiotic resistance rates in hospitals low.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-024-01460-y.
Keywords: ESBL, Prevalence, Rectal carriage, Molecular typing, Nosocomial transmission
Introduction
In Enterobacterales, extended-spectrum beta-lactamases (ESBL) cause resistance to most beta-lactam antibiotics, including third generation cephalosporins. Infections with ESBL producing Enterobacterales (ESBL-E) have been associated with higher morbidity and mortality rates as well as greater costs compared to infections with susceptible Enterobacterales [1–3]. The prevalence of ESBL-E is increasing worldwide [4–6], and the WHO lists ESBL-E as high-priority pathogens [7]. The prevalence of ESBL-E in the Netherlands is among the lowest worldwide [5, 8]. However, not much is known about trends in ESBL-E asymptomatic rectal carriage after 2016.
ESBL-E infections are often preceded by asymptomatic rectal carriage [9, 10], which can be widespread before outbreaks are noticed [11]. Therefore, knowledge of the reservoir of asymptomatic carriage can provide important information about the risks of nosocomial transmission and guide infection control policy [3].
In the present study, we describe the ESBL-E prevalence over the course of 10 years based on a series of annual point prevalence surveys (PPS) undertaken in a Dutch teaching hospital from 2013 to 2022. Secondly, we determine the ESBL-producing Enterobacterales species and the presence of different ESBL genes over time using whole-genome sequencing for genotypic characterisation. Lastly, we identify possible nosocomial transmission events based on whole genome cluster analysis.
Methods
Study design
This is a prospective cross-sectional study.
Setting
The study was performed in the Amphia hospital, a teaching hospital in Breda, the Netherlands. Nine one-day PPS were performed between 2013 and 2022, excluding the year 2018. The surveys were carried out between October – December in 2013–2017 and 2022 and in June in 2019–2021. More details of the hospital and infection control measures can be found in the supplementary methods.
Participants
The screening encompassed all inpatients, including the dialysis department and the day-care clinic. Patients admitted at the psychiatric ward or emergency department were excluded.
Sample collection and selective culturing
Culturing of the obtained peri-anal swabs was done according to the protocol previously described by Willemsen et al. [12]. In short, after the swab was plated on a blood agar plate to serve as a growth control, the liquid Amies eluent was inoculated in selective tryptic soy broth, containing cefotaxime (0.25 mg/L) and vancomycin (8 mg/L) (TSB-VC) and subsequently plates on EbSA agar plates (AlphaOmega, ‘s-Gravenhage, Netherlands) after one day of incubation. Species identification of all morphologically distinct oxidase-negative Enterobacterales was performed using MALDI-TOF MS (2013–2020 bioMerieux, Marcy l’Etoile, France; 2021–2022 Bruker Daltonics, Bremen, Germany), followed by susceptibility testing via VITEK2 AST (bioMerieux, Marcy l’Etoile, France). Phenotypic confirmation of ESBL production was performed of isolates exhibiting a MIC of > 1 mg/L for ceftazidime and/or cefotaxime using the combination disc method for cefotaxime, ceftazidime, and/or cefepime with or without clavulanic acid (MastGroup, Bootle, UK) in accordance with the EUCAST protocol (7).
Molecular typing
One isolate per species per patient was sequenced using Nextera XT chemistry on a MiSeq sequencer (Illumina, San Diego, CA, USA). Error-correction and de novo genome assembly processes were executed using CLC genomic workbench 21.0 (Qiagen, Germantown, MD, USA). The following quality control criteria were used: coverage ≥ 20; number of scaffolds ≤ 1,000; N50 ≥ 15,000 bases; maximum scaffold length ≥ 50,000 bases; used reads > 90% and expected genome size 90–115% of the reference. ESBL genes were identified using Abricate v1.0.1 based on the AMRFinderPlus database with a coverage and identity of at least 90%. Multi-Locus Sequence Typing (MLST) and clonal relatedness was analysed using Ridom SeqSphere + version 8.0 (Ridom GmgH, Munich, Germany). Clonal relatedness between all isolates was determined based on pairwise comparisons of alleles within species-specific whole genome MLST (wgMLST) schemes. Missing values were excluded from the pairwise comparisons. Clonally related isolates were defined as two isolates with the number of allelic differences below the cut-off of clonal relatedness. The following cut-offs based on percentage of different alleles were used: Escherichia coli 0.0095; Klebsiella pneumoniae 0.0045; Citrobacter spp. 0.0030; Enterobacter cloacae complex 0.0035 [13].
Data collection and analysis
Patients were considered ESBL-E positive when at least one of the isolates contained an ESBL gene (defined as such in the NCBI curated database (October 2023)). Cultures with a negative growth control or error in the culture procedure were excluded. Patient demographics were systematically gathered and subsequently anonymized upon integration with the culture data. Patient demographics collected were age, gender, treating specialty, admission date and hospital admission location. When patients transferred during the day of the PPS, only the first culture and corresponding demographics were included. The length of stay was defined as at the time from the start of admission to the time of testing.
Epidemiological analysis
When clonally related isolates were identified in two patients admitted to the same ward simultaneously, nosocomial transmission was deemed likely. Nosocomial transmission was considered possible, when clonally related isolates were detected in patients admitted at the same time to different wards within the same hospital location or when clonally related isolates were detected in patients admitted to the same ward not at the same time, but within a 60-day window from each other [14]. When clonally related isolates were identified in patients where the conditions for likely or possible nosocomial transmission were not met, nosocomial transmission was deemed unlikely.
Statistical analysis
The prevalence of ESBL-E carriage was calculated for each year and used as outcome variable in logistic regression analyses using Statistical Package for Social Science software (IBM SPSS Statistics for Windows, Version 24.0. Armonk, NY: IBM Corp.). The time trend over the years was calculated using logistic regression analysis correcting for potential confounders: continuous linear yearly time trend, patient’s age (years), length of stay (days), and ICU admission (yes/no). Those were chosen a-priori based on an association with ESBL-E carriage in previous studies [3, 12]. Statistical significance was accepted at a p-value < 0.05 for the time trend over the years. Additionally, for each year a multiple logistic regression model on the ESBL prevalence was trained with the ESBL carriage status as dependent variable and with the above-mentioned potential confounders. For each year, the ESBL-E prevalence was predicted based on the estimated logistic model of the total of the other eight years, given the average values of the confounders in the left-out year. The difference between this predicted ESBL prevalence and the observed prevalence was calculated and tested for statistical significance. P-values below 0.0064 per year were considered statistically significant, coinciding with a total experiment wise two-sided significance level alpha of 0.05 across the nine years of which eight were considered mutually independent (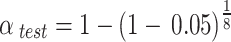 ). A more detailed description of the statistical analysis can be found in the supplementary methods.
). A more detailed description of the statistical analysis can be found in the supplementary methods.
Results
During the nine PPS, 5,494 patients were eligible for ESBL-E screening, of which 4,165 (75.8%) underwent screening resulting in 4,119 (75.0%) evaluable cultures (Table 1). A total of 46 cultures were excluded. Baseline characteristics of the included patients are shown in Table 1.
Table 1.
Baseline characteristics of screened patients in the nine conducted point prevalence surveys
| Year | 2013 | 2014 | 2015 | 2016 | 2017 | 2019 | 2020 | 2021 | 2022 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Period | Nov | Oct | Oct | Nov | Dec | June | June | June | Nov | |
| Hospitalised patients, n | 626 | 653 | 655 | 613 | 597 | 664 | 583 | 559 | 544 | 5494 |
| Patients screened, n (%) | 516 (82%) | 568 (87%) | 565 (86%) | 512 (84%) | 397 (69%) | 425 (64%) | 430 (74%) | 380 (68%) | 372 (68%) | 4165 (76%) |
| Excluded cultures, n | 0 | 10 | 5 | 10 | 4 | 0 | 5 | 7 | 5 | 46 (1%) |
| Evaluable cultures, n (%) | 516 (82%) | 558 (86%) | 560 (86%) | 502 (82%) | 393 (66%) | 425 (64%) | 425 (73%) | 373 (67%) | 367 (68%) | 4119 (75%) |
| Age (years), median, (IQR) | 65,5 (50-75.8) | 66 (50–76) | 67 (47,3–76) | 67 (54–78) | 71 (58.5–80) | 68 (53–76) | 68 (56–78) | 71 (57–78) | 70 (58–78) | 68 (53–77) |
| Length of stay (days), median, (IQR) | 2 (1–6) | 2 (0–6) | 2 (0–5) | 3.5 (1–8) | 4 (1–9) | 2 (0–7) | 3 (1-7.5) | 4 (1–8) | 3 (1–8) | 3 (1–7) |
| Sex, n (%) female | 250 (48%) | 290 (52%) | 290 (52%) | 271 (54%) | 172 (44%) | 204 (48%) | 221 (52%) | 174 (47%) | 180 (49%) | 2052 (50%) |
| Medical specialty, n (%) | ||||||||||
| Intensive care | 18 (4%) | 13 (2%) | 18 (3%) | 22 (4%) | 18 (5%) | 19 (5%) | 20 (5%) | 20 (5%) | 14 (4%) | 162 (4%) |
| Internal medicine | 107 (21%) | 105 (20%) | 106 (19%) | 108 (22%) | 55 (14%) | 87 (21%) | 87 (21%) | 71 (19%) | 102 (28%) | 828 (20%) |
| General surgery | 81 (16%) | 83 (15%) | 85 (15%) | 80 (16%) | 70 (18%) | 75 (18%) | 80 (19%) | 58 (16%) | 38 (10%) | 650 (16%) |
| Other | 310 (60%) | 357 (64%) | 351 (63%) | 292 (59%) | 250 (64%) | 244 (57%) | 238 (56%) | 224 (60%) | 213 (58%) | 2479 (60%) |
ESBL-E prevalence
In total, 214 Enterobacterales suspected of ESBL production were found in 206 patients. However, in 18 of these isolates the phenotypical ESBL test was considered false positive as no ESBL gene was identified (Table S1). This resulted in an ESBL-E rectal carriage prevalence of 5.0% based on phenotypical tests for ESBL production and in a prevalence of 4.6% (n = 188) based on WGS data (Table 2).
Table 2.
ESBL prevalence over time, including bacterial species and identified ESBL genes of the ESBL-E
| Year | 2013 | 2014 | 2015 | 2016 | 2017 | 2019 | 2020 | 2021 | 2022 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Period | Nov | Oct/Nov | Oct | Nov | Dec | June | June | June | Nov | |
| Evaluable cultures, n | 516 | 558 | 560 | 502 | 393 | 425 | 425 | 373 | 367 | 4119 |
| ESBL positive patients based on phenotype, n (%) | 28 (5.4%) | 39 (7.0%) | 33 (5.9%) | 16 (3.2%) | 21 (5.3%) | 25 (5.9%) | 9 (2.1%) | 18 (4.8%) | 17 (4.6%) | 206 (5.0%) |
| ESBL positive patients based on genotype, No. | 25 (4.8%) | 37(6.6%) | 32 (5.7%) | 14 (2.8%) | 18 (4.6%) | 24 (5.6%) | 9 (2.1%) | 15 (4.0%) | 14 (3.8%) | 188 (4.6%) |
| Patients with > 1 ESBL producing isolate, No. | 4 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | 8 |
| ESBL producing isolates | ||||||||||
| Genetically confirmed ESBL isolates, No. | 30 | 37 | 34 | 14 | 18 | 24 | 11 | 15 | 15 | 198 |
| Escherichia coli, No. | 25 | 29 | 30 | 12 | 15 | 18 | 10 | 10 | 9 | 158 (79.8%) |
| Klebsiella pneumoniae, No. | 5 | 4 | 2 | 2 | 2 | 2 | 0 | 2 | 3 | 22 (11.1%) |
| Enterobacter cloacae complex, No. | 0 | 3 | 2 | 0 | 1 | 4 | 0 | 3 | 2 | 15 (7.6%) |
| Other species, No. | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 (1.5%) |
| ESBL genes | ||||||||||
| Isolates with > 1 ESBL gene, No. | 3 | 4 | 3 | 0 | 1 | 2 | 1 | 1 | 2 | 17 (8.6%) |
| Total ESBL genes identified, No. | 33 | 41 | 37 | 14 | 19 | 26 | 12 | 16 | 17 | 215 |
| CTX-M family, No. | 30 | 36 | 32 | 11 | 15 | 21 | 10 | 14 | 14 | 183 (85.1%) |
| SHV family, No. | 2 | 5 | 4 | 2 | 2 | 3 | 1 | 1 | 3 | 23 (10.7%) |
| TEM family, No. | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 6 (2.8%) |
| Other, No. | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 (0.5%) |
Factors associated with ESBL-E prevalence
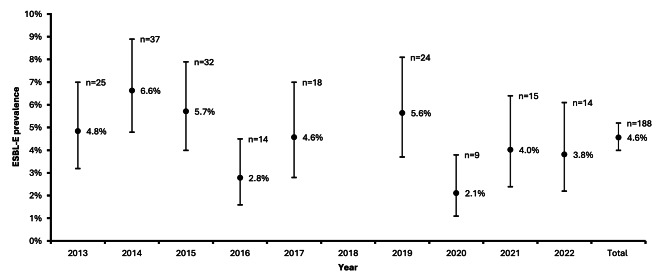
The ESBL-E prevalence ranged from 2.1 to 6.6% over the years (Fig. 1). The ESBL-prevalence decreased on average 5.5% per year (unadjusted odds ratio 0.945, 95% CI 0.899–0.994, p = 0.028; adjusted odds ratio 0.945, 95% CI 0.901–0.990, p = 0.017). A prolonged length of stay was associated with a 1.9% increased probability of ESBL-E carriage on average per extra day of hospitalization (unadjusted odds ratio 1.017, 95% IC 1.004–1.030, p = 0.011; adjusted odds ratio 1.019, 95% CI 1.006–1.032, p = 0.005). Age and ICU admission had no significant effect on ESBL-E carriage: a 10 year higher age was on average associated with a 1% higher ESBL-E prevalence (odds ratio 1.01, 95% CI 0.951–1.073, ns) and ICU-admission was on average associated with a 26% lower ESBL-E prevalence (odds ratio 0.74, 95% CI 0.32–1.71, ns). Post-hoc testing revealed that the ESBL-E prevalence in 2016 and 2020 was notably lower compared to other years and this observation persisted after adjusting for time trend and possible confounders (Table S2). In 2016 the ESBL-E prevalence was 2.8%, while a prevalence of 5.1% was predicted based on the other years (p = 0.0026, significance accepted at < 0.0064). Similarly, in 2020 the observed prevalence was 2.1% with a predicted prevalence of 4.2% based on the other years (p = 0.0072, significance accepted at < 0.0064).
Fig. 1.
ESBL-E prevalence from 2013 to 2022, excluding 2018, with adjusted Wald 95% confidence interval. The numbers of ESBL-E found per year are presented above the corresponding prevalence data
Identified ESBL producing species and sequence types
Eight of the 188 ESBL-E carrying patients carried two or more different isolates resulting in 198 ESBL-positive isolates. In total, 79.8% (n = 158) of the identified ESBL-E were E. coli, followed by 11.1% (n = 22) of K. pneumoniae (Table 2). Among the E. coli isolates, ST131 was the most common sequence type detected (n = 36, 22.8%). For K. pneumoniae isolates, no predominant MLST was observed (Table S3).
Identified ESBL genes
Collectively, the 198 ESBL-E harboured 215 ESBL genes, with a maximum of two ESBL genes per isolate, resulting in 9.7% (n = 19) of the ESBL-E isolates harbouring two EBSL genes. Most of the identified genes belonged to the CTX-M family (n = 182, 84.7%), with CTX-M-15 being the most prevalent (n = 85, 40.0%) (Table S3).
Clonal relatedness
Based on wgMLST analysis, 36 of the 198 EBSL-E isolates (18.2%) were clonally related to at least one other ESBL-E isolate. Of those, 25 were E. coli, corresponding to 15.8% of all 158 E. coli isolates found. The others were 7 of the 15 E. cloacae isolates (46.7%) and 4 of the 22 K. pneumoniae (18.2%). In 2020 no related isolates were found that were clonally related to any other ESBL-E isolate, and in 2016 only one isolate was clonally related to another isolate, which was from a different year (Table 3). Apart from the years 2016 and 2020–2022, clonal relatedness was detected between isolates derived from the same year as well as cross-annual (Table 3). ESBL-E isolates were never clonally related to more than one isolate detected in the same year. However, cross-annual analysis revealed an E. coli isolate clonally related to nine other isolates. For E. cloacae this maximum was two other isolates and for K. pneumoniae no cross-annual clonality was found.
Table 3.
Identified clonally related isolates each year within the same year and the total number of clonally related isolates including cross-annual clonal relatedness
| Year | ESBL-E isolates, No. | Total clonally related isolates within the same year, No. (%) | Total clonally related isolates incl. cross-annual, No. (%) |
|---|---|---|---|
| 2013 | 30 | 4 (13.3%) | 6 (20.0%) |
| 2014 | 37 | 4 (10.8%) | 6 (16.2%) |
| 2015 | 34 | 4 (11.8%) | 7 (20.6%) |
| 2016 | 14 | 0 | 1 (7.1%) |
| 2017 | 18 | 4 (22.2%) | 4 (22.2%) |
| 2019 | 24 | 2 (8.3%) | 7 (29.2%) |
| 2020 | 11 | 0 | 0 |
| 2021 | 15 | 2 (13.3%) | 2 (13.3%) |
| 2022 | 15 | 0 | 3 (20.0%) |
| Total | 198 | n.v.t. | 36 (18.8%) |
Nosocomial transmission events
A total of 38 out of 12,739 pairwise comparisons (consisting of 12.403 comparisons between the 158 E. coli isolates, 231 comparisons between the 22 K. pneumoniae isolates and 105 comparisons between the 15 E. cloacae isolates). revealed clonally related ESBL-E isolates from different patients. In 27 (71.1%) clonally related ESBL-E pairs no epidemiological link was found between the patients (Table S4), while in 11 pairs a link could be found deeming nosocomial transmission likely (n = 5) or possible (n = 6) (Table 4). There were two possible or likely nosocomial transmission events per year between patients enrolled in the same year in 2013, 2014, 2015, and 2017, and one per year in 2019 and 2021 (Table 4). Potential nosocomial transmission events were found in 8 of the 31 pairwise comparisons (25.8%) involving patients with clonally related E. coli isolates. As opposed to 20% (1/5) and 100% (2/2) of the comparisons between patient with clonal E. cloacae and K. pneumoniae isolates respectively. Overall, 5.9% (11/188) of ESBL-E rectal carriage cases could be attributed to potential nosocomial transmission, with 2.7% (5/188) considered likely.
Table 4.
Pairwise comparisons between two patients with clonally related ESBL-E isolates for which nosocomial transmission was deemed possible or likely based on the epidemiological link between those patients
| Year of collection | Species | MLST | ESBL genes | Epidemiological link | Likelihood nosocomial transmission |
|---|---|---|---|---|---|
| 2013 | Klebsiella pneumoniae | ST107 | CTX-M-14 | Same time, same location, same ward | Likely |
| 2013 | Escherichia coli | ST131 | 1 isolate CTX-M-32 1 isolate CTX-M-27 | Same time, other location of the hospital | Possible |
| 2014 | Escherichia coli | ST38 | CTX-M-15 | Same time, same location, different ward | Possible |
| 2014 | Klebsiella pneumoniae | ST4283 | CTX-M-15 | Same time, same location, same ward | Likely |
| 2015 | Escherichia coli | ST616 | CTX-M-15 | Same time, other location of the hospital | Possible |
| 2015 | Escherichia coli | ST131 | CTX-M-27 | Same time, same location, different ward | Possible |
| 2017 | Escherichia coli | ST131 | CTX-M-27 | Same time, other location of the hospital | Possible |
| 2017 | Escherichia coli | ST131 | CTX-M-15 | Same time, same location, same ward | Likely |
| 2017/2019 | Escherichia coli | ST131 | CTX-M-15 | Same time, same location, different ward | Possible |
| 2019 | Enterobacter cloacae complex | ST50 | SHV-12 | Same time, same location, different ward | Likely |
| 2021 | Escherichia coli | ST44 | CTX-M-15 | Same time, same location, same ward | Likely |
Discussion
We found an ESBL-E rectal carriage prevalence of 4.6% ranging from 2.1 to 6.6% based on WGS, measured in nine point prevalence surveys from 2013 to 2022. The ESBL-E prevalence declined on average 5.5% each year. Potential nosocomial transmission events were rarely observed in this study as only 5.9% of ESBL-E cases could be attributed to nosocomial transmission.
The observed decrease in ESBL-E prevalence over time is in line with NethMap and EARS-NET reports on the ESBL-E prevalence in the Netherlands and Europe, respectively, showing a stable or (slightly) decreasing prevalence of third generation cephalosporin resistance over the years in clinical isolates [8, 15]. Consistent with other studies, E. coli, and particularly ST131, predominated [4–6, 11, 12, 16, 17]. Among the ESBL genes, CTX-M genes and more specifically CTX-M-15, were most abundant [11, 12, 16]. Interestingly, we found that ST131 comprised only 22.8% of the E. coli isolates, but 80.6% of the clonally related pairwise comparisons between E. coli and 62.5% of the potential nosocomial transmission events. This indicates either relatively more genetic similarity between ST131 isolates or enhanced transmissibility of E. coli ST131. The latter could be a partial explanation of the remarkably fast global expansion of ESBL-producing ST131 of which the mechanism is still unknown [18].
Furthermore, we may have underestimated the number of nosocomial transmission events as indirect epidemiological links might be missed. For clonally related K. pneumoniae isolates, we could always identify a direct epidemiological link between patients. This was different for clonally related E. coli and E. cloacae complex isolates, for which a direct epidemiological link was only present in 25.8% and 20%, respectively. It could be that we missed patients that link these epidemiologically unrelated patients together or that patients share epidemiological links outside the hospital, indicating community transmission of E. coli and E. cloacae complex - and not of K. pneumoniae - which is detected in the hospital. This is in concordance with another study that identified transmission in the healthcare setting as important driver for ESBL producing K. pneumoniae, but not for spread of ESBL producing E. coli [19]. Missed indirect links between E. coli and E. cloacae isolates within the hospital rather than community transmission would imply that the community ESBL-E carriage prevalence is lower than in the hospital. However, the ESBL-E carriage prevalence observed in a Dutch study from 2014 to 2016 was similar to the in-hospital prevalence that we observed (≈ 5%) [20]. This makes missed indirect in-hospital links unlikely and would support community transmission as major player in E. coli carriage dynamics.
The lowest prevalence rates were observed in 2016 and 2020. Of particular interest is the year 2020, coinciding with the onset of the COVID-19 pandemic in the Netherlands, prompting the implementation of nationwide measures from March 2020 onwards to mitigate SARS-CoV-2 transmission. Subsequent extra attention for standard precautions to minimalize pathogen transmission across patients could explain the observed decrease in ESBL-E prevalence in 2020. This hypothesis is supported by the fact that no clonally related isolates and thus potential nosocomial transmission events were detected in 2020. Additionally, the nationwide measures to combat COVID-19 included travel restrictions, implemented a few months before the PPS in 2020 was conducted. It could also have resulted in lower ESBL-E prevalence rates compared to previous years as recent travel abroad is also established as a risk factor for ESBL-E colonization [11, 16]. Unfortunately, our dataset lacks comprehensive information regarding patients’ travel history and antibiotic usage. While the COVID-19 measure could explain the decline in 2020, the persistently low ESBL-E prevalences in 2021 and 2022 suggest the decline in ESBL-E prevalence belong the COVID-19 pandemic.
Beside leaking data on the patients’ travel history and antibiotic usage, limitations of our study are the lack of data on the year 2018 and the variation in the timing of PPS across different years. Some were conducted in June (2019–2021) and others in October-December (2013–2017, 2022). Despite this, our analysis did not reveal a difference in ESBL-E prevalence between seasons. While seasonal effects on ESBL carriage are not well-established, we considered the possibility of increased summer travel and higher incidences in enteric infectious diseases influencing prevalence [20, 21]. To investigate this, we conducted a PPS in both June and November 2021, but found no significant difference in ESBL-E rectal carriage prevalence (data not shown).
A notable strength of the present study is the consistent use of sensitive and selective ESBL-E culture methods each year. In 2021 the revised Dutch guideline for detection of highly resistant micro-organisms states that the use of enrichment broths for detection of ESBL carriage is favoured, because this increases the sensitivity of the ESBL-E cultures substantially [22, 23]. In this study, enrichment broths were already used from the study’s inception in 2013 ensuring reliable data comparison over the years.
Over a ten-year period, PPS provided continuous monitoring for the ESBL-E prevalence and transmission rates in our hospital informing and refining our infection control policies. Notably, when the hospital transitioned to a building with exclusively single-bed rooms, contact precaution measures for ESBL-E carriers were lifted, under the condition that periodic assessment of ESBL-E carriage rates was conducted. This strategy ensured that significant ESBL-E outbreaks were not overlooked and validated that standard precautions were sufficient to contain ESBL-E in a hospital environment consisting of single rooms with private sanitary facilities. This may also apply to multiple-bed rooms, as evidence indicates that multiple-bed rooms are non-inferior to single-bed rooms in containing ESBL-E when contact precautions for known ESBL-E carriers are implemented [24]. Nevertheless, further research is required to confirm this hypothesis. Annual PPS not only track the impact of changes in infection control policies, but also provide yearly insights into the efficacy of the hospital’s infection control measures and facilitate the early detection of potential ESBL-E outbreaks. Nonetheless, the implementation of prospective WGS on selected ESBL-E isolates could offer even more immediate data on potential outbreaks [14].
Conclusions
We observed a low ESBL-E prevalence with a declining trend over time with the COVID-19 pandemic and subsequent measures as possible explanations for the observed decrease in 2020. However, the persistently low ESBL-E prevalences in 2021 and 2022 indicate that the overall ESBL-E prevalence is declining over the years. Continuous monitoring of ESBL-E prevalence and transmission rates can aid infection control policy to keep antibiotic resistance rates in hospitals low.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank those who contributed to the completion of this research. Thanks to all the infection control practitioners of the Amphia hospital who gathered all the epidemiological data. We are also thankful to our colleagues of the Microvida NGS team who sequenced all the ESBL-E isolates and performed bioinformatical analyses. Lastly, we would like to thank Ina Willemsen who was one of the founders of this project in 2010.
Author contributions
Conceptualization—KH, VW, JK, and JS; methodology—KH, VW, WB, PM and JS; validation—KH, VW, WB and JS; formal analysis—KH, VW, MT and PM; investigation—KH and VW; resources—VW, WB, MR, JV, JM and JS; data curation—KH and MT; writing—original draft—KH, VW and JS; writing—review and editing—all authors; visualization—KH; supervision—VW and JS; project administration—KH, VW, WB, MR, JV, JM and JS; funding acquisition—VW, WB, JK, MR and JS.
Funding
This study received funding from the Amphia Fund for Science in 2019.
Data availability
Genomic sequences that support the findings of this study have been deposited in the European Nucleotide Archive with the primary accession code PRJEB78530.
Declarations
Ethics approval and consent to participate
This study was conducted as part of the hospital’s standard infection control policy. Ethical and medical approvals were not deemed necessary for this non-invasive procedure, as it aligned with Dutch regulations pertaining to research involving human subjects. Patients were provided the option to decline participation (opt-out) if they chose.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Phungoen P, Sarunyaparit J, Apiratwarakul K, Wonglakorn L, Meesing A, Sawanyawisuth K. The association of ESBL Escherichia coli with mortality in patients with Escherichia coli bacteremia at the emergency department. Drug Target Insights. 2022;16:12–6. 10.33393/dti.2022.2422. 10.33393/dti.2022.2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteve-Palau E, Solande G, Sánchez F, Sorlí L, Montero M, Güerri R, et al. Clinical and economic impact of urinary tract infections caused by ESBL-producing Escherichia coli requiring hospitalization: a matched cohort study. J Infect. 2015;71:667–74. 10.1016/J.JINF.2015.08.012. 10.1016/J.JINF.2015.08.012 [DOI] [PubMed] [Google Scholar]
- 3.Blom A, Ahl J, Månsson F, Resman F, Tham J. The prevalence of ESBL-producing Enterobacteriaceae in a nursing home setting compared with elderly living at home: a cross-sectional comparison. BMC Infect Dis. 2016;16. 10.1186/s12879-016-1430-5. [DOI] [PMC free article] [PubMed]
- 4.Chang K, Rattanavong S, Mayxay M, Keoluangkhot V, Davong V, Vongsouvath M, et al. Bacteremia caused by extended-spectrum beta-lactamase-producing enterobacteriaceae in Vientiane, Lao PDR: a 5-year study. Am J Trop Med Hyg. 2020;102:1137–43. 10.4269/AJTMH.19-0304. 10.4269/AJTMH.19-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bezabih YM, Sabiiti W, Alamneh E, Bezabih A, Peterson GM, Bezabhe WM, et al. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother. 2021;76:22–9. 10.1093/JAC/DKAA399. 10.1093/JAC/DKAA399 [DOI] [PubMed] [Google Scholar]
- 6.Raphael E, Glymour MM, Chambers HF. Trends in prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolated from patients with community- and healthcare-associated bacteriuria: results from 2014 to 2020 in an urban safety-net healthcare system. Antimicrob Resist Infect Control. 2021;10. 10.1186/s13756-021-00983-y. [DOI] [PMC free article] [PubMed]
- 7.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018;18. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed]
- 8.European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2021. Stockholm: ECDC; 2022. [Google Scholar]
- 9.Souverein D, Euser SM, Herpers BL, Kluytmans J, Rossen JWA, Den Boer JW. Association between rectal colonization with highly resistant gram-negative rods (HR-GNRs) and subsequent infection with HR-GNRs in clinical patients: a one year historical cohort study. PLoS ONE. 2019;14:e0211016. 10.1371/journal.pone.0211016. 10.1371/journal.pone.0211016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorrie CL, Mirc Eta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, et al. Gastrointestinal carriage is a Major Reservoir of Klebsiella pneumoniae infection in Intensive Care patients. Clin Infect Dis. 2017;65:208–15. 10.1093/cid/cix270. 10.1093/cid/cix270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolivet S, Vaillant L, Poncin T, Lolom I, Gaudonnet Y, Rondinaud E, et al. Prevalence of carriage of extended-spectrum β-lactamase-producing enterobacteria and associated factors in a French hospital. Clin Microbiol Infect. 2018;24:1311–4. 10.1016/j.cmi.2018.03.008. 10.1016/j.cmi.2018.03.008 [DOI] [PubMed] [Google Scholar]
- 12.Willemsen I, Oome S, Verhulst C, Pettersson A, Verduin K, Kluytmans J. Trends in Extended Spectrum Beta-Lactamase (ESBL) producing enterobacteriaceae and ESBL genes in a Dutch teaching hospital, measured in 5 yearly point prevalence surveys (2010–2014). PLoS ONE. 2015;10. 10.1371/journal.pone.0141765. [DOI] [PMC free article] [PubMed]
- 13.Den Kluytmans-Van MFQ, Rossen JWA, Bruijning-Verhagen PCJ, Bonten MJM, Friedrich AW, Vandenbroucke-Grauls CMJE, et al. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing enterobacteriaceae. J Clin Microbiol. 2016;54:2919–27. 10.1128/JCM.01648-16. 10.1128/JCM.01648-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherry NL, Gorrie CL, Kwong JC, Higgs C, Stuart RL, Marshall C, et al. Multi-site implementation of whole genome sequencing for hospital infection control: a prospective genomic epidemiological analysis. Lancet Reg Health West Pac. 2022;23:100446. 10.1016/j.lanwpc.2022.100446. 10.1016/j.lanwpc.2022.100446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.SC de Greeff E, Kolwijck AF, Schoffelen NM. 2023. Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands in 2022 / MARAN 2023. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2022. National Institute for Public Health and the Environment 2023.
- 16.Reuland EA, Al Naiemi N, Kaiser AM, Heck M, Kluytmans JAJW, Savelkoul PHM, et al. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother. 2016;71:1076–82. 10.1093/jac/dkv441. 10.1093/jac/dkv441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDanel J, Schweizer M, Crabb V, Nelson R, Samore M, Khader K, et al. Incidence of extended-spectrum β-Lactamase (ESBL)-producing Escherichia coli and klebsiella infections in the United States: a systematic literature review. Infect Control Hosp Epidemiol. 2017;38:1209–15. 10.1017/ice.2017.156. 10.1017/ice.2017.156 [DOI] [PubMed] [Google Scholar]
- 18.Pitout JDD, Finn TJ. The evolutionary puzzle of Escherichia coli ST131. Infection. Genet Evol. 2020;81. 10.1016/j.meegid.2020.104265. [DOI] [PubMed]
- 19.Duval A, Obadia T, Boëlleid PY, Fleury E, Herrmann JL, Guillemot D, et al. Close proximity interactions support transmission of ESBL-K. Pneumoniae but not ESBL-E. Coli in healthcare settings. PLoS Comput Biol. 2019;15. 10.1371/journal.pcbi.1006496. [DOI] [PMC free article] [PubMed]
- 20.van den Bunt G, van Pelt W, Hidalgo L, Scharringa J, de Greeff SC, Schürch AC, et al. Prevalence, risk factors and genetic characterisation of extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae (ESBL-E and CPE): a community-based cross-sectional study, the Netherlands, 2014 to 2016. Eurosurveillance. 2019;24. 10.2807/1560-7917.ES.2019.24.41.1800594. [DOI] [PMC free article] [PubMed]
- 21.Wielders CCH, Van Duijkeren E, Van Den Bunt G, Meijs AP, Dierikx CM, Bonten MJM, et al. Seasonality in carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in the general population: a pooled analysis of nationwide cross-sectional studies. Epidemiol Infect. 2020. 10.1017/S0950268820000539. 10.1017/S0950268820000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluytmans-Van Den Bergh MFQ, Verhulst C, Willemsen LE, Verkade E, Bonten MJM, Kluytmans JAJW. Rectal carriage of extended-spectrum-beta-lactamase-producing enterobacteriaceae in hospitalized patients: selective preenrichment increases yield of screening. J Clin Microbiol. 2015;53. 10.1128/JCM.01251-15. [DOI] [PMC free article] [PubMed]
- 23.Murk JLAN, Heddema ER, Hess DLJ, Bogaards JA, Vandenbroucke-Grauls CMJE, Debets-Ossenkopp YJ. Enrichment broth improved detection of extended-spectrum-beta-lactamase- producing bacteria in throat and rectal surveillance cultures of samples from patients in intensive care units. J Clin Microbiol. 2009;47. 10.1128/JCM.01406-08. [DOI] [PMC free article] [PubMed]
- 24.den Kluytmans-van MFQ, Bruijning-Verhagen PCJ, Vandenbroucke-Grauls CMJE, de Brauwer EIGB, Buiting AGM, Diederen BM, et al. Contact precautions in single-bed or multiple-bed rooms for patients with extended-spectrum β-lactamase-producing Enterobacteriaceae in Dutch hospitals: a cluster-randomised, crossover, non-inferiority study. Lancet Infect Dis. 2019;19:1069–79. 10.1016/S1473-3099(19)30262-2. 10.1016/S1473-3099(19)30262-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences that support the findings of this study have been deposited in the European Nucleotide Archive with the primary accession code PRJEB78530.